Significance
Gliomas exhibit high proportions of glioma stem cells (GSCs), anoikis resistance, increased brain parenchyma invasion, and resistance to therapy with high recurrence. GSCs display protective autophagy, a self-mediated lysosomal degradation process that balances sources of energy at critical times of stress. Protective autophagy in GSCs promotes resistance to anoikis, programmed death resulting from growth in an anchorage-independent manner. MDA-9 is critical in maintaining protective autophagy in GSCs, thereby contributing to anoikis-resistance. MDA-9 regulates critical molecules, including BCL2 and EGFR, which control autophagy. A link between MDA-9 and protective autophagy and anoikis-resistance identifies an Achilles’ heel of glioblastoma multiforme that may be exploited to define enhanced therapies with improved prognosis and decreased recurrence. Accordingly, targeting MDA-9 may represent a viable therapeutic strategy for glioblastoma multiforme.
Keywords: glioma stem cells, autophagy, anoikis resistance, MDA-9/Syntenin, cell death
Abstract
Glioma stem cells (GSCs) comprise a small subpopulation of glioblastoma multiforme cells that contribute to therapy resistance, poor prognosis, and tumor recurrence. Protective autophagy promotes resistance of GSCs to anoikis, a form of programmed cell death occurring when anchorage-dependent cells detach from the extracellular matrix. In nonadherent conditions, GSCs display protective autophagy and anoikis-resistance, which correlates with expression of melanoma differentiation associated gene-9/Syntenin (MDA-9) (syndecan binding protein; SDCBP). When MDA-9 is suppressed, GSCs undergo autophagic death supporting the hypothesis that MDA-9 regulates protective autophagy in GSCs under anoikis conditions. MDA-9 maintains protective autophagy through phosphorylation of BCL2 and by suppressing high levels of autophagy through EGFR signaling. MDA-9 promotes these changes by modifying FAK and PKC signaling. Gain-of-function and loss-of-function genetic approaches demonstrate that MDA-9 regulates pEGFR and pBCL2 expression through FAK and pPKC. EGFR signaling inhibits autophagy markers (ATG5, Lamp1, LC3B), helping to maintain protective autophagy, and along with pBCL2 maintain survival of GSCs. In the absence of MDA-9, this protective mechanism is deregulated; EGFR no longer maintains protective autophagy, leading to highly elevated and sustained levels of autophagy and consequently decreased cell survival. In addition, pBCL2 is down-regulated in the absence of MDA-9, leading to cell death in GSCs under conditions of anoikis. Our studies confirm a functional link between MDA-9 expression and protective autophagy in GSCs and show that inhibition of MDA-9 reverses protective autophagy and induces anoikis and cell death in GSCs.
Glioblastoma multiforme (GBM) is the most frequent and aggressive glial tumor, which consists of a small population of unique therapy-resistant cells, glioma stem cells (GSCs) (1). Current dogma suggests that tumor regrowth originates from GSCs (2), and these unique cells contribute to resistance to therapy, poor prognosis, and recurrence (3). These traits make GSCs an attractive yet challenging target for novel treatment approaches (1, 4). Nonadherent glioma neurosphere cultures are enriched in GSCs; however, their nonstem progeny undergoes anoikis, programmed cell death occurring when adherent cells grow detached from the extracellular matrix (ECM) (5). These observations imply that GSCs are inherently anoikis-resistant, and cell adhesion is not mandatory for their survival.
In nonstem cells, adhesion to ECM activates a number of prosurvival pathways via several key regulatory molecules. These critical molecules also function as master regulators of anoikis-resistance (6). The prosurvival pathways triggered by these agents promote expression and activation of antiapoptotic proteins. Loss of attachment to the ECM can incite distinct changes in cellular and molecular signaling that are not compatible with survival of these detached cells (7). The nonadherent cells show a substantial down-regulation in focal adhesion kinase (FAK) (8) and EGFR (6) signaling, which significantly contributes to inhibition of prosurvival pathways. GSCs, however, are anoikis-resistant (5) and can evade these changes when detached from the ECM, resulting in survival. Resistance to anoikis can be achieved through (i) constitutive activation of prosurvival signaling and (ii) by deregulating and adapting metabolism, through protective functions of autophagy (6).
Autophagy is a (patho-)physiological process occurring in both healthy and malignant cells and can function as either a tumor-suppressing or tumor-promoting factor (1, 9–11). Autophagy can prevent healthy cells from developing into cancer cells and can promote death in tumor cells. In contrast, autophagy induction is often observed during the progression of various human cancers to metastasis (12). Autophagy may play a key role at almost every stage of the metastatic cascade (12). More specifically, autophagy has been shown to be unambiguously involved in cancer stem cell viability, differentiation, as well as anoikis-resistance (12). Autophagy can provide a protective mechanism enabling cells to respond to stress. Protective autophagy in anoikis-resistant cells may compensate for the loss of extrinsic signals promoting nutrient and energy metabolism (13), but the details of this mechanism in GSCs is largely unknown.
Defining the mechanisms by which GBM cells resist therapy is mandatory for developing rational approaches to clinically manage this invariably fatal cancer. MDA-9 (syntenin; syndecan-binding protein/SDCBP), is an evolutionary conserved cytosolic protein (14, 15) implicated in the progression of multiple cancer types and is frequently highly expressed in these cancers (15–20). Expression of melanoma differentiation associated gene-9/Syntenin (MDA-9) correlates with advancing tumor grade in melanoma, breast cancer, glioma, and urothelial cell carcinoma, and is also overexpressed in gastric and breast cancer (15–19). Additionally, expression of MDA-9 is increased in more aggressive cell lines of multiple cancer types, unlike their less-invasive, less-aggressive, and normal counterparts.
MDA-9 is a regulator of GBM invasion, angiogenesis, and tumor progression (19), as well as GSC survival and stemness (21). Gene-expression analysis using publically available databases, such as the Cancer Genome Atlas (TCGA), revealed a correlation between high levels of MDA-9, astrocytoma grade, poor prognosis, and reduced survival (18), with highest expression in GBM (18). Suppression of MDA-9 sensitized GBM to radiation by inhibiting radiation-induced invasion gains and signaling changes (22). MDA-9 is comprised of two tandem PDZ domains, which facilitate the interaction and formation of c-Src–FAK complexes, which are crucial for cancer progression (14, 15).
