Significance
Glioblastoma (GBM) cells develop intrinsic or acquired insensitiveness to BET bromodomain inhibitors (BBIs) yet develop persistent BET protein dependency. Selective degradation of BET proteins by a next-generation chemical compound undermines the BET protein dependency and exerts superior antineoplastic effects over inhibition of BET bromodomain. Given the significant difference between bromodomain dependency and BET protein dependency in GBM cells, chemically induced degradation of BET proteins serves as a promising strategy to overcome anticipated clinical BBIs resistance.
Keywords: glioma, BRD2, BRD3, BRD4, E2F
Abstract
Competitive BET bromodomain inhibitors (BBIs) targeting BET proteins (BRD2, BRD3, BRD4, and BRDT) show promising preclinical activities against brain cancers. However, the BET protein-dependent glioblastoma (GBM)-promoting transcriptional network remains elusive. Here, with mechanistic exploration of a next-generation chemical degrader of BET proteins (dBET6), we reveal a profound and consistent impact of BET proteins on E2F1- dependent transcriptional program in both differentiated GBM cells and brain tumor-initiating cells. dBET6 treatment drastically reduces BET protein genomic occupancy, RNA-Pol2 activity, and permissive chromatin marks. Subsequently, dBET6 represses the proliferation, self-renewal, and tumorigenic ability of GBM cells. Moreover, dBET6-induced degradation of BET proteins exerts superior antiproliferation effects compared to conventional BBIs and overcomes both intrinsic and acquired resistance to BBIs in GBM cells. Our study reveals crucial functions of BET proteins and provides the rationale and therapeutic merits of targeted degradation of BET proteins in GBM.
Bromodomain and extraterminal (BET) family proteins are readers for histone lysine acetylation and key coactivators for oncogenic transcriptional programs in cancer (1). BET proteins (including BRD2, BRD3, BRD4, and BRDT) emerge as promising anticancer targets along with a growing list of competitive BET bromodomain inhibitors (BBIs) (e.g., JQ1, I-BET151, OTX015, and CPI203) (2–5). These BBIs block the acetyl-lysine binding activity of BET proteins and show broad anticancer effects. However, efficacy of BBIs is counteracted by emergence of both primary and acquired resistance in various types of cancers (6–8). Recent development of small molecules that redirect E3 ubiquitin ligases to degrade BET proteins shows promise for the treatment of leukemia, lymphoma, and breast cancer (9–11). Nevertheless, the efficacy of these BET protein degraders against BBI-resistant cancer cells remains to be explored.
Glioblastoma (GBM) is a therapy-refractory tumor with dismal prognosis that is awaiting new therapeutic innovations (12). Overexpression of BRD2 and BRD4 has been reported in human GBM (13). To date, despite heterogeneous dose responses across GBM cell lines, BBIs (JQ1, I-BET151, and OTX015) have been shown to trigger G1 cell cycle arrest and to retard xenografted tumor growth, partially through induction of p21 and inhibition of Bcl-xL and HOTAIR (4, 13–16). While BET proteins are important for gene expression, transcriptomic response of GBM cells to BBI and the genome-wide occupancy of BET proteins in these cells remain poorly studied. Therefore, the mechanism and the vulnerability of BET protein dependency in GBM cells need to be characterized further.
In this study, we report that GBM cells develop both intrinsic and acquired resistance to BBIs. By employing genetic and chemical approaches to selectively deplete BET proteins, we discovered a persistent growth dependency of GBM cells on BET proteins regardless of their sensitivities to BBIs. dBET6, a CRBN-dependent BET protein degrader, showed superior anti-GBM activities to conventional BBIs, as well as a strong capacity to overcome BBI resistance. dBET6-mediated depletion of BET proteins showed distinct transcriptional and cellular responses compared with bromodomain inhibition. Moreover, dBET6 exerted a profound impact on RNA-Pol2 function and histone modification. Integrative ChIP followed by sequencing (ChIP-seq) and RNA-sequencing (RNA-seq) analysis demonstrated E2F-dependent transcriptional program and cell cycle-related genes as primary downstream targets of BET proteins in both GBM cell lines and patient-derived GBM spheres. A core dBET6-responsive gene signature was identified further to be able to stratify pathological grades and patient prognosis in three independent glioma cohorts, strongly suggesting that glioma patients can benefit from therapeutic degradation of BET proteins. Thus, our studies not only provide therapeutic and mechanistic insights into the BET protein dependency of GBM, but also uncover targeted degradation of BET proteins as a promising alternative to overcome anticipated clinical BBI resistance.
Results
BET Proteins Are Functional Requisites for GBM Cell Growth.
Overexpression of BRD2, BRD3, and BRD4 (BRD2/3/4) is prevalent in human GBM samples (SI Appendix, Fig. S1A), while the level of BRDT remains low and unaltered in this disease. High expression of BRD2 and BRD4 predicts a worse prognosis for GBM patients (SI Appendix, Fig. S1B). To explore potential involvement of BRD2/3/4 in human GBM, we performed shRNA-mediated silencing of these genes and revealed their essential functions for GBM cell viability, anchorage-independent growth, and tumorigenicity (SI Appendix, Fig. S2 A–D). Surprisingly, the responsiveness of GBM cells to BET silencing was decoupled from their sensitivity to BBIs (e.g., JQ1, I-BET151, OTX015, and CPI203) (SI Appendix, Fig. S2 E–H).
