Abstract
Background
Blueberry cultivars require a fixed quantity of chilling hours during winter endo-dormancy for vernalization. In this study, transcriptome analysis using RNA sequencing data from nonchilled, chilled, and late pink buds of southern highbush blueberry ‘Legacy’ was performed to reveal genes associated with chilling accumulation and bud break.
Results
Fully chilled ‘Legacy’ plants flowered normally whereas nonchilled plants could not flower. Compared to nonchilled flower buds, chilled flower buds showed differential expression of 89% of flowering pathway genes, 86% of MADS-box genes, and 84% of cold-regulated genes. Blueberry orthologues of FLOWERING LOCUS T (FT) did not show a differential expression in chilled flower buds (compared to nonchilled flower bud) but were up-regulated in late-pink buds (compared to chilled flower bud). Orthologoues of major MADS-box genes were significantly up-regulated in chilled flower buds and down-regulated in late-pink buds. Functional orthologues of FLOWERING LOCUS C (FLC) were not found in blueberry. Orthologues of Protein FD (FD), TERMINAL FLOWER 1 (TFL1), and LEAFY (LFY) were down-regulated in chilled flower buds and in late-pink buds compared to nonchilled flower bud.
Conclusions
The changes from nonchilled to chilled and chilled to late-pink buds are associated with transcriptional changes in a large number of differentially expressed (DE) phytohormone-related genes and DE flowering pathway genes. The profile of DE genes suggests that orthologues of FT, FD, TFL1, LFY, and MADS-box genes are the major genes involved in chilling-mediated blueberry bud-break. The results contribute to the comprehensive investigation of the vernalization-mediated flowering mechanism in woody plants.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1311-8) contains supplementary material, which is available to authorized users.
Keywords: Chilling requirement, Cold hardness, Flowering time control, Freezing tolerance, Vaccinium corymbosum, Vernalization, Woody plant
Background
Winter dormancy (endo-dormancy) is essential for deciduous fruit crop survival [1, 2]. Under inductive low temperatures in the fall, deciduous woody fruit and nut crops are acclimated to develop freezing tolerance; meanwhile, accumulation of effective chilling hours is stimulated [3]. Sufficient chilling accumulation gives plants full vernalization, which is a prerequisite for bud-break in the spring.
Climate change in the last 40 years has caused earlier shifts in the onset of the growing season for trees (e.g., 2.3 days/decade in temperate Europe) and increased temperature fluctuation [4]. Early onset of the growing season causes insufficient chilling hours and prevents bud-break in fruit trees. Increased temperature fluctuation during plant bloom turns early season frosts into a danger, with freezing injuries to flowers and young fruits [5]. Plant breeding to manipulate chilling requirements and develop improved freeze tolerant cultivars are considered to be long-term solutions to mitigate reduced winter chill, decrease freezing damages, and secure deciduous fruit production [6].
Seasonal flowering plays a significant role in a plant’s life cycle and is controlled by a network of flowering pathway genes [7–9]. FLOWERING LOCUS C (FLC) is a key regulator in the vernalization pathway of winter-annual Arabidopsis thaliana ecotypes [7]. In winter wheat and barley, VERNALIZATION2 (VRN2) is a major regulator of vernalization-mediated flowering [8]. FLC and VRN2 analogs are recorded in peach (Prunus persica). These analogs are the DORMANCY ASSOCIATED MADS-box (DAM) genes which are a cluster of six MADS-box transcription factors. The loss of all or part of the DAMs resulted in the non-vernalized peach evergrowing mutant [10, 11]. The DAM genes are considered alternatives to FLC in regulating vernalization-mediated chilling requirement and flowering [10, 12]. However, the DAM genes show high similarities to A. thaliana AGAMOUS-LIKE 24 (AGL24) and SHORT VEGETATIVE PHASE (SVP) genes [12, 13]. Additionally, functional analysis of DAMs to reveal their roles in chilling-mediated flowering through reverse genetics has not been reported in peach. To date, neither a functional FLC-LIKE nor a VRN2-LIKE gene has been verified in Vaccinium plants [14].
Blueberries and cranberries are the most important Vaccinium fruits due to their high antioxidant and anti-inflammatory capacities [15]. Deciphering the mechanism of vernalization/chilling-mediated flowering will facilitate molecular breeding of blueberry cultivars for low-chilling requirement. To investigate flowering responses under nonconducive conditions, functional analysis of a blueberry FLOWERING LOCUS T (VcFT) gene has been conducted in highbush blueberry (Vaccinium corymbosum L.) [16–18]. Overexpression of VcFT (about 2900-fold increase in leaf tissues) caused continuous and precocious flowering in in vitro shoots and in one-year old ‘Aurora’ plants [16]. However, Overexpression of VcFT was not capable of fulfilling all chilling requirements in blueberry [17, 18]. Over 80% of the flower buds in two- and three-year old VcFT-overexpressing plants could not bloom under greenhouse conditions without a chilling period. To discover vernalization/chilling-responsive genes, transcriptome analyses were conducted with blueberry flower buds. The profile of differentially expressed (DE) genes will facilitate our understanding of the role of vernalization/chilling in blueberry bud-break.
Results
Identification of DE transcripts in chilled flower buds
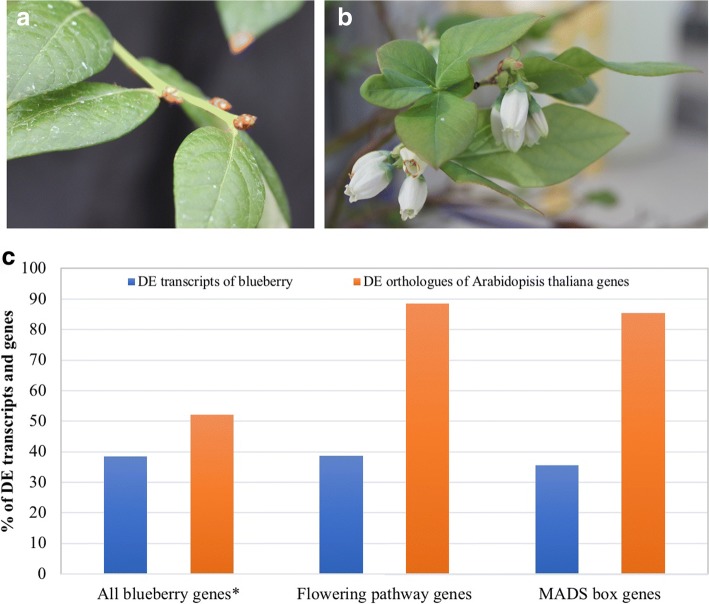
Nonchilled flower buds from the southern highbush blueberry ‘Legacy’ were grown in a heated greenhouse through the winter season. These buds did not flower in the following spring. In contrast, flower buds fully chilled under natural winter conditions flowered normally (Fig. 1). Transcriptome comparison of the chilled to nonchilled flower buds using Trinity and a non-annotated transcriptome reference (Reftrinity) of tetraploid blueberry in GenBank (Accession number: SRX2728597) [19, 20] revealed 37,000 differentially expressed genes (DEGs) and 47,000 DE transcripts.
Fig. 1.
Effect of vernalization of blueberry flowering. a Nonchilled flower buds could not bloom. b Early bloom of fully chilled flower buds. c Percentages of differentially expressed (DE) genes and transcripts in chilled flower buds (in comparison to nonchilled flower buds). *DE genes including A. thaliana genes and genes annotated to other species
To conduct transcriptome analysis, Reftrinity (180,000 genes and 250,000 isoforms) was annotated using Trinotate [20]. The annotation resulted in 14,000 known genes from 30,000 genes and 55,000 isoforms of Reftrinity. With this annotated reference, 64% of known blueberry genes and isoforms showed differential expression in the comparison of chilled to nonchilled flower buds (herein referring to DEGs/DE transcripts in chilled flower buds). Chilling affected expression of numerous genes simultaneously in blueberry flower buds.
