Significance
Generating mAbs against the native extracellular epitopes of multispanning membrane proteins is challenging, and as a result, few nonpeptidic mAbs against transporters have ever been isolated. Our approach here using virus-like particles and divergent host species for immunizations provides a means to overcome these challenges. The specific mAbs isolated here recognize native GLUT4 on the cell surface and can distinguish its different conformational states, thus representing some of the only state-specific mAbs ever isolated against any transporter. Epitope mapping of these mAbs revealed their binding sites as well as the mechanisms by which amino acids control the inward-open and outward-open states of GLUT4. Our studies demonstrate a valuable platform to isolate functional mAbs against important multispanning membrane proteins.
Keywords: lipoparticles, GLUT4, monoclonal antibody, chicken, phage display
Abstract
The insulin-responsive 12-transmembrane transporter GLUT4 changes conformation between an inward-open state and an outward-open state to actively facilitate cellular glucose uptake. Because of the difficulties of generating conformational mAbs against complex and highly conserved membrane proteins, no reliable tools exist to measure GLUT4 at the cell surface, follow its trafficking, or detect the conformational state of the protein. Here we report the isolation and characterization of conformational mAbs that recognize the extracellular and intracellular domains of GLUT4, including mAbs that are specific for the inward-open and outward-open states of GLUT4. mAbs against GLUT4 were generated using virus-like particles to present this complex membrane protein in its native conformation and using a divergent host species (chicken) for immunization to overcome immune tolerance. As a result, the isolated mAbs recognize conformational epitopes on native GLUT4 in cells, with apparent affinities as high as 1 pM and with specificity for GLUT4 across the human membrane proteome. Epitope mapping using shotgun mutagenesis alanine scanning across the 509 amino acids of GLUT4 identified the binding epitopes for mAbs specific for the states of GLUT4 and allowed the comprehensive identification of the residues that functionally control the GLUT4 inward-open and outward-open states. The mAbs identified here will be valuable molecular tools for monitoring GLUT4 structure, function, and trafficking, for differentiating GLUT4 conformational states, and for the development of novel therapeutics for the treatment of diabetes.
GLUT (SLC2A) proteins are a large family of glucose transporters, with 14 unique members identified in humans (reviewed in ref. 1). GLUT proteins are highly conserved, structurally complex molecules of ∼500 aa, each possessing 12 transmembrane (TM) domains, a large intracellular loop between helices VI and VII, and six small extracellular loops. The well-characterized glucose transporter isoforms GLUT1–4 have distinct regulatory and kinetic properties that reflect their specific roles in cellular and physiological glucose homeostasis. GLUT4 is one of the best-studied proteins of the GLUT family, reflecting its vital role in glucose metabolism, its complex mechanism of regulation by the hormone insulin, and the established links between disruption of GLUT4 regulation and prevalent human diseases, including diabetes and obesity (2).
GLUT4 is primarily expressed in adipose and muscle tissues. A complex network of signaling pathways and membrane-trafficking events downstream of the insulin receptor is responsible for the temporal regulation of GLUT4 expression at the plasma membrane. Under basal conditions, less than 1% of total GLUT4 is present at the plasma membrane, with the remainder localizing to intracellular organelles and GLUT4 storage vesicles. In response to insulin, GLUT4 is rapidly mobilized to the cell surface, where it increases the rate of glucose uptake into the cell, thereby contributing to the maintenance of acceptable glucose levels (3). Type 2 diabetes is, in part, defined by the inability of insulin to stimulate GLUT4 mobilization (insulin resistance).
Studying GLUT4 trafficking to the cell surface remains difficult, in part because of the scarcity of tools available to measure wild-type GLUT4 protein (3, 4). Translocation of GLUT4 to the plasma membrane has been studied primarily by using fluorescently tagged GLUT4. For example, by using an HA epitope tag inserted into the first external loop of GLUT4 and a GFP tag at the C terminus, the amount of HA tag exposed on the surface can be used to measure GLUT4 surface translocation (5, 6). Such approaches, although valuable, are limited to monitoring tagged exogenous proteins expressed in vitro. Antibodies that recognize endogenous GLUT4 have proven valuable as research tools (7–10), but all these antibodies recognize intracellular domains and linear epitopes of the protein (7), limiting their utility for studying GLUT4 on the cell surface.
Complicating its detection further, GLUT4 is known to cycle between several different conformational states that together facilitate glucose transit, including an inward-open state and an outward-open state (11). Pharmacological compounds such as cytochalasin B and phloretin can lock GLUT4 in each of these states, as can select point mutations such as E345Q and Y309I (12, 13). mAbs that can differentiate these states are not yet available for GLUT4—or for most other transporters—but state-specific mAbs have led to unprecedented insights into the structure, function, and signaling mechanisms of other membrane proteins, such as GPCRs and ion channels (14–16).
A key challenge in creating antibodies against the native extracellular regions of GLUT4 is maintenance of the native structure of the antigen for immunization and mAb isolation. Multispanning membrane proteins such as GLUT4 require the lipid membrane to maintain their structure, making antigen preparation difficult. Current commercial antibodies against GLUT4 all recognize short peptidic epitopes that are useful for applications such as Western blotting but are ineffective for applications that rely on natively folded epitopes, such as flow cytometry (17). Most successful approaches to raising mAbs that bind extracellular regions of multispanning membrane proteins have used the full-length protein in a membrane context by using whole cells, liposome-reconstituted protein, or DNA (18). Even then, however, the relatively small extracellular loops of GLUT4 (five of the six loops are 8–12 aa) form a small target antigen for immune reactivity. Compounding the difficulty of isolating mAbs, GLUT4 is highly conserved among mammals (95–97% homology), making traditional animal hosts poor candidates for immunization.
Here we report the isolation and characterization of conformational mAbs that recognize the extracellular and intracellular domains of GLUT4, including mAbs that are specific for both the inward-open and outward-open states of GLUT4. mAbs against GLUT4 were generated using virus-like particles (VLPs) to present this multispanning membrane protein in its native conformation, and a divergent host species (chicken) was chosen for immunization to overcome immune tolerance. The mAbs isolated and characterized (LM043, LM048, LM052, and LM059) recognize conformational epitopes on native GLUT4 within intact human cells and demonstrate remarkable specificity within the GLUT family, reacting with a mouse ortholog but not with most other members of the human membrane proteome. Biosensor analysis using intact GLUT4 demonstrated the high affinity of the mAbs, with the strongest apparent affinity displayed by LM048 (Kd < 1 pM), which also inhibited the function of GLUT4 and stained endogenous GLUT4 on 3T3-L1 adipocytes. Surprisingly, binding of mAbs LM043 and LM048 was dependent on the inward-open and outward-open conformational states of GLUT4, respectively. Epitope mapping of the mAbs, performed using shotgun mutagenesis alanine scanning across the 509 aa of GLUT4, identified distinct sets of GLUT4 residues critical for binding by each mAb. Also comprehensively identified were residues buried in the TM domains that control the GLUT4 inward-open and outward-open states. To the best of our knowledge, LM043 and LM048 are the only state-specific mAbs ever isolated against a glucose transporter, and most of the residues that functionally control GLUT4 conformational states have not been previously identified.
Results
Use of VLPs for Immunization and Phage Display to Isolate mAbs Against Native GLUT4.
To generate conformationally sensitive mAbs against GLUT4, we used murine leukemia virus (MLV)-based VLPs containing human GLUT4 (lipoparticles) for immunization. Retaining the native structure of multispanning membrane proteins during immunization (and later phage-panning steps) is vital for eliciting functionally relevant mAbs, which usually recognize conformational structures on the extracellular membrane protein face. VLPs are noninfectious lipid-enveloped retroviral particles that can present intact multispanning membrane proteins on their surface with native topology and conformation (19–21). VLPs can capture high levels of structurally intact GPCRs, ion channels, and transporters when the viral particles bud from the plasma membrane and are potent immunogens due to their particulate structure and multivalent epitope organization (22, 23). The level of incorporation of GLUT4 into the VLPs was ∼300 pmol/mg total protein. For comparison, commercial membrane preparations (e.g., from PerkinElmer or Millipore-Sigma) generally contain 1–10 pmol/mg, and intact cells generally contain 0.1–1 pmol/mg, so the VLPs concentrated GLUT4 ∼10–100 fold (24, 25).
The conservation of GLUT4 informed our choice of chickens as the host for immunization with human GLUT4 (hsGLUT4) VLPs. GLUT4 is highly conserved among mammals, with hsGLUT4 sharing 95–97% sequence identity with mouse, rat, and rabbit orthologs (26). Generating antibodies against highly conserved proteins is difficult due to immune tolerance, which severely limits both the magnitude of the humoral response and the diversity of epitopes recognized. In contrast, birds have a large evolutionary distance from mammals and so are capable of generating immune responses to human proteins that are highly conserved in mammals. Specifically, chickens lack a GLUT4 ortholog (27, 28), and their closest paralog is GLUT1, which is only 65% identical to hsGLUT4, making them an excellent host for antibody generation. Also, chicken Ig (IgY) is highly similar to mammalian IgG but with only a single variable heavy chain (VH) and variable light chain (VL) framework, making phage library creation and downstream humanization even easier than for murine antibodies (29, 30).
