Significance
Barbels are important sensory organs for food seeking of teleosts, reptiles, and amphibians, but the molecular basis of barbel development is unknown. Here, we exploited the barbel-less bottlenose catfish as a natural model to determine the genomic basis for barbel development. Through a series of comparative analyses using genome and transcriptome datasets, a chemokine gene, ccl33, was identified as a key regulator of barbel development. Its knockout in zebrafish led to the loss of barbels, further supporting the roles of ccl33 for barbel development. These findings demand functional studies of chemokines as key developmental, as well as immune, regulators.
Keywords: barbel, chemokine, regeneration, zebrafish, catfish
Abstract
Barbels are important sensory organs in teleosts, reptiles, and amphibians. The majority of ∼4,000 catfish species, such as the channel catfish (Ictalurus punctatus), possess abundant whisker-like barbels. However, barbel-less catfish, such as the bottlenose catfish (Ageneiosus marmoratus), do exist. Barbeled catfish and barbel-less catfish are ideal natural models for determination of the genomic basis for barbel development. In this work, we generated and annotated the genome sequences of the bottlenose catfish, conducted comparative and subtractive analyses using genome and transcriptome datasets, and identified differentially expressed genes during barbel regeneration. Here, we report that chemokine C-C motif ligand 33 (ccl33), as a key regulator of barbel development and regeneration. It is present in barbeled fish but absent in barbel-less fish. The ccl33 genes are differentially expressed during barbel regeneration in a timing concordant with the timing of barbel regeneration. Knockout of ccl33 genes in the zebrafish (Danio rerio) resulted in various phenotypes, including complete loss of barbels, reduced barbel sizes, and curly barbels, suggesting that ccl33 is a key regulator of barbel development. Expression analysis indicated that paralogs of the ccl33 gene have both shared and specific expression patterns, most notably expressed highly in various parts of the head, such as the eye, brain, and mouth areas, supporting its role for barbel development.
Barbels are integumentary sense organs in fishes, reptiles, and amphibians. Barbel structures are relatively simple, consisting of a central rod of cartilage, nerve trunk, artery, and taste buds. Although studies of barbels started over a century ago (1, 2), their origins, evolutions, and molecular basis of development have not been fully elucidated (3, 4). The cell types in barbels include skin cells, neural crest-derived pigment cells, circulatory vessels, taste buds, and sensory nerves, which are highly conserved in the teleost (5). Barbels are a useful system for studying angiogenesis, neural pathfinding, wound healing, scar formation, and other key processes in vertebrate physiology (5). Studies have been conducted to understand gene regulation during barbel regeneration. Many FGFs and Wnt pathway members, as well as myelin basic proteins, are differentially expressed during early barbel regeneration of zebrafish, but the mechanism of molecular control of barbel development, or its regeneration, is unknown (6).
Catfish (Siluriformes) are one of the most diverse vertebrate orders, with ∼4,000 species, and are most characteristic of possession of whisker-like barbels (7). However, barbel-less catfish species do exist in nature. Catfish in the genera Ageneiosus and Tympanopleura lack mandibular barbels and have maxillary barbels greatly reduced in size and lying in a groove at the corner of the mouth above the upper lip (8). As such, they are excellent natural systems for studying the molecular basis of barbel development. We previously generated the reference genome sequence of the channel catfish (Ictalurus punctatus) (9), a catfish with extremely abundant and long barbels. In this study, we generated the whole-genome sequences of the bottlenose catfish (Ageneiosus marmoratus), a barbel-less catfish. The opportunity lies in comparative subtraction of gene contents from barbeled and barbel-less fish species. The genome-level analysis, when coupled with transcriptome-level analysis of differentially expressed genes during barbel regeneration and gene knockout of the potential candidate gene(s), allows identification of both genes involved in barbel development and the key regulators of barbel formation and regeneration. Here, we report that a chemokine C-C motif ligand gene, ccl33, is present in genomes of barbeled fish but absent from genomes of barbel-less fish; it is highly expressed in the head, especially the mouth area; it is differentially expressed during barbel regeneration; and its knockout led to the loss of barbels in zebrafish, suggesting that ccl33 is a key regulator of barbel development in teleost fish.
Results and Discussion
Genome Sequencing, Assembly, and Annotation.
Paired-end and mate-pair Illumina reads of 180-fold genome coverage from a single adult male bottlenose catfish were generated, assembled, and annotated (Table 1). After trimming of low-quality reads, 455 million reads in the pair-end library and 825 million reads in the mate-pair libraries were assembled into 1.02-Gb genome sequences, with 16,063 scaffolds and a scaffold N50 (length of the smallest scaffold in the set that contains the fewest scaffolds whose combined length represents at least 50% of the assembly) of 223.1 kb (Table 1). The guanine-cytosine content of the bottlenose catfish genome was 39.0%. A total of 23,367 protein-coding genes were annotated from the bottlenose catfish genome. Approximately 265 Mb of repetitive elements (25.9% of the genome) was identified, of which interspersed repeats were the most abundant, accounting for 23.3% of the genome, followed by simple sequence repeats, accounting for 2.3% of the genome. Class II transposons were the most abundant elements of the interspersed repeat elements, making up 20.2% of the genome. Of the class II transposable elements, Tc1 transposons were the most abundant, representing 15.3% of the genome. Class I transposons (long interspersed nuclear elements, short interspersed nuclear elements, and long terminal repeats) were scarce, making up only 0.8% of the genome.
Table 1.
Assembly and annotation of the bottlenose catfish (A. marmoratus) genome sequences
| Field | Parameter | Contents |
| Genome sequencing | Paired-end reads (125 bp per read) | 498.5M (62.2-fold) |
| Mate-pair reads (library 1, 100 bp per read) | 586.7M (58.7-fold) | |
| Mate-pair reads (library 2, 100 bp per read) | 591.0M (59.1-fold) | |
| Total reads | 1,676.2M (180-fold) | |
| Genome assembly | Total length of genome assembly | 1,023 Mb |
| GC content | 39.0% | |
| Number of scaffolds | 16,063 | |
| N50 | 223,139 bp | |
| Average scaffold size | 64,122 bp | |
| Genome annotation | Number of predicted genes | 23,367 |
| Interspersed repeats | 238.4 Mb (23.3%) | |
| Satellites | 0.8 Mb (0.08%) | |
| Simple repeats | 23.7 Mb (2.3%) | |
| Low complexity | 2.1 Mb (0.2%) |
GC, guanine-cytosine; N50, length of the smallest scaffold in the set that contains the fewest scaffolds whose combined length represents at least 50% of the assembly.
Comparison of Gene Contents of Bottlenose Catfish and Channel Catfish.