Effect of chilling accumulation on flowering pathway genes
Differential expression was detected in chilled blueberry flower buds for 89% of flowering pathway orthologues of known A. thaliana flowering pathway genes (Table 1; Fig. 1). One of the top two orthologues of FT (VcFT: hereafter Vc before any A. thaliana gene refers to the blueberry orthologue gene) showed a slight decrease while the other showed a slight increase. Similar results were observed for CONSTANS-LIKE2 orthologues (Table 1).
Table 1.
Differentially expressed major flowering pathway genes in both chilled flower buds (CB) [vs nonchilled flower buds (NB)] and late-pink buds (LPB) (vs CB) in ‘Legacy’. LogFC for chilled buds: Log2(CB/NB). LogFC for late-pink buds: Log2(LPB/CB)
| Arabidopsis_ID|Vc_transcript_ID | LogFC for chilled buds | LogFC for late-pink buds | FDR for chilled buds | FDR for late-pink buds | e-value_to_blast | Gene_name | Annotation_by_Trinotate | Query_PmDAMs |
|---|---|---|---|---|---|---|---|---|
| AT5G60910.1|c77424_g2_i2 | 1.0 | 2.2 | 6E-03 | 3E-19 | 9E-44 | FUL, AGL8 | AGL11_ARATH | |
| AT1G69120.1|c92021_g1_i1 | −0.6 | −1.8 | 4E-02 | 5E-15 | 1E-80 | AP1, AGL7 | AGL8_SOLTU | |
| AT5G60910.1|c88116_g1_i1 | −0.4 | 0.8 | 4E-02 | 1E-10 | 5E-91 | FUL, AGL8 | AGL8_SOLTU | |
| AT1G69120.1|c88116_g8_i1 | 3.2 | −5.4 | 1E-16 | 2E-59 | 4E-94 | AP1, AGL7 | AGL8_SOLTU | |
| AT1G69120.1|c81830_g1_i1 | 3.5 | −2.1 | 2E-42 | 4E-24 | 2E-52 | AP1, AGL7 | AGL9_PETHY | |
| AT1G69120.1|c88116_g7_i1 | 3.6 | −2.7 | 3E-33 | 7E-41 | 3E-90 | AP1, AGL7 | AP1_SINAL | |
| AT5G15840.1|c77980_g1_i1 | −0.5 | 0.7 | 2E-03 | 3E-10 | 1E-66 | CO, FG | COL2_ARATH | |
| AT5G15840.1|c76265_g1_i1 | 2.3 | −2.5 | 2E-09 | 6E-14 | 4E-30 | CO, FG | COL2_ARATH | |
| AT5G60910.1|c79125_g1_i1 | −0.3 | −1.1 | 4E-02 | 2E-13 | 2E-35 | FUL, AGL8 | DEFA_ANTMA | |
| AT5G60910.1|c67980_g1_i1 | 3.4 | −4.6 | 7E-46 | 2E-70 | 5E-36 | FUL, AGL8 | DEFA_ANTMA | |
| AT5G61850.1|c96427_g2_i1 | −2.4 | −5.7 | 4E-13 | 4E-24 | 7E-102 | LFY, LFY3 | FLO_ANTMA | |
| AT5G61850.1|c96427_g2_i2 | −2.1 | −6.7 | 2E-13 | 9E-31 | 3E-98 | LFY, LFY3 | FLO_ANTMA | |
| AT1G65480.1|c84088_g2_i3 | −0.9 | −4.5 | 2E-02 | 7E-12 | 5E-95 | FT | HD3A_ORYSJ | |
| AT1G65480.1|c84088_g2_i5 | 1.1 | −4.9 | 1E-03 | 9E-28 | 1E-94 | FT | HD3A_ORYSJ | |
| AT1G26310.1|c77146_g1_i1 | 4.9 | −3.1 | 2E-57 | 9E-37 | 2E-56 | CAL, AGL10 | MADS6_ORYSJ | |
| AT2G45660.1|c89673_g3_i1 | 2.6 | −4.0 | 1E-09 | 3E-15 | 3E-53 | SOC1, AGL20 | SOC1_ARATH | |
| AT2G45660.1|c86010_g2_i1 | 3.9 | −4.4 | 1E-08 | 2E-09 | 2E-54 | SOC1, AGL20 | SOC1_ARATH | |
| AT2G22540.1|c91377_g1_i7 | 1.0 | −1.6 | 3E-02 | 1E-03 | 5E-69 | SVP, AGL22 | SVP_ARATH | |
| AT2G22540.1|c90829_g2_i1 | 1.2 | −1.2 | 1E-03 | 5E-04 | 4E-77 | SVP, AGL22 | SVP_ARATH | |
| AT4G24540.1|c91377_g1_i7 | 1.0 | −1.6 | 3E-02 | 1E-03 | 1E-39 | AGL24 | SVP_ARATH | |
| AT5G10140.1|c77146_g1_i1 | 4.9 | −3.1 | 2E-57 | 9E-37 | 2E-31 | FLC, FLF, AGL25 | MADS6_ORYSJ | |
| AT5G10140.1|c88116_g6_i1 | 4.1 | −1.9 | 5E-91 | 1E-26 | 7E-31 | FLC, FLF, AGL25 | SEP1_ARATH | |
| AT4G18960.1|c77424_g2_i2 | 1.0 | 2.2 | 6E-03 | 3E-19 | 1E-72 | MAF2, AGL31 | AGL11_ARATH | PmDAM1 |
| AT2G14210.1|c80388_g1_i2 | 3.9 | 3.2 | 4E-02 | 5E-09 | 3E-79 | MAF2, AGL31 | MAD23_ORYSJ | PmDAM1 |
| AT5G60910.1|c77146_g1_i1 | 4.9 | −3.1 | 2E-57 | 9E-37 | 7E-54 | MAF2, AGL31 | MADS6_ORYSJ | PmDAM1 |
| AT4G22950.1|c77424_g2_i2 | 1.0 | 2.2 | 6E-03 | 3E-19 | 1E-42 | MAF3, AGL70 | AGL11_ARATH | PmDAM1 |
| AT4G18960.1|c88293_g3_i1 | 4.7 | −4.7 | 5E-21 | 5E-20 | 5E-59 | MAF4 | AG_TOBAC | |
| AT5G15800.2|c88116_g6_i1 | 4.1 | −1.9 | 5E-91 | 1E-26 | 2E-87 | MAF4 | SEP1_ARATH | PmDAM1 |
| AT1G24260.2|c68983_g1_i1 | 2.5 | 0.0 | 3E-06 | 0E + 00 | 3E-42 | MAF5, AGL68 | . | |
| AT5G15800.2|c81830_g1_i1 | 3.5 | −2.1 | 2E-42 | 4E-24 | 3E-89 | MAF5, AGL68 | AGL9_PETHY | PmDAM1 |
#N/A: No differential expression
Orthologues of seven major MADS-box genes in flowering pathway genes of blueberry buds, APETALA1 (AP1), FRUITFULL (FUL), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), CAULIFLOWER (CAL), FLC, AGL24, and SVP, were significantly up-regulated (Table 1). The orthologues of Protein FD (FD), TERMINAL FLOWER 1 (TFL1), and LEAFY (LFY), and ACTIN-RELATED PROTEIN6 (ARP6) were down-regulated genes. Both VcFLC and VcSVP showed a contrasting response of FLC to vernalization in A. thaliana. The down-regulated VcFD and up-regulated VcSVP support the DE VcFT results [7]. However, decreased expression of VcLFY contradicts the decreased VcTFL1 and increased VcAP1, VcAGL24, and VcSOC1 expressions. The unchanged expression of VcFT and decreased expression of VcLFY orthologues could prevent the flowering of chilled flower buds during plant vernalization.