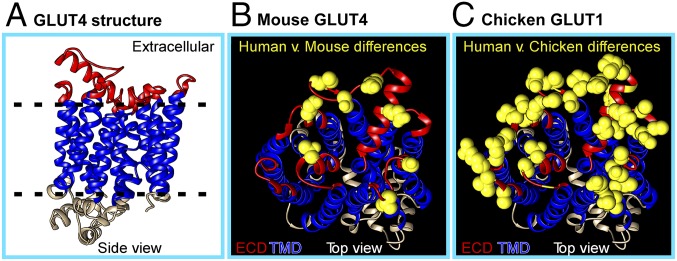
Although the structure of GLUT4 has not been solved, the crystal structure of hsGLUT1 (65% identity) (31) allows prediction of the position and size of the GLUT4 extracellular loops (Fig. 1A). GLUT4 has six predicted extracellular loops, all small (<12 aa) except for loop 1 (32 aa). A comparative alignment of the 80 amino acids in the extracellular regions of hsGLUT4 showed nine amino acid differences with mouse GLUT4 (mmGLUT4) but 29 amino acid differences with chicken GLUT1 (Fig. 1 B and C). Extracellular loop 1 has only four amino acid differences with mmGLUT4, but there are 11 amino acid differences with chicken GLUT1, as well as a 4-aa deletion, increasing the potential diversity of epitopes.
Fig. 1.
Homology-based model of hsGLUT4 structure. (A) hsGLUT4 is a 12-TM domain protein with both its C and N termini located in the cytosolic region. A homology-based structural model of hsGLUT4 was generated from the crystal structure of hsGLUT1 (PDB ID code 4pyp) by using Phyre2 to visualize antibody-accessible extracellular domains (in red). The six extracellular loops of hsGLUT4 are predicted to comprise 32, 8, 12, 11, 8, and 9 aa, respectively. (B) The extracellular amino acid residues on hsGLUT4 that are divergent in hsGLUT1and mmGLUT4 are shown in yellow. (C) The extracellular amino acid residues on hsGLUT4 that are divergent in hsGLUT4 and chicken GLUT1 (the closest ortholog) are shown in yellow.
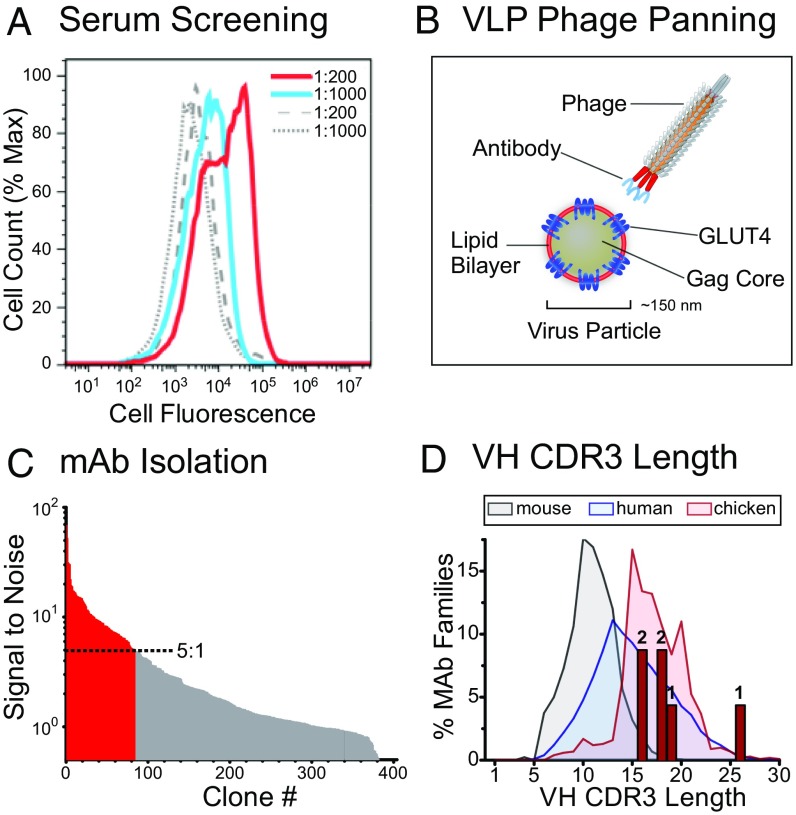
Based on this analysis, we selected chickens for immunization with hsGLUT4 VLPs to generate the most robust immune response against hsGLUT4. The sera of immunized chickens were screened by flow cytometry for reactivity with hsGLUT4 expressed in the avian cell line QT6 (Fig. 2A). B cells were collected from the animal with the most reactive serum, and antibody VH and VL gene fragments were amplified by PCR and used to construct a diverse (2 × 109) Ig single-chain variable fragment (scFv) library for phage display. The phage display library was panned in three consecutive rounds using hsGLUT4-VLPs (GLUT4 lipoparticles) (Fig. 2B), resulting in 364 reactive phage clones. Screening of these clones by ELISA identified 81 clones that bound to GLUT4 with a signal-to-background ratio of at least 5:1 (Fig. 2C). DNA sequencing of these clones and alignment of the VH genes identified 29 unique clones falling into six unrelated sequence families. Based on their sequence diversity and initial binding properties, four clones (LM043, LM048, LM052, and LM059) were selected for further characterization and were subcloned into a human IgG1 Fc fragment vector for mAb production and characterization. A fifth clone (LM040) was used in selected experiments as a control; the other clones isolated were not further characterized. LM052 and LM059 were from the largest VH sequence family and contained identical VH sequences but different VL genes. Interestingly, for the six families of isolated mAbs, the lengths of heavy chain hypervariable complementarity-determining region 3 (H-CDR3), the primary determinant of specificity for most mAbs, ranged from 16 to 26 aa (Fig. 2D and Table 1), reflecting the longer length of most chicken H-CDR3s compared with mouse or human (89% of chicken H-CDR3 sequences are between 15 and 23 residues) (32).
Fig. 2.
GLUT4 VLPs enable immunization and phage display for GLUT4-reactive mAbs. (A) Screening of serum from a GLUT4 VLP-immunized chicken for reactivity. QT6 cells expressing GLUT4 or a negative control were treated with serum samples followed by biotinylated bovine anti-IgY antibody and streptavidin PerCP and were analyzed by flow cytometry. Serum reactivity fluorescence profiles against hsGLUT4-expressing cells at 1:200 and 1:1,000 serum dilutions are indicated by red and blue traces, respectively. Serum reactivity fluorescence profiles against negative control-expressing cells are shown as dashed and solid gray lines at 1:200 and 1:1,000 serum dilutions, respectively. (B) A phage scFv library was created using B cells from a chicken with the highest reactive serum. The resulting phage library was used to pan against VLPs displaying the native form of hsGLUT4 on their surface. (C) scFvs isolated from the phage clones were tested for hsGLUT4 target selectivity in 384-well ELISA plates coated with hsGLUT4 VLPs or with null VLPs (VLPs produced without a specific receptor) as a negative control. Individual clones displaying more than a 5:1 signal-to-noise ratio for hsGLUT4 are shown as red bars, while the clones with a <5:1 ratio are shown as gray bars. Twenty-nine of these positive clones represented unique H-CDR3 sequences. (D) The distribution of murine, human, and chicken H-CDR3 length is shown, as reported in ref. 32. The six families of Glut4 mAbs isolated here are plotted by their H-CDR3 length (red bars). Numbers above each bar indicate the number of mAb families represented.
Table 1.
Key GLUT mAb characteristics
| Property | LM043 | LM048 | LM052 | LM059 |
| Apparent affinity | Not determined | 1.0E-12M | 1.8E-9M | 5.6E-10M |
| VH CDR3 length | 18 aa | 26 aa | 16 aa | 16 aa |
| hsGLUT4 binding | Yes | Yes | Yes | Yes |
| mmGLUT4 binding | Yes | Yes | Yes | Yes |
| hsGLUT1 binding | Yes | No | No | No |
| Epitope topology | Cytoplasmic | Extracellular | Extracellular | Extracellular |
| Epitope conformation | Inward-open | Outward-open | All | All |
| Epitope domain | ICL3, ICL5 | ECL4, ECL6 | ECL1 | ECL1 |
| Epitope critical residues | R265, G414 | E315, G446 | L61, G65, P66 | G65 |
Identification of mAbs That Bind Selectively to hsGlut4 and mmGLUT4.
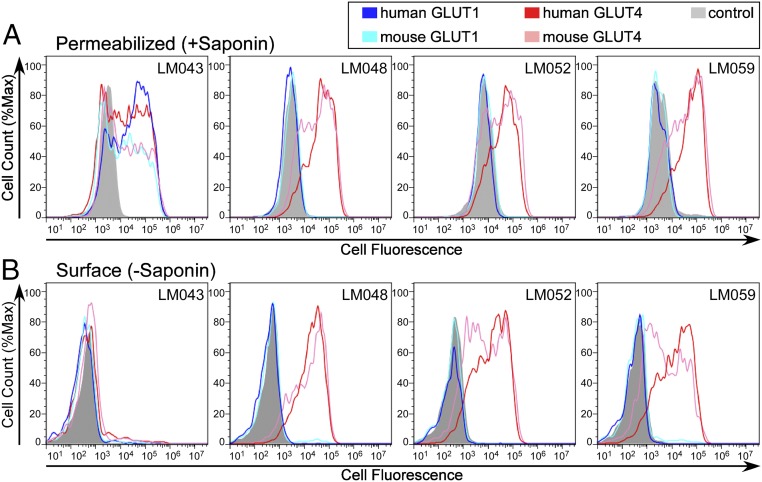
To confirm the reactivity of the mAbs against native GLUT4 on human cells, we first assessed mAb binding by flow cytometry after permeabilizing HEK-293T cells transfected with GLUT4. All four mAbs demonstrated strong reactivity with GLUT4 (Fig. 3A). All mAbs also demonstrated reactivity with mmGLUT4, which is important because murine cell lines such as 3T3-L1 adipocytes are routinely used to study GLUT4 trafficking. mAb LM048 reacted with endogenous GLUT4 on the surface of 3T3-L1 adipocytes in an insulin-dependent manner (SI Appendix, Fig. S1A), and mAbs LM043, LM048, and LM059 demonstrated various levels of reactivity with endogenous GLUT4 within permeabilized 3T3-L1 adipocytes (SI Appendix, Fig. S1B).