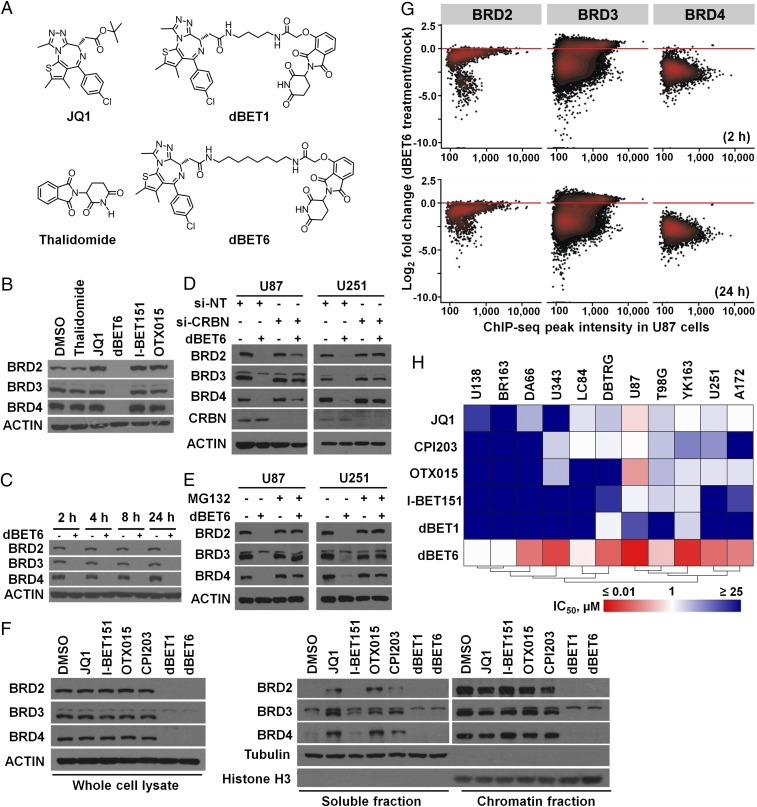
To address the aforementioned discrepancy, we employed dBET6, a compound developed from its molecular lead dBET1 (Fig. 1A) (10), to induce selective degradation of BET proteins in GBM cells. dBET6 degraded BRD2, BRD3, and BRD4 within 2 h via CRBN-dependent proteasomal pathway (Fig. 1 B–E), in contrast to BBI-induced displacement from chromatin (Fig. 1F). Interestingly, the protein level of CRBN tended to correlate positively with dBET6 sensitivity in GBM cells (SI Appendix, Fig. S3A). The cellular responsiveness to dBET6 was blunted by CRBN knockdown, which could be restored and enhanced upon overexpression of exogenous CRBN (SI Appendix, Fig. S3 B–F). Furthermore, disruption of the CRLCRBN complex via depletion of either RBX1 or DDB1 phenocopied the effect of CRBN knockdown (SI Appendix, Fig. S3 G and H), suggesting that the cellular activity of CRBN E3 ligase complex predisposes GBM cells to sensitivity to dBET6. ChIP-seq analysis revealed that dBET6 efficiently depleted the genome-wide chromatin occupancy of BRD2/3/4 (Fig. 1G). Comparative study among three BET degraders (dBET1, dBET6, and ARV-825) showed further that equimolar dBET6 was more efficient than the other two to reduce BRD4 (SI Appendix, Fig. S3I). We then screened and compared the antiproliferation efficacy of dBET6, dBET1, and BBIs against a panel of established and patient-derived GBM cell lines (Fig. 1H). Notably, although about half of the cell lines were intrinsically insensitive to both BBIs and dBET1 (IC50 > 10 µM), dBET6 exhibited submicromolar IC50 activities in 9 of 11 cell lines, suggesting that GBM cells are more susceptible to protein depletion than bromodomain inhibition of BRD2/3/4.
Fig. 1.
dBET6 induces efficient degradation of BET proteins and inhibits the proliferation of GBM cells. (A) Chemical structures of JQ1, dBET1, thalidomide, and dBET6. dBET6 was developed based on dBET1 by extending the linker. (B) dBET6 selectively depleted BET proteins. U87 GBM cells were treated with eqimolar concentrations (0.5 µM) of thalidomide, BBIs, and dBET6 for 24 h. (C) dBET6 depleted BET proteins within 2 h after treatment. (D) siRNA-mediated silencing of CRBN abolished dBET6-induced degradation of BET proteins. U87 cells were transfected with either si-NT or si-CRBN for 72 h before dBET6 treatment (0.5 µM, 2 h). (E) Proteasome inhibitor MG132 restored the expression of BET proteins in dBET6-treated cells. MG132 (10 µM) and dBET6 (0.5 µM) were added into culture medium for 2 h before cell harvest. (F) Effects of BBIs and BET protein degraders (dBET1 and dBET6) on the expression and chromatin loading of BRD2, BRD3, and BRD4. U87 cells were treated with indicated compounds (1 µM, 2 h) before chromatin fractionation. (G) Genome-wide depletion of BET protein occupancy as determined by time-course ChIP-seq of BRD2/3/4 in U87 cells treated with dBET6 (0.5 µM). Each point on the plot represents an individual peak of indicated BET protein. ChIP-seq peak intensity of BRD2/3/4 in mock-treated U87 cells was set as baseline control for comparison. (H) Hierarchical clustering of mean IC50 values of growth inhibition of four BBIs (JQ1, CPI203, OTX015, and I-BET151) and two BET degraders (dBET1 and dBET6) in 11 GBM cell lines. BR163, DA66, LC84, and YK163 were primary GBM explants. IC50 values were determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (96-h treatment), repeated independently at least three times.
The BET Degrader dBET6 Exerts Superior Anti-GBM Activities to BBIs.