The MADS-box transcription factor and flowering repressor, FLC, and FRIGIDA (FRI) are the major genes regulating the vernalization of A. thaliana. FRI activates FLC, but vernalization represses FLC [7]. Both VcFRI and VcFLC in blueberry are present in the annotated Reftrinity. VcFRI showed a high similarity to FRI (e = − 98) while two VcFLC transcripts showed a lower similarity to FLC (e = − 31). These two VcFLC transcripts were annotated to SEP1 of A. thaliana and MADS6 of rice, respectively (Table 1). In chilled flower buds, VcFRI did not show differential expression whereas expression of VcFLC was increased about 25-fold for the top-two VcFLC candidate genes (Table 1). When all 26 potential VcFLC candidate genes (e < − 20) were included in the analysis, four candidates showed a down-regulation with a fold-change of 0.6–0.8. The other 22 candidate genes showed an up-regulation with an average of 4.9-fold (Additional file 1: Table S2). The inconsistency between VcFLC and FLC response to chilling/vernalization suggests that a different or more complicated mechanism is involved in vernalization-mediated flowering in tetraploid, blueberry plants. Additionally, all other DEGs involved in known vernalization-mediated flowering pathway were up-regulated in chilled flower buds with the exception of VcARP6 (c49456_g2, Log2FC = − 7.7) (Additional file 2: Table S1).
Identification of DE transcripts in late-pink buds
Fully chilled blueberry flower buds remained dormant under chilling conditions in January until continuous warm conditions in April drove dormancy release and bloom. To investigate the effect of dormancy release on gene expression, RNA sequencing data was obtained from late-pink buds. Comparative transcripts analysis in late-pink buds to those in chilled flower buds (hereafter referring to DEGs/DE transcripts in late-pink buds) resulted in 28,000 DE isoforms, which were annotated to 11,000 known genes.
Expression of flowering pathway genes in late-pink buds
The major DE flower pathway genes in chilled flower buds were compared to those in late-pink buds (Additional file 2: Table S1). The major flowering pathway genes VcFT, VcSOC1, VcAP1, VcFUL, VcFLC, VcSVP, and VcLFY showed decreased expression in late-pink buds. In A. thaliana, FT, SOC1, AP1, FUL, and LFY promote flowering while SVP and FLC are flowering repressors [7, 21]. The expression changes of VcFD and VcTFL1 in late-pink buds were similar to those in chilled flower buds (Table 1). The decreased VcFD expression was related, at least in part, to VcFT down-regulation. Decreased VcTFL1 expression in late-pink buds was associated with a down-regulation of VcAP1 and VcLFY. Additionally, decreased VcSVP and VcFLC expression were associated with a decrease in VcFT expression.
Expression of MADS-box genes
Blueberry DE MADS-box and DAMs genes were identified using A. thaliana MADS-box genes and Japanese apricot (Prunus mume) DAMs (PmDAMs) (Additional file 1: Table S2; Table 1). Orthologues of 62 A. thaliana MADS-box gene were identified in blueberry (unpublished data). DE orthologues of 53 and 44 MADS-box genes were detected in chilled flower buds and late-pink buds, respectively. These orthologues include the major flowering pathway genes VcFLC, VcSOC1, VcSVP, VcAP1, VcFUL, VcCAL, and VcAGL24 (Table 1). The annotated VcSOC1 (c86010_g2_i1) showed high similarities to 25 A. thaliana MADS-box genes. Similarly, the annotated VcFLC homologues were similar to 23 A. thaliana MADS-box genes (Table 1). The results suggest that VcSOC1 and VcFLC could have multiple functions in blueberry.
Three DE PmDAM orthologues (VcPmDAM1, VcPmDAM2, and VcPmDAM5) were identified and showed high similarities to four A. thaliana MADS-box genes, MADS AFFECTING FLOWERING 2 (MAF2), MAF4, MAF5 and FOREVER YOUNG FLOWER. In A. thaliana, MAF2, MAF4, and MAF5 are FLC paralogs. MAF2, MAF5 and FLC are down-regulated and MAF5 is up-regulated during vernalization [22]. In contrast, the blueberry orthologues VcFLC, VcMAF1, VcMAF2, VcMAF4, and VcMAF5 were up-regulated while VcMAF3 was repressed in chilled flower buds (Table 1; Additional file 1: Table S2). Additionally, three DE VcPmDAMs were annotated to the homologues VcSOC1, VcSVP, VcAP1, and VcSEP1. The up-regulated VcPmDAM1 homologues were the only DE orthologue in chilled flower buds. In late-pink buds, 75% of DE VcPmDAM1 homologues and all DE VcPmDAM5 homologues were down-regulated while DE VcPmDAM2 was up-regulated (Additional file 1: Table S2). These blueberry MADS-box genes showed significant changes in response to both chilling and flower bud breaking (Table 1). However, during vernalization, the responses of VcFLC, VcMAFs, and VcPmDAMs diverges from FLC’s response to vernalization in A. thaliana.
Response of phytohormone-related genes in chilled and late-pink buds
For both chilled buds and late-pink buds, DE transcripts showed high similarities to the pathway genes for five major phytohormones (Additional file 3: Table S3). Over 50% of the DE blueberry orthologues were related to abscisic acid (ABA), ethylene, auxin, and gibberellin (GA) genes while 25% were related to cytokinin genes (Fig. 2). The late-pink bud showed more DE phyotohormone orthologues than the chilled buds (Fig. 2). The DE phytohormone genes suggest the potential involvement of these phytohormones during chilling and flowering.
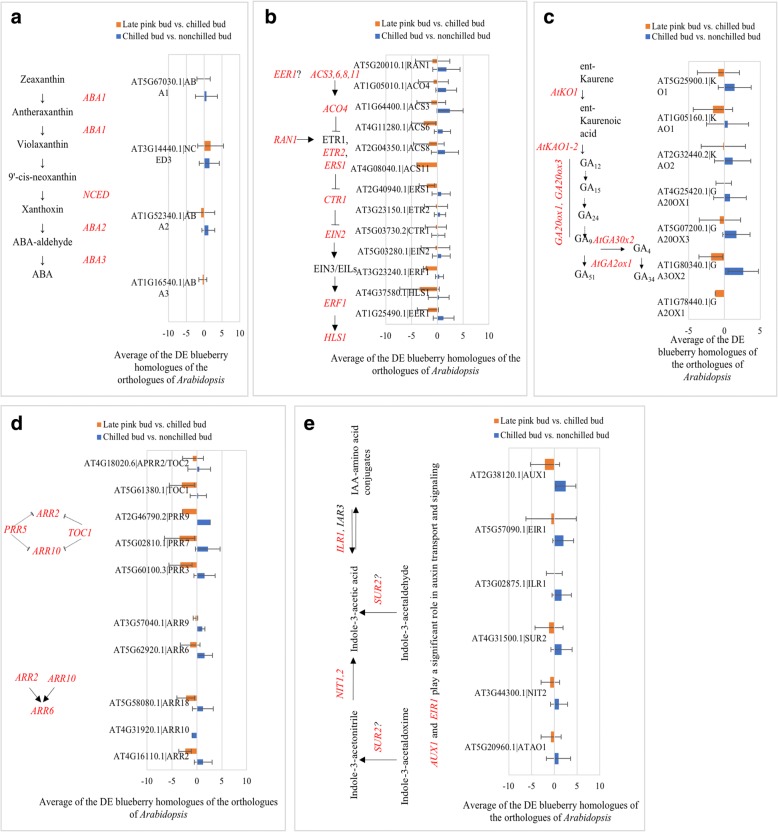
Fig. 2.