Fig. 3.
Reactivity of selected mAb clones against intracellular and surface GLUT4. Flow cytometry histograms for saponin-permeabilized (A) or surface-stained (B) HEK-293T cells expressing hsGLUT4, mmGLUT4, hsGLUT1, mmGLUT1, or a negative control protein and stained with one of four mAbs (LM043, LM048, LM052, or LM059). All GLUT proteins tested here contained a C-terminal myc epitope tag that was used to verify expression.
Reactivity was next assessed against surface-expressed hsGLUT4 and mmGLUT4. Three of the four tested mAbs (LM048, LM052, and LM059) were highly reactive against hsGLUT4 and mmGLUT4 on the cell surface (Fig. 3B). The remaining mAb, LM043, exhibited minimal reactivity against hsGLUT4 or mmGLUT4 on the surface of nonpermeabilized cells but reacted strongly when cells were permeabilized using saponin, suggesting binding to an intracellular domain of GLUT4. To confirm that LM043 recognizes an intracellular epitope, transfected cells were labeled with propidium iodide (PI), a nuclear-staining reagent that is not permeant to live cells, to differentiate naturally permeabilized (dead) and nonpermeabilized (live) cell populations. PI staining confirmed that LM043 binding to GLUT4 occurs exclusively in the permeabilized cell population (SI Appendix, Fig. S2, PI+ population). As positive controls, a commercial mAb (1F8) that binds an intracellular peptide on GLUT4 (33) and another mAb that we isolated (LM040) that binds to a similar intracellular epitope also bound exclusively to the naturally permeabilized (PI+) cell population. These results indicate that LM043 binds an intracellular epitope of GLUT4. Although the use of VLPs biases antibody discovery toward extracellular epitopes, VLPs that break open clearly allow the discovery of antibodies against intracellular epitopes as well.
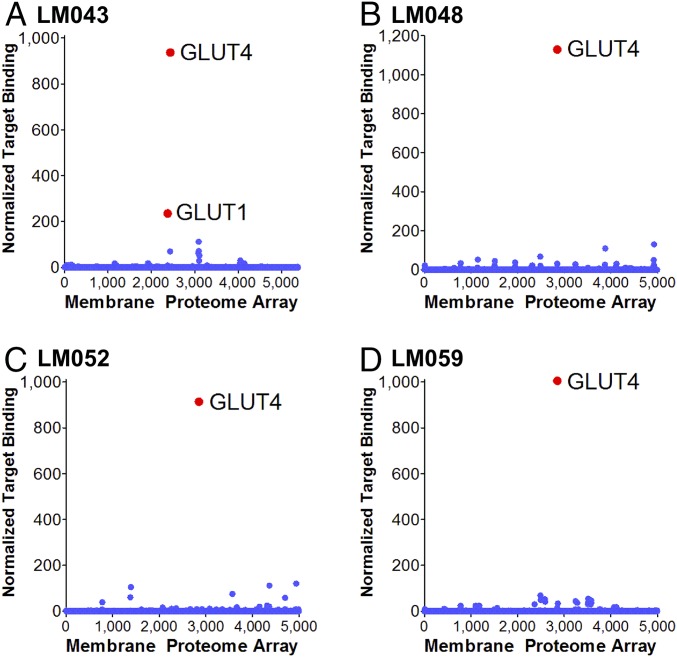
We next tested the specificity of the mAbs against other GLUT family members. mAbs LM048, LM052, and LM059 demonstrated no cross-reactivity with hsGLUT1, hsGLUT2, hsGLUT3, or mmGLUT1 (Fig. 3 and SI Appendix, Fig. S3), representing the GLUT family members with the highest homology to hsGLUT4. mAb LM043 was able to bind hsGLUT1 and mmGLUT1 (Fig. 3A), which can be explained by a common epitope (discussed below). To further determine if these mAbs displayed cross-reactivity with any other human membrane proteins, the binding specificities of LM043, LM048, LM052, and LM059 were tested for reactivity against an array of 4,571 human membrane proteins expressed in cells (the membrane proteome array; MPA) (Fig. 4), which includes GLUT1 to GLUT13. Each antibody was added to the MPA, and binding across the protein library was determined by flow cytometry. Three mAbs (LM048, LM052, and LM059) demonstrated very high specificity for GLUT4, with binding to GLUT4 at levels that were 10- to 100-fold above the other membrane proteins tested. LM043 bound to both GLUT4 and GLUT1 but did not bind well to any other membrane proteins. Interestingly, the very low-level reactivity of some mAbs with other membrane proteins can be explained by a common epitope (discussed below).
Fig. 4.
Membrane proteome-wide specificity testing of GLUT4 mAbs. The Integral Molecular Membrane Proteome Array (MPA) was used to determine the binding specificity of LM043 (A), LM048 (B), LM052 (C), and LM059 (D) across 4,571 different human membrane proteins expressed in duplicate in a cell-based array. Two hundred eight positive (Fc-binding) and negative (empty vector) control wells were also included in the screen. Antibodies were tested at a concentration of 1 μg/mL, followed by detection with flow cytometry using a fluorescently labeled secondary antibody. The resulting binding values are normalized to yield a single quantitative value for binding of the antibody against each target protein (normalized target binding).
mAbs Bind with Picomolar to Nanomolar Affinities to Native GLUT4.
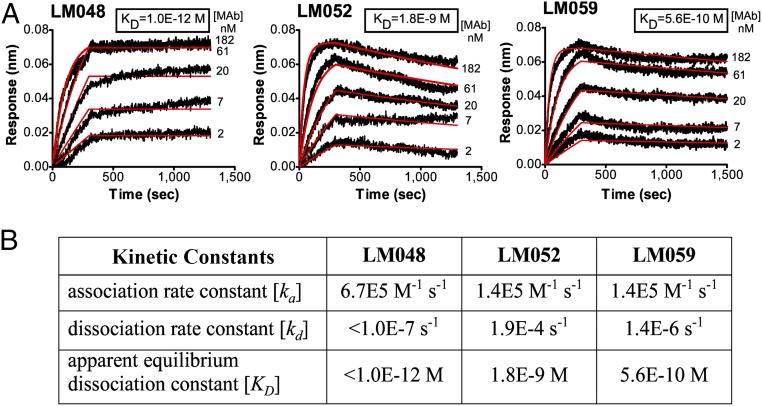
To further characterize the binding activities of the selected GLUT4 mAbs, the kinetics of mAb binding to conformationally native GLUT4 were assessed using biosensor analysis. For this, biotinylated hsGLUT4-VLPs were immobilized onto biosensor tips, and the binding affinity and kinetics of mAbs LM048, LM052, and LM059 were measured using biolayer interferometry (Fig. 5A). mAb titration experiments using bivalent scFv-Fc clones of each mAb (which results in apparent kinetics) demonstrated that LM048, LM052, and LM059 all have strong binding affinity to GLUT4, characterized by rapid association and slow dissociation (Fig. 5B). Of the three mAbs, LM048 exhibited the strongest apparent binding affinity to GLUT4 (Kd < 1 pM), primarily due to its extremely slow dissociation rate. The binding kinetics of LM043 could not be measured because it binds an intracellular epitope and is therefore inaccessible in immobilized VLPs that contain an intact lipid bilayer.
Fig. 5.
Isolated GLUT4 mAbs bind with picomolar to nanomolar apparent affinity. (A) Binding of LM048, LM052, and LM059 to native hsGLUT4 was detected by using biotinylated VLPs immobilized on biosensor tips. Kinetics of mAb binding to hsGLUT4 was assessed by fitting data to a 1:1 binding model to determine the rate constants. mAb titration experiments used bivalent scFv-Fc clones of each mAb, which results in apparent kinetics. Black curves represent the raw data curves of mAb binding, and red curves are fitted traces. For LM048, the experiment was run out to 45 min without measurable dissociation. (B) Table showing the association rate constant (ka), dissociation rate constant (kd), and apparent equilibrium dissociation constant (KD) for LM048, LM052, and LM059.
GLUT4 mAbs LM043 and LM048 Are Conformational and State Dependent.
Because conformationally intact GLUT4 was used for mAb isolation, we tested whether the resulting mAbs were reactive against conformational or linear epitopes of GLUT4. HEK-293T cells were transfected with a GLUT4-V5 construct, and lysates were assayed by Western blotting with the four selected mAbs. None of the GLUT4 mAbs showed reactivity against denatured GLUT4 by Western blot (SI Appendix, Fig. S4). GLUT4-V5 expression was confirmed by using an antibody against the V5 epitope tag included on its C terminus. These results suggest that all four GLUT4 mAbs recognize conformational epitopes formed by the 3D GLUT4 structure presented during VLP immunization.