We now demonstrate that MDA-9 plays a pivotal and decisive role in regulating protective autophagy in anoikis-resistant GSCs. We show that MDA-9 can influence protective autophagy, by triggering FAK/PKC/BCL2 and EGFR signaling. In the absence of MDA-9 expression, the protective function of autophagy is deregulated and the GSCs undergo toxic autophagy, resulting in cell death. These observations highlight the nodal role of MDA-9 in survival of GSCs and support the targeting of this molecule as a therapeutic strategy for GBM and GSCs.
Results
Autophagy Is Required for Survival and Protects Anoikis-Resistant GSCs.
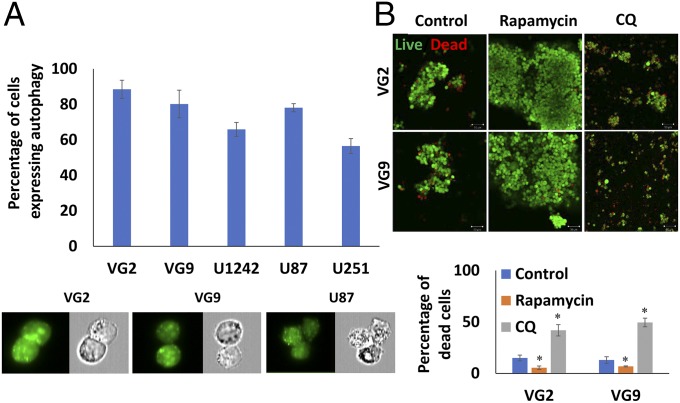
GSCs from multiple clinical samples and cell lines growing under anoikis conditions were evaluated for autophagy. The majority of GSCs in neurospheres were anoikis-resistant and expressed high basal levels of autophagy (Fig. 1A and Fig. S1A). Autophagy was evident in GSCs from clinical samples, VG2 (88% of total population of GSCs) and VG9 (80%), which were generally higher than in cell lines, U1242 (66%), U87 (78%), and U251 (57%), when grown as GSC neurospheres in nonadherent conditions. Quantification of autophagic vesicle markers by flow cytometry in viable GSCs cultured in nonadherent conditions indicated protective autophagy, with variable basal levels of multiple autophagy markers expressed in multiple GBM clinical samples and GBM cell lines. In VG2, U87, U87vIII, U1242, VG9, and VG10, the expression of ATG5 ranged from 7 to 60%; LC3B ranged from 23 to 65%; lysosome-associated membrane protein 1 (Lamp1) ranged from 8 to 55% (Table 1 and Table S1). The levels of these autophagy markers are similar to those observed in other studies of protective autophagy and anoikis-resistance (6, 12, 23). Induction of autophagy in neurospheres by treatment with rapamycin (10 µM) increased spheroid size, spheroids were more compact, and cell viability increased (Fig. 1B and Fig. S1B). Conversely, when autophagy was inhibited by chloroquine (CQ) (20 µM), spheroids showed significant loss of cell viability with decreased spheroid size (Fig. 1B and Fig. S1B). These results indicate that autophagy is protective in anoikis-resistant GSCs.
Fig. 1.
Autophagy is essential for survival of GSCs under anoikis conditions. (A) Percentage of autophagy in anoikis-resistant GSCs determined by flow cytometry. Fluorescence and phase-contrast images captured of the same cells [autophagic vacuoles (green) and blebbing/punctae]. (Magnification: 60×.) (B) Live/dead assay of neurospheres and GSC viability under conditions of autophagy induction (rapamycin treatment) and inhibition (CQ treatment) (green cells are viable). (Magnification: 100×.) Confocal images quantified for cell death and graphically represented. Error bars indicate ±SD, *P < 0.05.
Table 1.
Expression of EGFR, pEGFR, autophagy markers (ATG5, Lamp1, LC3), and pBCL2 in shcon and shmda-9 GSCs
| Cell line | % Expression in shcon GSCs | % Expression in shmda-9 GSCs |
| VG2 | ||
| EGFR | 46.9 ± 2.60 | 44.5 ± 3.80 |
| pEGFR | 39.6 ± 2.04 | 10.8 ± 0.39 |
| ATG5 | 15.1 ± 0.52 | 32.5 ± 1.30 |
| Lamp1 | 16.3 ± 0.78 | 20.7 ± 1.01 |
| LC3 | 23.4 ± 2.22 | 56.3 ± 2.47 |
| pBCL2 | 18.4 ± 0.27 | 1.4 ± 0.03 |
| U87 | ||
| EGFR | 35.7 ± 5.02 | 30.5 ± 7.95 |
| pEGFR | 21.8 ± 1.83 | 2.6 ± 0.05 |
| ATG5 | 13.5 ± 0.15 | 36.7 ± 2.58 |
| Lamp1 | 13.3 ± 1.41 | 37.9 ± 2.13 |
| LC3 | 27.5 ± 1.59 | 53.9 ± 4.03 |
| pBCL2 | 2.9 ± 0.06 | 0.3 ± 0.01 |
| U87 VIII | ||
| EGFR | 68.2 ± 9.10 | 65.9 ± 7.68 |
| pEGFR | 78.2 ± 6.24 | 22.3 ± 1.30 |
| ATG5 | 7.1 ± 0.26 | 35.8 ± 4.16 |
| Lamp1 | 7.6 ± 0.65 | 43.2 ± 2.46 |
| LC3 | 25.4 ± 0.29 | 47.9 ± 4.12 |
| pBCL2 | 20.6 ± 1.06 | 0.5 ± 0.02 |
MDA-9 Expression Correlates with Anoikis-Resistance and Protective Autophagy.