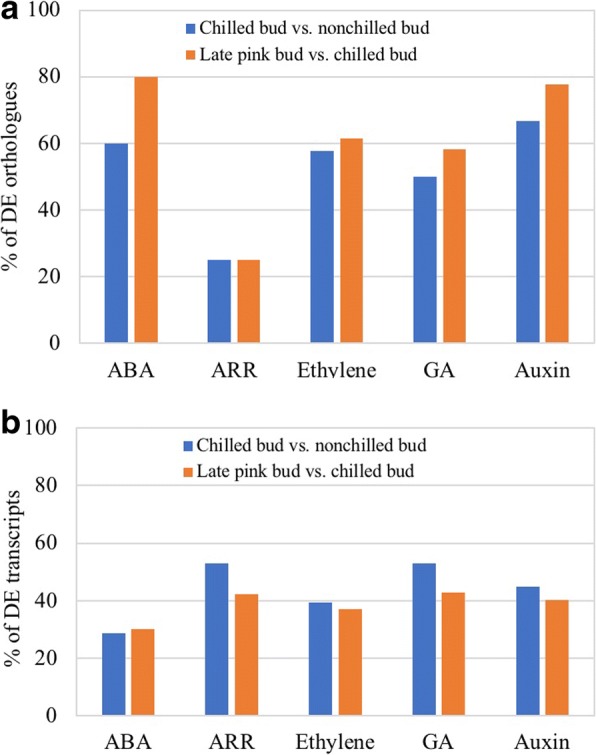
Response of phytohormone-related genes and transcripts in chilled flower buds (vs. nonchilled flower buds) and late-pink buds (vs. chilled flower buds). a Percentage of differentially expressed (DE) orthologues of A. thaliana genes (The number of DE A. thaliana genes ÷ total number of A. thaliana genes × 100). b Percentage of DE transcripts of transcripts of blueberry phytohormone-related genes (The number of DE transcripts ÷ total number of transcripts × 100)
Due to the tetraploid nature of ‘Legacy’, orthologues of each A. thaliana gene used for query often have more than one homologue (Fig. 2). Thus, an average of the changes (Log2Fold Change) for all the DE transcripts that are orthologues derived from a single A. thaliana query gene was used to represent the overall change of each phytohormone-related gene (Fig. 3). Increased expression of ABA1, ABA2 and NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 3 (NCED3) in the ABA biosynthesis pathway were seen in chilled flower buds. ABA1 and ABA2 continued to increase and NCED3 decreased in late-pink buds (Fig. 3). The increased expression of these orthologues indicates a potential increase in ABA biosynthesis during vernalization. Regardless of decreased ABA1 expression, increased NCED3 expression suggests that there is an increase in ABA biosynthesis during floral bud break (Fig. 3).
Fig. 3.
Average fold changes (Log2FC) of differentially expressed homologues for each of the phytohormone-related orthologue of A. thaliana in chilled flower buds (CB) [vs. nonchilled flower buds (NB)] and late-pink buds (LPB) [vs. chilled flower buds (CB)]. LogFC for chilled buds: Log2(CB/NB). LogFC for late-pink buds: Log2(LPB/CB). a Abscisic acid biosynthesis pathway genes [46]. b Ethylene biosynthesis and signaling pathway genes [45, 47]. c Gibberellin biosynthesis pathway genes [48]. d Two-component ARABIDOPSIS RESPONSE REGULATORS (ARR) [49, 50]. e Auxin biosynthesis pathway genes [51]. The bars represent standard deviation
DE orthologues of ethylene signaling pathway genes were all up-regulated in chilled buds and down-regulated in flowering buds (Fig. 3). These orthologues are considered regulators for freezing tolerance in A. thaliana. Indole-3-acetic acid (IAA) and GA biosynthesis pathway orthologues were up-regulated in chilled flower buds and decreased in late-pink buds (Fig. 3). GIBBERELLIN 3-BETA-DIOXYGENASE 2 (GA3OX2) in the GA pathway and AUX1 in the IAA pathway were major DEGs with high expression changes. The DE ARABIDOPSIS RESPONSE REGULATORS (ARRs) orthologues included two A-type, two B-type and five ARR-like genes in chilled buds and flowering buds. One B-type orthologue (ARR10) was suppressed only in chilled buds (Fig. 3).
Gene networks of DEGs in chilled flower buds and late-pink buds
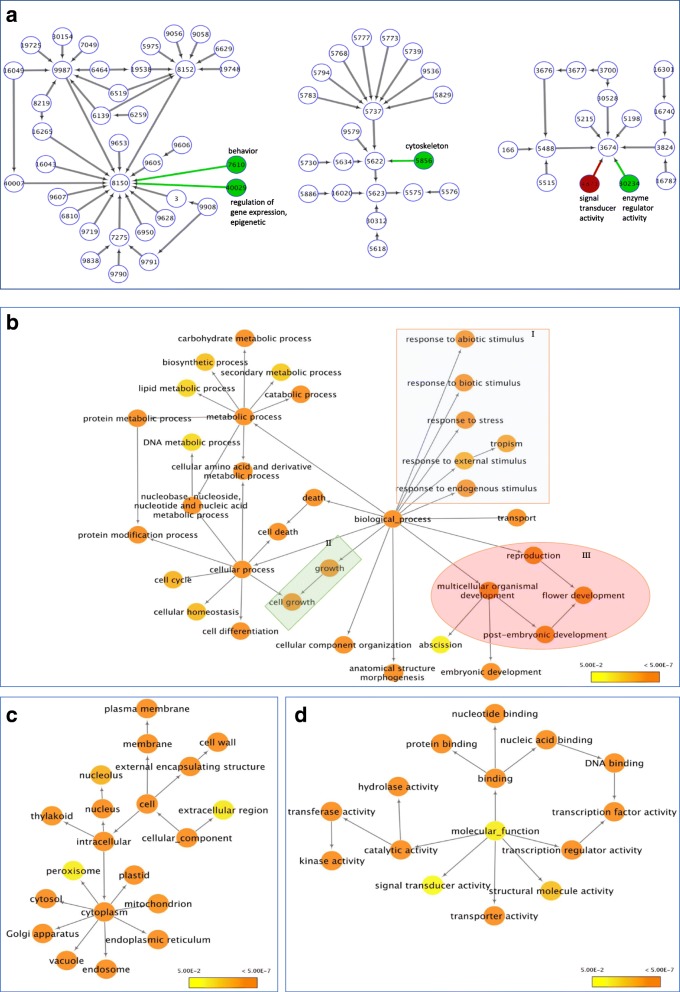
Over-represented Gene Ontology (GO) terms (P < 0.05) were grouped to visualize gene networks of the annotated DE transcripts using the GOslim_Plant as the selected GO file and A. thaliana annotation as the reference. The DE transcripts were classified in 70 and 73 over-represented GO terms for chilled flower buds and late-pink buds, respectively (Fig. 4). The over-represented GO terms for chilled flower buds and late-pink buds were identical except for two GO terms (Fig. 4), suggesting that the same transcripts responded to temperature changes in these buds. The difference in “biological_process” was two additional over-represented GO terms (GO:0007610-behavior and GO:040029-regulation of gene expression, epigenetic) found in late-pink buds but not in chilled flower buds. (Fig. 4).
Fig. 4.
Gene networks of differentially expressed genes (DEGs) in chilled flower buds and late-pink buds. DEGs in chilled flower buds were identified in comparison to nonchilled flower buds while DEGs in late-pink buds were identified in comparison to chilled flower buds. The ontology file of GOSlim_Plants in BiNGO was used to identify over-represented GO terms (P < 0.05). a Comparison of the gene network in chilled flower buds to late-pink buds; white nodes and black edges are present in both gene networks; red nodes and edges are present only in the chilled buds; and green nodes and edges are present only in the late-pink buds. The number in each circle is a GO identity number. A gene network in chilled flower buds (b “Biological_process” c “Cellular component” d “Molecular function”). I, II, and III in b show GO terms related to stress, plant growth, and reproduction, respectively
Over-represented GO terms in chilled flower buds revealed the impact of chilling on GO terms in three categories (Fig. 4). The over-represented GO terms in “biological_process” revealed that vernalization/chilling affected expression of genes in multiple GO terms related to growth, response to stress, and reproduction (Fig. 4). The gene network based on over-represented GO terms facilitate our understanding of the role DEGs in both chilled flower buds and late-pink buds (Fig. 4).
Validation of the expression of selected genes
In chilled flower buds and late-pink buds, qRT-PCR were used to validate DE transcripts of VcFD, VcTFL1, and VcARP6 (Fig. 5). The results suggested high-reliability of the RNA-seq data.
Fig. 5.
