Significance
Store-operated Orai1 channels mediate transcriptional, proliferative, and effector-cell programs in many cells. Mutations in Orai1 that block channel activation or evoke constitutive channel activity are known to cause debilitating diseases in humans such as immunodeficiency, autoimmunity, myopathy, and thrombocytopenia. However, our understanding of the underlying molecular mechanisms of these diseases is limited by fundamental gaps in how Orai1 channels are gated. Here, we map key functional interactions between the transmembrane domains of Orai1 and identify several contacts that are critical for conveying the STIM1 gating signal to the pore. Our findings illuminate important allosteric interactions between topologically distinct domains of Orai1 and help elucidate the molecular underpinnings of disease-causing mutations.
Keywords: CRAC channels, Orai1, STIM1, calcium, store-operated calcium entry
Abstract
Store-operated Orai1 channels are activated through a unique inside-out mechanism involving binding of the endoplasmic reticulum Ca2+ sensor STIM1 to cytoplasmic sites on Orai1. Although atomic-level details of Orai structure, including the pore and putative ligand binding domains, are resolved, how the gating signal is communicated to the pore and opens the gate is unknown. To address this issue, we used scanning mutagenesis to identify 15 residues in transmembrane domains (TMs) 1–4 whose perturbation activates Orai1 channels independently of STIM1. Cysteine accessibility analysis and molecular-dynamics simulations indicated that constitutive activation of the most robust variant, H134S, arises from a pore conformational change that opens a hydrophobic gate to augment pore hydration, similar to gating evoked by STIM1. Mutational analysis of this locus suggests that H134 acts as steric brake to stabilize the closed state of the channel. In addition, atomic packing analysis revealed distinct functional contacts between the TM1 pore helix and the surrounding TM2/3 helices, including one set mediated by a cluster of interdigitating hydrophobic residues and another by alternative ridges of polar and hydrophobic residues. Perturbing these contacts via mutagenesis destabilizes STIM1-mediated Orai1 channel gating, indicating that these bridges between TM1 and the surrounding TM2/3 ring are critical for conveying the gating signal to the pore. These findings help develop a framework for understanding the global conformational changes and allosteric interactions between topologically distinct domains that are essential for activation of Orai1 channels.
Ca2+ release-activated Ca2+ (CRAC) channels mediate store-operated Ca2+ entry, an essential mechanism for Ca2+ influx in many cells that is triggered in response to depletion of endoplasmic reticulum (ER) Ca2+ stores (1). Opening of CRAC channels evokes local and global increases in intracellular Ca2+, which not only refills ER Ca2+ stores but also drives numerous effector functions such as gene expression, cell proliferation, secretion of inflammatory mediators, and cell migration (1, 2). The importance of CRAC channels for human health is highlighted by mutations in CRAC channel proteins that give rise to severe immunodeficiency, autoimmunity, muscle weakness, skin and tooth defects, and thrombocytopenia (3, 4).
Activation of CRAC channels by store depletion occurs through a unique inside-out mechanism whose broad contours are now well established. In the first step of the process, depletion of ER Ca2+ stores mobilizes the ER Ca2+ sensor STIM1 into its active state, which exposes a catalytic domain in its cytoplasmic region, known as the CRAC activation domain (CAD) (5) or STIM1-Orai1 activating region (SOAR) (6). STIM1 then oligomerizes and migrates from the bulk ER to the ER–plasma membrane junctions (1). In the next step, the CAD/SOAR domain of STIM1 directly binds to the CRAC channel pore subunit Orai1, setting into motion conformational changes in the Orai1 protein that culminate in the opening of the channel gate. At the structural level, each Orai1 subunit of the hexameric CRAC channel contains four transmembrane helices (TMs) arranged in concentric layers, with the centrally located TM1 helices lining the pore, TM2 and TM3 in the next layer, and TM4 forming the most external, lipid-exposed segment (7). Recent studies indicated that the channel gate is formed, at least in part, by the combination of two hydrophobic pore residues, F99 and V102, which regulate the closed–open transition (8–11). STIM1 binding is proposed to open this gate by inducing a modest rotation of the pore helix to move the bulky F99 side chains away from the pore axis to lower the energy barrier for ion conduction (11).
A major unresolved question in this process is the molecular mechanism by which the STIM1 gating signal is communicated to the Orai1 pore. Although not considered in most early studies, there is increasing evidence from genetic disease association studies in human patients that the transmembrane domains of Orai1 may be critically involved in this process. In particular, several reports have described patients with tubular aggregate myopathy, thrombocytopenia, and congenital miosis caused by gain-of-function (GOF) Orai1 mutations located within the TM domains (12–15). In addition, recent structure–function analysis of several GOF mutations including that of a putative “hinge” at the base of TM4 (16), a Pro residue in TM4 (17), and several residues cataloged in a cancer genomics database (18), provide more direct support for involvement of the non–pore-lining TMs in Orai1 gating. These findings suggest that, as seen in many ligand-gated channels (19), local conformational changes at the agonist binding site (16, 20, 21) are allosterically relayed to the distally located channel gate in the pore (8, 11) through an unknown pathway.
The discovery of pathological human mutations, along with the increasing evidence from the structure–function studies described above, prompted us to examine the structural basis of how the non–pore-lining TMs regulate channel activation. By using scanning mutagenesis, state-dependent accessibility analysis, molecular-dynamics (MD) simulations, and atomic packing analysis, we generated a functional map of key interactions between the transmembrane domains of Orai1 that are critical for conveying the gating signal to the CRAC channel pore.
Results
Scanning Mutagenesis of Orai1 TMs Reveals Numerous Constitutively Active CRAC Channels.
Several recent studies have described GOF missense Orai1 mutations in TMs 2–4, with some linked to tubular aggregate myopathy with additional symptoms including thrombocytopenia and miosis (12–15). The identification of GOF mutations in the non–pore-lining segments suggests that these domains of Orai1 may play a greater role in channel activation than previously appreciated. To begin addressing this question, we started our study with an unbiased mutagenesis screen to identify residues in the TMs that could regulate Orai1 channel activation. There are 130 transmembrane residues in the human Orai1 sequence based on comparison with the crystal structure of highly homologous Drosophila melanogaster Orai (dOrai) protein [Protein Data Bank (PDB) ID code 4HKR)] (7), with three endogenous Cys residues (C126, C145, C196) in TM2 and TM3. We sequentially mutated each of the non-Cys Orai1 transmembrane residues to generate 127 DNA constructs, each containing a single Cys mutation. These mutants were transfected into HEK293 cells in the absence of STIM1 and tested by whole-cell mode patch clamping. Cys substitutions were used for this analysis because, as a moderately hydrophobic residue, Cys is generally well tolerated in TMs (22). In addition, this approach allowed us to take advantage of some previously generated TM1 and TM3 Cys mutants (23) for this work.
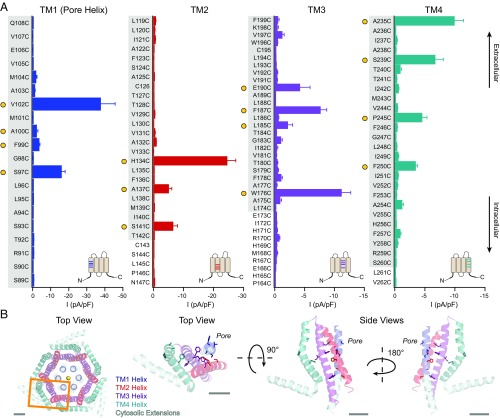
To our surprise, this screen revealed numerous constitutively active GOF mutations in Orai1 with varying levels of current density and ion selectivity (Figs. 1A and 2 and SI Appendix, Table S1). When mapped onto the crystal structure of dOrai (7) (Fig. 1B), the mutations showed broad distribution in the membrane-spanning regions of all four TM domains, including four in the TM4 segment that is farthest from the pore at a distance of 20–25 Å. Many GOF mutants exhibited current–voltage (I–V) relationships and a positive reversal potential (Vrev) resembling that of WT Orai1 channels activated by STIM1 (Vrev > 35 mV; Fig. 2 and SI Appendix, Table S1). These included two loci that have been linked to tubular aggregate myopathy with congenital miosis—S97C on the non–pore-lining face of TM1 (13) and P245C in TM4 (12)—as well as H134C in TM2, F187C in TM3, and A235C in TM4 (Figs. 1 and 2 and SI Appendix, Table S1). Others, such as W176C (24) and E190C on TM3, however, displayed substantial outward currents at positive potentials (Fig. 2 D and E). The wide variation in current amplitude and ion selectivity among the GOF mutants suggests that they stabilize Orai1 in different open conformations.