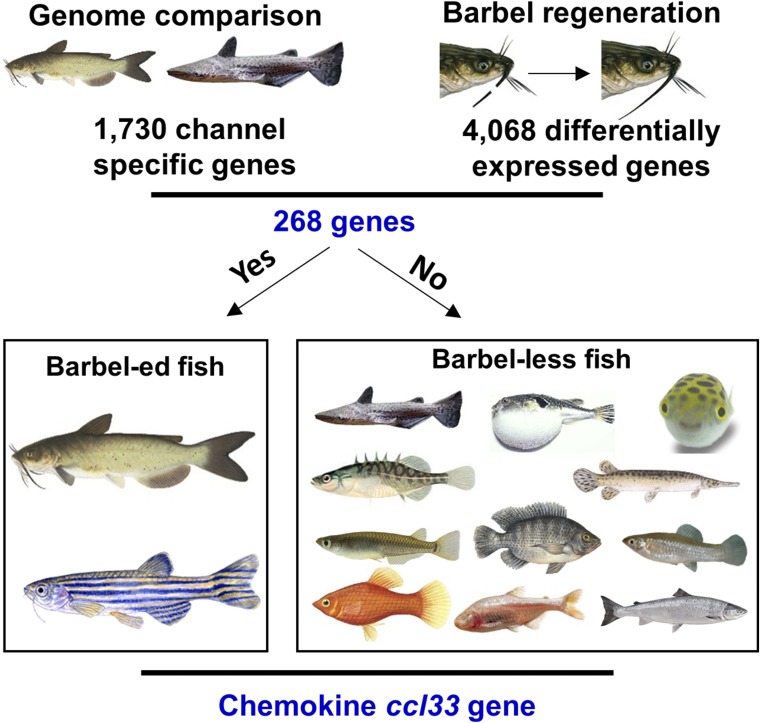
Bottlenose catfish and channel catfish both belong to the order Siluriformes and are the most closely related among those fish with sequenced genomes (SI Appendix, Fig. S1), but they have a sharp contrast when compared for barbels: Channel catfish have four pairs of barbels, while bottlenose catfish are generally regarded as barbel-less, with only one pair of ossified maxillary barbels that are greatly shortened (SI Appendix, Fig. S2). This trait makes them good natural models for understanding the genomic basis for barbel development. We first compared the gene contents of the bottlenose catfish genome and the channel catfish genome (9) to categorize the genomic differences between them. A total of 1,730 genes were identified in the channel catfish genome but absent from the bottlenose catfish genome (Dataset S1); conversely, 2,362 genes were found in the bottlenose catfish genome but absent from the channel catfish genome. Obviously, bottlenose catfish and channel catfish differ in many different traits, not just in their barbels. A large number of genes could be involved in barbel formation and growth. Therefore, we limited our analysis to those genes that are lost in barbel-less genomes. We reasoned that genes important for barbel development should perhaps be differentially expressed during barbel regeneration, and the genes that satisfy the following two conditions could be more likely involved in barbel development: (i) The gene(s) are present in barbeled fish but absent in barbel-less fish, and (ii) the gene(s) should be differentially expressed during barbel regeneration.
Differentially Expressed Genes During Barbel Regeneration in Channel Catfish.
We conducted a barbel regeneration experiment with channel catfish to identify the differentially expressed genes during barbel regeneration. After amputation of the maxillary barbels, the blastema covering the amputation site was visible 3 d after amputation. Then, the tissues began to accumulate on one side of the barbel. The barbels then gradually regenerated, and 1 mo later, the amputated barbels were regenerated to approximately half of the amputated portions. Gene expression profiles were determined during the first 2 wk during barbel regeneration. RNA-sequencing (RNA-Seq) was conducted for samples collected immediately after amputation, and at days 1, 2, 3, 7, and 14 after amputation. RNA-Seq reads were mapped to the channel catfish reference genome sequences to determine differentially expressed genes by comparing expression immediately after amputation at each of the postamputation time points. The largest number of differentially expressed genes was observed on day 1 after barbel amputation, with 2,723 differentially expressed genes. This was probably due to the induction of a large number of wound response genes after amputation of the barbels. The number of differentially expressed genes at day 2 was drastically decreased, with 1,212 genes being differentially expressed. Similarly, the number of differentially expressed genes on days 3, 7, and 14 were 1,013, 1,593, and 1,442 respectively (Dataset S2). When all of the differentially expressed genes at various time points were combined, a total of 4,068 genes were identified to be differentially expressed during barbel regeneration at least at one time point (Dataset S2).
Enrichment analysis of differentially expressed genes at each time point was conducted. The gene ontology (GO) terms that were most significantly enriched during barbel regeneration are listed in SI Appendix, Table S1. One day after amputation, the most significant enriched GO terms were “cellular respiration” and “wound healing.” Two days later, they were shifted to “organism development” and “single-organism developmental process.” Three days later, they became “anatomical structure development” and “embryonic organ development.” At 7 d and 14 d after amputation, the most significant GO terms were “cell adhesion” and “biological adhesion.” These most enriched GO terms were consistent with the observed characterizations during barbel regeneration.
Chemokine ccl33 as a Candidate Gene for Barbel Development.
To narrow down the list of genes potentially involved in the regulation of barbel development, the 1,730 genes present in the channel catfish genome but absent in the bottlenose catfish genome were compared with the 4,068 genes that were differentially expressed during barbel regeneration. A set of 268 genes was identified to be present in the channel catfish genome and absent from the bottlenose catfish genome, and also differentially expressed during barbel regeneration (SI Appendix, Table S2). These 268 genes were potential candidates important for barbel development. We further reasoned that if these genes are indeed essential for barbel development, they should be present in barbeled fish, such as channel catfish and zebrafish, but absent from barbel-less fish. An analysis of gene contents from genomes of 13 fish species present in the Ensembl database was conducted (Fig. 1), with two species, the channel catfish and zebrafish, representing barbeled fish, and 11 species [fugu (Takifugu rubripes), tetraodon (Tetraodon nigroviridis), stickleback (Gasterosteus aculeatus), tilapia (Oreochromis niloticus), spotted gar (Lepisosteus oculatus), medaka (Oryzias latipes), cave fish (Astyanax mexicanus), Amazon molly (Poecilia formosa), platyfish (Xiphophorus maculatus), Atlantic salmon (Salmo salar), and, of course, the bottlenose catfish] representing barbel-less fish. Such gene content analysis led to the identification of only one gene (GenBank GenInfo Identifier: 108268439) that was present in channel catfish and zebrafish, but absent in all of the barbel-less fish, and was differentially expressed in barbel regeneration. This gene was annotated as C-C motif chemokine 5-like at the National Center for Biotechnology Information (NCBI), but a recent comprehensive study of the chemokine gene family reannotated this gene as chemokine C-C motif ligand 33.7 (ccl33.7) (10). As such, the chemokine ccl33 is likely a candidate gene important for the regulation of barbel development and regeneration.
Fig. 1.
Chemokine ccl33 was identified to be present in channel catfish and zebrafish and absent in barbel-less fish, and it was differentially expressed during barbel regeneration of channel catfish. (Clockwise from Top Right) Image courtesy of © Joseph R. Tomelleri (artist); artwork courtesy of Texas Parks and Wildlife Department © 2004; image courtesy of Michi Tobler (photographer); image courtesy of Alamy Stock Photo/Paul R. Sterry; image courtesy of Dreamstime/Verastuchelova; image courtesy of Encyclopaedia Britannica, Inc., copyright 2010, used with permission; image courtesy of Amiidae/Ben Lee; image courtesy of Alamy Stock Photo/PF-(usna1).
Phylogenetic and Syntenic Analysis of ccl33.
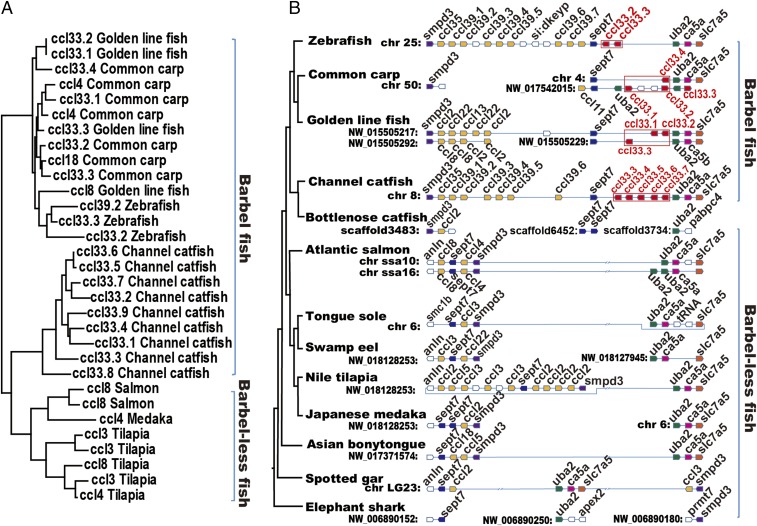
The chemokine ccl33 was identified as a candidate gene for barbel development, making it necessary to further determine its identity and status in various fish species. Chemokines are a group of small, secreted molecules that signal through G protein-coupled receptors to promote cell survival and proliferation and to provide directional guidance to migrating cells (11). They are divided into CC, CXC, CX3C, and XC according to the arrangements of the first two cysteine residues (12). Chemokine ccl33 is a member of the CC subfamily (10, 13). Phylogenetic analysis confirmed the identity of ccl33 (Fig. 2A and SI Appendix, Fig. S3). We further conducted syntenic analysis with fish species whose genome sequences are available to confirm that barbeled fish harbor ccl33, while barbel-less fish lack the ccl33 gene in their genomes. The conserved syntenic regions were then identified from barbeled fish, zebrafish, the common carp (Cyprinus carpio), and the golden line fish (Sinocyclocheilus grahami), and ccl33 genes were identified in the conserved syntenic regions (Fig. 2B). In the channel catfish, nine distinct transcripts of ccl33 were identified (10). In the zebrafish genome, two copies of ccl33 were identified. In the common carp and golden line fish genomes, four and three copies, respectively, of ccl33 were identified (SI Appendix, Table S3).