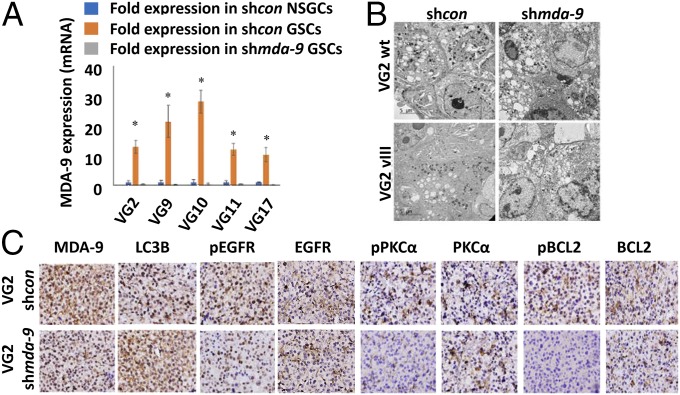
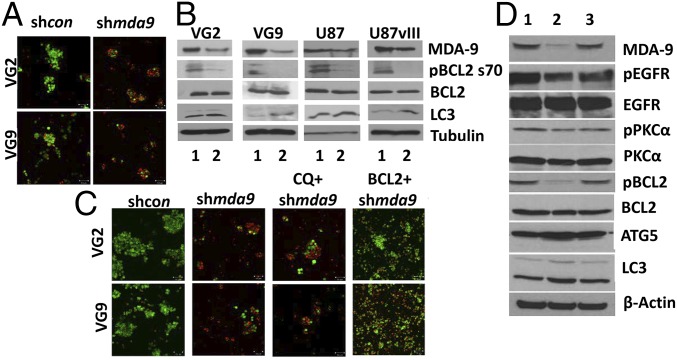
Anoikis-resistant GSCs expressed significantly higher levels of MDA-9 vs. anoikis-sensitive nonstem glioma cells (NSGCs) (Fig. 2A and Fig. S1C). When MDA-9 expression was suppressed in GSCs, the population of anoikis-resistant cells was greatly diminished, and there was a significant decrease in spheroid size (Fig. 3A). Flow cytometric analysis of shmda-9 GSCs cultured in nonadherent conditions indicated that autophagy was toxic, with elevated levels of multiple autophagy markers expressed in all clinical samples and cell lines. In patient (VG2, VG9, VG10) and cell line (U87, U87vIII, U1242) neurospheres, suppression of mda-9 expression with shmda-9 elevated expression of autophagic vesicles (Fig. 2B). This finding was corroborated by the elevated expression of autophagy proteins in shmda-9 GSCs, relative to shcon GSCs: ATG5 ranged from 32 to 73%; LC3B ranged from 48 to 87%; and Lamp1 ranged from 21 to 73% (Figs. 2C and 3B, Table 1, and Table S1). No significant change was observed in Beclin-1 expression (Fig. S1D). To rule out possible off-target effects, we used an alternate small interfering (si)RNA sequence for knockdown experiments and an mda-9/syntenin construct designed to be resistant to Ad.5/3–shmda-9. These studies confirmed our previous results in GSCs indicating that treatment with an siRNA targeting mda-9 (Fig. S1E) significantly increases LC3B expression compared with consi GSCs. Inhibition of autophagy by CQ (10 µM) in shmda-9 GSCs was not protective and did not restore viability in these cells (Fig. 3C). In contrast, when GSCs were treated with an shRNA-resistant mda-9 plasmid, there was no significant change compared with shcon GSCs (Fig. 3D). This indicated that autophagy alone was not responsible for anoikis-resistance, and that MDA-9 is crucial in maintaining GSC viability under anoikis conditions. These results indicate further that inhibition of MDA-9 promotes toxic autophagy and anoikis-sensitivity, whereas MDA-9 expression maintains protective autophagy and anoikis-resistance in nonadherent GSCs.
Fig. 2.
MDA-9 is crucial for maintaining protective autophagy in GSCs growing in anoikis conditions and loss of MDA-9 causes autophagy to become toxic. (A) MDA-9 expression in shcon NSGCs, shcon GSCs, and shmda-9 GSCs. Error bars indicate ±SD, *P < 0.05. (B) Electron microscopy images of shcon and shmda-9 GSCs. (Magnification: 2000×.) (C) VG2-luc shcon and shmda-9 GSCs injected intracranially into nude mice. Brain tumors isolated and sectioned. Expression of MDA-9, LC3B, EGFR, pEGFR, PKCα, pPKCα, BCL2, and pBCL2 in in vivo tumors. (Magnification: 400×.)
Fig. 3.
MDA-9 regulates GSC survival and autophagy through BCL2 in nonadherent conditions. (A) Viability of shcon and shmda-9 GSCs. (Magnification: 100×.) (B) Effect of MDA-9 suppression on BCL2, pBCL2, and LC3B as shown by Western blotting. 1, shcon; 2, shmda-9. (C) shcon, shmda-9 GSCs, and shmda-9 GSCs treated with CQ, or overexpressing BCL2, by live/dead assay, where green cells represent viable cells, and red cells represent dead cells. (Magnification: 100×.) (D) shmda-9–resistant mda-9 plasmid abrogates mda-9 silencing-induced molecular changes. 1, shcon GSCs; 2, shmda-9 GSCs; 3, shmda-9 GSCs treated with shmda-9–resistant mda-9 plasmid.
MDA-9 Expression Regulates EGFR Activation and PKCα Signaling-Mediated Antiapoptotic BCL2 Protein Phosphorylation.
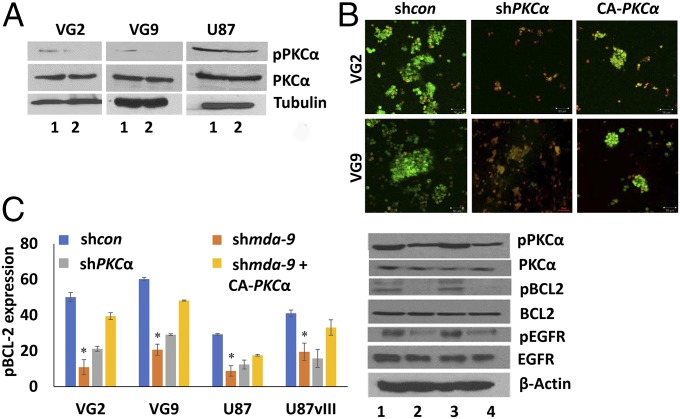
Protein expression analysis of nonadherent shcon and shmda-9 GSCs by flow cytometry and Western blotting indicated that EGFR and PKCα phosphorylation was significantly decreased in shmda-9 GSC neurospheres, both in vitro and in vivo in intracranial glioma xenografts (Figs. 2C and 4 and Table 1). While there was no significant change in total EGFR expression, pEGFR (Tyr-1068) expression decreased ∼25%, 29%, 56%, 35%, 29%, and 67% in shmda-9 GSCs from VG2, U87, U87vIII, U1242, VG9, and VG10 cells, respectively (Fig. 2C, Table 1, and Table S1). No significant change was observed in total PKCα; however, shmda-9 GSCs had decreased pPKCα (Thr-638) expression both in vitro and in vivo (Figs. 2C and 4). A decrease in the antiapoptotic protein pBCL2 (s70) was evident in shmda-9 GSCs (Figs. 3B and 4) and shPKCα GSCs (Fig. 4), suggesting that BCL2 is downstream of PKCα and MDA-9. Similar results were obtained in GSCs treated with an alternate siRNA targeting mda-9 (Fig. S1E), compared with consi GSCs. In contrast, when the GSCs were treated with an shRNA resistant mda-9 plasmid there was no significant change compared with shcon GSCs (Fig. 3D). Overexpression of a constitutively active PKCα (CA-PKCα) (Fig. 4 and Fig. S2) and BCL2 (Fig. 3C and Fig. S3) rescued pBCL2 expression and promoted survival in MDA-9–inhibited GSCs. These data support the hypothesis that MDA-9 regulates survival in anoikis-resistant GSCs through the PKCα/BCL2 axis as well as through EGFR signaling.