Fig. 1.
A cysteine screen of Orai1 TMs reveals several constitutively active mutants in all four TMs. (A) Current densities of Orai1 cysteine mutants in the absence of STIM1. For comparison, the current densities of WT Orai1 without and with STIM1 are −0.2 ± 0.01 pA/pF and −48 ± 8 pA/pF, respectively. Constitutively active mutants (defined as >2 pA/pF) are marked with filled yellow circles. Gray shaded areas on the labels indicate the boundaries of the membrane as represented in the dOrai crystal structure (7). (Insets) Approximate positions of the mutations on a topology diagram of Orai1 (n = 4–16 cells; values are mean ± SEM). (B) Top-down view of the crystal structure of dOrai with one subunit outlined in an orange box. A top-down view and two side views of one dOrai subunit are also shown. TMs 1–4 are colored in blue, red, purple, and teal, respectively, with the positions of constitutively open cysteine mutants represented as sticks. (Scale bars: 10 Å.)
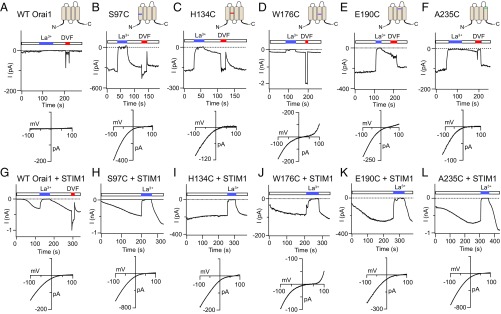
Fig. 2.
Channel properties of Orai1 GOF cysteine mutants. (A–F) Constitutive activity of the indicated Orai1 variants. (Top) Traces show time course of current in the absence of STIM1 following whole-cell break-in (t = 0). (Bottom) Graphs depict I–V relationships of the indicated mutants measured in 20 mM extracellular Ca2+ solution. (G–L) Store-operated activation of WT and mutant Orai1 channels in STIM1-coexpressing cells. In contrast to all other mutants, H134C does not show additional current increase following whole-cell break-in. (Bottom) Graphs show I–V relationships of the different mutants. Note that STIM1 coexpression shifts the reversal potential of the mutant channels in all cases (SI Appendix, Table S1).
Because Orai1 has three endogenous Cys residues at positions 126, 143, and 195, it is possible that some of the GOF phenotypes arise from disulfide bond formation with the introduced Cys residues. However, replacing the native cysteines with serines (i.e., Cys3S construct) did not affect the GOF phenotypes of the tested open mutants (SI Appendix, Fig. S1), indicating that the constitutive activity is not dependent on the endogenous cysteines. Further, the open channel phenotype does not arise from enhanced binding to the endogenous pool of STIM1, as the introduction of an additional L273D mutation which abrogates STIM1 binding (25) did not affect the constitutive activity of the tested mutants (SI Appendix, Fig. S1). Finally, consistent with the well-established requirement of STIM1 for Ca2+-dependent fast inactivation (CDI) in Orai1 (26–28), the constitutively open cysteine mutants did not exhibit CDI (SI Appendix, Fig. S1). Taken together, these results indicate that the cysteine mutations described here earlier activate Orai1 channels independently of STIM1 binding, likely by disrupting endogenous interactions between amino acids that are involved in keeping the channel closed.
Interestingly, adding a K85E mutation, which has been shown to be essential for Orai1 channel gating (29–31), or deleting the N terminus of Orai1 (Orai1 ∆2–85), which harbors a putative CAD binding site (5, 30, 32), eliminated the constitutive activity of the GOF mutations (SI Appendix, Fig. S2). The loss of function (LOF) of these mutants was not caused by defects in channel expression or plasma membrane localization (SI Appendix, Fig. S2). These findings are in line with recent studies that have also observed that K85E and other N-terminal mutations abolish the constitutive activity of some GOF Orai1 mutations (16, 33) and suggest that the Orai1 N terminus, and specifically K85, are essential for maintaining channel function independently of interactions with STIM1.
Differential Modulation of GOF Orai1 Cysteine Mutants by STIM1.
Although constitutively active in the absence of STIM1 coexpression, the majority of the open mutants described here earlier displayed increases in current amplitude and became significantly more Ca2+-selective when overexpressed with STIM1 (Fig. 2 H–L). As a consequence, the macroscopic mutant channel properties in the presence of STIM1 including I–V relationship, reversal potentials, and CDI were, in most cases, indistinguishable from those of WT Orai1 channels activated by STIM1 (Fig. 2 and SI Appendix, Table S1). For example, E190C, which was only moderately Ca2+-selective in the absence of STIM1 (Fig. 2E and SI Appendix, Table S1), became significantly more Ca2+-selective when coexpressed with STIM1 (Fig. 2K and SI Appendix, Table S1). Even W176C, which displayed an outwardly rectifying I–V relationship at positive potentials similar to 2-APB–gated Orai3 currents (24, 34, 35) (Fig. 2D), showed a robust rightward shift in Vrev in the presence of STIM1 (Fig. 2J). This modulation of ion selectivity is consistent with previous findings indicating that STIM1 not only activates Orai1 channels, but also concomitantly boosts its Ca2+ selectivity in a dose-dependent manner (8). Overall, these findings demonstrate that the constitutively open mutant channels are not maximally active but instead adopt one or more open states that can be further activated by STIM1.
Modulation of channel activity and Ca2+ selectivity by STIM1, however, varied among the different mutants. Plots of the increase in current, or Vrev, showed an inverse relationship with the amount of constitutive activity (SI Appendix, Fig. S3), suggesting that the efficacy of STIM1 to further activate a mutant and enhance its Ca2+ selectivity diminishes with increasing baseline activity. Notably, these plots showed that H134C, which has the largest constitutive current and a very positive Vrev, exhibits the smallest STIM1-mediated increases in current and Vrev (SI Appendix, Fig. S3). Based on these criteria, we conclude that among all of the preactivated mutants identified from the cysteine screen, H134C is the closest in its properties to physiological STIM1-gated Orai1 channels.
H134 Regulates Orai1 Gating Through a Steric Mechanism.
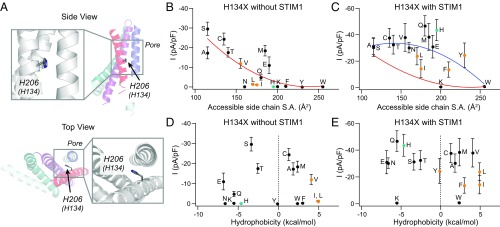
The robust GOF phenotype of H134C on TM2 (Fig. 3A) prompted us to further probe the mechanism by which this mutation induces constitutive channel activation. To address this question, we mutated H134 to all other amino acids and tested them in the absence and presence of STIM1. This analysis revealed that substitutions of H134 to smaller, or moderately sized but flexible, residues (S/A/C/T/V/Q/E/M) produced GOF channels that were open without STIM1, whereas mutation to larger, more rigid residues failed to evoke constitutive activity (Fig. 3B). A plot of the current density vs. side-chain surface area revealed a trend toward spontaneous channel activation with diminishing side-chain size (Fig. 3B). By contrast, no obvious dependence of constitutive channel activation with respect to hydrophobicity was observed (Fig. 3D). A recent study has proposed that H134 forms hydrogen bonds with two serine residues on the back of TM1 (S93 and S97) to maintain the closed channel state and that disrupting these hydrogen bonds evokes constitutive channel activity (18). However, our data argue against this hypothesis because H134S and H134T channels, which have hydroxyl groups presumably available for hydrogen bonding, are robustly open without STIM1 (Fig. 3 B and D and SI Appendix, Fig. S4).