Fig. 2.
Chemokine ccl33 genes were identified to be specific in barbeled fish. (A) Phylogenetic clade containing chemokine ccl33 genes was only present in barbeled fish. (B) Conserved syntenic region that included the ccl33 (in red box) genes was specific in barbeled fish.
In contrast, the ccl33 gene was not found in the conserved syntenic regions of the Atlantic salmon, half-smooth tongue sole (Cynoglossus semilaevis), swamp eel (Monopterus albus), Nile tilapia, medaka, and spotted gar, all of which are barbel-less fish. The syntenic analyses provided additional evidence that ccl33 was indeed present in barbeled fish but absent from barbel-less fish. Analysis of ccl33 and its copy numbers strongly supported the notion that its presence or absence in fish genomes is correlated with the presence or absence of barbels.
Expression of ccl33 Genes in Zebrafish and Channel Catfish.
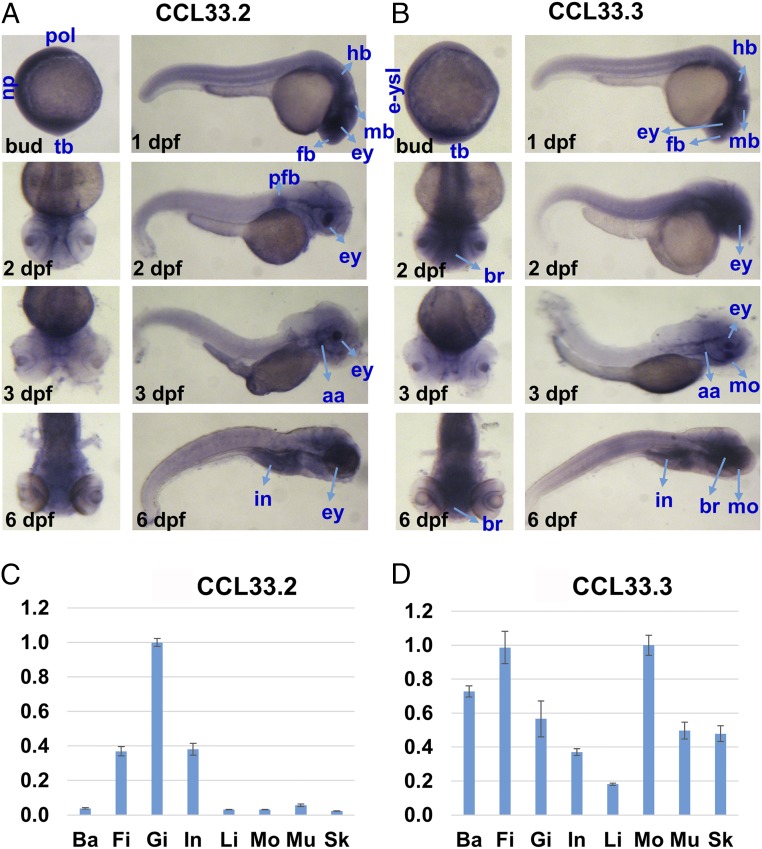
The expression of ccl33.2 and ccl33.3 in zebrafish embryos at different developmental stages, including the bud stage, 1 d postfertilization (dpf), 2 dpf, 3 dpf, and 6 dpf, was analyzed by in situ hybridization. At the bud stage, ccl33.2 was expressed at high levels in the polster, neural plate, and tail bud. At 1 dpf, ccl33.2 was expressed highly in the eye, forebrain, midbrain, and hindbrain. At 2 dpf, ccl33.2 was expressed highly in the eye and pectoral fin bud. At 3 dpf, ccl33.2 was expressed highly in the eye and aortic arches. At 6 dpf, ccl33.2 was expressed highly in the eye and intestine (Fig. 3A).
Fig. 3.
Expression of ccl33 genes in zebrafish. (A and B) In situ hybridization of ccl33.2 and ccl33.3 in zebrafish embryos of different developmental stages, including the bud, 1 dpf, 2 dpf, 3 dpf, and 6 dpf. aa, aortic arches; br, brain; ey, eye; e-ysl, external yolk syncytial layer; fb, forebrain; hb, hindbrain; mb, midbrain; mo, mouth; np, neural plate; pfb, pectoral fin bud; pol, polster; tb, tail bud. (C and D) Expression of ccl33.2 and ccl33.3 in adult zebrafish organs using real-time PCR analysis. The y axis represents the relative expression value. Ba, barbel; Fi, caudal fin; Gi, gill; In, intestine; Li, liver; Mo, mouth; Mu, muscle; Sk, skin.
In contrast, ccl33.3 was expressed at high levels in the tail bud and external yolk syncytial layer during the bud stage. At 1 dpf, ccl33.3 was also expressed highly in the eye, forebrain, midbrain, and hindbrain. At 2 dpf, ccl33.3 showed high levels of expression in the eye and brain. At 3 dpf, ccl33.3 was expressed highly in the mouth area and eye. At 6 dpf, ccl33.2 was expressed highly in the mouth area, brain, and intestine (Fig. 3B). It is apparent that ccl33.2 and ccl33.3 have both shared and specific expression patterns, most notably expressed highly in various parts of the head, such as the eye, brain, and mouth areas, supporting its role for barbel development. This is particularly true with ccl33.3, which is expressed highly in the mouth area where barbels are generated (Fig. 3B). In addition, these genes are expressed highly in the brain, eye, and intestine, suggesting their roles in other processes beyond barbel development.
The expression of ccl33.2 and ccl33.3 in adult zebrafish organs, including the barbel, caudal fin, gill, intestine, liver, mouth, muscle, and skin, was analyzed by real-time PCR. Expression of ccl33.2 was preferentially high in the gill, caudal fin, and intestine of adult zebrafish but very low in the babel, liver, mouth, muscle, and skin (Fig. 3C). In comparison, expression of ccl33.3 was high in most of the tested tissues, with the highest expression in the mouth, followed by the caudal fin, barbels, gill, muscle, skin, intestine, and liver of adult zebrafish (Fig. 3D).