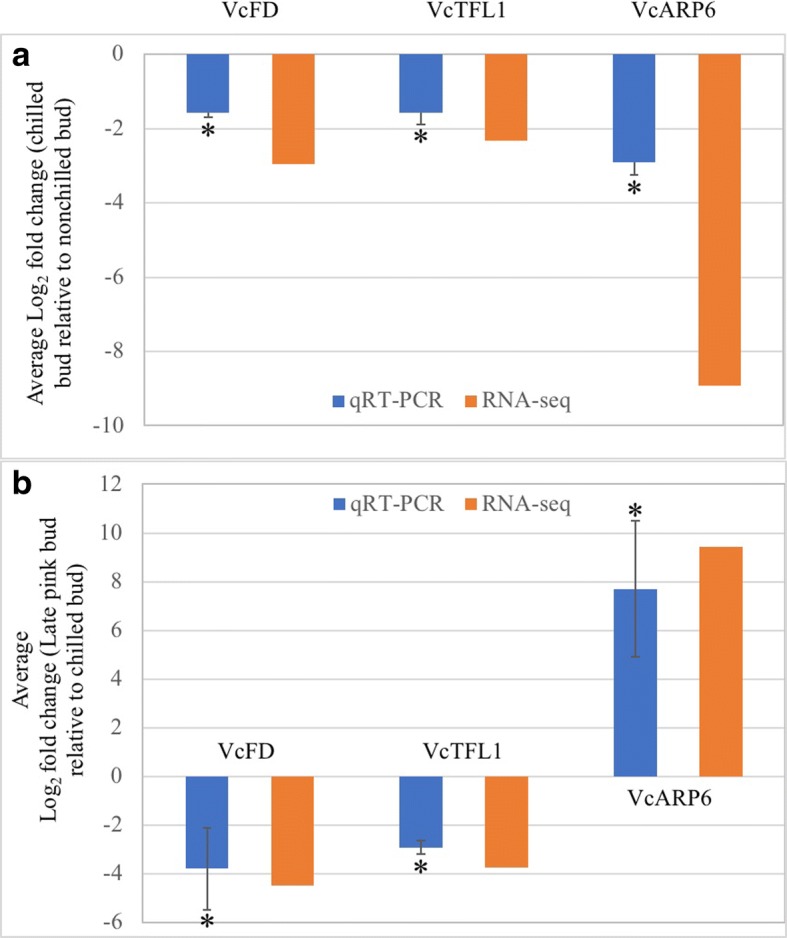
Comparison of RNA sequencing and qRT-PCR analysis of three differentially expressed genes in (a) chilled flower buds (compared to nonchilled flower buds) (b) and late-pink buds (compared to chilled flower buds). Eukaryotic translation initiation factor 3 subunit H is the internal control. Log2fold-change was calculated by -∆∆Ct = − [(CtGOI – Ctnom) tissue 1 – (CtGOI – Ctnom) tissue 2]. Average Log2fold-change ± standard deviation of three biological replicates. Significant average fold-change determined using a Student’s t-test is denoted. An asterisk (*) indicates p < 0.001
Discussion
Transcriptome analysis is an effective approach to study flowering pathway genes [23, 24]. Using this approach, DEGs in response to vernalization have been identified in Japanese pear (Pyrus pyrifolia Nakai) and oriental lily [24]. For blueberries, expressed sequence tags have been generated from blueberry flower buds [25]. However, comparative transcriptome analyses of different stage floral buds have not been documented.
The roles of VcFT, VcLFY and VcARP6 in vernalization-mediated blueberry flowering
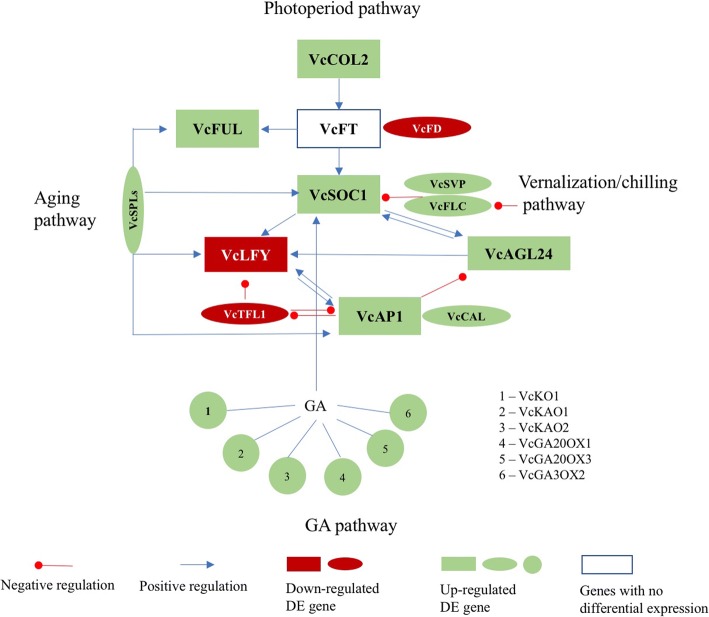
Overexpression of VcFT (expression level > 2000-fold in leaf tissues) resulted in precocious flowering [16, 17]. However, the high expression level did not completely reverse the need for chilling for normal plant flowering which suggests that chilling requirement is not replaceable by VcFT manipulation. When the results were aligned to the flowering pathway of A. thaliana [7], the acquired chill did not change VcFT expression in blueberry flower buds (Fig. 6). This result was also observed in woody pear but not in herbaceous lily [24]. The inactive VcFT expression in response to chilling may be one major reason that chilled flower buds remain dormant prior to exposure to bud-breaking temperatures since VcFT increased in late-pink buds (Table 1).
Fig. 6.
Response of major flowering pathway genes in chilled flower buds (compared to nonchilled flower buds). The relationships among the listed genes are drawn according to the diagram for A. thaliana by Fornara et al. 2010 [7], although not all DE genes of blueberry align perfectly with the correlations proposed for A. thaliana. All the listed genes in this diagram showed down-regulation in late-pink buds (compared to chilled flower buds) (Additional file 2: Table S1)
Overexpression of VcFT in leaves promoted expression of downstream genes VcSOC1, VcFUL, VcAP1, and VcLFY [17]. In this study, expression of VcSOC1, VcFUL, and VcAP1 were up-regulated but VcLFY was repressed regardless of VcFT expression in flower buds (Table 1: Fig. 5). The expression of VcFT-downstream genes was regulated independently of VcFT in chilled flower buds. Additionally, repressed VcLFY response in chilled flower buds is similar to results observed in grapevine (Vitis vinifera) [26]. These results suggest that VcLFY supression may play a role in chilling-mediated flowering by maintaining bud dormancy before bud break.
The interaction of FD and FT promotes flowering in A. thaliana while TFL1 is a negative regulator of FT [27, 28]. In this study, reduced VcFD and VcTFL1 expressions did not changed VcFT expression in chilled flower buds. However, increased VcFD and decreased VcTFL1 in late-pink buds were associated with increased VcFT. TFL1 was considered a repressor of both LFY and AP1 in A. thaliana until recent evidence suggest that TFL1 transcription was suppressed by AP1 but promoted by LFY [29, 30]. For chilled blueberry flower buds, increased expression of VcAP1 and decreased expression of VcLFY were associated with decreased expression of VcTFL1. This result is consistent with the recent report about the interactions among these three genes [30]. During flower bud break, repressed VcTFL1 associated with decreased VcLFY and VcAP1 supports the theory that TFL1 represses LFY [30]. Although some DEGs of flowering pathway genes in blueberry match the proposed interactions in A. thaliana [7], VcFD and VcTFL1 seem to be playing a more significant role in blueberry (Fig. 4).
FLC interacts with SVP and both are repressed during vernalization in A. thaliana [19]. In chilled blueberry flower buds, both VcFLC and VcSVP homologues showed decreased expression but increased in late-pink buds (Table 1). In A. thaliana, ARP6 activates FLC, MAF4, and MAF5, which are all repressors of plant flowering in vernalization pathway [31]. In blueberry, decreased VcARP6 in chilled flower buds was not associated with decreased expression of VcFLC, VcMAF4, or VcMAF5. However, increased VcARP6 (c49456_g2, Log2FC = 8.1) was associated with an increase in these genes in late-pink buds (Table 1; Additional file 2: Table S1). Due to ARP6’s role in A. thaliana vernalization, DE VcARP6 may significantly contribute to chilling-mediated flowering of blueberry flower buds.