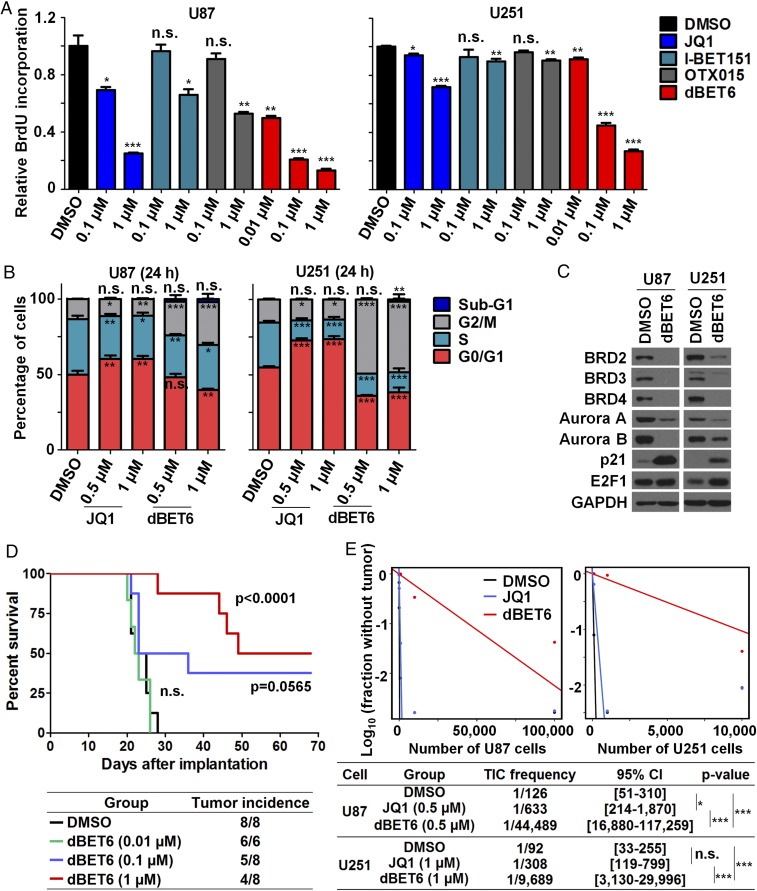
Next, we explored differential responses of GBM cells to dBET6 and BBIs. Exposure of GBM cells to dBET6 inhibited BrdU incorporation robustly and to a greater extent than BBIs (Fig. 2A). Different from BBI-induced G1/G0 arrest, dBET6 triggered a marked G2/M arrest of GBM cells, with concordant dysregulation of key cell cycle-related genes (Fig. 2 B and C and SI Appendix, Fig. S3J). However, 24-h treatment of dBET6 did not induce obvious sub-G1 population and apoptosis (Fig. 2B and SI Appendix, Fig. S3 K and L). Therefore, acute dBET6 treatment inhibits GBM cell viability mainly through its antiproliferation effect. Remarkably, short-term exposure of dBET6 elicited a profound impact on GBM tumor initiation in vivo. In an intracranial xenograft model, pretreatment of U87 cells with dBET6 for 24 h significantly prolonged murine survival and reduced tumor incidence in a dose-dependent manner (Fig. 2D). In vivo limiting dilution assay confirmed further the advantage of dBET6 over JQ1 to suppress the tumorigenic ability of GBM cells (Fig. 2E). These observations prompted us to examine the effect of dBET6 against tumor spheres (mixture of stem and progenitor cells) isolated from GBM patients (17, 18). dBET6 exerted a potent antiproliferative activity against GBM spheres and mitigated their sphere-forming efficiency (Fig. 3 A–C). Moreover, transcriptome profiling of NNI-24 tumor spheres after dBET6 treatment revealed a strong suppression of gene sets associated with stemness and neural stem cell proliferation (SI Appendix, Fig. S3M), suggesting that dBET6 inhibits both propagation and self-renewal of GBM initiating cells. Notably, 24-h exposure of patient-derived GBM spheres to dBET6 was able to induce durable antiproliferative effect on the intracranial tumor formation and expansion, resulting in prolonged survival of recipient mice (Fig. 3 D–G). Interestingly, BET proteins in orthotopic GBM xenografts can be down-regulated within 2 h post i.p. injection of dBET6 (SI Appendix, Fig. S3N), although intracranial concentration of dBET6 remained undetectable in healthy brain. Collectively, these data reveal the encouraging antiproliferative potential of dBET6 in GBM cells.
Fig. 2.
dBET6 exerts superior anti-GBM activities over BBIs. (A) dBET6 showed a stronger inhibitory effect on BrdU incorporation of GBM cells than BBIs; mean ± SD, n = 3. (B) Cell cycle analysis of GBM cells in response to dBET6 and JQ1 treatment; mean ± SD; n = 3. (C) Levels of cell cycle-related proteins in U87 (0.5 µM) and U251 (1 µM) GBM cells after dBET6 treatment (24 h). (D) Short-term dBET6 pretreatment impaired tumor formation of U87 cells in an intracranial model and prolonged nude mice survival postimplantation. U87 cells were treated with indicated concentration of dBET6 for 24 h before stereotaxic implantation. Log-rank test was applied for survival analysis; n = 6 or 8. (E) Effects of DMSO, JQ1, and dBET6 exposure (24 h) on GBM cell tumorigenicity in NOD/SCID gamma mice. Tumor-initiating cell (TIC) frequency was estimated by in vivo limiting tumor-formation assay. P values in E are shown to indicate the pair-wise differences in active cell frequency between groups; χ2 likelihood ratio test was applied. *P < 0.05; **P < 0.01; ***P < 0.001. n.s., not significant.