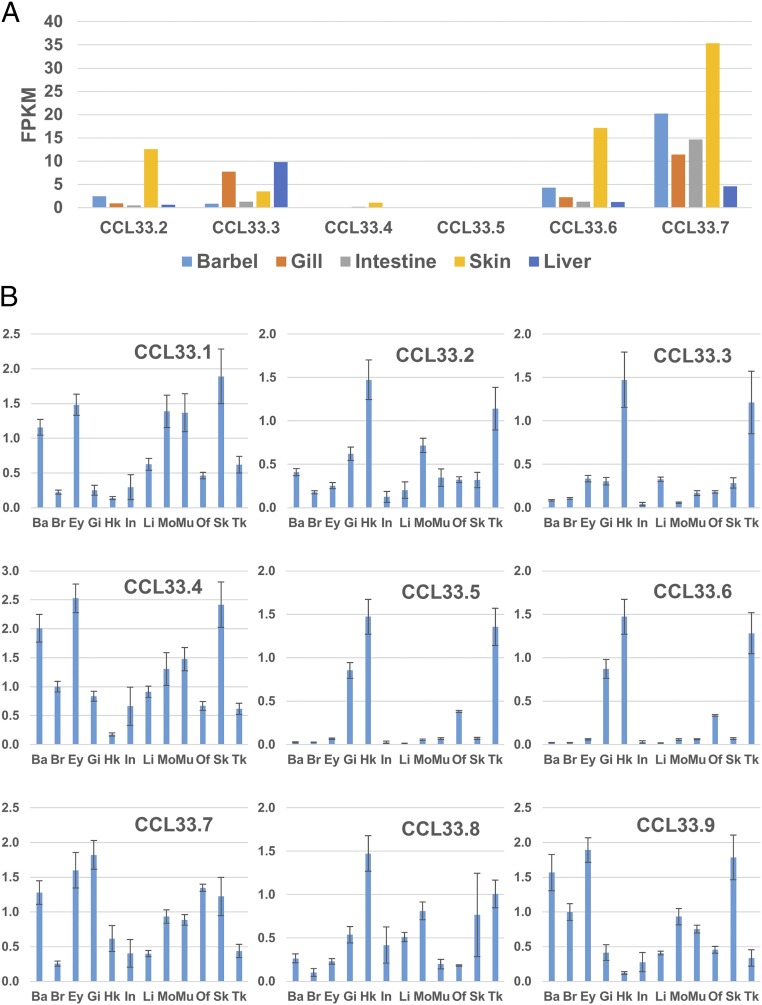
The expression of ccl33 genes in channel catfish was determined through meta-analysis of RNA-Seq datasets from various tissues, including the barbel, gill, intestine, skin, and liver. Of the nine copies of the channel catfish ccl33, six of them (ccl33.2–ccl33.7) were mapped to the reference genome sequence, while the other three copies (ccl33.1, ccl33.8, and ccl33.9) were on unmapped sequence tags (SI Appendix, Table S3). After mapping the RNA-Seq reads to the channel catfish genome, the expression values (fragments per kilobase of transcript per million mapped reads, FPKM) of the six copies of ccl33 were generated (Fig. 4A). Most of the ccl33 genes (ccl33.2, ccl33.4, ccl33.6, and ccl33.7) were expressed highly in skin, followed by the barbel tissue. Because barbels are a skin appendage, they may share similar structures and functions. The ccl33.3 gene was expressed highly in the liver and gill, while the ccl33.5 gene was not expressed in all of the five tissues.
Fig. 4.
Expression analysis of ccl33 in different tissues of channel catfish. (A) Expression of ccl33 genes in the barbel, gill, intestine, skin, and liver of channel catfish using meta-analysis. (B) Expression of ccl33 genes in 12 tissues of channel catfish using real-time PCR analysis. The y axis represents the relative expression value. Ba, barbel; Br, brain; Ey, eye; Gi, gill; Hk, head kidney; In, intestine; Mo, mouth; Mu, muscle; Of, olfactory organ; Sk, skin; Tk, trunk kidney.
The expression pattern of the nine copies of ccl33 in channel catfish was further determined by RT-PCR in 12 tissues, including the barbel, brain, eye, gill, head kidney, intestine, liver, mouth, muscle, olfactory organ, skin, and trunk kidney. As shown in Fig. 4B, the ccl33.1, ccl33.4, ccl33.7, and ccl33.9 genes were expressed highly in the skin, eye, and barbel, suggesting their roles in sensory organs. The ccl33.2, ccl33.3, ccl33.5, ccl33.6, and ccl33.8 genes were expressed highly in the head kidney and trunk kidney (Fig. 4B), suggesting their potential functions in immune responses.
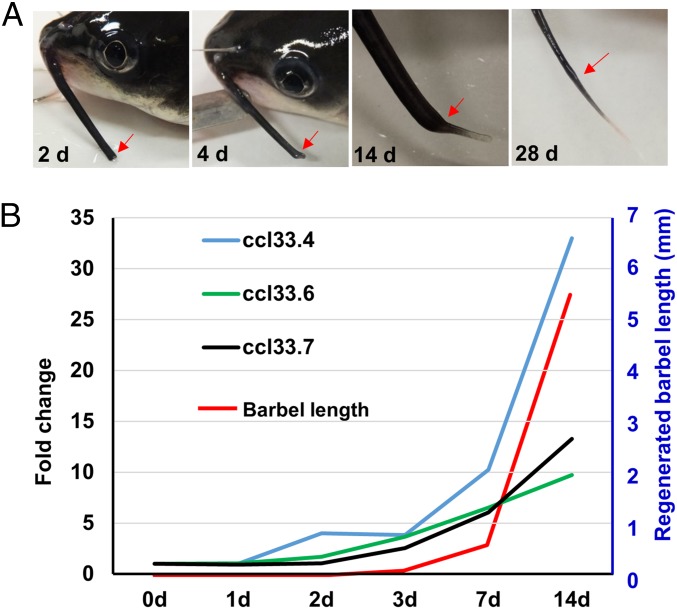
Of all of the nine ccl33 genes in channel catfish, three (ccl33.4, ccl33.6, and ccl33.7) were differentially expressed during barbel regeneration. Their expression during barbel regeneration was approximately synchronized with barbel regeneration (Fig. 5). At the beginning of barbel regeneration, their expression was low; significantly increased expression started at day 3 and then drastically increased after 1 wk during barbel regeneration. Interestingly, the increase in regenerated barbel lengths was well correlated with increased expression of the ccl33 genes (Fig. 5B). Correlated expression of ccl33.4, ccl33.6, and ccl33.7 with barbel regeneration provided additional evidence for their involvement in the regulation of barbel development.
Fig. 5.
Barbel regeneration of channel catfish and expression of ccl33 genes. (A) Morphology of the channel catfish maxillary barbel at different days after amputation. Red arrows indicate the amputation site. (B) Expression of ccl33 genes was timed concordant with the timing of barbel regeneration.
Chemokine ccl33 Knockout in Zebrafish.
We deployed zebrafish knockouts to test the roles of ccl33 for barbel development. In zebrafish, two ccl33 genes exist: ccl33.2 and ccl33.3. The ccl33.2 and ccl33.3 in zebrafish were knocked out by using the CRISPR/Cas9 system. We targeted three regions each for ccl33.2 and ccl33.3 (SI Appendix, Table S4). After injection, the morphology of the zebrafish embryo and larvae was continuously observed and recorded. A barbel blastema bud in the wild-type zebrafish was observed 28 d after hatching (SI Appendix, Fig. S4), similar to previous reports (5, 14). We terminated the experiments at 56 d after hatching as zebrafish became sexually mature and began to breed.