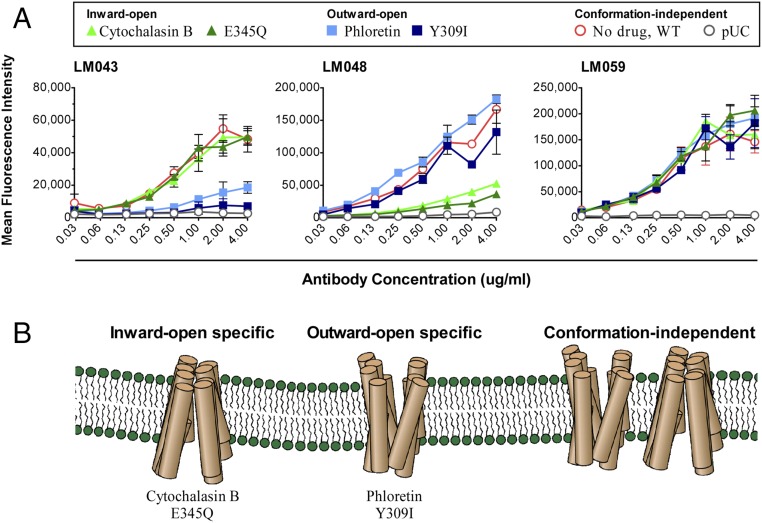
Glucose transporters undergo large ligand-induced conformational changes during their transport cycle and so can exist in several distinct states (11, 31, 34, 35). To test if binding of any of the isolated mAbs was state dependent, we next assessed mAb binding to GLUT4 in distinct conformational states induced by either chemical compounds or point mutations. GLUT4 can be locked into an inward-open conformation by either the transport inhibitor cytochalasin B (12) or an E345Q mutation (36). Alternatively, GLUT4 can be locked into an outward-open conformation by the GLUT4 inhibitor phloretin or a Y309I mutation (12, 13). Unexpectedly, two of the four mAbs tested demonstrated state-specific binding. LM043 showed strong binding to GLUT4 in the inward-open conformation induced by both cytochalasin B and E345Q (Fig. 6). In contrast, locking GLUT4 into the outward-open conformation with phloretin or the Y309I mutation essentially abolished binding of LM043 to GLUT4. These results indicate that LM043 binding to GLUT4 is specific for the inward-open conformation of GLUT4.
Fig. 6.
mAbs LM043 and LM048 show state-specific binding to GLUT4. (A) Wild-type hsGLUT4 (WT) expressed on HEK-293T cells was treated with either cytochalasin B or phloretin or was mutated (E345Q or Y309I) to induce specific GLUT4 conformational states. The treated or mutated hsGLUT4 proteins were then assayed for binding by mAbs LM043, LM048, or LM059 followed by an Alexa Fluor 488-conjugated goat anti-human secondary antibody. Cells transfected with a pUC vector were used as a negative control. (B) The mAbs isolated are sensitive to GLUT4 conformations induced by the presence of cytochalasin B or E345Q (inward open) or by phloretin or Y309I (outward open) or are conformation independent.
mAb LM048 also demonstrated state-dependent binding to GLUT4 but in the opposite manner. LM048 showed strong binding to GLUT4 in the outward-open conformation induced by both phloretin and Y309I. In contrast, locking GLUT4 into the inward-open conformation with cytochalasin B or the E345Q mutation essentially abolished binding of LM048 to GLUT4, indicating that LM048 binding is dependent on the outward-open conformation of GLUT4.
Binding to GLUT4 by the other mAbs, LM052 and LM059, was unaffected by phloretin or cytochalasin B treatment or by the two point mutations. Therefore, LM052 and LM059 appear to bind to a GLUT4 epitope whose availability is independent of the inward-open or outward-open state of GLUT4.
State-specific GLUT4 mAbs are expected to act as inhibitors of GLUT4-mediated glucose transport. We therefore tested the ability of antibodies to inhibit the ability of GLUT4 to transport a glucose analog using a bioluminescent assay. As expected, LM048 inhibited the ability of GLUT4 to transport 2-deoxyglucose, while neither LM059 (state independent) or LM043 (intracellular) showed inhibition at any concentration tested (SI Appendix, Fig. S5).
Mapping of mAb Epitopes and Residues That Control GLUT4 State Dependence.
Because of the state dependence of the isolated mAbs, we next wished to understand the mechanism by which each mAb could recognize GLUT4 in its different conformations. The residues required for binding of each mAb were determined using shotgun mutagenesis comprehensive alanine scanning (37). All residues of hsGLUT4 were individually mutated to alanine, with existing alanines changed to serines. Mutations were obtained for 508 of the 509 residues of GLUT4 (excluding the initiating methionine). The entire mutation library was transfected into human HEK-293T cells in a 384-well array format (one clone per well) and was assessed for immunoreactivity using high-throughput flow cytometry.
mAb epitopes were obtained by testing each mAb against the entire mutation library and identifying residues whose mutation to alanine impaired binding relative to wild-type GLUT4 (SI Appendix, Table S1). Residues critical for each GLUT4 mAb epitope were identified as those where GLUT4 mutations resulted in less than 20% reactivity for the mAb of interest (relative to wild-type GLUT4) but greater than 70% wild-type binding by a reference mAb. Residues were further validated as critical by comparing their reactivities across all mAbs tested to verify that the mutation did not globally disrupt the binding of diverse mAbs. The location and exposure of the identified residues were also taken into account as an indication of their potential for direct interaction with each mAb. Identified residues are hot spots that generally represent the most energetically important amino acids contributing to the binding of each mAb (38). As a control, epitope mapping of the commercial mAb 1F8 identified distal C-terminal amino acids D506 and N508 as its critical epitope residues (SI Appendix, Fig. S6A), consistent with prior localization of the 1F8 epitope to a linear peptide from the distal C terminus of GLUT4 (7). Epitope mapping of our own mAb LM040 identified a binding site overlapping that of 1F8 (SI Appendix, Fig. S6B).
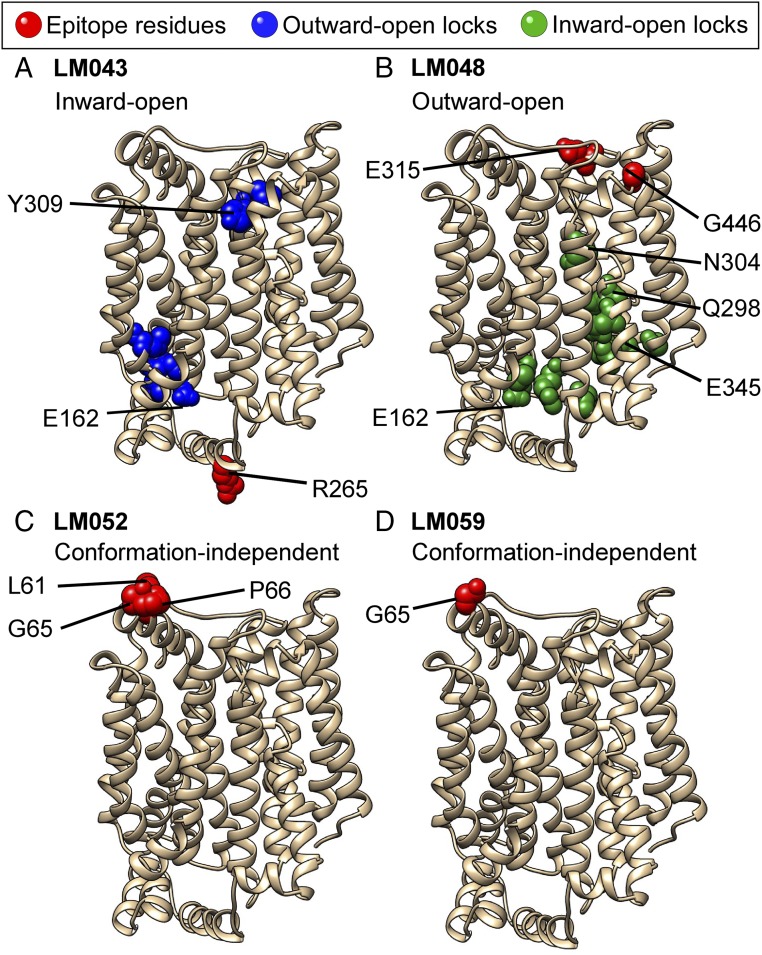
The critical residues comprising the binding epitopes for mAbs LM043, LM048, LM052, and LM059 are shown on the structural model of hsGLUT4 (Fig. 7, red residues). Mapping of mAb LM059 identified a single residue, G65, that, when mutated to alanine, eliminated LM059 binding. G65 is located at the apex of the largest GLUT4 extracellular loop (ECL1, 32 aa), so binding of LM059 at this site is consistent with its location and exposure. Other GLUT4 residues likely also contact LM059 but do not sufficiently contribute to the energetics of the interaction for a single alanine substitution to disrupt binding.
Fig. 7.
Residues that define the epitopes of GLUT4 mAbs and control the conformation of GLUT4. The critical residues comprising the epitopes for each of the mAbs [LMO43 (A), LMO48 (B), LMO52 (C), and LMO59 (D)] are shown in red on a structural model of hsGLUT4 generated from the crystal structure of human GLUT1 (PDB ID code 4pyp) using Phyre2 homology modeling. Residues that are buried in the TM domains, but that are critical for the binding of mAbs LM043 or LM048, are identified as outward-open locks (blue in A) or inward-open locks (green in B), respectively. Select residues discussed in the text are labeled.