Fig. 3.
dBET6 inhibits the propagation and stemness of patient-derived gliomaspheres. (A) Relative cell viability of patient-derived gliomaspheres treated with the indicated concentrations of dBET6 for 5 d. IC50 values (mean, n = 3) of dBET6 are shown. (B) Levels of cell cycle-related proteins in gliomaspheres after dBET6 treatment (50 nM, 24 h). (C) In vitro limiting-dilution assay showing the effect of dBET6 treatment (14 d) on gliomasphere formation. Sphere-forming cell (SFC) frequency was estimated. P values in C (Lower) were shown to indicate the pair-wise differences in active cell frequency between groups; χ2 likelihood ratio test was applied. (D–G) Short-term dBET6 pretreatment impaired tumor formation of NNI-21 cells (D–F) and NNI-31 cells (G) in an intracranial model and prolonged the survival of recipient NOD/SCID gamma mice. GBM propagating cells were treated with indicated concentration of dBET6 for 24 h before stereotaxic implantation. Log-rank test was applied for survival analysis; n = 8. (D) H-score and (E) representative images showing immunohistochemistry staining of Ki67 signals in end point tumors harvested from individual animals in F. (Scale bar, 100 μm.) Data of D represent mean ± SD; n = 3. **P < 0.01; ***P < 0.001. n.s., not significant.
dBET6 Overcomes Acquired Resistance of GBM Cells to BBIs.
Acquired resistance to BBIs has been identified in various cancer types (6–8, 19). We showed that GBM cells developed adaptive tolerance to JQ1 after chronic exposure and became less sensitive to other BBIs (Fig. 4 A and B). Based on the observations from (−)-JQ1 (an inactive enantiomer of JQ1), the anti-GMB activity of JQ1 at a dose exceeding 10 µM was complicated by off-target effect other than BET bromodomain inhibition (SI Appendix, Fig. S4A). We hereby designated these GBM cells with acquired tolerance to JQ1 as “BBI-resistant cells”. BBI-resistant U87 (U87R) cells alleviated the JQ1-responsive transcriptional network and exhibited a distinct gene expression pattern compared with parental cells (Fig. 4C and SI Appendix, Fig. S4 B and C). Interestingly, genes within chromosome 5p15, a region showing concurrent copy number gain, were significantly up-regulated in U87R cells (SI Appendix, Fig. S4 D and E). No common gatekeeper or secondary mutation was identified between the two BBI-resistant lines (U87R and U251R) (SI Appendix, Table S1). Notably, we discovered that the growth of U87R and U251R cells still remained dependent on BRD2/3/4 expression. Depletion of BET proteins by either shRNAs or dBET6 robustly inhibited the proliferation of BBI-resistant cells (Fig. 4 D–F). Moreover, RNA-seq uncovered that JQ1 and dBET6 triggered distinct transcriptional responses in U87 cells (Fig. 4G) and that the impact of dBET6 on its target genes was largely unaffected by acquired BBI resistance (Fig. 4H). Together, these data demonstrate the strength of dBET6 to overcome acquired insensitiveness to BBIs by undermining the persistent BET protein dependency of GBM cells.
Fig. 4.
dBET6 and BET protein depletion overcome acquired resistance to BBIs in GBM cells. (A) Schematic diagram showing the establishment of JQ1-resistant U87 and U251 GBM cells (designated U87R and U251R, respectively) by chronic exposure to increasing doses of JQ1. (B) Resistance of U87R and U251R to JQ1, OTX015, and I-BET151. Cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (96-h treatment); mean ± SD, n ≥ 3. (C) Transcriptome responses of parental U87 cells and U87R cells to JQ1 treatment (1 µM, 24 h). JQ1-responsive genes (FDR < 0.1; absolute log2 fold change > 1.2) were identified based on parental cells. Their amplitudes of differential expression in parental and resistant cells were then compared using paired t test. (D) Responses of U87R, U251R, and their parental cells to shRNA-mediated silencing of BRD2/3/4. Cell viability was determined by MTT assay (96 h postseeding); mean ± SD, n = 3. All sh-BRD2/3/4 groups were significantly different from sh-NT control group (P < 0.001, Student’s t test). (E and F) Effects of dBET6 on cell viability (E) and BrdU incorporation (F) of U87R, U251R, and their parental cells; mean ± SD, n = 3. (G) Principle component (PC) analysis of parental U87 and U87R cells in response to DMSO, JQ1, and dBET6 treatment. After JQ1 withdrawal for 48 h, U87R cells were treated with indicated compounds (1 µM, 24 h). (H) Heat map showing the transcriptome responses of parental U87 cells and U87R cells to dBET6 treatment (1 µM, 24 h). Genes that were sensitive to dBET6 treatment (FDR < 0.1; absolute log2 fold change > 1.2) were identified in parental cells and used for plotting. *P < 0.05; ***P < 0.001. n.s., not significant.
dBET6 Blocks RNA-Pol2 Function and Active Histone Marks.