Fig. 3.
The constitutive activity of the H134X mutations depends on the size of the introduced amino acid. (A) Top-down and side views of H134 in the context of a dOrai monomer. H134 is located on TM2, and its imidazole ring faces the non–pore-lining side of TM1. (B and C) Current densities of H134X mutants without and with STIM1 coexpression plotted against side-chain surface area. Red and blue lines represent polynomial fits to the data (without and with STIM1, respectively) to enable visualization of the overall trends. H134G and H134P were excluded from this analysis because of the propensity of glycine and proline residues to break α-helical secondary structure (45). H134R Orai1 did not express to the membrane and is not included. The native His residue is represented in teal. Orange points indicate mutants with nonselective currents (Vrev < 20 mV). (D and E) Current density of H134X mutants vs. the solvation energies (in kilocalories per mole) of the substituted amino acid as a measure of its hydrophobicity. There is a strong inverse correlation between amino acid size and current amplitude in the absence of STIM1, but no clear dependence on hydrophobicity (n = 5–12 cells; values are mean ± SEM).
When overexpressed with STIM1, the H134X channels exhibited several different phenotypes. Like H134C, H134A/S/T channels were also nearly maximally active at baseline, and did not display increases in current over time following whole-cell break-in (SI Appendix, Fig. S4 and Table S2). By contrast, H134V/E/M/Q channels were only partially active without STIM1 (Fig. 3 B and D), and STIM1 coexpression boosted current amplitude and Ca2+ selectivity of these mutants (SI Appendix, Fig. S4 and Table S2). H134N was the only mutant that was closed at rest and activated by STIM1 with kinetics indistinguishable from WT Orai1 (SI Appendix, Fig. S4). Several mutants additionally displayed LOF phenotypes with defects in STIM1-dependent gating. For example, H134W and H134K channels showed very little current with STIM1 (Fig. 3 C and E) despite seemingly normal membrane expression, suggesting that these bulky side chains essentially block channel activation by STIM1. On the contrary, H134L/I/F/Y exhibited “reluctant” gating in the presence of STIM1, with small, nonselective currents with reversal potentials of 8–14 mV compared with 50–60 mV in WT channels (Fig. 3 C and E and SI Appendix, Fig. S4 and Table S2).
These results indicate that the constitutive activation of H134X channels arises from a reduction in the bulk of the native His at this position. We hypothesize that a decrease in the side-chain volume releases a “brake” at the TM1–TM2 domain interface, enabling the channel to open (Discussion). Conversely, increasing the volume of the side chain inhibits STIM1’s ability to gate the channel. Although no straightforward dependence of current density with respect to hydrophobicity of the introduced amino acid at H134 was observed (Fig. 3D), nonpolar substitutions tended to yield channels that were less selective for Ca2+ when activated by STIM1 in comparison with polar substitutions (Fig. 3E and SI Appendix, Table S2). This feature suggests that hydrophobicity at the TM1–TM2 interface may play a role in orienting TM1 helices into a Ca2+-selective pore, a possibility meriting further study.
The GOF H134S Orai1 Pore Configuration Mimics STIM1-Gated Channels.
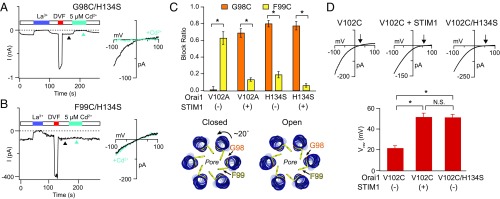
Given that H134S strongly activates Orai1, we used this mutant as a tool to further dissect the mechanism of constitutive channel activation. Previous studies have shown that the outer region of the Orai1 pore harbors a hydrophobic gate that encompasses the residues V102 and F99 (8, 10, 11). STIM1 activates the channel, at least in part, by triggering rotation of the pore helix to displace F99 away from the pore axis and lower the energy barrier for ion conduction. One operational consequence of this displacement is that the coordination of metal ions such as Cd2+ and Zn2+ by a Cys introduced at F99 (F99C) sharply declines upon STIM1 binding (11). By contrast, G98, which is located on the opposite face of the pore helix (Fig. 4C), becomes accessible to pore-applied divalent metal ions in the presence of STIM1, resulting in enhancement of Cd2+ coordination by G98C and, consequently, channel blockade (11). We hypothesized that, if the GOF mutations represent a STIM1-like activated state of Orai1 with a rotated pore helix, there should also be weak Cd2+ accessibility at F99 but strong blockade at G98. By contrast, a “leaky” gate phenotype as in V102A or F99Y Orai1 mutants resulting from destruction of the channel gate should show strong coordination at F99, but not G98 (11). Consistent with the former scenario, we observed very little Cd2+ block at F99 (19 ± 4% blockade in F99C/H134S) but strong blockade at G98 (80 ± 4% blockade in G98C/H134S), indicating that F99C is positioned away from the pore axis in the H134S mutant (Fig. 4). This pattern of Cd2+ blockade is akin to the pattern observed in V102A channels in the presence of STIM1 (11), indicating that the orientation of F99 and G98 is similar to that of STIM1-gated channels (Fig. 4C).
Fig. 4.
Cd2+ accessibility studies suggest that the pore conformation of H134S Orai1 is similar to that of STIM1-gated channels. (A and B) Cd2+ (5 µM) strongly blocks G98C, but not F99C, residues in the H134S mutant. I–V relationships were measured at times indicated by the arrowheads. (C) Summary of Cd2+ blockade at G98C or F99C in the H134S mutant (n = 4–6 cells; values are mean ± SEM). For comparison, Cd2+ blockade at these positions is also illustrated in the constitutively conducting V102A channels in the presence or absence of STIM1 (data from ref. 11). The pattern of blockade in the H134S mutant resembles that seen in STIM1-bound V102A channels, suggesting similar pore conformations. A schematic illustrating the open and closed states of the Orai1 pore showing rotation of the pore helix induced by STIM1 binding (adapted from ref. 46). (D) I–V relationships of the V102C mutant alone, or in the presence of either STIM1 or an additional H134S mutation. The lower graph summarizes the Vrev under these conditions. As seen with STIM1, H134S confers Ca2+ selectivity to V102C channels. Arrowheads indicate reversal potentials (n = 6–9 cells; values are mean ± SEM; *P < 0.05).
In a second test to examine whether the H134S channel conformation resembles that evoked by STIM1, we asked whether this mutation increases the Ca2+ selectivity of the poorly Ca2+-selective, constitutively conducting Orai1 variant V102C. We have previously shown that gating evoked by STIM1 enhances the Ca2+ selectivity of the V102C mutant channel (8). In a similar fashion, introducing the H134S mutation boosted the Ca2+ selectivity of V102C Orai1, as evidenced by a significant rightward shift in the Vrev of V102C/H134S channels (Fig. 4D). Taken together, these results indicate that the H134S activating mutation assumes a pore configuration close to that of STIM1-gated Orai1 channels.
Molecular Simulations Show That H134S Induces Rotation of TM1 and Increased Pore Hydration.