For mAb LM052, which shares the same VH as LM059 but demonstrates a weaker affinity, G65 was also identified as a critical residue, along with adjacent residues L61 and P66. Mutation G356A reduced the binding of LM052 to just below 20%, but its location in the middle of TM9 suggests that this mutation may have an allosteric effect on the LM052 epitope. Notably, although both LM052 and LM059 bind nearly sequential residues, neither recognizes denatured GLUT4 by Western blot (SI Appendix, Fig. S4), suggesting that the epitope structure recognized by these mAbs forms a 3D conformation that requires the entire protein to be natively presented and is disrupted by protein unfolding. Interestingly, the epitope of LM052 also explains at least some of its low-level reactivity with other membrane proteins. For example, at very high concentrations LM052 reacts with the membrane protein Notch1 at levels that are above background (albeit well below GLUT4 levels) (SI Appendix, Fig. S7). Remarkably, the critical epitope residues recognized by LM052 (61-LGXXGP-66) are mirrored almost exactly by a disulfide-constrained amino acid loop in Notch1 (91-LGXXGP-96), despite GLUT4 and Notch1 having no significant (<7%) overall sequence identity. These data suggest that off-target binding of antibodies to genetically and structurally unrelated proteins needs to be tested on a case-by-case basis across the proteome.
mAb LM048 binds to GLUT4 on the cell surface, and binding is specific for GLUT4 in its outward-open conformational state. Screening of the GLUT4 mutation library identified 13 critical residues with less than 20% binding of LM048. Residues E315, located on extracellular loop 4 (ECL4), and G446, on ECL6, were the only extracellular-accessible residues identified as likely to form the critical binding amino acids for LM048. The locations of E315 and G446 explain the state-specific binding shown by LM048. Comparisons between structures of GLUT proteins demonstrate that TM7 undergoes extensive local structural changes in the shift from the outward-open to the inward-open conformation (31, 34, 39), which changes the relative positions of E315 and G446 at the GLUT4 extracellular surface and eliminates the conformational epitope formed by these residues (SI Appendix, Fig. S8).
Interestingly, additional residues critical for the binding of mAb LM048 were also identified but are located within the TM region and are not sufficiently exposed to allow mAb binding (Fig. 7B, green residues). These residues, when individually mutated to alanine, abolish the binding of mAb LM048, which binds only the outward-open state of GLUT4. Remarkably, many of these residues have been implicated in controlling the conformational rearrangement of GLUT4 during glucose transport (SI Appendix, Table S2), including residue E345, the site of the E345Q mutation that locks GLUT into the inward-open state (36). This cluster also includes two residues (Q298 and N304) that are part of the binding site for cytochalasin B (40), which locks GLUT4 into the inward-open state and abolishes LM048 binding. Cumulatively, these data suggest that this group of residues in the TM region locks GLUT4 in an inward-open state.
mAb LM043 binds to GLUT4 only in permeabilized cells, and binding is specific for GLUT4 in its inward-open conformational state. Screening of the GLUT4 mutation library identified seven critical residues with less than 20% binding by LM043. Residue R265, located on intracellular loop 3 (ICL3), was the only intracellular-accessible residue identified and so is likely the critical binding contact for LM043. Residue G414, located on ICL5, was the only other poorly reactive amino acid located intracellularly and so is likely also bound by LM043 but did not quite meet our threshold (23% reactivity with LM043). Interestingly, the locations of R265 and G414 may explain the state-specific binding shown by LM043. Comparisons between the structures of GLUT proteins demonstrate that ICL3 (proposed as a “door closer” that determines the extent of inward opening) undergoes extensive local structural changes in the shift from the outward-open to the inward-open conformation (31, 34, 39), which changes the relative positions of R265 (and G414) on the GLUT4 intracellular loops and eliminates the conformational epitope formed by these residues (SI Appendix, Fig. S8). These epitope residues also explain LM043’s cross-reactivity with GLUT1, as both R265 and G414 are conserved within GLUT1, but at least one is changed in each of the other human GLUT family members.
Similar to the analysis of LM048 binding, two additional clusters of residues critical for the binding of mAb LM043 were identified, but these clusters are located within the TM region and are not sufficiently exposed to allow mAb binding (Fig. 7A, blue residues). These residues, when individually mutated to alanine, abolish the binding of mAb LM043, which binds only the inward-open state of GLUT4. Remarkably, many of these residues have been implicated in controlling the conformational rearrangement of GLUT4 during glucose transport (SI Appendix, Table S2), including residue Y309, the site of the Y309I mutation that locks GLUT into the outward-open state (13). The extracellular-most cluster (most of the residues from 306 to 314) forms the top of TM7B that is also known to partly unwind and bend to form the outward-open state of GLUT4 (31, 34, 40). Cumulatively, these data suggest that this group of residues in the TM region locks GLUT4 in an outward-open state.
Interestingly, mutation E162A was identified as disrupting both the inward-open state and the outward-open state (i.e., eliminating the ability of both LM043 and LM048 to bind). E162 is structurally located toward the cytoplasmic face of GLUT4 at the interface between the cluster of outward-open locking residues and inward-open locking residues (SI Appendix, Fig. S8, aqua residue). These data suggest that E162 may be centrally involved in the transition between the outward-open and inward-open conformational states of GLUT4.
Discussion
GLUT4, one of the most studied members of the GLUT family of glucose transporters, plays the central role in maintaining glucose homeostasis at cellular and physiological levels. Insulin induces GLUT4 translocation from intracellular storage vesicles to the cell surface where it drives postprandial glucose uptake and maintenance of euglycemia. Disruption of GLUT4 regulation by insulin resistance results in the development of human diseases, including diabetes and obesity. Monitoring the GLUT4 translocation process not only is useful for basic biochemical and cellular studies but also has been used to screen for small-molecule insulin mimetics that can induce GLUT4 trafficking to the cell surface (41) and could be a valuable diagnostic tool to screen patients for early-stage insulin resistance before disease onset. However, to date, there are no tools available to help understand, detect, and visualize the native trafficking and conformational states of GLUT4 or most other transporters. The mAbs isolated in this study, therefore, will be valuable as tools to study human diseases that impact glucose homeostasis as well as for the development of assays to screen for small-molecule drugs that modify GLUT4 transport.
Generating mAbs against the native extracellular epitopes of multispanning membrane proteins is challenging due to multiple issues, including the difficulty in maintaining the membrane-dependent structure during immunization and panning/screening, poor immunogenicity of small extracellular loops, and immune tolerance due to the high sequence identity among mammals that limits the antibody response. As a result, there have been no mAbs that recognize the conformational structure of GLUT4, and, more generally, few mAbs against native epitopes for any multispanning membrane proteins, including most GPCRs, ion channels, and transporters (42). Here, we have isolated conformationally sensitive mAbs against one of the most challenging multispanning membrane proteins, GLUT4, containing 12 TM domains and six small, highly conserved extracellular loops.
The biological characteristics of retroviral VLPs were critical for both immunization and mAb isolation by phage panning. Multispanning membrane proteins such as GLUT4 require the continued presence of a lipid membrane to maintain native extracellular epitopes. Retroviral VLPs are produced by budding from the plasma membrane. Therefore, GLUT4 was maintained in its native cellular membrane at all stages of production and purification, eliminating the need to extract protein from the membrane with the attendant risk of denaturation and misfolding. Membrane proteins are also often difficult to express at high levels due to poor translation, folding, and trafficking. We were able to incorporate high levels of GLUT4 in VLPs (∼300 pmol/mg), ∼10- to 100-fold more concentrated than in cells or traditional membrane preparations, contributing to the success of both immunization and phage panning. Immunogenicity was likely further improved by the size (∼150 nm) and particulate nature of VLPs, which are efficiently targeted by dendritic cells for antigen processing and presentation (22). Although we were primarily interested in generating mAbs against extracellular epitopes, we also identified mAbs against intracellular regions. Intracellular epitopes are likely exposed when the VLP immunogen is broken open during immunization, antigen presentation, and phage panning, although the conformation-dependent nature of the isolated mAbs suggests that GLUT4 within VLPs still maintained its native structure throughout these manipulations. Based on the state-dependent mAbs isolated, VLPs are also able to present integral membrane proteins in multiple biologically relevant states.
To avoid problems associated with immune tolerance, chickens were chosen as the immunization host species, because the most similar GLUT homolog in chickens is only 65% identical to hsGLUT4, whereas hsGLUT4 and mmGLUT4 are 95% identical. In many respects, chickens are very similar to, and in many respects even better than, mammalian hosts in terms of antibody generation and isolation, and produce an Ig, IgY, that is highly similar to mammalian IgG. Chicken antibodies have a single VH and VL framework, which simplifies the molecular biology of phage library construction, minimizes VH/VL incompatibility when building a large scFv library, and makes humanization of any desired antibodies much easier than murine humanization (29, 30).
The anti-GLUT4 scFv phage display library created here was constructed using VH and VL genes derived from B cells from a single seropositive chicken. After panning on hsGLUT4 VLPs, 29 unique clones were identified representing six unrelated VH-CDR3 sequence families. Given the relatively modest extent of our screen, it is likely that further screening of additional phage clones and under different conditions would identify additional unique GLUT4 antibodies. For example, similar campaigns we have since conducted against other multispanning membrane proteins have identified hundreds of unique family members after more extensive screening. Future discovery campaigns could also utilize conformationally locked GLUT4 to bias the recovery of mAbs with desired binding properties. LM052 and LM059 have distinct VL genes but have identical VH genes, overlapping epitopes, and similar apparent affinities, reflecting the dominance of VH in mAb binding. Chicken VH genes appear to have an especially dominant role in antigen interactions since they have relatively long VH CDR3s (29, 32, 43). Long VH CDR3s may also be especially useful for binding to multispanning membrane proteins, which often have functionally important binding pockets into which long CDRs may reach.