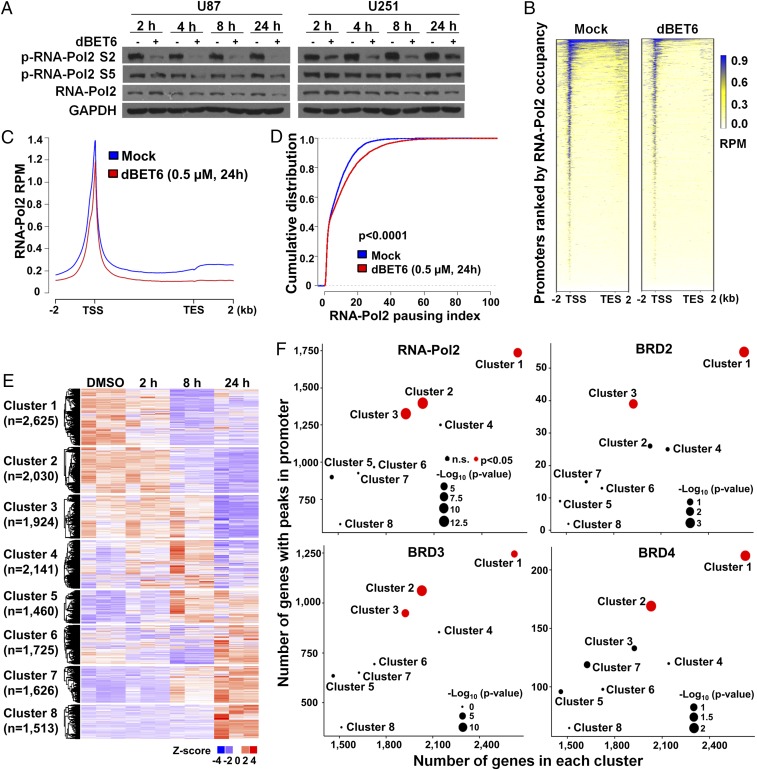
To gain insight into BET protein dependency of GBM cells, we assessed the consequence of dBET6-induced BET protein depletion on RNA-Pol2 function and epigenetic modifications. Recruitment of BET proteins to hyperacetylated chromatin potentiates active transcription (20, 21). Herein, we observed that dBET6 inhibited phosphorylation of RNA-Pol2 (especially Ser2 at the C-terminal domain) with a moderate decrease of total RNA-Pol2 (Fig. 5A). RNA-Pol2 ChIP-seq analysis revealed that dBET6 treatment reduced the average RNA-Pol2 occupancy in both promoter/transcription start site (TSS) and gene body, and resulted in a shift of the average RNA-Pol2 peak center toward the gene body (Fig. 5 B and C and SI Appendix, Fig. S5A). Further, dBET6 treatment substantially increased the RNA-Pol2 pausing indexes of genes whose promoter/TSS regions did not show drastic reduction of RNA-Pol2 binding (Fig. 5D). These data indicate that dBET6 inhibits both RNA-Pol2 loading and elongation. In parallel to defective transcription, dBET6 treatment reduced overall levels of active histone marks (H3K27ac and H3K4me3), whereas it moderately elevated the repressive histone marks (H3K27me3 and H3K9me3) (SI Appendix, Fig. S5B), implying that initial recruitment of BET proteins contributes to the maintenance of a permissive chromatin state.
Fig. 5.
dBET6 impairs RNA-Pol2 activity and BET protein-dependent transcription. (A) Effect of dBET6 treatment on RNA-Pol2 phosphorylation. U87 and U251 GBM cells were incubated with dBET6 (0.5 µM and 1 µM, respectively) for indicated durations. (B) Heat map for the ChIP-seq signal of RNA-Pol2 across the gene body in mock-treated and dBET6-treated U87 cells. The x axis was extended to 2 kb upstream of the TSS and 2 kb downstream of the transcription end site (TES). Color density reflects enrichment of ChIP signal. (C) Metagenes showing average RNA-Pol2 ChIP-seq signals across active RefSeq genes with basal RNA-Pol2 peaks in their promoter/TSS regions. Blue, mock treatment; red, dBET6 treatment. Units are mean tags per 20-bp bin per million reads (RPM) across the transcribed region of each gene with 2-kb upstream and downstream flanking regions. (D) Effect of dBET6 treatment on RNA-Pol2 pausing index. RefSeq genes showing no or moderate reduction of RNA-Pol2 signal in TSS region after dBET6 treatment were included in this analysis. Mann–Whitney U test was applied for statistical comparison of RNA-Pol2 pausing indexes under mock condition (shown in blue) and dBET6 treatment (shown in red). (E) Heat map showing the time-course transcriptomic response of U87 cells to dBET6 treatment (0.5 µM). RNA-seq was performed in triplicate, and data were subjected to model-based clustering analysis by MCLUST. (F) Enrichment analyses of genes with RNA-Pol2, BRD2, BRD3, and BRD4 peaks in their promoter/TSS regions in each cluster. Average frequency of each protein’s occupancy at the promoter/TSS of all clusters was used as reference. Clusters with P values smaller than 0.05 are highlighted in red; diameter of the circle represents the P value of Fisher’s exact test (one-sided).
dBET6 Impairs the Transcriptional Program Coactivated by BET Proteins and E2F1 in GBM Cells.
To determine BET protein-dependent transcriptional networks in GBM, we performed a time-course transcriptome analysis of vehicle- and dBET6-treated U87 cells (Fig. 5E). Genes with RNA-Pol2 and BET protein occupancy around their promoter/TSS regions (−1,000 bp to +200 bp from the TSS) were actively transcribed and subsequently down-regulated by dBET6 (Fig. 5F). Notably, genes with their promoters bound with BRD3; BRD2 and 3; BRD3 and 4; and BRD2, 3, and 4 were hypersensitive to dBET6 (SI Appendix, Fig. S6A). Gene set enrichment analysis (GSEA) showed a robust suppression of pathways related to mitotic cell cycle in both dBET6-treated U87 cells and NNI-24 gliomaspheres (SI Appendix, Fig. S6 B and C). In line with prior cell cycle analysis, genes with well-implicated functions in both mitotic cell cycle and gliomagenesis (e.g., AURKA, AURKB, and PLK1) were extensively repressed by dBET6 (SI Appendix, Fig. S6D) (22–24). We then sought to identify potential transcription factors cooperating with BET proteins. Genes with E2F binding motifs in their promoters were particularly sensitive to dBET6 treatment and were significantly down-regulated in both U87 cells and NNI-24 tumor spheres (Fig. 6A). In addition, we performed GSEA of multiple functionally defined E2F target gene sets in these cells and found that E2F targets were consistently suppressed after 24-h dBET6 treatment (SI Appendix, Fig. S7A). Importantly, we verified the E2F1 dependency of GBM cells by genetic silencing assays (SI Appendix, Fig. S7 B–D) (25). E2F1 deficiency strongly mitigated the soft-agar colony-forming and xenograft tumor-forming abilities of U87 cells. Earlier works have suggested that BRD2 physically interacts with E2F1 and that BRD4 cooperates with E2F1 in transcriptional activation (26–28). In this study, we found that dBET6 attenuated E2F1-dependent oncogenic transcription without inhibiting E2F1 protein expression (Figs. 2C and 3B), supporting the notion that BET proteins are transcriptional coactivators of E2F1 in GBM cells. ChIP-seq analysis unveiled a preferential E2F1 binding at active promoters with coenrichment of BET proteins, RNA-Pol2, and active histone marks (Fig. 6B). Similar to BET proteins, promoter-bound E2F1 was a robust marker positively correlated with both high basal expression and early responsiveness to dBET6 treatment (Fig. 6C and SI Appendix, Figs. S7 E and F and S8). Moreover, we refined a genuine set of promoter-bound E2F1 targets based on our E2F1 ChIP-seq and verified the significant down-regulation of E2F1 targets in both U87 and NNI-24 cells upon dBET6 treatment (Fig. 6D). Therefore, our data suggest that dBET6 primarily impairs the E2F1 and BET protein coactivated transcriptional program in GBM cells.