Expression of blueberry MADS-box genes in chilled flower buds and late-pink buds
The major flowering pathway genes SOC1, FLC, AP1, FUL, SVP, and AGL24 are MADS-box genes encoding MIKCc (classical MIKC) proteins [7]. Similar to A. thaliana, multiple blueberry MADS-box genes are present and activated at different flowering stages. VcSOC1, VCAP1 and VcFUL are responsive to VcFT overexpression [17]. Additionally, constitutively expressed VcSOC1 or Keratin-like (K) domain of VcSOC1 promoted blueberry flowering [32]. In this study, the functional orthologues of FLC and AGL24 were not detected in blueberry, suggesting that the vernalization/chilling-mediated flowering pathway of blueberry is different from A. thaliana. VcSVP showed differential expression in chilled and late-pink buds (Table 1).
In woody fruit crops, functional FLC orthologues have not been identified. The peach DAM genes mimic FLC response in A. thaliana under dormancy [10]. The DAM genes are the orthologues of A. thaliana AGL24 and SVP genes [12, 33]. In this study, the DE DAMs showed similarity to several MADS-box genes (VcAP1, VcSVP, VcSOC1, and VcSPL3) (Table 1). Therefore, it is possible that the interaction of multiple MADS-box genes co-regulates chilling-mediated flowering in blueberry as well as other woody plants.
Response of phytohormone genes during vernalization and devernalization
Phytohormones are involved in plant flowering and dormancy. In A. thaliana, cold acclimation, dormancy, and plant flowering are affected by phytohormone gene expression [7, 34, 35]. The gibberellin pathway interacts with the flowering pathway through SOC1 [7, 36]. Ethylene signaling pathway genes are considered regulators for freezing tolerance in A. thaliana. In this study, DE phytohormone genes were identified in both chilled buds and late-pink buds of blueberry (Fig. 2; Additional file 3: Table S3). These DE phytohormone genes reveal their potential roles in cold acclimation, dormancy, freezing tolerance, and chilling-mediated flowering in blueberries. For example, chilled blueberry buds showed higher freezing tolerance than nonchilled buds and flower tissue [37]. Increased DE orthologues of ethylene genes in the chilled blueberry buds were responsible for the enhanced freezing tolerance in chilled buds while decreased expression of DE ethylene orthologues in late-pink buds reduced freezing tolerance (Fig. 3).
Conclusions
The changes from nonchilled to chilled and chilled to late-pink buds are associated with transcriptional changes in a large number of DE phytohormone-related genes and DE flowering pathway genes. The DE flowering pathway genes suggest that orthologues of FT, FD, TFL1, LFY, and MADS-box genes are the major genes involved in chilling-mediated blueberry bud-break. The DE phytohormone genes reveal the potential roles of phytohormone genes in cold acclimation, dormancy, freezing tolerance, and chilling-mediated flowering in blueberries. The results contribute to the comprehensive investigation of the chilling-mediated flowering mechanism in woody plants.
Method
Plant materials
The tetraploid southern highbush blueberry ‘Legacy’ needs over 800 chilling units (CU) for normal flowering. Twelve 4-year old ‘Legacy’ plants were obtained through micropropagation of in vitro cultured shoots. All plants were grown in 4-gal pots in a secured courtyard under natural light conditions at Michigan State University, East Lansing, Michigan (latitude 42.701847, longitude − 84.482170). The average low and high temperatures in January 2016 were − 11 °C and − 1.8 °C, respectively (http://www.usclimatedata.com/climate/east-lansing/michigan/united-states/usmi0248). In September 2015, six plants were moved to a heated greenhouse with a 12-h photoperiod and a minimum temperature of 23 °C in order to keep the plants from any chilling hour accumulation. The remaining six plants were kept in the secured courtyard. In November, three plants were selected from the greenhouse and 30–50 flower buds were harvested per plant. These flower buds did not receive any chilling temperatures and were labeled as nonchilled flower buds. At the end of January 2016, three plants were selected from the courtyard and 30–50 flower buds were harvested per plants. These flower buds experienced natural chilling conditions through mid-winter and were labeled as chilled flower buds. In April, 20–30 flower buds per plant were obtained from a second harvest of the same three plants in the courtyard. These flower buds experienced natural chilling conditions and began to flower in early spring. The buds selected were at early-pink-stage and were labeled as late-pink buds. All tissues collected were frozen immediately in liquid nitrogen and stored at − 80 °C. Three plants for each bud type were used as the three biological replicates for transcriptome analysis.
RNA preparation, sequencing, and de novo transcriptome assembly
Total RNA of each blueberry sample (from individual plants) was isolated from 200 mg of bud tissues using a separate CTAB method [38] and was purified using RNeasy Mini Kit (Qiagen, Valencia, CA, USA). All RNA samples were purified using On-Column DNase digestion with the RNase-free DNase Set (Qiagen). The integrity of the RNA samples was assessed using the Agilent RNA 6000 Pico Kit (Agilent Technologies, Inc., Germany). All samples had an RNA quality score greater than 8.0 prior to submission for sequencing (100-bp pair end reads) using the Illumina HiSeq2500 platform at the Research Technology Support Facility at Michigan State University (East Lansing, Michigan, USA). The FastQC program (www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to assess the quality of sequencing reads for the per base quality scores ranging from 30 to 40.
Differential expression analysis and transcriptome annotation
RNA-seq reads of three biological replicates for nonchilled, chilled, and late-pink buds were analyzed. Two technical replicates were sequenced for each biological replicate and were combined together for analysis. The paired reads, two sets for each biological replicate, were aligned to the transcriptome reference Reftrinity developed for ‘Legacy’ [17] and the abundance of each read was estimated using the Trinity command “align_and_estimate_abundance.pl”. The Trinity command “run_DE_analysis.pl --method edgeR” was used for differential expression analysis. The DE transcripts with false discovery rate (FDR) values below 0.05 were used for further analyses. Comparison of transcriptome in chilled to nonchilled flower buds resulted in DE transcripts/genes in chilled flower buds. Comparison of transcriptome in late-pink buds to chilled flower buds resulted in DE transcripts/genes in late-pink buds. DE transcripts in chilled buds were annotated using Trinotate_v2.0 (https://trinotate.github.io).
Gene network construction
Annotated transcripts were imported to Cytoscape 3.5.0 under BiNGO’s default parameters with selected ontology file ‘GOSlim_Plants’ and selected organism A. thaliana [39, 40].
Identification of the selected pathway genes
Representative protein sequences of selected genes of A. thaliana were downloaded from the TAIR server (https://www.arabidopsis.org/tools/bulk/sequences/index.jsp). The retrieved sequences were used to search the blueberry transcriptome reference (refTrinity) using the tblastn command of BLAST+. The resultant transcripts that show e-value lower than − 20 were used to screen the DE transcript list of nonchilled floral buds.
The blueberry floral genes identified in the previous study [17] were used to analyze flowering pathway genes. The pathway genes of major phytohormones (gibberellin [41], abscisic acid [42], cytokinin/Arabidopsis Responsive Regulator [43], indole-3-acetic acid [44], and ethylene [45]) in A. thaliana were retrieved from TAIR_10 server based on published gene identities (Additional file 3: Table S3). Additionally, sequences of A. thaliana MADS-box proteins were used to analyze blueberry MADS-box genes. Percentages of DE phyotohormone genes were calculated based either on the number of orthologues to A. thaliana genes or on the number of DE blueberry transcripts.
RT-PCR of DE transcripts
Reliability of DE genes or transcripts identified through RNA-seq was evaluated through qRT-PCR analysis of six selected transcripts (Additional file 4: Table S4). These transcripts are from the representative DE genes in auxin, ethylene, cytokinin, and gibberellin pathways. They have high fold changes (> 2) and sequence specificity (based on alignment result of different isoforms) for PCR amplification. Eukaryotic translation initiation factor 3 subunit H was the internal control (Additional file 4: Table S4).