To develop a detailed understanding of how the mAbs bind, LM043, LM048, LM052, and LM059 were characterized for affinity, specificity, and the nature of the epitopes that they recognize (summarized in Table 1). Given the 95% identity between hsGLUT4 and mmGLUT4, it was not surprising that all of the mAbs were able to recognize both proteins. However, generating mouse antibodies able to cross-react across human and mouse orthologs is often difficult using traditional immunization host species because of immune-tolerance mechanisms. Using chickens as the host animal enabled us to easily isolate human–mouse cross-reactive antibodies. While hsGLUT1, the closest paralog to hsGLUT4, is only 65% identical, one mAb, LM043, was also able to cross-react with hsGLUT1, explained by the identified LM043 epitope residues being conserved between hsGLUT1 and hsGLUT4.
Unexpectedly, of the four mAbs characterized, we identified two state-specific mAbs, LM043 and LM048, with specificity for opposing inward-open and outward-open conformations of GLUT4. Locking GLUT4 into defined inward- or outward-open conformations, using either pharmacological agents or mutations, enabled or eliminated mAb binding. Additionally, binding of LM048 was able to inhibit the transport function of GLUT4, consistent with its ability to bind GLUT4 extracellularly and in a state-specific fashion. Consistent with their state specificity, the epitopes of LM043 and LM048 each spanned two loops of GLUT4 that demonstrate relative structural rearrangement between the two states, whereas the epitopes for the conformationally insensitive mAbs LM052 and LM059 were contained in a single loop. To the best of our knowledge, LM043 and LM048 are the only state-specific mAbs ever isolated against a glucose transporter and are some of the only state-specific mAbs against any transporter. One other antibody was recently reported that inhibits the eukaryotic transporter ABCG2 from entering its inward facing conformation (44). mAbs specific for the conformational states of other complex membrane proteins, such as eukaryotic GPCRs and ion channels (14–16), as well as the lactose permease prokaryotic transporter (45), have proven critical for understanding the structure and function of these proteins. We expect that the availability of state-specific mAbs against GLUT4 will be helpful in detecting transient conformational changes of GLUT4 during glucose transport and for future structural studies of GLUT4 stabilized in state-specific conformations. These mAbs may also be useful as tools for identifying small molecules that preferentially bind specific states of GLUT4. Finally, the ability of mAbs to functionally modulate transporter function (e.g., as antagonists or agonists) opens potential new avenues of therapeutic intervention for diseases such as diabetes and cancer.
The epitopes of the GLUT4 mAbs were determined using comprehensive alanine scanning shotgun mutagenesis, as we have used previously for epitope mapping of over 500 mAbs against other complex membrane proteins, including GPCRs, ion channels, and viral envelopes (37). Two mAbs, LM052 and LM059, bound the apex of ECL1, the largest extracellular loop (32 aa) and the most accessible extracellular site on GLUT4. LM048 bound across ECL4 and ECL6, which are each only ∼10 residues long and are recessed in a cavity on the surface of GLUT4. Interestingly, LM048 contains the longest VH CDR3 (26 residues) among the characterized mAbs, which may enable it to bind in the cavity formed by ECL4 and ECL6 and sense the outward-open state of GLUT4. A mAb with a CDR3 of this length would not likely be found using traditional immunization hosts, but chicken CDR3s are generally longer than either mouse or human CDR3s and so make such mAbs more likely (Fig. 2D). We note that the epitope data for LM048 (and LM043) do not enable us to distinguish between states of GLUT4 with or without bound ligand, and we have not seen differences in binding in the presence or absence of glucose. Studies using nontransportable sugars such as maltose may enable determination of the exact conformational state bound by these antibodies.
Because binding of the state-dependent mAbs LM043 and LM048 was tested against alanine mutations across the entire GLUT4 sequence, we were also able to comprehensively identify mutations that lock GLUT4 into inward-open or outward-open conformations. Remarkably, the sites of these mutations were clustered around many positions known to influence the conformational state of GLUT4, including previously defined locking mutants and the binding site for cytochalasin. Still, it is possible that these residues could be affecting GLUT4 and mAb binding in alternative ways, such as allosterically affecting the mAb-binding sites.
Interestingly, mutation of residue E162 eliminated both LM043 and LM048 reactivity (although not LM052 or LM059 reactivity, GLUT4 surface trafficking, or GLUT4 expression). E162 is located at the interface between the outward-open (blue) and inward-open (green) locking residue clusters that we identified (SI Appendix, Fig. S8, in aqua) and was previously identified as essential for conformational rearrangement (36, 46). Its location, effect on GLUT4 function, and effect on GLUT4 conformation (stabilizing a conformation of GLUT4 that is neither inward-open nor outward-open) suggest that residue E162 plays a critical role in the transition between the inward-open and outward-open conformations.
The results of this study highlight the utility of VLPs and divergent host animals for the isolation of conformational mAbs against complex multispanning membrane proteins. mAbs specific for extracellular epitopes, intracellular epitopes, and the two conformational states of GLUT4 were readily isolated using the strategies outlined here. These mAbs represent unique reagents that can be used for monitoring native GLUT4 function and trafficking, studying the structure of GLUT4, differentiating different GLUT4 epitopes and states, and high-throughput screening of novel diabetes therapeutics.
Materials and Methods
Isolation of GLUT4 mAbs.
VLPs displaying GLUT proteins (commercially referred to as “lipoparticles”) were produced by cotransfection of HEK-293T cells with plasmids carrying hsGLUT1 or hsGLUT4 genes and the retroviral (MLV) Gag protein, as previously described (19, 47). Null VLPs were produced the same way but without transfection of a specific receptor.
An scFv phage display library was constructed from B cells of a hsGLUT4 VLP-immunized chicken that showed serum reactivity with GLUT4 by flow cytometry, as previously described (29). For panning, the phage library (2E9) was allowed to bind to wells coated with GLUT4 VLPs (for positive selection) or with null VLPs (for deselection). Bound phage was trypsin-eluted and amplified through three rounds of panning before the clones were screened for binding to GLUT4. Individual scFv peripreps were prepared from single colonies by induction with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 28 °C overnight followed by extraction of the periplasmic fraction by freeze–thaw and then were screened for hsGLUT4-specific binding by ELISA using VLPs.
For screening phage clones by periprep ELISA, purified hsGLUT4-displaying VLPs were coated on a 96-well white, flat-bottomed microtiter plate overnight at 4 °C using 0.25 µg of protein per well in 0.1 M sodium bicarbonate buffer, pH 8.6. The wells were washed with PBS and blocked with 4% PBS without calcium or magnesium (PBS−/−) with 4% milk (PBSM) for 1 h. scFv in 4% PBSM were added to each well, and the plate was incubated for 1 h at 37 °C with gentle agitation. The scFv solution was discarded, and the plate was washed three times with PBS plus 0.01% Tween 20. To detect bound scFv, a 1:5,000 dilution of anti-human Fd conjugated with HRP (SouthernBiotech) in 4% PBSM was added to the wells, and the plate was incubated at room temperature (22 °C) for 30 min with gentle agitation. The plate was washed three times with PBS plus 0.01% Tween 20 and was developed according to the manufacturer’s instructions (Super Signal West Pico; Thermo Scientific). Negative controls included a buffer blank (no antigen) and a nonspecific antigen.
Candidate scFv were converted to the human IgG1-Fc format for production in HEK-293T cells. Briefly, the scFv region was PCR amplified and cloned using infusion cloning (Clontech) in-frame with a leader sequence and the Fc fragment of human IgG1 to create an scFv-Fc gene. scFv-Fc constructs were transfected into HEK-293T cells by calcium phosphate precipitation. Secreted scFv-Fc were purified from the culture medium at 48–72 h posttransfection by protein A chromatography, followed by concentration and buffer exchange against PBS. Quantification of the purified scFv-Fc was performed using a bicinchoninic acid (BCA) assay (Thermo Scientific).
Biosensor-Binding Kinetics.
All biosensor studies were performed in PBS buffer supplemented with 1 mg/mL BSA (PBS-B) at 25 °C using a ForteBio Octet Red biosensor system (Pall-ForteBio, Inc.). Streptavidin biosensor tips were loaded with biotinylated hsGLUT4 VLPs (diluted to 20 µg/mL in PBS-B) for 45 min and were allowed to stabilize for 10 min. Antibody binding and dissociation kinetics were determined for serial dilutions of antibody in PBS-B starting at 20 µg/mL. Antibody association was measured for 5 min followed by dissociation for up to 45 min in buffer. Nonspecific binding was assessed by using sensor tips with VLPs containing only endogenous proteins (null VLPs). Data analysis was performed with the Octet data analysis program (v8.1; ForteBio) using a standard 1:1 binding model.
Flow Cytometry.
HEK-293T cells were transfected by CaPO4 with plasmids in six-well culture plates (Falcon) at 750,000 cells per well. At 48 h post transfection, cells were stained with anti-V5 and anti-GLUT4 mAbs (purified scFv, 2 µg/mL) followed by biotinylated goat anti-mouse or human antibody (1:500) and streptavidin PerCP (Biolegend, 1:500). Fluorescence was detected using an Intellicyt high-throughput flow cytometer (HTFC). For cell permeabilization, cells were treated with saponin at 0.1% in PBS for 20 min. To discriminate intact (live) and permeabilized (dead) cells without the use of detergent, PI was added to HEK-293T cells at 1 μg/mL just before flow cytometry. Where indicated, cytochalasin B (Sigma) was used at a final concentration of 10 μM, and phloretin (Sigma) was used at a final concentration of 100 μM, each added in 10% normal goat serum (NGS) for 30 min before antibody staining as well as included with the primary antibody.