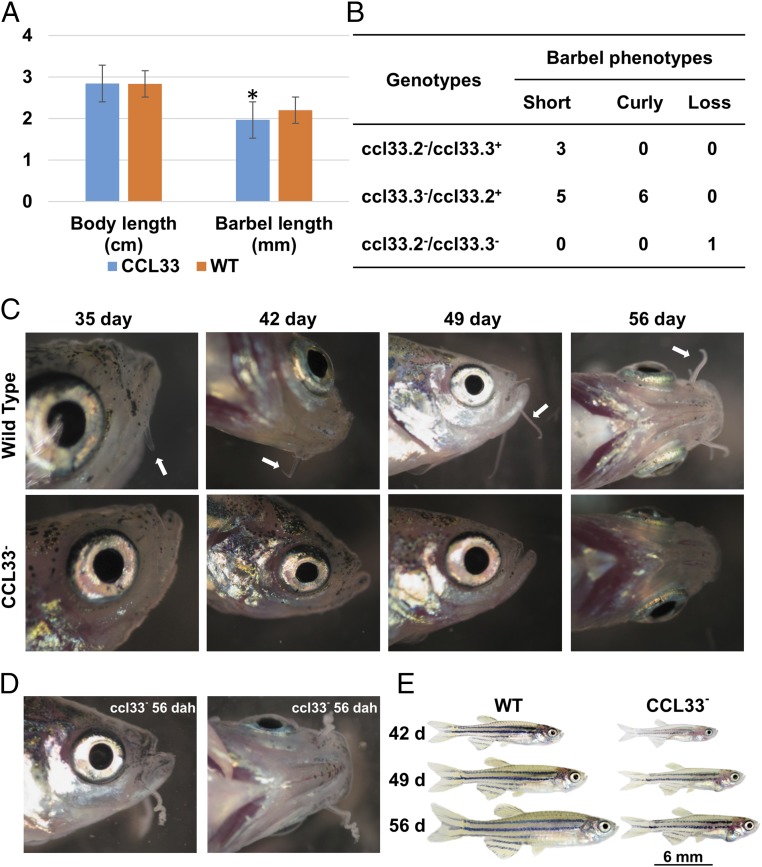
The body length and barbel length were measured from 124 injected zebrafish and 21 noninjected control zebrafish. Body length between the injected fish and the control fish was not statistically different (P < 0.05) using an ANOVA test; however, the barbel length of the injected fish was significantly smaller (P = 0.019) than that of the control fish (Fig. 6A). In addition to the reduced barbel sizes, abnormal barbel phenotypes, including complete loss of barbels and curly barbels, were observed among the injected fish, but not among the control fish (Fig. 6 B–D).
Fig. 6.
Knockout of ccl33.2 and ccl33.3 in zebrafish using the CRISPR/Cas9 system. (A) Body length of injected fish and wild-type (WT) fish was not statistically different; however, the barbel length of the injected fish was significantly smaller (P = 0.019) than that of the WT fish. (B) Number of individuals with different phenotypes in the ccl33− group and their associated genotypes. (C) Head region of WT and ccl33− zebrafish at different days after hatching. White arrows indicate the barbels of WT zebrafish. No barbels were observed in the ccl33− individual. (D) Individual with curly barbels in the ccl33− group. dah, days after hatching. (E) Gross morphology of zebrafish from the WT group and the individual with double knockout of ccl33.2 and ccl33.3.
The genotypes at the ccl33 loci were determined from the injected fish. The target regions from selected individuals with different barbel phenotypes were amplified, and sequenced. A total of 39 fish among the 124 injected fish were observed with abnormal barbels: 23 fish had reduced barbel lengths, 15 fish had curly barbels, and a single fish completely lost its barbels. Of the 39 fish with barbel abnormalities, genotypes were obtained from 15 fish (eight with short barbels, six with curly barbels, and one with lost barbels). The genotypes and phenotypes of the 15 fish are summarized in Fig. 6B: The fish that completely lost its barbels carried a double knockout of both ccl33.2 and ccl33.3, fish with curly barbels carried a single knockout of ccl33.3, and fish with short barbels carried a single knockout of either ccl33.2 or ccl33.3 (Fig. 6B and SI Appendix, Fig. S5). Apparently, double knockout at both ccl33.2 and ccl33.3 loci was rare in our experiments. Interestingly, this individual with double knockout was smaller in body size (Fig. 6E), but this observation was made with only a single individual obtained from the experiment. Future studies are warranted to determine if the double knockouts also have a significantly reduced body size. It is possible that the double knockouts may have pleiotropic effects, including reduced survival and reduced body size. However, the pleiotropic effects on body size, if validated, could be a direct effect of the double knockouts or an indirect effect as a consequence of the barbel loss that may have had an effect on food searching and feeding. Taken together, these results suggested that ccl33 is a key regulator of barbel development. In addition, it might have other functions related to growth and development in general.
Although the genotypes of the knockout fish supported the observed barbel abnormalities, determination of causation of the observed phenotypes requires future experimentation because the action of Cas9 can cause other nonspecific mutations that were not determined in our study. However, we do believe that the phenotypes of parental generation injected fish provided strong evidence for the involvement of ccl33 in the regulation of barbel formation and growth, because all 15 genotyped fish with mutations in ccl33 loci exhibited barbel abnormalities. Future research is warranted to produce pure knockout lines of ccl33 loci for the determination of dose effects of single and double knockouts. However, it is unknown at present if breeding of fish with knockouts is possible, especially with double knockouts. The very low rate of double knockouts in our study (only one fish), along with reduced body size, suggested potential difficulties for the production of pure lines of double knockouts.
The primary functions of homeostatic chemokines in development include organogenesis (15). Homeostatic chemokines have an important role in several steps, including directing the migration of a committed stem cell to the site where an organ is going to develop (16, 17), promoting angiogenesis (18), maintaining stem cells in a niche site until they are needed to produce progeny cells (19), providing a framework or cellular organization to produce the organ (20, 21), and direct proliferative effects on cells (22). The functions of several chemokines and their receptors in directing organogenesis or regeneration have been well studied in zebrafish. For example, Cxcl12, also known as stromal cell-derived factor 1 (Sdf1), is the guidance cue for the lateral line primordium in zebrafish (23). The cxcl12a and cxcr4 genes were essential for heart regeneration in zebrafish (24). Cxcr4 guides gonadal stem cells to their proper site in the developing embryo (25). Cxcr4 is also involved, along with Cxcr7, in modulating the development of the central nervous system (26, 27). Pathways activated by members of the CXC family of chemokines play important roles in the mechanisms of liver repair and regeneration through their effects on hepatocytes (28). The CXC chemokines regulate the hepatic proliferative response and subsequent liver regeneration (29). Despite the knowledge available about the involvement of chemokines or their receptors in organogenesis, no information was available for the function of ccl33. In this work, we identified ccl33 as a candidate gene for barbel development in teleost fish. Its correlated expression during barbel regeneration and its knockout provided strong support for its candidacy as a key regulator of barbel development. In addition, its broad effects on growth and development suggested that ccl33 may have important functions for normal development and growth in zebrafish. Although its knockout was not lethal, the knockout fish were generally smaller and weak in performance, suggesting additional roles of ccl33 beyond controlling barbel development. Although analysis of other effects of ccl33 was not the objective of this study, it is possible that the loss of barbels or their reduced size may have a secondary effect on growth because of the roles of barbels in sensing food, and thereby the effects on feeding. In addition, we must keep in mind that the mainstream of chemokine functions is their involvement in immune responses; the ccl33 knockout could have other effects, such as reduced immunity against infections. This research therefore opens a venue for increased understanding of the functions of chemokines in development, as well as in the defense response against infectious agents.
Materials and Methods
Ethics Statement.
All of the procedures involved in handling and treatment of fish during this study were approved by the Auburn University Institutional Animal Care and Use Committee before the initiation of the study. Tissue samples were collected after euthanasia. All animal procedures were carried out according to the Guide for the Care and Use of Laboratory Animals (30) and the Animal Welfare Act in the United States.
DNA Extraction and Genome Sequencing of Bottlenose Catfish.
The bottlenose catfish used in this study was bought from an online ornamental shop. The morphology indicated that it was a male. After arriving, the fish was anesthetized with MS-222 (Sigma–Aldrich). Blood was drawn from the fish using a 1-mL syringe and stored in cell lysis solution for DNA extraction. Genomic DNA was extracted by using standard protocols. In general, the blood sample stored in cell lysis solution was incubated at 55 °C overnight before extraction. The blood sample was lysed in cell lysis solution containing proteinase K. The protein in the solution was eliminated by adding protein precipitation solution. DNA was precipitated by isopropanol and collected by brief centrifugation. The collected DNA was washed twice with 70% ethanol and then air-dried and resuspended in TE buffer (Thermo Scientific). The quantity and purity of the DNA was measured using a Nanodrop instrument (Thermo Scientific). The collected genomic DNA was send to the Genomics Services Laboratory at HudsonAlpha for sequencing.