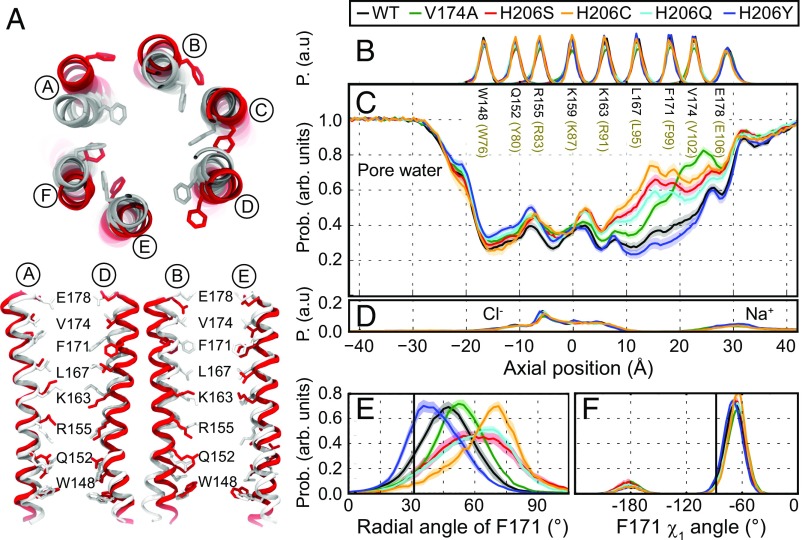
To understand how the H134S mutation alters the energetics of channel activation, we performed MD simulations using models constructed from the Drosophila Orai structure (SI Appendix, Fig. S5 and Movies S1 and S2). We examined the degree of pore hydration and associated conformational fluctuations of WT, V174A, and H206S/C/Q/Y mutations corresponding to human Orai1 V102A and H134S/C/Q/Y. As previously observed (11), the pore helices of WT and V174A dOrai displayed spontaneous counterclockwise rotations, with the angular position of F171 shifted by 17 ± 1° and 22 ± 1°, respectively, relative to the crystallographic dOrai structure (Fig. 5E). This spontaneous relaxation/rotation was strongly enhanced (P < 0.001) in the dOrai H206S and H206C mutants, in which the average pore helix rotations were 30 ± 1° and 34 ± 2°, respectively (Fig. 5 A and E and Movies S1 and S2). Further, the extent of helix rotation in H206S/C was accompanied by a striking increase in the number of water molecules in the hydrophobic stretch of the pore (Fig. 5C). Notably, enhanced hydration was observed over a significantly larger stretch of the pore encompassing more of the hydrophobic zone (L167–V174) than in the constitutively permeant V174A mutant.
Fig. 5.
MD simulations demonstrate increased pore helix rotation and pore hydration in GOF mutations H206S/C. (A) Superposition of snapshots at t = 350 ns from MD simulations of WT (gray) and H206S (red) as viewed from the top. (Bottom) Pairs of diagonal subunits viewed from the side. (B) Distributions of the axial position of Cα atoms for all pore-lining residues. The residues corresponding to human Orai1 are shown in brown. Data in A–F were computed from simulations of WT (black) and of V174A (green), H206C (orange), H206S (red), H206Q (aqua), and H206Y (navy) mutant channels. Average distribution of water oxygen atoms (C) and Na+ and Cl− ions (D) along the pore axis. The water occupancies of V174A and H206S/C/Q mutants deviate from those of WT and H206Y dOrai most significantly in the hydrophobic stretch of the pore. Bound Cl− and Na+ counter ions are found in the basic region of the inner pore and near E178 side chains, respectively. (E) Average distribution of the radial angle of residue 171 defined as the angle between the pore axis, the center of mass of the two helical turns centered at residue 171, and the Cα atom of residue 171. The means of these distributions over all simulation repeats in degrees are 47 ± 1° for WT, 53 ± 1° for V174A, 61 ± 1° for H206S, 65 ± 2° for H206C, 59 ± 1° for H206Q, and 42 ± 1° for H206Y. The radial angle in the crystallographic structure (31°) is shown as a black vertical bar for reference. (F) Average distribution of side-chain torsion χ1 of residue 171 in WT and V174A and H206S/C/Q/Y mutants. The χ1 of residue 171 in the crystallographic structure (−88°) is shown as a black vertical bar for reference. The traces depict values ± SEM.
To understand how these features compare with other mutants with differing levels of channel activity, we also modeled the H206Q (hOrai1 H134Q) mutant, which, as described earlier, yields partially active channels in the absence of STIM1 (Fig. 3 and SI Appendix, Fig. S4 and Table S2). In line with this intermediate level of constitutive channel activity, MD simulations of H206Q showed slightly increased pore helix rotation (29 ± 1°) and pore hydration (Fig. 5 C and E) compared with WT channels. By contrast, hOrai1 H134Y is not constitutively open and exhibits impaired STIM1-mediated gating with low Ca2+ selectivity (SI Appendix, Fig. S4 and Table S2). Consistent with this phenotype, MD simulations showed a lower extent of pore helix rotation as well as pore hydration in H206Y compared with WT (P < 0.001), with an average F171 Cα rotation of only 11 ± 1°, and a modest decrease in pore hydration compared with WT dOrai (Fig. 5 C and E). Thus, H206S/C and H206Y elicit opposite effects on pore helix rotation and pore hydration. The rotamer populations of F171 were comparable across all variants, indicating that lateral displacement of F171 occurred as a result of pore helix rotation rather than side-chain fluctuations. Interestingly, the simulations also showed that the activated mutants (H206S/C/Q and V174A) exhibit noticeable pore dilation compared with WT channels as measured by HOLE (36) analysis, especially in the hydrophobic stretch encompassing F99 (SI Appendix, Fig. S6). Moreover, pore dilation at F99 was reduced in the LOF H206Y mutant, suggesting that the extent of dilation is correlated with the “openness” of the mutants (SI Appendix, Fig. S6).
Several interesting insights can be gleaned from this analysis. First, the close correlation between the constitutive activity of the variants as assessed by electrophysiology and pore hydration and pore helix rotation (Fig. 5 C and E) indicates that the molecular simulations are able to recapitulate some key aspects of the experimentally detectable structural changes induced by the mutations. Second, the simulations reveal that changes in pore hydration are limited nearly exclusively to the hydrophobic section encompassing V102 and F99, with little to no change seen in the rest of the pore (Fig. 5C), reaffirming the importance of the hydrophobic zone for Orai1 gating (8–11). The simulations also revealed hitherto unappreciated pore dilation in the hydrophobic stretch that is well correlated with the degree of helix rotation and especially with pore hydration (SI Appendix, Fig. S6). This observation raises the possibility that, along with the experimentally detectable displacement of the F99 residues, widening of the pore in the hydrophobic zone may also contribute to lowering of the free energy barrier for ion conduction. However, because current electrophysiological methods do not have the resolution needed to resolve the small changes in pore radius seen in the simulations, these predictions await experimental validation with alternative approaches.
Atomic Packing and Hydrophobicity Analysis Reveals Distinct Functional Contacts Between the TM1 and TM2/3 Helices.