The same RNA samples used for RNA-sequencing, including samples of three biological replicates, were used for cDNA preparation. Reverse transcription of RNA to cDNA was performed using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The resulting cDNA of one micro gram of RNA was diluted (volume 1: 4) in water and a 1 μl/sample (25 ng) was used for PCR reactions.
Integrated DNA Technologies, Inc. (https://www.idtdna.com/Primerquest/Home/Index) provided the online tool for primer design and synthesized the primers (Additional file 4: Table S4). Three qRT-PCR analyses were performed on an Agilent Technologies Stratagene Mx3005P (Agilent Technologies, Santa Clara, CA) using the SYBR Green system (Life Technologies, Carlsbad, CA). In each 25 μl reaction mixture, 25 ng of cDNA, 200 nM of primers, and 12.5 μl of 2× SYBR Green master mix were included. The reaction conditions for all primer pairs were 95 °C for 10 min, 40 cycles of 30 s at 95 °C, 60 s at 60 °C and 60 s at 72 °C, and followed by one cycle of 60 s at 95 °C, 30 s at 55 °C and 30 s at 95 °C. The specificity of the amplification reaction for each primer pair was determined by the melting curve. Transcript levels within samples were normalized to the eukaryotic translation initiation factor 3 subunit H. Fold changes were calculated by -∆∆Ct = − [(CtGOI – Ctnom) tissue 1 – (CtGOI – Ctnom) tissue 2] (n = 3).
Additional files
Table S2. DE MADS-box genes in chilled flower buds (CB) [vs nonchilled flower buds (NB)] and late-pink buds (LPB) (vs CB) in ‘Legacy’. LogFC for chilled buds: Log2(CB/NB). LogFC for late-pink buds: Log2(LPB/CB). Except #N/A (no differential expression), all the rest are DE genes. (XLSX 23 kb)
Table S1. DE floral genes in chilled flower buds (CB) [vs nonchilled flower buds (NB)] and late-pink buds (LPB) (vs CB) in ‘Legacy’. LogFC for chilled buds: Log2(CB/NB). LogFC for late-pink buds: Log2(LPB/CB). #N/A: no differential expression. (XLSX 140 kb)
Table S3. DE phytohormones in chilled flower buds(CB) [vs nonchilled flower buds (NB)] and late-pink buds (LPB) (vs CB) in ‘Legacy’. LogFC for chilled buds: Log2(CB/NB). LogFC for late-pink buds: Log2(LPB/CB). Except #N/A (no differential expression), all the rest are DE genes. (XLSX 401 kb)
Table S4. Primers used in this study. (DOCX 54 kb)
Acknowledgements
The authors would thank Dr. Wayne H. Loescher for reviewing this manuscript, Dr. Jeff Landgraf and Mr. Kevin Carr at Michigan State University Research Technology Support Facility for RNA sequencing. This research is partially supported by AgBioResearch Project GREEEN of Michigan State University (http://www.canr.msu.edu/research/plant-agriculture/project_greeen/).
Funding
This research was partially supported by AgBioResearch of Michigan State University (http://agbioresearch.msu.edu/programs/info/project_greeen).
Availability of data and materials
Our blueberry transcriptome reference Reftrinity has been deposited in GenBank (Accession number: SRX2728597). Datasets from the current study are available from the corresponding author on request.
Abbreviations
- ABA
abscisic acid
- DE
Differentially expressed
- FDR
False discovery rate
- GA
gibberellin
- GO
Gene ontology
- IAA
indole-3-acetic acid
- qRT-PCR
Quantitative reverse transcriptase polymerase chain reaction
Authors’ contributions
GS conceived and supervised the study; QC and GS conducted the experiments; GS analyzed the data and wrote the manuscript. Both authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1311-8) contains supplementary material, which is available to authorized users.
References
- 1.Anderson JV. Advances in plant dormancy. Switzerland: Springer International Publishing; 2015. [Google Scholar]
- 2.Zinn KE, Tunc-Ozdemir M, Harper JF. Temperature stress and plant sexual reproduction: uncovering the weakest links. J Exp Bot. 2010;61(7):1959–1968. doi: 10.1093/jxb/erq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ouellet F, Charron J-B. eLS. Chichester: John Wiley & Sons, Ltd; 2013. Cold acclimation and freezing tolerance in plants. [Google Scholar]
- 4.Chuine IC, Bonhomme M, Legave J-M, De Cortázar-atauri I, Charrier G, Lacointe A, Améglio T. Can phenological models predict tree phenology accurately in the future? The unrevealed hurdle of endodormancy break. Glob Chang Biol. 2016;22:17. doi: 10.1111/gcb.13383. [DOI] [PubMed] [Google Scholar]
- 5.Luedeling E, Girvetz EH, Semenov MA, Brown PH. Climate change affects winter chill for temperate fruit and nut trees. PLoS One. 2011;6(5):e20155. doi: 10.1371/journal.pone.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkinson CJ, Brennan RM, Jones HG. Declining chilling and its impact on temperate perennial crops. Environ Exp Bot. 2013;91:48–62. doi: 10.1016/j.envexpbot.2013.02.004. [DOI] [Google Scholar]
- 7.Fornara F, de Montaigu A, Coupland G. SnapShot: control of flowering in Arabidopsis. Cell. 2010;141(3):550, 550.e1–2. [DOI] [PubMed]
- 8.Greenup A, Peacock WJ, Dennis ES, Trevaskis B. The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann Bot. 2009;103(8):1165–1172. doi: 10.1093/aob/mcp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Higgins JA, Bailey PC, Laurie DA. Comparative genomics of flowering time pathways using Brachypodium distachyon as a model for the temperate grasses. PLoS One. 2010;5(4):e10065. doi: 10.1371/journal.pone.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielenberg DG, Wang Y, Li ZG, Zhebentyayeva T, Fan SH, Reighard GL, Scorza R, Abbott AG. Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet Genomes. 2008;4(3):495–507. doi: 10.1007/s11295-007-0126-9. [DOI] [Google Scholar]
- 11.Wang Y, Georgi LL, Reighard GL, Scorza R, Abbott AG. Genetic mapping of the evergrowing gene in peach [Prunus persica (L.) Batsch] J Hered. 2002;93(5):352–358. doi: 10.1093/jhered/93.5.352. [DOI] [PubMed] [Google Scholar]
- 12.Sasaki R, Yamane H, Ooka T, Jotatsu H, Kitamura Y, Akagi T, Tao R. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot. Plant Physiol. 2011;157(1):485–497. doi: 10.1104/pp.111.181982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jimenez S, Reighard GL, Bielenberg DG. Gene expression of DAM5 and DAM6 is suppressed by chilling temperatures and inversely correlated with bud break rate. Plant Mol Biol. 2010;73(1–2):157–167. doi: 10.1007/s11103-010-9608-5. [DOI] [PubMed] [Google Scholar]
- 14.Wilkie JD, Sedgley M, Olesen T. Regulation of floral initiation in horticultural trees. J Exp Bot. 2008;59(12):3215–3228. doi: 10.1093/jxb/ern188. [DOI] [PubMed] [Google Scholar]
- 15.Ehlenfeldt MK, Prior RL. Oxygen radical absorbance capacity (ORAC) and phenolic and anthocyanin concentrations in fruit and leaf tissues of highbus blueberry. J Agr Food Chem. 2001;49(5):2222–2227. doi: 10.1021/jf0013656. [DOI] [PubMed] [Google Scholar]
- 16.Song GQ, Walworth A, Zhao DY, Jiang N, Hancock JF. The Vaccinium corymbosum FLOWERING LOCUS T-like gene (VcFT): a flowering activator reverses photoperiodic and chilling requirements in blueberry. Plant Cell Rep. 2013;32(11):1759–1769. doi: 10.1007/s00299-013-1489-z. [DOI] [PubMed] [Google Scholar]