Western Blot.
HEK-293T cells were plated in six-well plates and were transfected with plasmids at 8 µg per well via a calcium phosphate transfection. After 48 h, cells were lysed using 1 mL of radioimmunoprecipitation assay (RIPA) lysis buffer, and lysates were collected and spun at 17,000 × g for 20 min at 4 °C. Protein concentration in the supernatant was determined using a BCA assay (Pierce). Thirty micrograms of cell lysates and 2 µg of VLPs were further solubilized in sample buffer (62.5 mM Tris⋅HCl, 5% glycerol, 2% SDS, 0.0025% Bromophenol blue, and 25 mM DTT). Samples were run on a 10% urea gel at 110 V for 1.33 h (Bio-Rad). Proteins were transferred onto a PVDF membrane (Thermo Fisher) overnight at 4 °C at 22 V (Bio-Rad). Membrane was blocked using 5% milk in blocking buffer (PBS with 0.2% Tween-20) for 0.5 h and then was stained with primary antibodies at a concentration of 2–5 µg/mL in blocking buffer for 1 h. The appropriate HRP secondary antibody (either anti-human or anti-mouse; SouthernBiotech) was used at a 1:5,000 dilution in 5% milk in blocking buffer for 0.5 h. Blots were developed using West Femto chemiluminescent substrate (Thermo Scientific) and were imaged on an Alpha Innotech FluorChem system.
MPA Specificity Testing.
The MPA is Integral Molecular’s cell-based platform of 4,571 different human membrane proteins, each overexpressed in live cells from expression plasmids that are individually transfected in separate wells of a 384-well plate (48). The entire library is arrayed in duplicate in a matrix format to facilitate testing and analysis. For testing here, the array of plasmids in the MPA was expressed in HEK-293T cells for 24 h. Before testing on the array, primary mAb concentrations were determined using an independent immunofluorescence titration curve against wild-type hsGLUT4 to ensure that the signal-to-background ratio was optimal for target detection. Cells were permeabilized with 0.1% saponin, each antibody was added to the MPA at 1 μg/mL, and binding across the protein library was measured by Intellicyt HTFC using a fluorescent secondary antibody. Each array plate contains both positive (Fc-binding) and negative (empty vector) controls to ensure plate-by-plate data validity. Any identified targets were confirmed in a second flow cytometry experiment using serial dilutions of antibody, and the target identity was re-verified by sequencing.
Shotgun Mutagenesis Epitope Mapping of GLUT4 mAbs.
An hsGLUT4 expression construct (Uniprot accession no. P14672) was subjected to high-throughput alanine scanning mutagenesis to generate a comprehensive mutation library. Primers were designed to mutate each residue to alanine, with alanine codons mutated to serine. In total, 508 hsGLUT4 mutants were generated across the entire GLUT4 sequence (residues 2–509), the mutations were sequence confirmed, and the mutants were arrayed into 384-well plates at one mutant per well.
The GLUT4 mutation library, arrayed in 384-well microplates, was transfected into HEK-293T cells and allowed to express for 22 h. Cells were stained with purified mAbs LM043 (0.5 µg/mL), LM048 (1 µg/mL), LM052 (0.5 µg/mL), or LM059 (0.5 µg/mL) diluted in 10% NGS (Sigma-Aldrich). Primary mAb concentrations were determined using an independent immunofluorescence titration curve against wild-type GLUT4 to ensure that the signals were within the linear range of detection. For LM043, binding to the library was measured in the presence of 10 μM cytochalasin B. mAbs were detected using an Alexa Fluor 488-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) at 3.75 µg/mL in 10% NGS. Cells were washed twice in PBS−/− and were resuspended in Cellstripper solution (CELLGRO) with 0.1% BSA (Sigma-Aldrich). Mean cellular fluorescence was detected using Intellicyt HTFC. mAb reactivities against each mutant GLUT4 clone relative to the reactivity against wild-type GLUT4 protein were calculated by subtracting the signal from mock-transfected controls and normalizing the signal to the signal from wild-type GLUT4-transfected controls.
Mutated residues were identified as being critical to the mAb epitope if they did not support the reactivity of the test mAb but did support the reactivity of a reference GLUT4 mAb and additional GLUT4 mAbs. This counterscreen strategy facilitates the exclusion of GLUT4 mutants that are locally misfolded or that have an expression defect (37, 49). Residues constituting the mAb epitope were visualized on the homology-based structural model of GLUT4 generated from the crystal structure of GLUT1 [Protein Data Bank (PDB) ID code 4pyp] using Phyre2 (50).
3T3-L1 Adipocyte Immunofluorescence.
3T3-L1 preadipocytes were grown and differentiated into the adipocyte phenotype as previously described (51). Adipocytes between 7–8 d post differentiation were serum-starved for 3 h in DMEM with 220 mM bicarbonate and 20 mM Hepes (pH 7.4) before stimulation for 20 min at 37 °C with or without 170 nM human insulin (Sigma Aldrich) prepared in the same medium with 2% BSA. Adipocytes were washed twice with ice-cold PBS with calcium and magnesium (PBS+/+) and remained on ice for all subsequent steps.
Adipocytes were blocked with 10% NGS in PBS+/+ for 30 min on ice before the addition of test antibodies at 10 μg/mL in 10% NGS. Primary antibodies were incubated for 2 h and then were washed twice with PBS+/+. Cells were subsequently fixed for 5 min in 4% paraformaldehyde, washed twice with PBS+/+, and then stained with secondary antibody (AF488-conjugated goat anti-human; Jackson ImmunoResearch) diluted 1:200 in 10% NGS. Secondary antibody was incubated for 1 h after washing times with PBS+/+. For intracellular staining, cells were fixed before the blocking step and were permeabilized with 10% NGS containing 0.2% saponin. The 1F8 antibody was used at 1 µg/mL and was detected using AF647-conjugated goat anti-mouse secondary antibody diluted 1:400 (Jackson ImmunoResearch). Alexa Fluor 488 was visualized using the B-2EC cube with an exposure time of 1.5 s. Alexa Fluor 647 was visualized using the Cy5 cube with an exposure time of 2 s.
2-Deoxyglucose Uptake Assay.
HEK-293T cells were transfected with hsGLUT4 expression plasmid or pUC19 control plasmid in poly-l-lysine–coated, black 384-well plates with clear bottoms (Costar) and were incubated at 37 °C for 22 h. The growth medium was removed, and cells were washed three times with PBS+/+ to remove residual glucose. Cells were incubated at 25 °C for 2 h in PBS+/+ containing 0.5% BSA, 25–100 μg/mL scFv-Fc, and 10 nM BAY-876 (Sigma-Aldrich) to block endogenous GLUT1 channels. Glucose uptake was initiated by the addition of 1 mM 2-deoxyglucose and was allowed to proceed for 20 min at 37 °C. Cellular 2-deoxyglucose uptake was measured in total cell lysates using the Glucose Uptake-Glo kit (Promega) according to the manufacturer’s instructions. Luminescence was measured using an Envision (PerkinElmer), and nonspecific luminescence was measured in lysates from cells pretreated with 10 mM cytochalasin B.
Supplementary Material
Acknowledgments
We thank Samantha Gilman, Mayda Hernandez, Soma Banik, Anu Thomas, Andrew Ettenger, Chris Bryan, Bernard Lieberman, Natasha Kushnir, and Srikar Reddy for valuable technical assistance and scientific advice; Tim McGraw at Cornell University for valuable discussions; and Morrie Birnbaum for advice and cells. This work was supported by NIH Grants DC010105 and GM113556.
Footnotes
Conflict of interest statement: S.H.W., B.J.D., and J.B.R. are shareholders of Integral Molecular.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716788115/-/DCSupplemental.