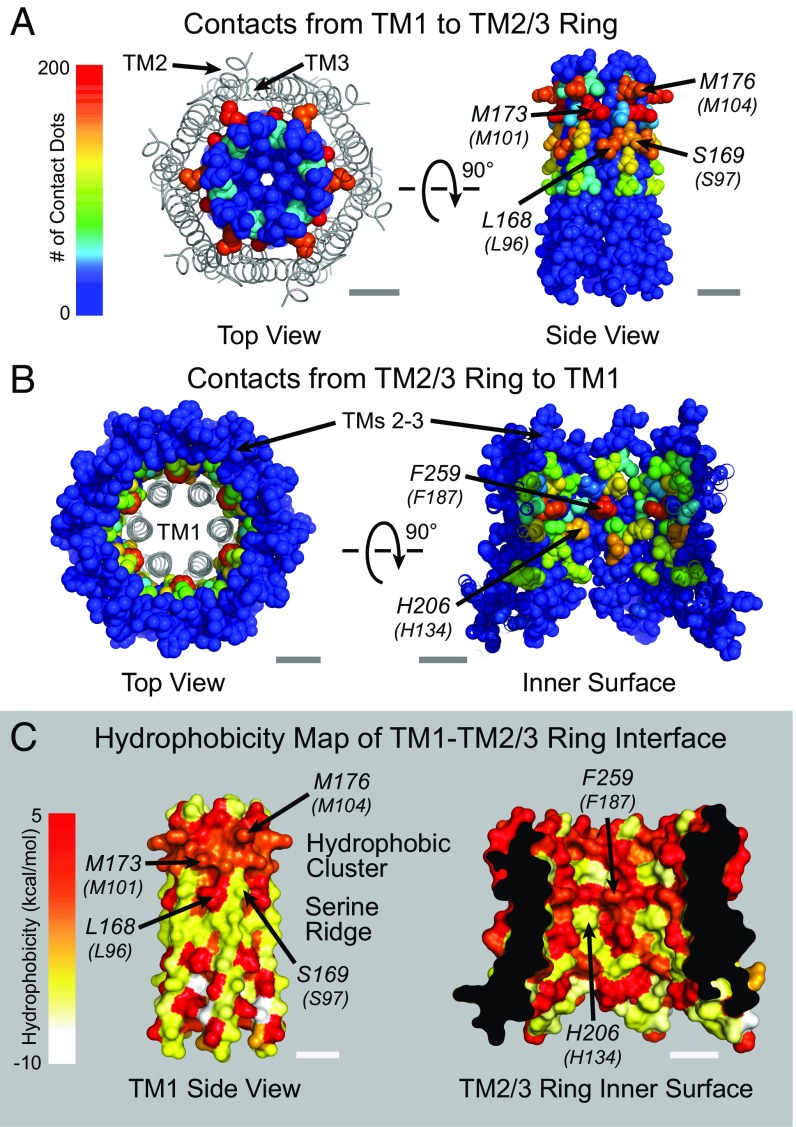
The results discussed thus far imply that the non–pore-lining TMs play crucial roles in Orai1 channel gating by STIM1. To understand the structural basis of how the individual TM amino acids mediate these effects, we next mapped the packing densities of interhelical surfaces by using a small-probe contact dots protocol (37, 38), which provides a rapid and computationally facile means to evaluate packing interactions on a residue-by-residue basis. We complemented this analysis with maps of the hydrophobicity of the residues at the TM1–TM2/3 interface to assess the contribution of hydrophobic interactions to this interface. As the enthalpic contribution to the stabilization of packing interactions should be proportional to the atomic surface areas in contact, residues exhibiting extensive interactions likely contribute more to the stability of an interface, and to the coupling between structural elements, than residues with less interaction surface. As a consequence, tightly packed regions in proteins are generally more rigid, whereas loosely packed regions more readily undergo conformational change (37, 38).
Applying this analysis to the residues in direct contact with the pore helix revealed several intriguing structural features. First, the contact dots analysis of dOrai shows that the atomic packing density varies significantly across and within the TM1–TM4 bundle of helices (SI Appendix, Fig. S7). Interestingly, the TM2/3 helix pair displays substantially more contacts and a more uniform packing density across its interfaces than do TM1 or TM4 (SI Appendix, Fig. S7). With both intra- and intersubunit surfaces lined mostly by large hydrophobic residues, TMs 2 and 3 have the appearance of an interlocked ring situated between TMs 1 and 4. Second, the analysis reveals a prominent cluster of interdigitating hydrophobic residues lining the interface between TM1 and the TM2/3 ring near the extracellular region that forms a hydrophobic stack (Figs. 6 and 7). Third, moving toward the cytoplasmic side, there are alternating ridges of hydrophobic and polar residues on the back of TM1 that appear to interact with complementary hydrophobic and polar stripes on the inner interface of the TM2/3 ring (Figs. 6C and 8). Later, we address the functional roles of these structural elements for relaying the gating signal to the central pore.
Fig. 6.
Atomic packing analysis and hydrophobicity mapping of dOrai reveals two distinct interfaces between the TM2/3 and TM1 segments. (A and B) Top and side view space-filling representations of the interface between TM1 and the TM 2/3 ring colored by the number of contacts per residue. TM4 is hidden for clarity. In A, TMs 2 and 3 are shown as ribbons. (C) Surface representation of the non–pore-lining residues of TM1 and residues of the TM2/3 ring facing TM1 colored according to amino acid hydrophobicity (47). A cluster of hydrophobic amino acids towards the extracellular side and a “serine ridge” towards the cytosolic side are also labeled. (Scale bars: 10 Å.)
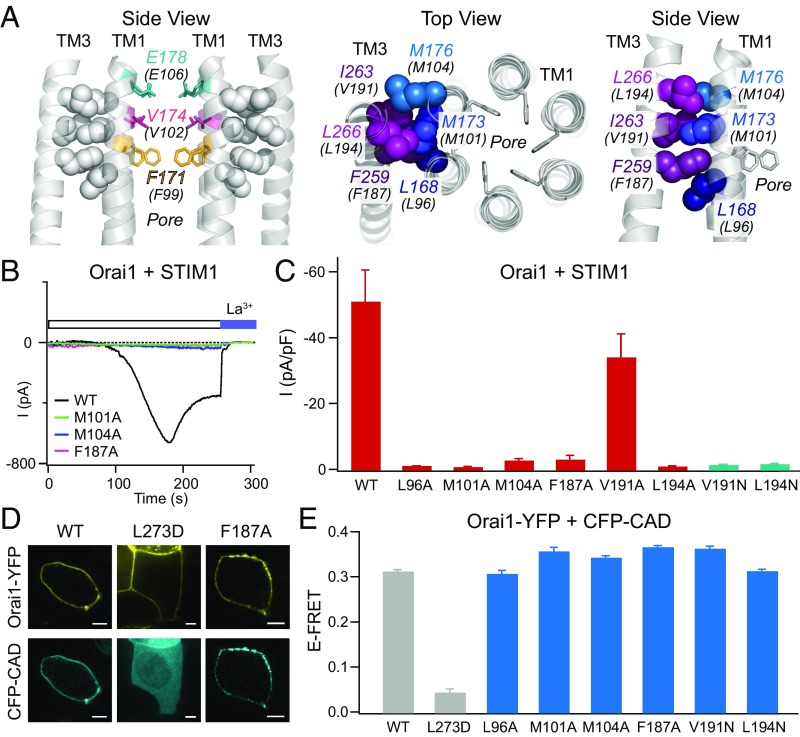
Fig. 7.
The TM3–TM1 hydrophobic cluster is essential for Orai1 gating by STIM1. (A) (Left) Side view of the hydrophobic contacts (gray spheres) between the TM1 pore helices with the TM3 segment. The selectivity filter (E106, teal) and hydrophobic gate residues (V102, magenta; F99, yellow) are also shown. Four TM1 helices and two TM3 helices are shown for simplicity. (Middle and Right) Top-down and side views of the six residues forming the hydrophobic stack. Amino acids are labeled with dOrai numbering with hOrai1 numbering shown in parentheses. (B) Point mutations of the hydrophobic stack abrogate Orai1 activation by STIM1. Representative traces of WT Orai1 and M101A, M104A, and F187A Orai1 mutants coexpressed with STIM1 following whole-cell break-in are shown. (C) Current densities of the indicated Orai1 mutations in the TM3–TM1 hydrophobic cluster coexpressed with STIM1 (n = 4–6 cells; values are mean ± SEM). With the exception of V191A, these mutations abrogate Orai1 activation by STIM1. (D) Confocal images of WT, L273D, and F187A Orai1-YFP constructs coexpressed with CFP-CAD showing normal CAD binding in the F187A mutant. (Scale bars: 5 μm.) (E) FRET between the indicated Orai1-YFP channels and CFP-CAD shows that the LOF mutations in the hydrophobic stack do not impair Orai1-STIM1 binding (n = 53–88 cells for each mutant; values are mean ± SEM).