Fig. 6.
dBET6 inhibits E2F protein-dependent transcriptional networks. (A) Hyperenrichment of down-regulated genes with E2F binding motifs in their promoters [Molecular Signature Database (MSigDB) C3]. U87 and NNI-24 cells were treated with 0.5 µM and 50 nM of dBET6, respectively. (B) Heat maps for the ChIP-seq signals of indicated antibodies ±2 kb from TSS in U87 cells. (C) Enrichment analysis of genes with E2F1 peaks in their promoter/TSS regions in each cluster. Average frequency of E2F1 occupancy at the promoter/TSS of all clusters was used as reference. Clusters with P values smaller than 0.05 are highlighted in red; diameter of the circle represents the P value of Fisher’s exact test (one-sided). (D) GSEA plots of a ChIP-seq–defined E2F1 target gene set in the U87 and NNI-24 GBM cells treated with DMSO versus dBET6. (E) Heat map showing the differential expression of genes within dBET6-responsive gene signature in response to dBET6 treatment in U87 and NNI-24 cells. (F) The dBET6-responsive gene signature stratified glioma molecular subtypes, pathological grades, and patient survival in three clinical glioma databases (TCGA, REMBRANDT, and Gravendeel). Log-rank test was applied to compare the Kaplan–Meier survival curves. Hazard ratio (HR) from the Cox proportional hazards model was reported. (G) Effects of JQ1 and dBET6 treatment (1 µM, 24 h) on BET proteins, phosphorylation of RNA-Pol2, and the expression of mesenchymal master regulators in U87 cells.
dBET6-Responsive Gene Signature Shows Prognostic Potential.
Furthermore, to explore the role of dBET6-impaired transcriptional program in patient prognosis, we generated a core dBET6-responsive gene signature based on integrative analysis of ChIP-seq (H3K27ac, RNA-Pol2, E2F1, and BET proteins) and differential expression upon dBET6 treatment (Fig. 6E and SI Appendix, Fig. S9 A and B). Among these seven signature genes, ARL4C was characterized further as an important downstream target of dBET6 with both strong prognostic potential and unrecognized GBM-promoting function (SI Appendix, Fig. S9 B–F). By using the Connectivity Map analysis (29), gliomas with their transcriptional patterns matching the cellular responses to dBET6 treatment were identified as the dBET6-positive group. dBET6 positivity was anticorrelated with mesenchymal subtype, while it was strongly associated with proneural subtype, lower tumor grades, and longer patient survival in three independent cohorts (Fig. 6F and SI Appendix, Tables S2 and S3) (30–32), suggesting that glioma patients can benefit from therapeutic degradation of BET proteins. In line with the association between dBET6-responsive transcription and GBM cell plasticity, dBET6 treatment undermined the mesenchymal-specific core transcriptional regulatory network involving CEBPB, RUNX1, FOSL2, and STAT3 (Fig. 6G and SI Appendix, Fig. S9G) (33). Together, these data reveal an enhanced BET protein dependency of mesenchymal GBM cells and establish the dBET6-responsive gene signature as a promising tool to stratify pathological grades and prognosis in glioma.
Discussion
Our study reveals the significant difference between BET bromodomain dependency and BET protein dependency in GBM cells. We also report on a BET protein degrader, dBET6, and demonstrate in-depth its potency against GBM cells. Since GBM cells with either intrinsic or acquired insensitiveness to BBIs remain vulnerable to genetic silencing of BET genes and chemically induced degradation of BET proteins, we propose that targeting BET protein dependency can be a promising strategy to circumvent anticipated clinical BBI resistance in this disease.
Competitive inhibitors of BET bromodomains have been investigated actively in a broad spectrum of human malignancies, including GBM. In line with previous reports that JQ1, I-BET151, and OTX015 showed positive preclinical efficacy in GBM models with diverse genetic backgrounds (4, 13–16), our data support the antiproliferative activities of these BBIs in some GBM cell lines (e.g., U87 cells). These BBIs consistently triggered p21 expression and cell cycle arrest at G1 phase. Surprisingly, we found that over 70% of the GBM cell lines showed intrinsic insensitiveness to at least one BBI, which may challenge the clinical effectiveness of BBIs in GBM patients. Moreover, we demonstrated that GBM cells developed adaptive tolerance and cross-resistance to BBIs after chronic exposure. We did not observe recurrent mutations or copy number variations between the two GBM cells with acquired resistance to BBIs based on whole exome sequencing analysis. In light of the results from BBI-resistant leukemia and breast cancer (6, 7), our data suggest that multiple mechanisms may confer the acquired BBI resistance on GBM cells. Transcriptome profiling of U87 and U87R cells identified 1,096 genes that were differentially expressed [false discovery rate (FDR) of less than 0.1]. GSEA analysis showed that 5p15 amplicon was the only gene signature significantly up-regulated in U87R cells. Notably, 5p15 was also focally amplified (copy number = 4) in U87R cells, while its amplification in gliomas is infrequent (<2%) according to the TCGA-database merged cohort of lower-grade gliomas and GBM (n = 794, via cBio Portal). Since TERT expression has been shown to be inhibited by JQ1 in GBM cells (15), its elevated expression in U87R cells may contribute to the tolerance of BBI treatment. Although detailed mechanisms underlying the BBI resistance in GBM cells await further investigation, our observations strongly urge the development of alternative approaches to target BET proteins.