Fig. 4.
MDA-9 mediates cell survival in anoikis-resistant GSCs through PKC and BCL2. (A) Effect of MDA-9 on PKC activation in shcon and shmda-9 GSCs. 1, shcon; 2, shmda-9. (B) Effect of PKC suppression and expression of constitutively active PKC on GSC survival. (Magnification: 100×.) (C) Effect of PKC suppression and expression of constitutively active PKC on pBCL2 expression by flow cytometry and Western blotting. 1, shcon; 2, shmda-9; 3, shmda-9+CA-PKCα; 4, shPKCα. Error bars indicate ±SD, *P < 0.05.
EGFR Signaling Plays an Important Role in MDA-9–Mediated Protective Autophagy.
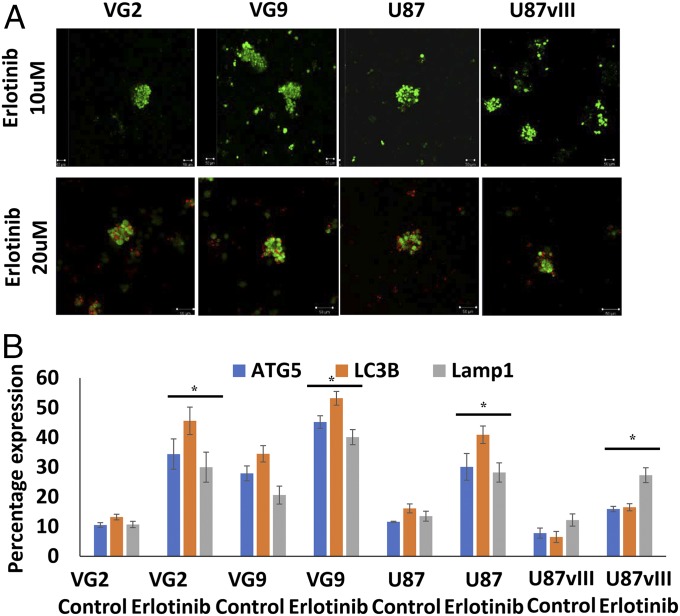
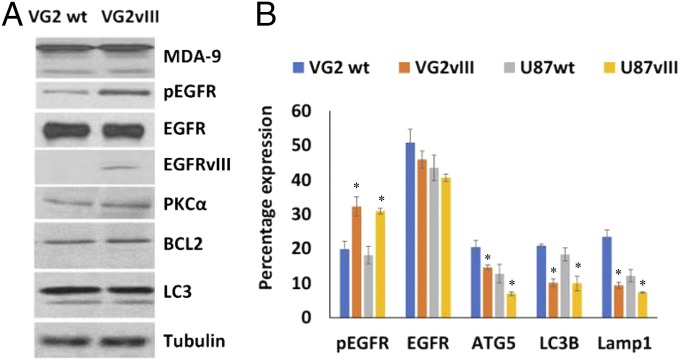
To assess the effect of EGFR signaling on protective autophagy, GSCs were treated with the EGFR tyrosine kinase inhibitor erlotinib, as well as by overexpressing a constitutively active EGFR variant III (EGFRvIII) in GSCs. Erlotinib treatment (20 µM) caused cell death in nonadherent GSCs (Fig. 5A), suggesting that EGFR contributes to GSC survival and anoikis-resistance, and a high dose of inhibitor induced cell death in otherwise anoikis-resistant GSCs. Erlotinib treatment at 10 µM led to a significant increase in autophagy (Fig. 5B and Figs. S4 and S5), which coincides with earlier reports (24–27). Erlotinib treatment approximately doubled the expression of autophagy markers in VG2 and U87 GSCs (Figs. S4 and S5). Overexpression of a constitutively active form of EGFR confirmed that both VG2wt and VG2vIII express similar levels of EGFR; however, only VG2vIII expresses EGFRvIII, along with decreased expression of autophagy markers (Fig. 6). Compared with the parental EGFRwt cells, the EGFRvIII GSCs showed significantly decreased expression of ATG5, LC3, and Lamp1. ATG5 expression was decreased by ∼25%, and 45%, LC3B expression was decreased ∼51% and 46%, and Lamp1 expression was decreased by ∼60% and 39% in VG2 and U87 EGFRvIII GSCs, respectively, compared with the WT cells (Fig. 6 and Fig. S5). Consequently, suppression of EGFR signaling increased autophagy, whereas increased EGFR signaling decreased autophagy. These results suggest that MDA-9–mediated EGFR signaling may regulate levels of autophagy. Loss of MDA-9 expression leads to increased autophagy, possibly due to the loss of the regulatory functions of EGFR.
Fig. 5.
EGFR activation plays an important role in regulating protective autophagy in anoikis-resistant GSCs. Inhibition of EGFR activation leads to increased autophagy. Effect of erlotinib on (A) GSC survival and (B) expression of autophagy markers ATG5, LC3B, and Lamp1. Error bars indicate ±SD, *P < 0.05. (Magnification: 100×.)
Fig. 6.
EGFR activation regulates protective autophagy in anoikis-resistant GSCs. Constitutive activation of EGFR decreases autophagy. (A) Effect of constitutive EGFR activation on EGFR, PKC, BCL2 signaling, and MDA-9, LC3B expression by Western blotting. (B) Effect of constitutive EGFR activation on EGFR, pEGFR, ATG5, Lamp1, and LC3B expression determined by flow cytometry. Error bars indicate ±SD, *P < 0.05.
FAK Regulates MDA-9–Mediated EGFR and PKC Signaling, Which Is Crucial for Protective Autophagy and Anoikis-Resistance.