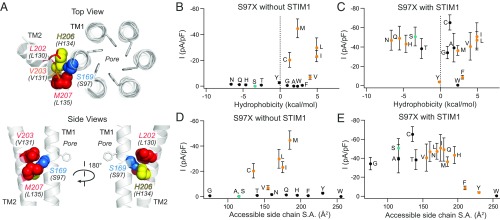
Fig. 8.
Hydrophobic substitutions at S97 evoke constitutive channel activity. (A) Top-down and side views of TMs 1 and 2 in the dOrai structure showing S169 (hOrai1 S97, blue), H206 (hOrai1 H134, yellow), and the nonpolar residues L202 (hOrai1 L130, red), V203 (hOrai1 V131, orange sticks), and M207 (hOrai1 L135, red). F99 side chains are shown as gray sticks. (B and C) The current densities of S97X mutants in the absence and presence of STIM1 coexpression are plotted against the solvation energies (in kilocalories per mole) of the substituted amino acids (47). The native Ser is depicted in teal. Substitutions that give rise to nonselective channels (Vrev < 35 mV) are shown as orange points. (D and E) Current densities of S97X mutants plotted against the accessible side-chain surface area of the introduced amino acid without and with STIM1. Mutant channels with small substitutions retain store-operated, Ca2+-selective WT behavior, whereas nonpolar or large substitutions cause the channel to become nonselective or unable to be activated by STIM1 (n = 4–11 cells; values are mean ± SEM).
A Cluster of TM3–TM1 Hydrophobic Contacts Is Critical for STIM1-Mediated Orai1 Gating.
The stack of interdigitating hydrophobic residues on the extracellular region of the channel is the locus of highest packing density at the TM1–TM2/3 ring interface (Fig. 6 A and B). This hydrophobic cluster is comprised of TM1 residues L168, M173, M176, and V177 (hOrai1 L96, M101, M104, V105) closely packed against the TM2/3 residues F195, F259, I263, and L266 (hOrai1 F123, F187, V191, L194; Fig. 7A). Intriguingly, this cluster of hydrophobic residues is located at the same depth as the selectivity filter (E106) and the hydrophobic gate (V102/F99), raising the tempting possibility that this structural transition zone between the pore domain and the TM2/3 ring is a natural site of communication for regulating pore opening and ion selectivity.
We examined this possibility by substituting each of the residues of the hydrophobic cluster to Ala, a small, less hydrophobic amino acid that is not expected to destabilize the local helical structure. Consistent with our prediction, the alanine scan revealed that decreasing the number of contacts and the hydrophobicity in this region resulted in channels that could no longer be gated by STIM1 (Fig. 7 B and C). The L96A, M101A, M104A, F187A, and L194A mutations all abrogated STIM1-mediated Orai1 activation (Fig. 7 B and C). The one exception to this trend was V191A, which retained store-operated gating (Fig. 7C), possibly reflecting the similar size and nonpolar nature of Val and Ala residues, and potentially a lesser role for V191 in the middle of the hydrophobic stack compared with its larger, more hydrophobic Met and Leu neighbors (Fig. 7A). However, even at this position, a V191N substitution impaired STIM1 gating (Fig. 7C). The loss of functional activity of these mutants is not a result of deficiencies in expression in the plasma membrane or STIM1 binding, as indicated by unaltered levels of FRET between CFP-CAD and Orai1-YFP and the recruitment of CFP-CAD to the plasma membrane (Fig. 7 D and E). Taken together, mutational analysis of the hydrophobic clamp indicates that disrupting hydrophobic interactions between TM3 and TM1 abolishes communication between the TM1 pore segment with the other helices and the ability of STIM1 to open the pore.
The Disease Mutation S97C Activates Orai1 Through a Hydrophobic Switching Mechanism.
In striking contrast to the tight packing in the hydrophobic cluster toward the extracellular region, the cytoplasmic region of the non–pore-facing TM1 surface exhibits relatively low packing density with the TM2/3 ring (Fig. 6A). This is because the central residues at the interface arise from a ridge of serine side chains S154, S161, S162, S165, and S169 (hOrai1 S82, S89, S90, S93, S97) (7), which, in addition to being more loosely packed than the hydrophobic clamp, are also polar (Fig. 6 A and C). A band of alternating hydrophobic and polar residues formed by L96 and S97 features prominently in this zone (Fig. 6C). This pattern of polar-hydrophobic regions on TM1 appear to pair with complementary polar-hydrophobic stripes on the surface of the TM2/3 ring (Fig. 6C), suggesting that mutations that increase the hydrophobicity of the serine ridge may force an interaction with a hydrophobic region on TM2/3 ring instead of its usual polar interaction partners. This possibility is intriguing because a recent report has described a GOF human Orai1 mutation S97C that causes a Stormorken-like syndrome with tubular aggregate myopathy and congenital miosis (13). The molecular basis of the GOF phenotype of this mutant is currently not understood, but such a switching mechanism could potentially explain its constitutive activity.
Consistent with this scenario, hydrophobic substitutions at S97 including Cys, Val, Leu, Ile, and Met yielded Orai1 channels that were partially open in the absence of STIM1 (Fig. 8B). By contrast, mutation of S97 to similarly sized, polar residues such as Asn, Gln, and Thr did not produce constitutively open channels (Fig. 8D); they instead remained store-operated. In the dOrai crystal structure, S97 is located on the non–pore-lining face of TM1 surrounded by three hydrophobic TM2 residues (L130, V131, L135) and the bulky, polar H134 (Fig. 8A). Although speculative, this structural feature raises the possibility that nonpolar substitutions at S97 promote hydrophobic interactions with residues V131 and L135 on the TM2/3 ring, thereby moving this residue away from the polar H134 (Fig. 8A). The net effect is predicted to induce a slight counterclockwise rotation of the TM1 helix, which should lower the free energy barrier at the V102/F99 region in the pore to allow ion conduction.
Interestingly, with the exception of S97G/A/T mutants, which exhibited WT store-operated behavior, introduction of hydrophobic or large amino acids at position 97 yielded nonselective currents when gated by STIM1 (Fig. 8 C and E and SI Appendix, Fig. S8 and Table S3). We postulate that hydrophobic substitutions disrupt the alternating polar/nonpolar interface with the TM2/3 ring, thereby creating a “sticky” hydrophobic patch that constrains the motions of the TM1 segment and traps the channel in a nonselective open state. Large amino acid substitutions (S97F/Y/W) likely reduce the conformational flexibility of the TM1 segment through steric effects (Fig. 8E), much like the LOF phenotypes of bulky H134X mutants (Fig. 3C). Overall, these data suggest that the small, polar nature of S97 as part of the serine ridge provides a degree of conformational flexibility at the TM1–TM2/3 ring interface that is essential for STIM1 gating and ion selectivity.
Discussion
Whereas previous studies have identified individual GOF and LOF mutations within the Orai1 TMs (12–17), our study systematically examines the roles of all four TMs in gating. The identification of GOF mutations encompassing all regions of Orai1 strongly suggests a concerted, global gating mechanism that involves the entire protein rather than a localized motion at the N terminus and TM1, consistent with a gating signal being transmitted through the lipid-spanning regions of the TMs. Notably, the large number of GOF mutations identified through the cysteine screen implies that the energy barrier to Orai1 pore opening is likely not large. We postulate that the constitutively open cysteine mutations activate the channel by removing inter-TM constraints that maintain the closed conformation. Because the mutants have varying degrees of current density and ion selectivity and almost all can be further gated by STIM1, several of the mutants likely represent trapped intermediate states along the STIM1 gating pathway. These mutants could serve as a critical resource for future drug-discovery studies intended for manipulating the function of Orai1 channels and for X-ray crystallography or cryo-EM studies aimed at elucidating the structure of the open channel.