Genome Assembly, Gene Prediction, and Annotation of Bottlenose Catfish.
The quality of the raw reads was checked using FastQC (31). Based on the results of FastQC, the adapters, primers, and low-quality bases in the reads were trimmed using Cutadapt (32). The trimmed high-quality reads were input into the ALLPATHS-LG (33) pipeline to assemble the bottlenose catfish genome de novo. The assembled contigs and the reads from the two mate-pair libraries were used in the scaffolding process in SSPACE (34). After scaffolding, five rounds of gap-filling processes were performed to fill the gaps using GapFiller (35).
After gap filling, de novo repeat family identification and modeling were performed to construct a custom repeat library using RepeatModeler (36). The repetitive elements, including low-complexity sequences and interspersed repeats in the scaffolds, were masked by alignment with the constructed custom repeat library using RepeatMasker (37). The masked scaffolds were input into AUGUSTUS (38) to predict the genes. The predicted genes were annotated by BLAST in the UniProt (39) and NCBI nonredundant (NR) databases.
Repetitive Element Analysis.
The derived repetitive sequences in the bottlenose catfish genome were searched against curated libraries and the repetitive DNA sequence databases Repbase (40) and Dfam (41) derived from the RepeatMasker package. All of the sequences were further queried and searched against the NCBI nucleotide collection database using Nucleotide-Nucleotide BLAST (BLASTN) to get deeper annotations (E-value < 1e-5).
Comparative Genome Analysis of Bottlenose Catfish and Channel Catfish.
The availability of the channel catfish genome (9) makes it possible to conduct a comparative genomic analysis between channel catfish and bottlenose catfish. An available pipeline was used in this study, with some modifications (9). The orthologs and orthogroups between bottlenose catfish and channel catfish were generated using OrthoFinder (42). To obtain the species-specific genes, Protein BLAST (BLASTP) was performed in which genes not included in the orthogroups were queried against the genes in the orthogroups within the same species, with a maximal E-value of 1e-10. A reciprocal BLASTP with a maximal E-value of 1e-5 was used to query genes with no hits from previous steps (9). The genes with no hits to any orthologs were considered as species-specific genes.
Phylogenomic Analyses.
Phylogenomic analyses were based on protein-coding sequence alignments. We obtained 372 single-copy ortholog sequences from 13 representative species [Elephant shark (Callorhinchus milii), human (Homo sapiens), mouse (Mus musculus), chicken (Gallus gallus), lizard (Anolis carolinensis), Xenopus tropicalis, zebrafish, bottlenose catfish, channel catfish, cave fish, fugu, medaka, and spotted gar] by alignment with MAFFT v7.310 (43). The alignments were combined in a supermatrix with FASconCAT v1.0 (44) and input into GBlocks v0.91b (45) to exclude possible misaligned regions. Phylogenetic reconstruction was carried out in RAxML v8.2.10 (46) with the Jones–Taylor–Thornton (JTT)+F+Γ4 model. Another round of phylogenetic reconstruction was performed using ExaBayes v1.5 (47). The posterior consensus tree was obtained with exclusion of the first 2,000 trees as burn-in and sampling every 500 generations from both chains. Node support was evaluated with 100 bootstrap replicates for RAxML and posterior probability for ExaBayes. To explore further the robustness of any given node of the reconstructed phylogeny, we calculated the internode certainty (IC) of each node and the internode certainty all (ICA) value. IC and ICA metrics were developed to inform the certainty of a phylogeny’s internode based on a given set of individual gene trees. The IC was calculated based on the most prevalent conflicting bipartition at the trees set, whereas all prevalent conflicting bipartitions were considered when calculating the ICA. The IC calculation was conducted on the JTT+F+Γ4 phylogeny using the individual genes’ phylogenies as the tree dataset to quantify incongruence. Individual gene phylogenies were estimated in RAxML using the same model as the one used in the main phylogeny. The 225 individual gene alignments that included sequence information for all 13 species were included in the calculation of IC values in RAxML.
Phylogenetic Analysis of Chemokine Ligand Genes.
Phylogenetic analysis was conducted using C-C and C-X-C motif chemokine ligand genes from fish and some representative species, including the channel catfish, bottlenose catfish, zebrafish, Atlantic salmon, tilapia, Amazon molly, spotted gar, medaka, platyfish, cave fish, fugu, stickleback, tetraodon, coelacanth (Latimeria chalumnae), lamprey (Petromyzon marinus), elephant shark, human, mouse, cattle (Bos taurus), Xenopus, and chicken. Multiple amino acid sequences were aligned by ClustalW (48). The phylogenetic analysis was conducted using MEGA6 (49) with the maximum likelihood method and JTT model. Bootstrapping with 1,000 replications was performed to evaluate the phylogenetic tree.
Syntenic Analysis.
Syntenic analysis was conducted to provide additional evidence for the identification of genes. The genomic region containing the candidate gene was retrieved from the NCBI genome database and the Ensembl database. The localization of candidate genes was compared.
Expression of ccl33 in Zebrafish Embryos by in Situ Hybridization Analysis.
The wild-type EK line of zebrafish was used for in situ hybridization analysis. Development of embryos was at 28 °C, and staging was determined by both hours postfertilization and morphological characteristics (50). Whole-mount RNA in situ hybridization was carried out as previously described (51, 52). Antisense probes for ccl33.2 and ccl33.3 were prepared from cDNA isolated using the primers listed in SI Appendix, Table S5. Amplicons were cloned in pJET1.2 (Thermo Scientific) vectors. Digoxigenin (DIG)-labeled antisense riboprobes were synthesized using a DIG Labeling Kit (Roche).
Expression of ccl33 in Different Tissues of Channel Catfish Using Meta-Analysis.
The RNA-Seq reads from different tissues of channel catfish were downloaded from the NCBI Sequence Read Archive database, including the gill (SRR493667), intestine (SRR357322), skin (SRR1986577), and liver (SRR917955, SRR917956, and SRR917957). The expression of ccl33 in different tissues was analyzed using the TopHat and Cufflinks pipeline (53).
Expression of ccl33 in Different Tissues of Zebrafish and Channel Catfish Using Real-Time PCR Analysis.
Expression of ccl33.2 and ccl33.3 in the adult zebrafish barbel, caudal fin, gill, intestine, liver, mouth, muscle, and skin was analyzed by real-time PCR. Then, expression of ccl33.1–ccl33.9 in the adult channel catfish barbel, brain, eye, gill, head, kidney, intestine, liver, mouth, muscle, olfactory organ, skin, and trunk kidney was also analyzed. Primers for the ccl33 genes in zebrafish (SI Appendix, Table S6) and channel catfish (SI Appendix, Table S7) were designed using NCBI Primer-BLAST. Reverse transcription was performed using a qScript cDNA Synthesis Kit (Quanta Bioscience) according to the manufacturer’s instructions. All of the cDNA products were diluted to 250 ng/μL and utilized for quantitative real-time PCR assay using PerfeCTa SYBR Green SuperMix (Quanta Bioscience) on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories). The profile of thermal cycling for PCR reactions invoked an initial denaturation at 95 °C for 30 s, 40 amplification cycles with a denaturation temperature at 95 °C for 5 s, an extension temperature at 58 °C for 5 s, and an additional temperature ramping step from 65 °C to 95 °C to produce melting curves of the reaction. Test PCR assays were performed in advance to ensure all of the genes were amplified with the expected PCR product sizes. Three replicates were performed for each sample. The generated raw fluorescent data were imported into Bio-Rad CFX Manager (version 1.6) for baseline correction and to estimate amplification efficiencies for each amplicon. Two potential endogenous reference genes (18S rRNA and b2m) (54), and four potential endogenous reference genes (18S rRNA, gusb, gapdh, and tuba) were tested for their expression stability. The calculated starting concentrations of these reference genes were used to rank the stability value and select the optimal endogenous reference genes. The normalized fold expression of the target genes was calculated.