Inspired by the strong growth dependency of GBM cells on BET gene expression (SI Appendix, Fig. S2 A–E) (13, 15, 16), we hypothesize that GBM cells are more vulnerable to BET protein depletion than BET bromodomain inhibition. Indeed, GBM cells with either intrinsic or acquired BBI resistance still relied on the presence of BET proteins for continuous growth. Likewise, wild-type BRD4 has been shown to be essential for JQ1-resistant triple-negative breast cancer cells (6), supporting the notion that depletion of BET proteins overrides acquired BBI resistance. To test our hypothesis, we employed dBET6, a JQ1-phthalimide–moiety hybrid compound developed from its chemical lead dBET1 by extending the length of linker (Fig. 1A) (10). dBET6 induced efficient degradation of BET proteins via CRBN E3 ubiquitin ligase, which can be reversed by either CRBN knockdown or proteasome inhibitor. The expression and the activity of CRBN E3 ligase complex serve as a promising predictive biomarker for cellular responsiveness to dBET6. Concordant with the decrease in total protein levels, dBET6 treatment diminished the majority of BRD2/3/4 ChIP-seq signals within 2 h. This chemical approach helped to illustrate a high-confidence map of BRD2/3/4 occupancy across the GBM genome. Moreover, dBET6 displayed much higher activity to inhibit GBM cell viability as compared to dBET1 and BBIs. Importantly, we demonstrated the potential of dBET6 to overcome both primary and acquired BBI resistance, supporting that GBM cells are vulnerable to chemically induced degradation of BET proteins. Although the desirable pharmacodynamics parameters have been reported recently (34), dBET6 concentration was below the detectable level in the crude lysate of normal brain tissues from mice receiving i.p. injection (up to 50 mg/kg). Nevertheless, i.p. injection of dBET6 can down-regulate the level of BET proteins in orthotopic GBM xenografts, suggesting that a trace amount of dBET6 can penetrate the blood brain barrier and modulate the expression of downstream targets. More efforts are needed to improve the bioavailability and brain delivery of dBET6 in vivo.
Based on integrated analysis of RNA-seq and ChIP-seq data, BET protein targets and E2F1 targets are particularly sensitive to dBET6 treatment in GBM cells. Similarly, BRD4 has been reported as a coactivator for E2F1-driven oncogenic transcriptional programs in diffuse large B cell lymphoma (27). As E2F1 is essential for GBM cell growth, our study suggests preventing the coactivator binding as a strategy to restrain the oncogenic potential of E2F1. In addition, we showed that equimolar dBET6 was more efficient than other BET degraders (e.g., dBET1 and ARV-825) to reduce BRD4, MYC, and phospho-RNA-Pol2 (SI Appendix, Fig. S3I) (10, 11). Despite the common strategy of targeted degradation of BET proteins by chemical ligands, our study not only expands the molecular toolbox of BET degraders, but also provides mechanistic insight into the BET protein dependency in GBM.
Of note, dBET6 and BBIs exerted distinct impacts on GBM cells. Apart from increased antiproliferative potency, dBET6 showed unique activity to induce G2/M cell cycle arrest, and suppress the self-renewal and tumorigenic potential of GBM initiating cells. Remarkably, dBET6 showed extraordinary strength against patient-derived GBM spheres composed of GBM stem and progenitor cells, with IC50 values ranging from 3 to 40 nM. Similar to a recent report of BET degraders in triple-negative breast cancer (9), our study observed drastic transcriptomic changes of GBM cells exposed to dBET6. dBET6 and JQ1 elicited different transcriptomic responses in GBM cells. dBET6-responsive network was largely unaffected by acquired resistance to BBIs, indicating that acquired resistance to BET bromodomain inhibition does not counteract the activity of the BET protein degrader. Therefore, our data suggest the functional differences between BET protein depletion and BET bromodomain inhibition.
Collectively, our work illustrates the genomic occupancy of BET proteins and reveals the BET protein-dependent transcriptional program in GBM cells. We also demonstrate the superior anti-GBM efficacy of the BET protein degrader dBET6 over BBIs, which provides a rationale for, and mechanistic insight into, inducing ligand-dependent degradation of BET proteins as an alternative therapeutic approach to combat both primary and acquired resistance to BBIs. We anticipate that a similar strategy can be applied to additional drug targets with acquired insensitiveness yet persistent protein dependency.
Materials and Methods
Extended materials and methods are provided in SI Appendix, Materials and Methods. Patient-derived GBM spheres were established from tumor specimens obtained with written informed consent, as part of a study protocol approved by the SingHealth Centralised Institutional Review Board A and the National Healthcare Group Domain Specific Review Board A. All animal work was performed according to protocols approved by the National Neuroscience Institute at Tan Tock Seng Hospital and the National University of Singapore Institutional Animal Care and Use Committees. Unless otherwise stated, two-tailed Student’s t test was used to analyze the potential statistical difference between two groups; with *P < 0.05, **P < 0.01, and ***P < 0.001. Log-rank test was used for survival analysis.