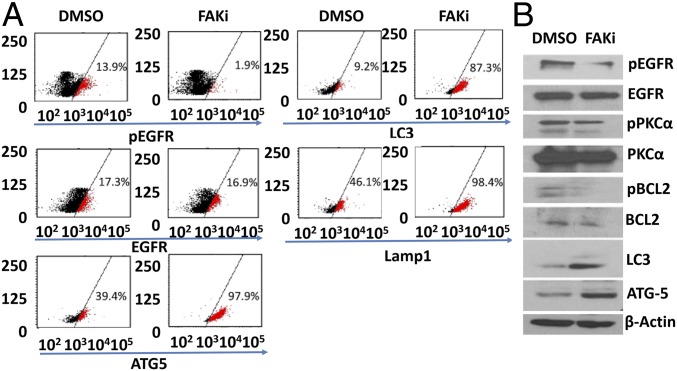
To assess whether MDA-9–mediated effects on autophagy were regulated by FAK, we treated GSCs cultured in nonadherent conditions with FAK inhibitor 14 (FAKi) (10 µM). Inhibition of FAK lead to decreased EGFR phosphorylation (Fig. 7) and up-regulation of autophagy markers ATG5, LC3B, and Lamp1 in both VG2 and VG9 GSCs (Fig. 7A). Western blotting analysis of the same lysates indicated similar up-regulation of autophagy markers (Fig. 7B).
Fig. 7.
Effect of FAK inhibitor 14 (FAKi) on EGFR, PKC and autophagy signaling as shown by (A) flow cytometry and (B) Western blotting.
Discussion
Autophagy is a lysosome-dependent process in which enzymatic degradation and recycling of cytosolic components is instigated following exposure of cells to stressful conditions (28). Anoikis, is a form of apoptosis, triggered when cells detach from the ECM (6, 7) and the catabolic process of autophagy imparts anoikis-resistance in solid tumors (29, 30). Autophagy in cancer stem cells provides a link between maintenance of stemness and metastasis-associated anoikis-resistance (31). However, the role of autophagy in cancer is complicated because autophagy can be a double-edged sword (i.e., a prosurvival or prodeath agent depending on the context and the stimuli), and the details of what regulates protective or toxic autophagy is not fully understood. In our studies, anoikis-resistant GSCs grown in nonadherent conditions display a basal level of autophagy, which is protective (Fig. 1, Table 1, Fig. S1 A and B, and Table S1). Inhibition of this autophagy caused a loss of anoikis-resistance and induced cell death, whereas autophagy induction promoted survival. The current study establishes that inhibition of MDA-9 in GSCs shifts autophagy from prosurvival to procell death, leading to loss of anoikis-resistance (Figs. 2 and 3, Table 1, Fig. S1C, and Table S1). In addition to observing these molecular changes in shmda-9–treated gliomas in vivo, increased survival was also evident in these mice, as we reported previously (21). MDA-9 is critical for maintaining protective autophagy, which is required for survival under anoikis-inducing conditions. This complex phenomenon is intricately orchestrated with the aid of molecules, such as ATG5, LC3B, Lamp1, EGFR, PKCα, and BCL2.
The ATG5 protein is essential in the early stages of autophagy (32). Autophagosome formation initiates at phagophore assembly sites, where several proteins required for autophagosome formation localize in a hierarchical manner close to the acceptor membrane, with ATG5 facilitating membrane binding (33). Inducing transcription and translation of the ATG RNA/protein, respectively, is essential for autophagy and sustaining survival following detachment (7, 34), but these changes can also contribute to autophagic cell death depending on the context (35). The ATG5-ATG12 complex further advances autophagosome formation and site of synthesis by helping in LC3 conjugation to phosphatidylethanolamine (36), also known as LC3-II, the membrane bound form (37, 38). LC3-II remains associated with autophagosomes even after fusion with lysososmes (called autolysosomes at this stage) until degradation (38). The autophagosomes, after fusing with the lysososmes, acquire Lamp1 (39). Hence, the right amount of ATG5, LC3, and Lamp1 expression may be critical in regulating the protective function of autophagy, and deregulated expression of these proteins can lead to loss of cell survival. When autophagy is deregulated and expression exceeds a threshold, cell damage exceeds the capacity for cell survival, resulting in self-digestion by autophagy and programmed cell death (40). This occurs in the shmda-9 GSCs cultured under anoikis-inducing conditions. These GSCs express aberrantly high levels of ATG5, LC3, and Lamp1 (Figs. 2 and 3, Table 1, and Table S1), and these cells lose viability, with protective autophagy shifting to toxic autophagy in the MDA-9 suppressed GSCs.
The epidermal growth factor receptor EGFR, a tyrosine kinase, is pivotal in glioma progression. EGFR is an 1,186 amino acid transmembrane receptor with three functional domains: extracellular (ECD), transmembrane and intracellular (ICD) (41). Tyrosine phosphorylation at Y1068 of EGFR is one of the major sites for EGFR autophosphorylation and indicates activation (42). FAK and PKCα regulate EGFR signaling (43, 44). The phosphorylation of tyrosine (Y) residue 1068, is responsible for regulating autophagy-induced cell death (42, 45). Several recent studies indicate that plasma membrane- and cytoplasm-located EGFR (pcEGFR) act as a tyrosine kinase regulating autophagy (45). Treatment of cancer cells with EGFR-tyrosine kinase inhibitors, such as gefitinib and erlotinib, induce autophagy (26), which we also observe in multiple GSCs (Fig. 5 and Figs. S4 and S5). In addition, suppression of MDA-9 decreases phosphorylation of EGFR at Y1068, causing loss of EGFR tyrosine kinase activation. These results indicate that regulation of EGFR activation by MDA-9 promotes maintenance of protective autophagy and anoikis-resistance in GSCs. Suppression of MDA-9 or EGFR signaling leads to increased autophagy beyond its beneficial threshold, thereby causing cell death.
In a considerable percentage of patients with GBM, EGFR signaling is constitutively active, because they contain a mutant EGFRvIII, thereby promoting tumor progression and poor prognosis (46). EGFRvIII has an in-frame deletion of 801 bp of coding sequence from exons 2–7 (46, 47), rendering it incapable of binding any known ligand. Despite this deletion, EGFRvIII displays low-level ligand-independent and constitutive receptor phosphorylation (47–49). We hypothesize that MDA-9–mediated EGFR signaling and autophagy regulation are important in GBM. To test this supposition, we analyzed autophagy levels in EGFRwt and EGFRvIII GSCs. EGFRwt and EGFRvIII anoikis-resistant GSCs have similar MDA-9 levels, VG2 (Fig. 6) and U87 (Fig. S1D). However, the EGFRvIII cells express less autophagy markers (Fig. 6, Table 1, and Table S1). Despite lower autophagy levels, when MDA-9 is suppressed in these GSCs, the autophagy still shifts the balance to toxicity (Fig. 3 and Table 1).