Barbel Regeneration of Channel Catfish.
The channel catfish barbel regeneration trial was conducted at E. W. Shell Fisheries Center, North Auburn Unit. A total of 175 healthy 2-y-old channel catfish were randomly selected. To document the regeneration of channel catfish barbel tissue, we surgically removed left-side maxillary barbels at 11 time points (immediately after amputation and 3 h, 6 h, 12 h, 1 d, 2 d, 3 d, 7 d, 14 d, 21 d, and 28 d after amputation) and observed the progress of regeneration. On the day of surgery, fish were lightly anesthetized in system water containing 0.015% buffered tricaine. Sterile fine forceps were used to elevate the maxillary barbel tissue. At the beginning of the experiment (0 h), one-third of the left-side maxillary barbels of the 175 channel catfish were amputated. The right-side barbels were kept as controls. At each of the following 10 time points, 15 individual fish were randomly picked out and another one-third of the left-side maxillary barbels were amputated. After the surgery, the fish were restocked in the tank and fed twice daily. The amputated barbel tissues from 15 individuals were randomly stored in three 1.5-mL tubes and were considered as three replicates. All of the barbel tissue samples were stored in dry ice temporarily before being moved into a −80 °C refrigerator.
A total of six samples (immediately after amputation and at day 1, day 2, day 3, day 7, and day 14 after amputation) were selected for RNA-Seq analysis. Total RNA was extracted from the collected barbel tissue using an RNeasy Plus Kit (Qiagen). The collected RNA samples were send to Genomics Services Laboratory at HudsonAlpha for sequencing. Poly(A) RNA-Seq libraries were prepared for RNA sequencing. After quantitation and dilution, the libraries were sequenced on an Illumina HiSeq 2000 instrument.
Identification of Differentially Expressed Genes During Channel Catfish Barbel Regeneration.
The TopHat and Cufflinks pipeline was used to identify differentially expressed genes during channel catfish barbel regeneration (53). Briefly, the adapters, low-quality sequences, and short sequences (length < 30 bp) in the raw RNA-Seq reads were trimmed using Trimmomatic (55). After trimming, the remaining high-quality reads were aligned to the channel catfish fish genome using TopHat (56). The aligned RNA-Seq reads and channel catfish transcript annotation file downloaded from the NCBI were input into Cufflinks to detect the differentially expressed genes (53). The sample collected immediately after amputation was used as the control. At each time point (day 1, day 2, day 3, day 7, and day 14 after amputation), the expression value of each gene (FPKM) was compared with that of the control. The q-value (false discovery rate-adjusted P value) of the test was calculated. Genes with expression differences fulfilling statistical significance criteria (q-value < 0.05, |fold change| > 2) at each time point compared with the control were regarded as differentially expressed genes.
Enrichment Analysis.
Enrichment analysis of differentially expressed genes at each time point was performed using the PANTHER classification system (57). Due to the relatively less information available about GO annotation for channel catfish, the zebrafish background was selected for further analysis. The orthologs of channel catfish genes in zebrafish were identified by aligning the channel catfish genes to the zebrafish genome. The identified zebrafish gene identification and zebrafish genome background were selected to perform the enrichment analysis.
Knockout of ccl33 in Zebrafish.
To knock out ccl33.2 and ccl33.3 in zebrafish, we followed a high-throughput, targeted mutagenesis pipeline using CRISPR/Cas9 technology (58). We used a cloning-free method to generate single-guide RNA (sgRNA) templates. Briefly, to generate sgRNA templates, an oligo consisting of the T7 promoter, 18- to 20-nt target sequences, and a 20-nt sequence that overlapped to a generic sgRNA template was designed. The targeting oligo was annealed with an 80-nt chimeric sgRNA core sequence. The annealed oligos were then filled in using Accuzyme polymerase (Bioline). The quality of the assembled oligos was checked on a 2.5% agarose gel. Approximately 8 μL of gRNA template was then used to transcribe RNA by in vitro transcription using a HiScribe T7 High Yield RNA Synthesis Kit (New England BioLabs) according to the manufacturer’s instructions. The sgRNAs were purified by passage through a Microspin S-200 HR Column (GE Healthcare).
For Cas9 mRNA, the zebrafish codon-optimized cas9 plasmid pCS-nCas9n was used as a template (59). The template DNA was linearized by XbaI and purified using a Zymo Clean and Concentrator purification column (Zymo Research). Five hundred to 1,000 ng of linearized template was used to synthesize capped RNA using an mMESSAGE mMACHINE sp6 Kit (Life Technologies) and precipitated using Phenol/Chloroform/Isoamyl Alcohol.
The sgRNAs and Cas9 RNA were coinjected into wild-type EK strain zebrafish embryos. A total volume of ∼2 nL of Cas9 RNA and sgRNAs was coinjected into one-cell-stage embryos at a concentration of 30 pg of sgRNA with about 150 pg of Cas9. Then, embryos were moved into 1-L glass beakers containing E3 zebrafish medium and incubated at 28 °C until hatching. Dead embryos were removed, and water was changed daily.
Genotyping Analysis.
Injected fish were grown to adulthood and screened for CRISPR-induced mutations. Mutagenesis was analyzed by cloning and sequencing. Genomic DNA was extracted from muscle tissue. Genotyping primers for zebrafish (SI Appendix, Table S8) were designed using NCBI Primer-BLAST. A short stretch of the genomic region flanking the target site was PCR-amplified from the genomic DNA. PCR amplification consisted of a 2-min denaturing step at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 s at 57 °C, and 30 s at 72 °C. The PCR amplicons were purified using a QIAquick PCR Purification Kit (Qiagen). The purified PCR products were cloned using a TA Cloning Kit (Invitrogen). The plasmids containing targeted regions were sequenced. The collected sequences were used for genotyping analysis.
Data Availability.
All sequence data that support the findings of this study have been deposited in the NCBI under BioProject accession no. PRJNA427361: SRR6425174, SRR6425175, and SRR6425176 for the bottlenose catfish genome sequence of the 220-bp, 3–5kbp, and 7–10kpb libraries, respectively, and SRR6414583–SRR6414594 for RNA-Seq reads from the channel catfish barbel transcriptome.
Supplementary Material
Acknowledgments
We thank Dr. Michael Miller (Auburn University Harrison School of Pharmacy) for help with microphotography and Dr. Jim Stoeckel and Karen Veverica for providing facilities for and assistance with holding the fish. This project was supported by Agriculture and Food Research Initiative Competitive Grants 2015-67015-22907 and 2017-67015-26295 from the US Department of Agriculture National Institute of Food and Agriculture Animal Genomics, Genetics, and Breeding Program. T.Z. is supported by a scholarship from the China Scholarship Council.
Footnotes
The authors declare no conflict of interest.
Data deposition: All sequence data reported in this paper have been deposited in the National Center for Biotechnology Information under BioProject accession no. PRJNA427361: SRR6425174, SRR6425175, and SRR6425176 for the bottlenose catfish genome sequence of the 220-bp, 3–5 kilobase pair (kbp), and 7–10 kbp libraries, respectively, and SRR6414583–SRR6414594 for RNA-sequencing reads from the channel catfish barbel transcriptome.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718603115/-/DCSupplemental.