References
- 1.Mueckler M, Thorens B. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med. 2013;34:121–138. doi: 10.1016/j.mam.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang S, Czech MP. The GLUT4 glucose transporter. Cell Metab. 2007;5:237–252. doi: 10.1016/j.cmet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Jaldin-Fincati JR, Pavarotti M, Frendo-Cumbo S, Bilan PJ, Klip A. Update on GLUT4 vesicle traffic: A cornerstone of insulin action. Trends Endocrinol Metab. 2017;28:597–611. doi: 10.1016/j.tem.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Klip A, Sun Y, Chiu TT, Foley KP. Signal transduction meets vesicle traffic: The software and hardware of GLUT4 translocation. Am J Physiol Cell Physiol. 2014;306:C879–C886. doi: 10.1152/ajpcell.00069.2014. [DOI] [PubMed] [Google Scholar]
- 5.Dawson K, Aviles-Hernandez A, Cushman SW, Malide D. Insulin-regulated trafficking of dual-labeled glucose transporter 4 in primary rat adipose cells. Biochem Biophys Res Commun. 2001;287:445–454. doi: 10.1006/bbrc.2001.5620. [DOI] [PubMed] [Google Scholar]
- 6.Lampson MA, Racz A, Cushman SW, McGraw TE. Demonstration of insulin-responsive trafficking of GLUT4 and vpTR in fibroblasts. J Cell Sci. 2000;113:4065–4076. doi: 10.1242/jcs.113.22.4065. [DOI] [PubMed] [Google Scholar]
- 7.James DE, Brown R, Navarro J, Pilch PF. Insulin-regulatable tissues express a unique insulin-sensitive glucose transport protein. Nature. 1988;333:183–185. doi: 10.1038/333183a0. [DOI] [PubMed] [Google Scholar]
- 8.Zorzano A, et al. Insulin-regulated glucose uptake in rat adipocytes is mediated by two transporter isoforms present in at least two vesicle populations. J Biol Chem. 1989;264:12358–12363. [PubMed] [Google Scholar]
- 9.Friedman JE, et al. Immunolocalization of glucose transporter GLUT4 within human skeletal muscle. Diabetes. 1991;40:150–154. doi: 10.2337/diab.40.1.150. [DOI] [PubMed] [Google Scholar]
- 10.Slot JW, Geuze HJ, Gigengack S, James DE, Lienhard GE. Translocation of the glucose transporter GLUT4 in cardiac myocytes of the rat. Proc Natl Acad Sci USA. 1991;88:7815–7819. doi: 10.1073/pnas.88.17.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohan S, Sheena A, Poulose N, Anilkumar G. Molecular dynamics simulation studies of GLUT4: Substrate-free and substrate-induced dynamics and ATP-mediated glucose transport inhibition. PLoS One. 2010;5:e14217. doi: 10.1371/journal.pone.0014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yano Y, May JM. Ligand-induced conformational changes modify proteolytic cleavage of the adipocyte insulin-sensitive glucose transporter. Biochem J. 1993;295:183–188. doi: 10.1042/bj2950183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori H, et al. Substitution of tyrosine 293 of GLUT1 locks the transporter into an outward facing conformation. J Biol Chem. 1994;269:11578–11583. [PubMed] [Google Scholar]
- 14.Steyaert J, Kobilka BK. Nanobody stabilization of G protein-coupled receptor conformational states. Curr Opin Struct Biol. 2011;21:567–572. doi: 10.1016/j.sbi.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ring AM, et al. Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staus DP, et al. Regulation of β2-adrenergic receptor function by conformationally selective single-domain intrabodies. Mol Pharmacol. 2014;85:472–481. doi: 10.1124/mol.113.089516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown MC, et al. Impact of immunization technology and assay application on antibody performance–A systematic comparative evaluation. PLoS One. 2011;6:e28718. doi: 10.1371/journal.pone.0028718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith AJ. New horizons in therapeutic antibody discovery: Opportunities and challenges versus small-molecule therapeutics. J Biomol Screen. 2015;20:437–453. doi: 10.1177/1087057114562544. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman TL, Canziani G, Jia L, Rucker J, Doms RW. A biosensor assay for studying ligand-membrane receptor interactions: Binding of antibodies and HIV-1 Env to chemokine receptors. Proc Natl Acad Sci USA. 2000;97:11215–11220. doi: 10.1073/pnas.190274097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endres MJ, et al. Targeting of HIV- and SIV-infected cells by CD4-chemokine receptor pseudotypes. Science. 1997;278:1462–1464. doi: 10.1126/science.278.5342.1462. [DOI] [PubMed] [Google Scholar]
- 21.Balliet JW, Bates P. Efficient infection mediated by viral receptors incorporated into retroviral particles. J Virol. 1998;72:671–676. doi: 10.1128/jvi.72.1.671-676.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludwig C, Wagner R. Virus-like particles-universal molecular toolboxes. Curr Opin Biotechnol. 2007;18:537–545. doi: 10.1016/j.copbio.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitoh R, et al. Viral envelope protein gp64 transgenic mouse facilitates the generation of monoclonal antibodies against exogenous membrane proteins displayed on baculovirus. J Immunol Methods. 2007;322:104–117. doi: 10.1016/j.jim.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Lai X. Reproducible method to enrich membrane proteins with high purity and high yield for an LC-MS/MS approach in quantitative membrane proteomics. Electrophoresis. 2013;34:809–817. doi: 10.1002/elps.201200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryk AH, Wiśniewski JR. Quantitative analysis of human red blood cell proteome. J Proteome Res. 2017;16:2752–2761. doi: 10.1021/acs.jproteome.7b00025. [DOI] [PubMed] [Google Scholar]
- 26.Zhao FQ, Keating AF. Functional properties and genomics of glucose transporters. Curr Genomics. 2007;8:113–128. doi: 10.2174/138920207780368187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kono T, et al. Characterisation of glucose transporter (GLUT) gene expression in broiler chickens. Br Poult Sci. 2005;46:510–515. doi: 10.1080/00071660500181289. [DOI] [PubMed] [Google Scholar]
- 28.Seki Y, Sato K, Kono T, Abe H, Akiba Y. Broiler chickens (Ross strain) lack insulin-responsive glucose transporter GLUT4 and have GLUT8 cDNA. Gen Comp Endocrinol. 2003;133:80–87. doi: 10.1016/s0016-6480(03)00145-x. [DOI] [PubMed] [Google Scholar]
- 29.Finlay WJ, Bloom L, Varghese S, Autin B, Cunningham O. Optimized generation of high-affinity, high-specificity single-chain Fv antibodies from multi-antigen immunized chickens. Methods Mol Biol. 2017;1485:319–338. doi: 10.1007/978-1-4939-6412-3_16. [DOI] [PubMed] [Google Scholar]
- 30.Finlay WJ, et al. Phage display: A powerful technology for the generation of high-specificity affinity reagents from alternative immune sources. Methods Mol Biol. 2017;1485:85–99. doi: 10.1007/978-1-4939-6412-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng D, et al. Crystal structure of the human glucose transporter GLUT1. Nature. 2014;510:121–125. doi: 10.1038/nature13306. [DOI] [PubMed] [Google Scholar]
- 32.Wu L, et al. Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids. J Immunol. 2012;188:322–333. doi: 10.4049/jimmunol.1102466. [DOI] [PubMed] [Google Scholar]
- 33.Thoidis G, Kotliar N, Pilch PF. Immunological analysis of GLUT4-enriched vesicles. Identification of novel proteins regulated by insulin and diabetes. J Biol Chem. 1993;268:11691–11696. [PubMed] [Google Scholar]
- 34.Deng D, et al. Molecular basis of ligand recognition and transport by glucose transporters. Nature. 2015;526:391–396. doi: 10.1038/nature14655. [DOI] [PubMed] [Google Scholar]
- 35.Quistgaard EM, Löw C, Moberg P, Trésaugues L, Nordlund P. Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters. Nat Struct Mol Biol. 2013;20:766–768. doi: 10.1038/nsmb.2569. [DOI] [PubMed] [Google Scholar]
- 36.Schürmann A, et al. Role of conserved arginine and glutamate residues on the cytosolic surface of glucose transporters for transporter function. Biochemistry. 1997;36:12897–12902. doi: 10.1021/bi971173c. [DOI] [PubMed] [Google Scholar]
- 37.Davidson E, Doranz BJ. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology. 2014;143:13–20. doi: 10.1111/imm.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 39.Nomura N, et al. Structure and mechanism of the mammalian fructose transporter GLUT5. Nature. 2015;526:397–401. doi: 10.1038/nature14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapoor K, et al. Mechanism of inhibition of human glucose transporter GLUT1 is conserved between cytochalasin B and phenylalanine amides. Proc Natl Acad Sci USA. 2016;113:4711–4716. doi: 10.1073/pnas.1603735113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lanzerstorfer P, et al. Identification of novel insulin mimetic drugs by quantitative total internal reflection fluorescence (TIRF) microscopy. Br J Pharmacol. 2014;171:5237–5251. doi: 10.1111/bph.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Douthwaite JA, Finch DK, Mustelin T, Wilkinson TC. Development of therapeutic antibodies to G protein-coupled receptors and ion channels: Opportunities, challenges and their therapeutic potential in respiratory diseases. Pharmacol Ther. 2017;169:113–123. doi: 10.1016/j.pharmthera.2016.04.013. [DOI] [PubMed] [Google Scholar]
- 43.Könitzer JD, et al. Generation of a highly diverse panel of antagonistic chicken monoclonal antibodies against the GIP receptor. MAbs. 2017;9:536–549. doi: 10.1080/19420862.2016.1276683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor NMI, et al. Structure of the human multidrug transporter ABCG2. Nature. 2017;546:504–509. doi: 10.1038/nature22345. [DOI] [PubMed] [Google Scholar]
- 45.Smirnova I, et al. Outward-facing conformers of LacY stabilized by nanobodies. Proc Natl Acad Sci USA. 2014;111:18548–18553. doi: 10.1073/pnas.1422265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mueckler M, Makepeace C. Cysteine-scanning mutagenesis and substituted cysteine accessibility analysis of transmembrane segment 4 of the Glut1 glucose transporter. J Biol Chem. 2005;280:39562–39568. doi: 10.1074/jbc.M509050200. [DOI] [PubMed] [Google Scholar]
- 47.Willis S, et al. Virus-like particles as quantitative probes of membrane protein interactions. Biochemistry. 2008;47:6988–6990. doi: 10.1021/bi800540b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huston-Paterson DJ, Banik SSR, Doranz BJ. Screening the membrane proteome. Genet Eng Biotechnol News. 2016;36:18–19. [Google Scholar]
- 49.Paes C, et al. Atomic-level mapping of antibody epitopes on a GPCR. J Am Chem Soc. 2009;131:6952–6954. doi: 10.1021/ja900186n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi SM, et al. Insulin regulates adipocyte lipolysis via an Akt-independent signaling pathway. Mol Cell Biol. 2010;30:5009–5020. doi: 10.1128/MCB.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.