To maintain protective autophagy, antiapoptotic proteins likely play an important role, to prevent autophagy from promoting programmed cell death. BCL2 functions to suppress apoptosis and single-site phosphorylation at serine 70 (S70) of this protein is required for its antiapoptotic function (50, 51). PKCα functions as a direct BCL2 kinase at S70 in the BCL2 protein (52), making active PKCα and BCL2 as key mediators in maintaining cell viability in the presence of autophagy. We show that MDA-9 is necessary for PKCα activation (Figs. 2 and 4), thereby further contributing to the antiapoptotic balance in autophagy. This was documented further by gain-of-function and loss-of function studies of PKCα and BCL2 (Fig. 4 and Fig. S2), confirming that PKCα and BCL2 are key mediators of survival in anoikis-resistant MDA-9–expressing GSCs. Multiple studies suggest that MDA-9, PKCα, FAK, and EGFR signaling may be functionally interconnected (22, 27, 43, 44, 53, 54). MDA-9 is capable of regulating PKC (53), as well as FAK signaling (14, 22, 54), and the same pathways are activated in anoikis-resistant GSCs. Furthermore, MDA-9 regulates EGFR, and PKCα activation through FAK in an interconnected and interdependent manner (Fig. 8). GSCs treated with FAKi display the same phenotype as shmda-9 GSCs with increased autophagy and cell death (Fig. 7).
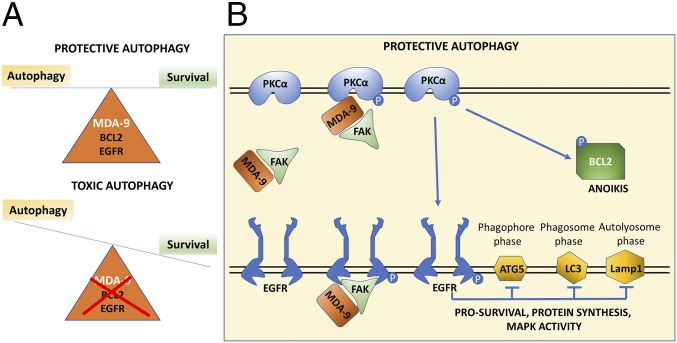
Fig. 8.
Schematic diagram of MDA-9-mediated protective autophagy in anoikis-resistant GSCs. (A) MDA-9 regulation of EGFR, PKC, and BCL2 activation maintains protective autophagy in anoikis-resistant GSCs. Loss of MDA-9 expression deregulates the balance, causing autophagy levels to exceed the threshold level, thereby shifting autophagy from protective to toxic. (B) Diagrammatic representation of the multiple pathways that MDA-9 directly and indirectly regulates to maintain protective autophagy in anoikis-resistant GSCs. MDA-9 regulates EGFR and PKCα activation through FAK in an interconnected and interdependent manner. EGFR activation decreases autophagy marker expression and PKCα activation leads to phosphorylation of BCL2, both pathways contributing to protective autophagy in anoikis-resistant GSCs.
In summation, our studies elucidate the protective function of autophagy in anoikis-resistant GSCs and mechanistically show that this process is maintained by MDA-9 through FAK/PKC/EGFR. PKC controls survival in GSCs, by regulating the antiapoptotic protein BCL2. EGFR maintains autophagy levels via regulation of ATG5, LC3, and Lamp1, so autophagy does not exceed threshold levels, which result in a shift from cell viability to toxicity. Because both MDA-9 and autophagy are involved in many important cancer-related cellular processes and signaling events, our study reinforces the potential use of MDA-9 suppression strategies in conjunction with other therapeutic strategies to promote GSCs death. Autophagy is often induced by several anticancer drugs, such as rapamycin, erlotinib, and so forth, and MDA-9 suppression in such therapeutic regimens could prove beneficial. In addition, these combinatorial strategies could potentially inhibit GSC-mediated resistance, relapse, and poor patient prognosis. GSCs and circulating stem cells utilize the mechanism of autophagy for their survival and resistance to a variety of stressful conditions.
The ability to develop selective inhibitors capable of targeting specific protein domains is coming of age (15), and these strategies can potentially be applied in developing small-molecule inhibitors of MDA-9’s PDZ domains. In addition, detailed elucidation of the role of MDA-9 in exosome biology could prove instructive in developing targeting strategies (15). We recently reported that radiation-induced glioblastoma invasion was inhibited by adenovirus-based genetic as well as pharmacological targeting of MDA-9. PDZ1i (113B7), a specific inhibitor of MDA-9 activity, crosses the blood–brain barrier, resulting in reduced invasion gains in GBM cells following radiation (22). MDA-9–targeted therapy could potentially be used clinically as part of a combinatorial approach with chemotherapy or radiotherapy, both of which often exploit protective autophagy in their resistance mechanisms. Genetic targeting of mda-9 has also been effective in melanoma, breast, gastric cancer, and urothelial cell carcinoma (15–17, 55). Accordingly, based on elevated MDA-9 expression, both genetic and pharmacological targeting strategies might be applicable to multiple cancer types. Considering all of the available data, MDA-9 inhibition could provide a promising approach for selectively targeting these chronically therapeutic-resistant cells, thereby improving therapeutic responses in patients with malignant gliomas and other cancers.
Materials and Methods
Reagents, Plasmids, Adenoviruses, and Stable Cell Lines.
shmda-9 plasmid and adenovirus construction are described in SI Materials and Methods (19, 21). Flag-BCL2 (Plasmid #18003) and Myr.PKCα.FLAG (Plasmid #10807) constructs were obtained from Addgene. EGFRwt and EGFRvIII plasmids were obtained from F.B.F. (56). PKC-α shRNA (HSH014706-LVRU6) was obtained from Genecopoeia.
Cell Lines, Cell Culture, and Chemicals.
Primary human malignant brain tumors were obtained from patients undergoing surgical removal of their tumors. Informed consent was obtained according to the research proposals approved by the Institutional Review Board at the Virginia Commonwealth University Tissue and Data Acquisition and Analysis Core. The patients were informed of the nature and requirements of the study and written consent was procured from them, which allowed them to donate their tissues for research purposes. Other human glioma cell lines used in this study are described in SI Materials and Methods. Erlotinib, rapamycin, CQ diphosphate, 3MA, and FAKi were obtained from Sigma. For animal studies, mice were maintained under pathogen-free conditions as approved by the American Association for Accreditation of Laboratory Animal Care, as well as in agreement with present regulations and standards of the US Department of Agriculture, US Department of Health and Human Services, and NIH.