A noteworthy feature of the constitutively active mutants is that their function is abrogated by mutations in the Orai1 N terminus (K85E and Δ2–85) that are known to impair STIM1-mediated gating (29, 39). Although residues 73–85 in this region of the N terminus are also thought to directly bind STIM1 (5), this result suggests that the loss of channel function in the Orai1 N-terminal mutations may not be directly related to lack of STIM1 binding to this site. This conclusion is consistent with other recent studies that have examined the contribution of the membrane-proximal N terminus for Orai1 gating (16, 31). We do not yet know how the Orai1 N terminus contributes to channel function, but possible explanations could include an interaction with another part of Orai1 in the open state (40) or a role in ion permeation (41).
Our results also suggest a potential mechanism by which H134 orchestrates Orai1 gating. The position and orientation of histidine side chains are defined by two torsion angles, χ1 and χ2. However, at the 3.35-Å resolution of the electron density map of 4HKR (7), the χ2 angle cannot be determined directly and must be inferred from the availability of potential hydrogen bonding partners and known rotameric positions available to histidine side chains in α-helices (42, 43). For H206 (hOrai1 H134) in 4HKR, the assigned χ1, χ2 angles of −82°, −60° orient the ND1 nitrogen toward the side-chain OH of S165 (hOrai1 S93; SI Appendix, Fig. S9), raising the possibility that a hydrogen bond between these residues plays a significant functional role (7, 18). However, a different side-chain rotamer is also fully consistent with the crystallographic electron density. This rotamer, which adopts χ1, χ2 angles of −82°, 80° (i.e., a ∼140° rotation about the Cβ-Cγ bond) is found to be selectively enriched in TMs (42) because it allows for intrahelical hydrogen bonding between the ND1 nitrogen of the imidazole ring and the carbonyl oxygen of the i-4 residue preceding it in the α-helix (SI Appendix, Fig. S9). We can propose then, with equal validity based on the crystallographic evidence, that the potentially significant interaction of H206 (hOrai1 H134) side chain is not with S165 of TM1 (hOrai1 S93), but instead with the backbone carbonyl of L202 (hOrai1 L130). This intrahelical interaction could stabilize the orientation of the histidine side chain such that it functions as a steric “brake” at the nexus of the interface between helices TM1, TM2, and TM3 that can be released through the substitution of the endogenous His with small and flexible amino acids. Intriguingly, an asparagine substitution at position 206 is isosteric for the four overlapping atoms with the His, predicting that this side chain should also readily adopt a rotameric position that allows formation of an intrahelical hydrogen bond of its ND2 nitrogen with the carbonyl oxygen of L202 (43) (SI Appendix, Fig. S9). Indeed, the hOrai1 H134N substitution is the one mutant that behaves like WT Orai1. Overall, these results suggest that the loss of the privileged intrahelical stabilization available for His and Asn side chains by small and flexible substitutions destabilizes the interface to release the brake. By contrast, larger substitutions at H134 would be expected to hinder the flexibility between the TM1 and TM2 segments needed for gating, leading to LOF phenotypes.
Based on our results, there are two possible ways in which Orai1 can conduct ions in the absence of STIM1: (i) because of a leaky gate caused by mutations of V102/F99 residues that comprise a hydrophobic channel gate (8, 10, 11) or (ii) as a result of activation gating that would involve displacement the F99 side chains away from the pore axis (11). Albeit through different mechanisms, both would be expected to diminish the free energy barrier for water and ions, thereby permitting ion conduction. In the Ca2+-selective H134S mutant channels where the hydrophobic gate itself is intact, our results indicate that this mutation mimics the activated state of Orai1 by configuring the pore into a state analogous to STIM1-activated channels. We believe that a similar mechanism involving activation of the hydrophobic channel gate underlies the GOF phenotypes of S97C and P245L, given the similarity of permeation properties of H134S with these disease-linked mutations (12, 13). It is also notable that in the MD simulations, the hydrophobic stretch is the only region undergoing changes in hydration between closed vs. open channel variants, reaffirming that this region is likely the primary regulator of channel gating.
The non–pore-lining surface of TM1 contains two qualitatively distinct functional interfaces identifiable on the basis of packing density and hydrophobic or hydrophilic character. Towards the extracellular side of the channel is a hydrophobic stack formed by residues of TM1 and the TM2/3 ring. Positioned close to the selectivity filter and the hydrophobic gate in the pore, this interdigitating stack of hydrophobic residues is optimally located to relay structural rearrangements from the TM2/3 ring to the channel gate and the selectivity filter, a prediction borne out in the STIM1-mediated gating defects seen in point mutations in this stack. By contrast, the cytoplasmic side of the TM1 helix is lined by a ridge of serine residues rendering this surface polar, flexible, and loosely packed at the interface, suggesting that this segment is allowed considerable conformational freedom in relation to the surrounding helical ring. Together, these characteristics optimize the communication of the gating signal between the TM2/3 ring and TM1 to drive STIM1-dependent pore opening and confer high Ca2+ selectivity.
Methods
CRAC currents were recorded from recombinant Orai1 channels overexpressed in HEK293 cells in the presence or absence of STIM1. Methods for whole-cell patch-clamp recordings of Orai1 currents and FRET measurements for monitoring Orai1–CAD interactions were as previously described (11, 44). MD simulations were carried out using the crystal structure of the dOrai channel (PDB ID code 4HKR) as previously described (11). Atomic packing analysis was performed by using PROBE (37, 38). A detailed description of study methods is provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank members of the M.P. laboratory for helpful discussions. We acknowledge the supercomputer resources provided by WestGrid (www.westgrid.ca) and Compute Canada Calcul Canada for the molecular simulations. This work was supported by National Institutes of Health (NIH) Grants NS057499 and GM114210 (to M.P.), Canadian Institutes of Health Research Grant MOP130461 (to R.P.), NIH Predoctoral Fellowships T32GM008382 and F31NS101830 (to P.S.-W.Y.), and the Medical Scientist Training Program (to P.S.-W.Y.). Northwestern University’s Center for Advanced Microscopy is supported by NIH Grant NCRR 1S10 RR031680-01.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718373115/-/DCSupplemental.