- 17.Walworth AE, Chai B, Song GQ. Transcript profile of flowering regulatory genes in VcFT-overexpressing blueberry plants. PLoS One. 2016;11(6):e0156993. doi: 10.1371/journal.pone.0156993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X, Walworth AE, Mackie C, Song GQ. Overexpression of blueberry FLOWERING LOCUS T is associated with changes in the expression of phytohormone-related genes in blueberry plants. Hortic Res. 2016;3:16053. doi: 10.1038/hortres.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song GQ, Gao X. Transcriptomic changes reveal gene networks responding to the overexpression of a blueberry DWARF AND DELAYED FLOWERING 1 gene in transgenic blueberry plants. BMC Plant Biol. 2017;17(1):106. doi: 10.1186/s12870-017-1053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mateos JL, Madrigal P, Tsuda K, Rawat V, Richter R, Romera-Branchat M, Fornara F, Schneeberger K, Krajewski P, Coupland G. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 2015;16:31. doi: 10.1186/s13059-015-0597-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents Vernalization by Short periods of cold. Plant Cell. 2003;15(5):1159–1169. doi: 10.1105/tpc.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen Z, Guo W, Li J, Lin H, He C, Liu Y, Zhang Q, Liu W. Comparative Transcriptomic analysis of Vernalization- and Cytokinin-induced floral transition in Dendrobium nobile. Sci Rep. 2017;7:45748. doi: 10.1038/srep45748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Liu X, Lu Y. Transcriptome comparison reveals key candidate genes in response to vernalization of oriental lily. BMC Genomics. 2016;17:664. doi: 10.1186/s12864-016-2955-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowland LJ, Alkharouf N, Darwish O, Ogden EL, Polashock JJ, Bassil NV, Main D. Generation and analysis of blueberry transcriptome sequences from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol. 2012;12:46. doi: 10.1186/1471-2229-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carmona MJ, Cubas P, Martinez-Zapater JM. VFL, the grapevine FLORICAULA/LEAFY ortholog, is expressed in meristematic regions independently of their fate. Plant Physiol. 2002;130(1):68–77. doi: 10.1104/pp.002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309(5737):1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- 28.Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309(5737):1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 29.Liljegren SJ, Gustafson-Brown C, Pinyopich A, Ditta GS, Yanofsky MF. Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell. 1999;11(6):1007–1018. doi: 10.1105/tpc.11.6.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goslin K, Zheng BB, Serrano-Mislata A, Rae L, Ryan PT, Kwasniewska K, Thomson B, O'Maoileidigh DS, Madueno F, Wellmer F, et al. Transcription factor interplay between LEAFY and APETALA1/CAULIFLOWER during floral initiation. Plant Physiol. 2017;174(2):1097–1109. doi: 10.1104/pp.17.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deal RB, Kandasamy MK, McKinney EC, Meagher RB. The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell. 2005;17(10):2633–2646. doi: 10.1105/tpc.105.035196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song GQ, Walworth A, Zhao DY, Hildebrandt B, Leasia M. Constitutive expression of the K-domain of a Vaccinium corymbosum SOC1-like (VcSOC1-K) MADS-box gene is sufficient to promote flowering in tobacco. Plant Cell Rep. 2013;32(11):1819–1826. doi: 10.1007/s00299-013-1495-1. [DOI] [PubMed] [Google Scholar]
- 33.Jimenez S, Lawton-Rauh AL, Reighard GL, Abbott AG, Bielenberg DG. Phylogenetic analysis and molecular evolution of the dormancy associated MADS-box genes from peach. BMC Plant Biol. 2009;9:81. doi: 10.1186/1471-2229-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi Y, Ding Y, Yang S. Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 2015;56(1):7–15. doi: 10.1093/pcp/pcu115. [DOI] [PubMed] [Google Scholar]
- 35.Kendall SL, Hellwege A, Marriot P, Whalley C, Graham IA, Penfield S. Induction of dormancy in Arabidopsis summer annuals requires parallel regulation of DOG1 and hormone metabolism by low temperature and CBF transcription factors. Plant Cell. 2011;23(7):2568–2580. doi: 10.1105/tpc.111.087643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Showk S, Ruonala R, Helariutta Y. Crossing paths: cytokinin signalling and crosstalk. Development. 2013;140(7):1373–1383. doi: 10.1242/dev.086371. [DOI] [PubMed] [Google Scholar]
- 37.Walworth AE, Rowland LJ, Polashock JJ, Hancock JF, Song GQ. Overexpression of a blueberry-derived CBF gene enhances cold tolerance in a southern highbush blueberry cultivar. Mol Breed. 2012;30(3):1313–1323. doi: 10.1007/s11032-012-9718-7. [DOI] [Google Scholar]
- 38.Zamboni A, Pierantoni L, De Franceschi P. Total RNA extraction from strawberry tree (Arbutus unedo) and several other woodyplants. Iforest. 2008;1:122–125. doi: 10.3832/ifor0465-0010122. [DOI] [Google Scholar]
- 39.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 41.Regnault T, Daviere JM, Heintz D, Lange T, Achard P. The gibberellin biosynthetic genes AtKAO1 and AtKAO2 have overlapping roles throughout Arabidopsis development. Plant J. 2014;80(3):462–474. doi: 10.1111/tpj.12648. [DOI] [PubMed] [Google Scholar]
- 42.Xiong L, Zhu JK. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003;133(1):29–36. doi: 10.1104/pp.103.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang D, Chen HC, Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002;129(2):500–515. doi: 10.1104/pp.005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Normanly J, Bartel B. Redundancy as a way of life - IAA metabolism. Curr Opin Plant Biol. 1999;2(3):207–213. doi: 10.1016/S1369-5266(99)80037-5. [DOI] [PubMed] [Google Scholar]
- 45.Wang KL, Li H, Ecker JR. Ethylene biosynthesis and signaling networks. Plant Cell. 2002;14(Suppl):S131–S151. doi: 10.1105/tpc.001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong T, Park Y, Hwang I. Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015;58:29–48. doi: 10.1042/bse0580029. [DOI] [PubMed] [Google Scholar]
- 47.Corbineau F, Xia Q, Bailly C, El-Maarouf-Bouteau H. Ethylene, a key factor in the regulation of seed dormancy. Front Plant Sci. 2014;5:539. doi: 10.3389/fpls.2014.00539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S. Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell. 2004;16(2):367–378. doi: 10.1105/tpc.018143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muller B, Sheen J. Advances in cytokinin signaling. Science. 2007;318(5847):68–69. doi: 10.1126/science.1145461. [DOI] [PubMed] [Google Scholar]
- 50.Greenham K, McClung CR. Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet. 2015;16(10):598–610. doi: 10.1038/nrg3976. [DOI] [PubMed] [Google Scholar]
- 51.Velasquez SM, Barbez E, Kleine-Vehn J, Estevez JM. Auxin and cellular elongation. Plant Physiol. 2016;170(3):1206–1215. doi: 10.1104/pp.15.01863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. DE MADS-box genes in chilled flower buds (CB) [vs nonchilled flower buds (NB)] and late-pink buds (LPB) (vs CB) in ‘Legacy’. LogFC for chilled buds: Log2(CB/NB). LogFC for late-pink buds: Log2(LPB/CB). Except #N/A (no differential expression), all the rest are DE genes. (XLSX 23 kb)
Table S1. DE floral genes in chilled flower buds (CB) [vs nonchilled flower buds (NB)] and late-pink buds (LPB) (vs CB) in ‘Legacy’. LogFC for chilled buds: Log2(CB/NB). LogFC for late-pink buds: Log2(LPB/CB). #N/A: no differential expression. (XLSX 140 kb)
Table S3. DE phytohormones in chilled flower buds(CB) [vs nonchilled flower buds (NB)] and late-pink buds (LPB) (vs CB) in ‘Legacy’. LogFC for chilled buds: Log2(CB/NB). LogFC for late-pink buds: Log2(LPB/CB). Except #N/A (no differential expression), all the rest are DE genes. (XLSX 401 kb)
Table S4. Primers used in this study. (DOCX 54 kb)
Data Availability Statement
Our blueberry transcriptome reference Reftrinity has been deposited in GenBank (Accession number: SRX2728597). Datasets from the current study are available from the corresponding author on request.