Isolation and Culture of Human GBM, Putative GSCs, and NSGCs.
Human GBM GSCs and NSGCs were isolated from GBM tissue from surgical samples and from established U87, U87vIII, U251, and U1242/luc-GFP GBM cells. GBM tissue samples were dissociated and GSCs were isolated using CD44 and CD133 markers as described previously (21).
Preparation of Whole-Cell Lysates and Western Blotting Analysis.
Preparation of cell lysates and subsequent Western blotting analysis of the lysates were performed as described previously (57). Details are briefly described in SI Materials and Methods.
Flow Cytometry Sorting and Analysis.
Flow cytometry was performed after 24 h of incubation, before any cell death was observed. Details are described in SI Materials and Methods.
Immunohistochemistry.
H&E staining and immunohistochemistry were performed as described previously (22). Details are described in SI Materials and Methods.
Statistical Analysis.
For all experiments, statistical analyses were conducted using Student’s t test and ANOVA (Microsoft Excel). The data are presented as the mean ± SD of the values from three or more independent determinations. Probability values < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
Support was provided by the National Foundation for Cancer Research (P.B.F. and W.K.C.), the Virginia Commonwealth University Institute of Molecular Medicine (P.B.F.), and the Genetics Enhancement Fund (to P.B.F., S.K.D., and L.E.). Microscopy was performed at the Virginia Commonwealth University Department of Anatomy & Neurobiology Microscopy Facility, supported in part by funding from NIH-National Institute of Neurological Disorders and Stroke Center Core Grant 5 P30 NS047463 and, in part, by funding from NIH-National Cancer Institute (NCI) Cancer Center Support Grant P30 CA016059. The Virginia Commonwealth University Massey Cancer Center (MCC) Flow Cytometry Shared Resource, supported in part with funding from NIH-NCI Cancer Center Support Grant P30 CA016059 (to P.B.F. and D.S.), generated services and products in support of the research project. Histology services and products in support of the research project were generated by the MCC Mouse Model Shared Resource, supported in part with funding from NIH-NCI Cancer Center Support Grant P30 CA016059 (to J.J.W., P.B.F., and D.S.). P.B.F. holds the Thelma Newmeyer Corman Chair in Cancer Research in the MCC.
Footnotes
Conflict of interest statement: P.B.F. and W.K.C. are cofounders of InVaMet Therapeutics, Inc. P.B.F., W.K.C., Virginia Commonwealth University, and the Sanford-Burnham-Prebys Medical Discovery Institute own stock in InVaMet Therapeutics, Inc.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721650115/-/DCSupplemental.
References
- 1.Talukdar S, Emdad L, Das SK, Sarkar D, Fisher PB. Evolving strategies for therapeutically targeting cancer stem cells. Adv Cancer Res. 2016;131:159–191. doi: 10.1016/bs.acr.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed AU, Auffinger B, Lesniak MS. Understanding glioma stem cells: Rationale, clinical relevance and therapeutic strategies. Expert Rev Neurother. 2013;13:545–555. doi: 10.1586/ern.13.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalkan R. Glioblastoma stem cells as a new therapeutic target for glioblastoma. Clin Med Insights Oncol. 2015;9:95–103. doi: 10.4137/CMO.S30271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlashi E, et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc Natl Acad Sci USA. 2011;108:16062–16067. doi: 10.1073/pnas.1106704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paoli P, Giannoni E, Chiarugi P. Anoikis molecular pathways and its role in cancer progression. Biochim Biophys Acta. 2013;1833:3481–3498. doi: 10.1016/j.bbamcr.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 7.Buchheit CL, Weigel KJ, Schafer ZT. Cancer cell survival during detachment from the ECM: Multiple barriers to tumour progression. Nat Rev Cancer. 2014;14:632–641. doi: 10.1038/nrc3789. [DOI] [PubMed] [Google Scholar]
- 8.Zouq NK, et al. FAK engages multiple pathways to maintain survival of fibroblasts and epithelia: Differential roles for paxillin and p130Cas. J Cell Sci. 2009;122:357–367. doi: 10.1242/jcs.030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhutia SK, et al. Autophagy: Cancer’s friend or foe? Adv Cancer Res. 2013;118:61–95. doi: 10.1016/B978-0-12-407173-5.00003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Debnath J. The evolving, multifaceted roles of autophagy in cancer. Adv Cancer Res. 2016;130:1–53. doi: 10.1016/bs.acr.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Bischof J, et al. Cancer stem cells: The potential role of autophagy, proteolysis, and cathepsins in glioblastoma stem cells. Tumour Biol. 2017;39:1010428317692227. doi: 10.1177/1010428317692227. [DOI] [PubMed] [Google Scholar]
- 12.Mowers EE, Sharifi MN, Macleod KF. Autophagy in cancer metastasis. Oncogene. 2017;36:1619–1630. doi: 10.1038/onc.2016.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenific CM, Thorburn A, Debnath J. Autophagy and metastasis: Another double-edged sword. Curr Opin Cell Biol. 2010;22:241–245. doi: 10.1016/j.ceb.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boukerche H, Su ZZ, Prévot C, Sarkar D, Fisher PB. mda-9/syntenin promotes metastasis in human melanoma cells by activating c-Src. Proc Natl Acad Sci USA. 2008;105:15914–15919. doi: 10.1073/pnas.0808171105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kegelman TP, et al. Targeting tumor invasion: The roles of MDA-9/syntenin. Expert Opin Ther Targets. 2015;19:97–112. doi: 10.1517/14728222.2014.959495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das SK, et al. MDA-9/syntenin and IGFBP-2 promote angiogenesis in human melanoma. Cancer Res. 2013;73:844–854. doi: 10.1158/0008-5472.CAN-12-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dasgupta S, et al. Novel role of MDA-9/syntenin in regulating urothelial cell proliferation by modulating EGFR signaling. Clin Cancer Res. 2013;19:4621–4633. doi: 10.1158/1078-0432.CCR-13-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bacolod MD, et al. Examination of epigenetic and other molecular factors associated with mda-9/syntenin dysregulation in cancer through integrated analyses of public genomic datasets. Adv Cancer Res. 2015;127:49–121. doi: 10.1016/bs.acr.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kegelman TP, et al. MDA-9/syntenin is a key regulator of glioma pathogenesis. Neuro-oncol. 2014;16:50–61. doi: 10.1093/neuonc/not157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarkar D, Boukerche H, Su Z-Z, Fisher PB. mda-9/syntenin: More than just a simple adapter protein when it comes to cancer metastasis. Cancer Res. 2008;68:3087–3093. doi: 10.1158/0008-5472.CAN-07-6210. [DOI] [PubMed] [Google Scholar]