References
- 1.Prakriya M, Lewis RS. Store-operated calcium channels. Physiol Rev. 2015;95:1383–1436. doi: 10.1152/physrev.00020.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feske S. ORAI1 and STIM1 deficiency in human and mice: Roles of store-operated Ca2+ entry in the immune system and beyond. Immunol Rev. 2009;231:189–209. doi: 10.1111/j.1600-065X.2009.00818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feske S. CRAC channelopathies. Pflugers Arch. 2010;460:417–435. doi: 10.1007/s00424-009-0777-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacruz RS, Feske S. Diseases caused by mutations in ORAI1 and STIM1. Ann N Y Acad Sci. 2015;1356:45–79. doi: 10.1111/nyas.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park CY, et al. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan JP, et al. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou X, Pedi L, Diver MM, Long SB. Crystal structure of the calcium release-activated calcium channel Orai. Science. 2012;338:1308–1313. doi: 10.1126/science.1228757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNally BA, Somasundaram A, Yamashita M, Prakriya M. Gated regulation of CRAC channel ion selectivity by STIM1. Nature. 2012;482:241–245. doi: 10.1038/nature10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong H, Fiorin G, Carnevale V, Treptow W, Klein ML. Pore waters regulate ion permeation in a calcium release-activated calcium channel. Proc Natl Acad Sci USA. 2013;110:17332–17337. doi: 10.1073/pnas.1316969110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudlur A, et al. STIM1 triggers a gating rearrangement at the extracellular mouth of the ORAI1 channel. Nat Commun. 2014;5:5164. doi: 10.1038/ncomms6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita M, et al. STIM1 activates CRAC channels through rotation of the pore helix to open a hydrophobic gate. Nat Commun. 2017;8:14512. doi: 10.1038/ncomms14512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nesin V, et al. Activating mutations in STIM1 and ORAI1 cause overlapping syndromes of tubular myopathy and congenital miosis. Proc Natl Acad Sci USA. 2014;111:4197–4202. doi: 10.1073/pnas.1312520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garibaldi M, et al. A novel gain-of-function mutation in ORAI1 causes late-onset tubular aggregate myopathy and congenital miosis. Clin Genet. 2017;91:780–786. doi: 10.1111/cge.12888. [DOI] [PubMed] [Google Scholar]
- 14.Böhm J, et al. ORAI1 mutations with distinct channel gating defects in tubular aggregate myopathy. Hum Mutat. 2017;38:426–438. doi: 10.1002/humu.23172. [DOI] [PubMed] [Google Scholar]
- 15.Endo Y, et al. Dominant mutations in ORAI1 cause tubular aggregate myopathy with hypocalcemia via constitutive activation of store-operated Ca2+ channels. Hum Mol Genet. 2015;24:637–648. doi: 10.1093/hmg/ddu477. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, et al. The STIM1-binding site nexus remotely controls Orai1 channel gating. Nat Commun. 2016;7:13725. doi: 10.1038/ncomms13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palty R, Stanley C, Isacoff EY. Critical role for Orai1 C-terminal domain and TM4 in CRAC channel gating. Cell Res. 2015;25:963–980. doi: 10.1038/cr.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frischauf I, et al. Transmembrane helix connectivity in Orai1 controls two gates for calcium-dependent transcription. Sci Signal. 2017;10:eaao0358. doi: 10.1126/scisignal.aao0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Changeux JP, Christopoulos A. Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell. 2016;166:1084–1102. doi: 10.1016/j.cell.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 20.Tirado-Lee L, Yamashita M, Prakriya M. Conformational changes in the Orai1 C-terminus evoked by STIM1 binding. PLoS One. 2015;10:e0128622. doi: 10.1371/journal.pone.0128622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palty R, Fu Z, Isacoff EY. Sequential steps of CRAC channel activation. Cell Rep. 2017;19:1929–1939. doi: 10.1016/j.celrep.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bogdanov M, Zhang W, Xie J, Dowhan W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM(TM)): Application to lipid-specific membrane protein topogenesis. Methods. 2005;36:148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNally BA, Yamashita M, Engh A, Prakriya M. Structural determinants of ion permeation in CRAC channels. Proc Natl Acad Sci USA. 2009;106:22516–22521. doi: 10.1073/pnas.0909574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srikanth S, Yee MK, Gwack Y, Ribalet B. The third transmembrane segment of orai1 protein modulates Ca2+ release-activated Ca2+ (CRAC) channel gating and permeation properties. J Biol Chem. 2011;286:35318–35328. doi: 10.1074/jbc.M111.265884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, et al. Graded activation of CRAC channel by binding of different numbers of STIM1 to Orai1 subunits. Cell Res. 2011;21:305–315. doi: 10.1038/cr.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mullins FM, Park CY, Dolmetsch RE, Lewis RS. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc Natl Acad Sci USA. 2009;106:15495–15500. doi: 10.1073/pnas.0906781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scrimgeour N, Litjens T, Ma L, Barritt GJ, Rychkov GY. Properties of Orai1 mediated store-operated current depend on the expression levels of STIM1 and Orai1 proteins. J Physiol. 2009;587:2903–2918. doi: 10.1113/jphysiol.2009.170662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derler I, et al. A Ca2(+)release-activated Ca2(+) (CRAC) modulatory domain (CMD) within STIM1 mediates fast Ca2(+)-dependent inactivation of ORAI1 channels. J Biol Chem. 2009;284:24933–24938. doi: 10.1074/jbc.C109.024083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lis A, Zierler S, Peinelt C, Fleig A, Penner R. A single lysine in the N-terminal region of store-operated channels is critical for STIM1-mediated gating. J Gen Physiol. 2010;136:673–686. doi: 10.1085/jgp.201010484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNally BA, Somasundaram A, Jairaman A, Yamashita M, Prakriya M. The C- and N-terminal STIM1 binding sites on Orai1 are required for both trapping and gating CRAC channels. J Physiol. 2013;591:2833–2850. doi: 10.1113/jphysiol.2012.250456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derler I, et al. The extended transmembrane Orai1 N-terminal (ETON) region combines binding interface and gate for Orai1 activation by STIM1. J Biol Chem. 2013;288:29025–29034. doi: 10.1074/jbc.M113.501510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, et al. STIM1 gates the store-operated calcium channel ORAI1 in vitro. Nat Struct Mol Biol. 2010;17:112–116. doi: 10.1038/nsmb.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derler I, et al. Authentic CRAC channel activity requires STIM1 and the conserved portion of the Orai N terminus. J Biol Chem. 2018;293:1259–1270. doi: 10.1074/jbc.M117.812206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindl R, et al. 2-aminoethoxydiphenyl borate alters selectivity of Orai3 channels by increasing their pore size. J Biol Chem. 2008;283:20261–20267. doi: 10.1074/jbc.M803101200. [DOI] [PubMed] [Google Scholar]
- 35.Yamashita M, Prakriya M. Divergence of Ca(2+) selectivity and equilibrium Ca(2+) blockade in a Ca(2+) release-activated Ca(2+) channel. J Gen Physiol. 2014;143:325–343. doi: 10.1085/jgp.201311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360, 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 37.Word JM. 2000. All-atom small-probe contact surface analysis: An information-rich description of molecular goodness-of-fit. PhD thesis (Duke University, Durham, NC)
- 38.Word JM, et al. Visualizing and quantifying molecular goodness-of-fit: Small-probe contact dots with explicit hydrogen atoms. J Mol Biol. 1999;285:1711–1733. doi: 10.1006/jmbi.1998.2400. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, et al. Mapping the interacting domains of STIM1 and Orai1 in Ca2+ release-activated Ca2+ channel activation. J Biol Chem. 2007;282:29448–29456. doi: 10.1074/jbc.M703573200. [DOI] [PubMed] [Google Scholar]
- 40.Fahrner M, et al. Communication between N terminus and loop2 tunes Orai activation. J Biol Chem. 2018;293:1271–1285. doi: 10.1074/jbc.M117.812693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong H, Klein ML, Fiorin G. Counterion-assisted cation transport in a biological calcium channel. J Phys Chem B. 2014;118:9668–9676. doi: 10.1021/jp5059897. [DOI] [PubMed] [Google Scholar]
- 42.Chamberlain AK, Bowie JU. Analysis of side-chain rotamers in transmembrane proteins. Biophys J. 2004;87:3460–3469. doi: 10.1529/biophysj.104.044024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lovell SC, Word JM, Richardson JS, Richardson DC. Asparagine and glutamine rotamers: B-factor cutoff and correction of amide flips yield distinct clustering. Proc Natl Acad Sci USA. 1999;96:400–405. doi: 10.1073/pnas.96.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navarro-Borelly L, et al. STIM1-Orai1 interactions and Orai1 conformational changes revealed by live-cell FRET microscopy. J Physiol. 2008;586:5383–5401. doi: 10.1113/jphysiol.2008.162503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blaber M, Zhang XJ, Matthews BW. Structural basis of amino acid alpha helix propensity. Science. 1993;260:1637–1640. doi: 10.1126/science.8503008. [DOI] [PubMed] [Google Scholar]
- 46.Yeung PS, Yamashita M, Prakriya M. Pore opening mechanism of CRAC channels. Cell Calcium. 2017;63:14–19. doi: 10.1016/j.ceca.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Radzicka A, Wolfenden R. Comparing the polarities of the amino-acids: Side-chain distribution coefficients between the vapor-phase, cyclohexane, 1-octanol, and neutral aqueous-solution. Biochemistry. 1988;27:1664–1670. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.