References
- 1.Pollard H. The “cirrhostomial” origin of the head in vertebrates. Anat Anz. 1894;9:349–359. [Google Scholar]
- 2.Pollard H. The oral cirri of siluroids and the origin of the head in vertebrates. Zool Jb Anat Ontog. 1895;8:379–424. [Google Scholar]
- 3.Fox H. Barbels and barbel-like tentacular structures in sub-mammalian vertebrates: A review. Hydrobiologia. 1999;403:153–193. [Google Scholar]
- 4.Diogo R, Chardon M. The structures associated with catfish (Teleostei: Siluriformes) mandibular barbels: Origin, anatomy, function, taxonomic distribution, nomenclature and synonymy. Neth J Zool. 2000;50:455–478. [Google Scholar]
- 5.LeClair EE, Topczewski J. Development and regeneration of the zebrafish maxillary barbel: A novel study system for vertebrate tissue growth and repair. PLoS One. 2010;5:e8737. doi: 10.1371/journal.pone.0008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duszynski RJ, Topczewski J, LeClair EE. Divergent requirements for fibroblast growth factor signaling in zebrafish maxillary barbel and caudal fin regeneration. Dev Growth Differ. 2013;55:282–300. doi: 10.1111/dgd.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferraris CJ. Checklist of Catfishes, Recent and Fossil (Osteichthyes: Siluriformes), and Catalogue of Siluriform Primary Types. Magnolia Press; Auckland: 2007. [Google Scholar]
- 8.Birindelli JLO. Phylogenetic relationships of the South American Doradoidea (Ostariophysi: Siluriformes) Neotrop Ichthyol. 2014;12:451–564. [Google Scholar]
- 9.Liu Z, et al. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat Commun. 2016;7:11757. doi: 10.1038/ncomms11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Q, et al. The chemokinome superfamily: II. The 64 CC chemokines in channel catfish and their involvement in disease and hypoxia responses. Dev Comp Immunol. 2017;73:97–108. doi: 10.1016/j.dci.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Wang J, Knaut H. Chemokine signaling in development and disease. Development. 2014;141:4199–4205. doi: 10.1242/dev.101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachelerie F, et al. New nomenclature for atypical chemokine receptors. Nat Immunol. 2014;15:207–208. doi: 10.1038/ni.2812. [DOI] [PubMed] [Google Scholar]
- 13.Fu Q, et al. The chemokinome superfamily in channel catfish: I. CXC subfamily and their involvement in disease defense and hypoxia responses. Fish Shellfish Immunol. 2017;60:380–390. doi: 10.1016/j.fsi.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Hansen A, Reutter K, Zeiske E. Taste bud development in the zebrafish, Danio rerio. Dev Dyn. 2002;223:483–496. doi: 10.1002/dvdy.10074. [DOI] [PubMed] [Google Scholar]
- 15.Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ-specific metastasis. Nat Rev Immunol. 2011;11:597–606. doi: 10.1038/nri3049. [DOI] [PubMed] [Google Scholar]
- 16.Doitsidou M, et al. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–659. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- 17.Knaut H, Werz C, Geisler R, Nüsslein-Volhard C. Tübingen 2000 Screen Consortium A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature. 2003;421:279–282. doi: 10.1038/nature01338. [DOI] [PubMed] [Google Scholar]
- 18.Cui K, et al. The CXCR4-CXCL12 pathway facilitates the progression of pancreatic cancer via induction of angiogenesis and lymphangiogenesis. J Surg Res. 2011;171:143–150. doi: 10.1016/j.jss.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Steinberg M, Silva M. Plerixafor: A chemokine receptor-4 antagonist for mobilization of hematopoietic stem cells for transplantation after high-dose chemotherapy for non-Hodgkin’s lymphoma or multiple myeloma. Clin Ther. 2010;32:821–843. doi: 10.1016/j.clinthera.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Mahabaleshwar H, Boldajipour B, Raz E. Killing the messenger: The role of CXCR7 in regulating primordial germ cell migration. Cell Adhes Migr. 2008;2:69–70. doi: 10.4161/cam.2.2.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valentin G, Haas P, Gilmour D. The chemokine SDF1a coordinates tissue migration through the spatially restricted activation of Cxcr7 and Cxcr4b. Curr Biol. 2007;17:1026–1031. doi: 10.1016/j.cub.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 22.Ehtesham M, Mapara KY, Stevenson CB, Thompson RC. CXCR4 mediates the proliferation of glioblastoma progenitor cells. Cancer Lett. 2009;274:305–312. doi: 10.1016/j.canlet.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perlin JR, Talbot WS. Signals on the move: Chemokine receptors and organogenesis in zebrafish. Sci STKE. 2007;2007:pe45. doi: 10.1126/stke.4002007pe45. [DOI] [PubMed] [Google Scholar]
- 24.Itou J, et al. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development. 2012;139:4133–4142. doi: 10.1242/dev.079756. [DOI] [PubMed] [Google Scholar]
- 25.Raz E, Mahabaleshwar H. Chemokine signaling in embryonic cell migration: A fisheye view. Development. 2009;136:1223–1229. doi: 10.1242/dev.022418. [DOI] [PubMed] [Google Scholar]
- 26.Thelen M, Thelen S. CXCR7, CXCR4 and CXCL12: An eccentric trio? J Neuroimmunol. 2008;198:9–13. doi: 10.1016/j.jneuroim.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 27.Dambly-Chaudière C, Cubedo N, Ghysen A. Control of cell migration in the development of the posterior lateral line: Antagonistic interactions between the chemokine receptors CXCR4 and CXCR7/RDC1. BMC Dev Biol. 2007;7:23. doi: 10.1186/1471-213X-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Sweringen HL, et al. CXC chemokine signaling in the liver: Impact on repair and regeneration. Hepatology. 2011;54:1445–1453. doi: 10.1002/hep.24457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson GC, et al. CXC chemokines function as a rheostat for hepatocyte proliferation and liver regeneration. PLoS One. 2015;10:e0120092. doi: 10.1371/journal.pone.0120092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 31.Andrews S. 2010 FastQC: A quality control tool for high throughput sequence data. Available at www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed September 9, 2015.
- 32.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 33.Maccallum I, et al. ALLPATHS 2: Small genomes assembled accurately and with high continuity from short paired reads. Genome Biol. 2009;10:R103. doi: 10.1186/gb-2009-10-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 35.Boetzer M, Pirovano W. Toward almost closed genomes with GapFiller. Genome Biol. 2012;13:R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smit A, Hubley R. 2010 RepeatModeler Open-1.0. Available at www.repeatmasker.org/RepeatModeler. Accessed August 3, 2016.
- 37.Smit AF, Hubley R, Green P. 2015 RepeatMasker Open-4.0. Available at www.repeatmasker.org. Accessed August 30, 2016.
- 38.Stanke M, Steinkamp R, Waack S, Morgenstern B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium TU. UniProt Consortium UniProt: A hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler TJ, et al. Dfam: A database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 2013;41:D70–D82. doi: 10.1093/nar/gks1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emms DM, Kelly S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada KD, Tomii K, Katoh K. Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics. 2016;32:3246–3251. doi: 10.1093/bioinformatics/btw412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kück P, Meusemann K. FASconCAT: Convenient handling of data matrices. Mol Phylogenet Evol. 2010;56:1115–1118. doi: 10.1016/j.ympev.2010.04.024. [DOI] [PubMed] [Google Scholar]
- 45.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 46.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aberer AJ, Kobert K, Stamatakis A. ExaBayes: Massively parallel bayesian tree inference for the whole-genome era. Mol Biol Evol. 2014;31:2553–2556. doi: 10.1093/molbev/msu236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2:Unit 2.3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 49.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 51.Pham VN, et al. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hauptmann G, Gerster T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 1994;10:266. doi: 10.1016/0168-9525(90)90008-t. [DOI] [PubMed] [Google Scholar]
- 53.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCurley AT, Callard GV. Characterization of housekeeping genes in zebrafish: Male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol Biol. 2008;9:102. doi: 10.1186/1471-2199-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varshney GK, et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015;25:1030–1042. doi: 10.1101/gr.186379.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data that support the findings of this study have been deposited in the NCBI under BioProject accession no. PRJNA427361: SRR6425174, SRR6425175, and SRR6425176 for the bottlenose catfish genome sequence of the 220-bp, 3–5kbp, and 7–10kpb libraries, respectively, and SRR6414583–SRR6414594 for RNA-Seq reads from the channel catfish barbel transcriptome.