- 21.Talukdar S, et al. Novel function of MDA-9/syntenin (SDCBP) as a regulator of survival and stemness in glioma stem cells. Oncotarget. 2016;7:54102–54119. doi: 10.18632/oncotarget.10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kegelman TP, et al. Inhibition of radiation-induced glioblastoma invasion by genetic and pharmacological targeting of MDA-9/syntenin. Proc Natl Acad Sci USA. 2017;114:370–375. doi: 10.1073/pnas.1616100114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JL, et al. Autophagy induction results in enhanced anoikis resistance in models of peritoneal disease. Mol Cancer Res. 2017;15:26–34. doi: 10.1158/1541-7786.MCR-16-0200-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han W, et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS One. 2011;6:e18691. doi: 10.1371/journal.pone.0018691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YY, Lam SK, Mak JC, Zheng CY, Ho JC. Erlotinib-induced autophagy in epidermal growth factor receptor mutated non-small cell lung cancer. Lung Cancer. 2013;81:354–361. doi: 10.1016/j.lungcan.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Fung C, Chen X, Grandis JR, Duvvuri U. EGFR tyrosine kinase inhibition induces autophagy in cancer cells. Cancer Biol Ther. 2012;13:1417–1424. doi: 10.4161/cbt.22002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei Y, et al. EGFR-targeted mAb therapy modulates autophagy in head and neck squamous cell carcinoma through NLRX1-TUFM protein complex. Oncogene. 2016;35:4698–4707. doi: 10.1038/onc.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine B. Cell biology: Autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, et al. Integration of autophagy and anoikis resistance in solid tumors. Anat Rec (Hoboken) 2013;296:1501–1508. doi: 10.1002/ar.22769. [DOI] [PubMed] [Google Scholar]
- 30.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biol Cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ojha R, Bhattacharyya S, Singh SK. Autophagy in cancer stem cells: A potential link between chemoresistance, recurrence, and metastasis. Biores Open Access. 2015;4:97–108. doi: 10.1089/biores.2014.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima N, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 33.Romanov J, et al. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avivar-Valderas A, et al. PERK integrates autophagy and oxidative stress responses to promote survival during extracellular matrix detachment. Mol Cell Biol. 2011;31:3616–3629. doi: 10.1128/MCB.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pyo JO, et al. Essential roles of Atg5 and FADD in autophagic cell death: Dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 36.Rubinsztein DC, Shpilka T, Elazar Z. Mechanisms of autophagosome biogenesis. Curr Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 37.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 39.Eskelinen EL. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol Aspects Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Tang Y, et al. Obatoclax and lapatinib interact to induce toxic autophagy through NOXA. Mol Pharmacol. 2012;81:527–540. doi: 10.1124/mol.111.076851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrett TP, et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, et al. Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy. 2016;12:1029–1046. doi: 10.1080/15548627.2016.1164357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones G, Machado J, Jr, Merlo A. Loss of focal adhesion kinase (FAK) inhibits epidermal growth factor receptor-dependent migration and induces aggregation of nh(2)-terminal FAK in the nuclei of apoptotic glioblastoma cells. Cancer Res. 2001;61:4978–4981. [PubMed] [Google Scholar]
- 44.Stewart JR, O’Brian CA. Protein kinase C-α mediates epidermal growth factor receptor transactivation in human prostate cancer cells. Mol Cancer Ther. 2005;4:726–732. doi: 10.1158/1535-7163.MCT-05-0013. [DOI] [PubMed] [Google Scholar]
- 45.Li H, You L, Xie J, Pan H, Han W. The roles of subcellularly located EGFR in autophagy. Cell Signal. 2017;35:223–230. doi: 10.1016/j.cellsig.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 46.Gan HK, Cvrljevic AN, Johns TG. The epidermal growth factor receptor variant III (EGFRvIII): Where wild things are altered. FEBS J. 2013;280:5350–5370. doi: 10.1111/febs.12393. [DOI] [PubMed] [Google Scholar]
- 47.Zadeh G, Bhat KP, Aldape K. EGFR and EGFRvIII in glioblastoma: Partners in crime. Cancer Cell. 2013;24:403–404. doi: 10.1016/j.ccr.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 48.Nishikawa R, et al. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Sci Signal. 2009;2:re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 50.Deng X, Gao F, Flagg T, May WS., Jr Mono- and multisite phosphorylation enhances Bcl2’s antiapoptotic function and inhibition of cell cycle entry functions. Proc Natl Acad Sci USA. 2004;101:153–158. doi: 10.1073/pnas.2533920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blagosklonny MV. Unwinding the loop of Bcl-2 phosphorylation. Leukemia. 2001;15:869–874. doi: 10.1038/sj.leu.2402134. [DOI] [PubMed] [Google Scholar]
- 52.Ruvolo PP, Deng X, May WS. Phosphorylation of Bcl2 and regulation of apoptosis. Leukemia. 2001;15:515–522. doi: 10.1038/sj.leu.2402090. [DOI] [PubMed] [Google Scholar]
- 53.Hwangbo C, Kim J, Lee JJ, Lee JH. Activation of the integrin effector kinase focal adhesion kinase in cancer cells is regulated by crosstalk between protein kinase Calpha and the PDZ adapter protein mda-9/syntenin. Cancer Res. 2010;70:1645–1655. doi: 10.1158/0008-5472.CAN-09-2447. [DOI] [PubMed] [Google Scholar]
- 54.Das SK, et al. Raf kinase inhibitor RKIP inhibits MDA-9/syntenin-mediated metastasis in melanoma. Cancer Res. 2012;72:6217–6226. doi: 10.1158/0008-5472.CAN-12-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Menezes ME, et al. MDA-9/syntenin (SDCBP) modulates small GTPases RhoA and Cdc42 via transforming growth factor β1 to enhance epithelial-mesenchymal transition in breast cancer. Oncotarget. 2016;7:80175–80189. doi: 10.18632/oncotarget.13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mukherjee B, et al. EGFRvIII and DNA double-strand break repair: A molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69:4252–4259. doi: 10.1158/0008-5472.CAN-08-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emdad L, et al. Astrocyte elevated gene-1 (AEG-1) functions as an oncogene and regulates angiogenesis. Proc Natl Acad Sci USA. 2009;106:21300–21305. doi: 10.1073/pnas.0910936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.