Supplementary Material
Acknowledgments
We thank Nathanael S. Gray, Jinhua Wang, Tinghu Zhang, Lavina Tay, Shwu-Yuan Wu, and Cheng-Ming Chiang for reagent sharing; and Hazimah Binte Mohd Nordin for help with mouse work. We are grateful to Bing Ren, Sudhakar Jha, Fang Hu, and members of the H.P.K. laboratory for kind discussions and suggestions. This work is funded by the National Research Foundation Singapore under the Singapore Translational Research Investigator Award NMRC/STaR/0021/2014 (to H.P.K.); the Singapore Ministry of Education Academic Research Fund Tier 2 (MOE2013-T2-2-150 and MOE2017-T2-1-033); the Singapore Ministry of Health’s National Medical Research Council (NMRC) Centre Grant awarded to National University Cancer Institute of Singapore (NCIS), the National Research Foundation Singapore, and the Singapore Ministry of Education under its Research Centres of Excellence initiatives; and is additionally supported by a Seed Funding Program within the NCIS Centre Grant, an NCIS Yong Siew Yoon Research Grant through donations from the Yong Loo Lin Trust and philanthropic donations from the Melamed family, Aaron Eshman, and Valerie Baker Fairbank who deeply inspires our work. B.T.A. and C.T. are supported by an NMRC Translational & Clinical Research Flagship Programme Grant (NMRC/TCR/016-NNI/2016). D.L.B. was supported by the Damon Runyon Cancer Research Foundation as a Merck Fellow (DRG-2196-14). The computational work for this article was partially performed on resources of the National Supercomputing Centre, Singapore (https://www.nscc.sg).
Footnotes
Conflict of interest statement: J.E.B. is an employee, shareholder, and executive of Novartis Pharmaceuticals. J.E.B. is also a Scientific Founder of Tensha Therapeutics, C4 Therapeutics, Syros Pharmaceuticals, SHAPE Pharmaceuticals, and Acetylon Pharmaceuticals. D.L.B. is an employee of the Novartis Institutes for BioMedical Research. None of these relationships constitutes a conflict of interest for the present work. The remaining authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: RNA-seq, and ChIP-seq data generated in this study have been deposited in the NCBI Gene Expression Omnibus database (accession no. GSE99183). Whole exome sequencing data are available in the NCBI Sequence Read Archive database (accession no. SRP107365).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712363115/-/DCSupplemental.
References
- 1.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12:465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berenguer-Daizé C, et al. OTX015 (MK-8628), a novel BET inhibitor, displays in vitro and in vivo antitumor effects alone and in combination with conventional therapies in glioblastoma models. Int J Cancer. 2016;139:2047–2055. doi: 10.1002/ijc.30256. [DOI] [PubMed] [Google Scholar]
- 5.Moros A, et al. Synergistic antitumor activity of lenalidomide with the BET bromodomain inhibitor CPI203 in bortezomib-resistant mantle cell lymphoma. Leukemia. 2014;28:2049–2059. doi: 10.1038/leu.2014.106. [DOI] [PubMed] [Google Scholar]
- 6.Shu S, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529:413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fong CY, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rathert P, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525:543–547. doi: 10.1038/nature14898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai L, et al. Targeted degradation of BET proteins in triple-negative breast cancer. Cancer Res. 2017;77:2476–2487. doi: 10.1158/0008-5472.CAN-16-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter GE, et al. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: From molecular pathology to targeted treatment. Annu Rev Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 13.Pastori C, et al. BET bromodomain proteins are required for glioblastoma cell proliferation. Epigenetics. 2014;9:611–620. doi: 10.4161/epi.27906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F, et al. EGFR mutation promotes glioblastoma through epigenome and transcription factor network remodeling. Mol Cell. 2015;60:307–318. doi: 10.1016/j.molcel.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Z, et al. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastori C, et al. The bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci USA. 2015;112:8326–8331. doi: 10.1073/pnas.1424220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan X, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 18.Chong YK, et al. Cryopreservation of neurospheres derived from human glioblastoma multiforme. Stem Cells. 2009;27:29–39. doi: 10.1634/stemcells.2008-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurimchak AM, et al. Resistance to BET bromodomain inhibitors is mediated by kinome reprogramming in ovarian cancer. Cell Reports. 2016;16:1273–1286. doi: 10.1016/j.celrep.2016.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 22.Xia Z, et al. AURKA governs self-renewal capacity in glioma-initiating cells via stabilization/activation of β-catenin/Wnt signaling. Mol Cancer Res. 2013;11:1101–1111. doi: 10.1158/1541-7786.MCR-13-0044. [DOI] [PubMed] [Google Scholar]
- 23.Diaz RJ, Golbourn B, Shekarforoush M, Smith CA, Rutka JT. Aurora kinase B/C inhibition impairs malignant glioma growth in vivo. J Neurooncol. 2012;108:349–360. doi: 10.1007/s11060-012-0835-2. [DOI] [PubMed] [Google Scholar]
- 24.Lee C, et al. Polo-like kinase 1 inhibition kills glioblastoma multiforme brain tumor cells in part through loss of SOX2 and delays tumor progression in mice. Stem Cells. 2012;30:1064–1075. doi: 10.1002/stem.1081. [DOI] [PubMed] [Google Scholar]
- 25.Alonso MM, et al. ICOVIR-5 shows E2F1 addiction and potent antiglioma effect in vivo. Cancer Res. 2007;67:8255–8263. doi: 10.1158/0008-5472.CAN-06-4675. [DOI] [PubMed] [Google Scholar]
- 26.Peng J, et al. Brd2 is a TBP-associated protein and recruits TBP into E2F-1 transcriptional complex in response to serum stimulation. Mol Cell Biochem. 2007;294:45–54. doi: 10.1007/s11010-006-9223-6. [DOI] [PubMed] [Google Scholar]
- 27.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denis GV, Vaziri C, Guo N, Faller DV. RING3 kinase transactivates promoters of cell cycle regulatory genes through E2F. Cell Growth Differ. 2000;11:417–424. [PMC free article] [PubMed] [Google Scholar]
- 29.Lamb J, et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 30.Madhavan S, et al. Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7:157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gravendeel LA, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 32.Verhaak RG, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter GE, et al. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol Cell. 2017;67:5–18.e19. doi: 10.1016/j.molcel.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.