Significance
Plant roots nurture a large diversity of soil microbes via exudation of chemical compounds into the rhizosphere. In turn, beneficial root microbiota promote plant growth and immunity. The root-specific transcription factor MYB72 has emerged as a central regulator in this process. Here, we show that MYB72 regulates the excretion of the coumarin scopoletin, an iron-mobilizing phenolic compound with selective antimicrobial activity that shapes the root-associated microbial community. Selected soil-borne fungal pathogens appeared to be highly sensitive to the antimicrobial activity of scopoletin, while two MYB72-inducing beneficial rhizobacteria were tolerant. Our results suggest that probiotic root-associated microbes that activate the iron-deficiency response during colonization stimulate MYB72-dependent excretion of scopoletin, thereby potentially improving their niche establishment and enhancing plant growth and protection.
Keywords: root metabolome, coumarin, induced systemic resistance, iron-deficiency response, microbiome assembly
Abstract
Plant roots nurture a tremendous diversity of microbes via exudation of photosynthetically fixed carbon sources. In turn, probiotic members of the root microbiome promote plant growth and protect the host plant against pathogens and pests. In the Arabidopsis thaliana–Pseudomonas simiae WCS417 model system the root-specific transcription factor MYB72 and the MYB72-controlled β-glucosidase BGLU42 emerged as important regulators of beneficial rhizobacteria-induced systemic resistance (ISR) and iron-uptake responses. MYB72 regulates the biosynthesis of iron-mobilizing fluorescent phenolic compounds, after which BGLU42 activity is required for their excretion into the rhizosphere. Metabolite fingerprinting revealed the antimicrobial coumarin scopoletin as a dominant metabolite that is produced in the roots and excreted into the rhizosphere in a MYB72- and BGLU42-dependent manner. Shotgun-metagenome sequencing of root-associated microbiota of Col-0, myb72, and the scopoletin biosynthesis mutant f6′h1 showed that scopoletin selectively impacts the assembly of the microbial community in the rhizosphere. We show that scopoletin selectively inhibits the soil-borne fungal pathogens Fusarium oxysporum and Verticillium dahliae, while the growth-promoting and ISR-inducing rhizobacteria P. simiae WCS417 and Pseudomonas capeferrum WCS358 are highly tolerant of the antimicrobial effect of scopoletin. Collectively, our results demonstrate a role for coumarins in microbiome assembly and point to a scenario in which plants and probiotic rhizobacteria join forces to trigger MYB72/BGLU42-dependent scopolin production and scopoletin excretion, resulting in improved niche establishment for the microbial partner and growth and immunity benefits for the host plant.
Plant roots exude a significant proportion of their photosynthetically fixed carbon into the rhizosphere (1). As a result, the rhizosphere nurtures one of the richest microbial ecosystems on Earth. The microbial community that inhabits the root–soil interface contains up to 1011 microbial cells per gram of root and collectively represents the root microbiome (2, 3). Root exudates greatly influence the composition of the root microbiome, a phenomenon called the “rhizosphere effect” (2, 4). Besides detrimental pathogens, the root microbiome also harbors beneficial members that promote plant growth or stimulate plant health (2, 5). Such mutualistic microbes serve plants in acquiring water and nutrients, fixing nitrogen, suppressing soil-borne pathogens, or stimulating plant immunity (2, 4).
Selected plant growth-promoting rhizobacteria (PGPR) can trigger an induced systemic resistance (ISR) that is effective against a broad range of foliar pathogens and even insect herbivores (6). ISR is well studied in the interaction between Arabidopsis thaliana and the PGPR Pseudomonas simiae WCS417 (7). WCS417-ISR in Arabidopsis functions independently of the defense hormone salicylic acid (SA) but instead requires a functional response to the plant hormones ethylene and jasmonic acid (JA) (6, 8). In the absence of a pathogen, WCS417-ISR–expressing leaves do not display abundant transcriptional changes (9). However, upon pathogen or insect attack, ISR-expressing leaves develop an accelerated, primed defense response that is associated with enhanced resistance (9–11). In contrast to foliar tissues, WCS417-colonized roots show abundant transcriptional changes (9, 11–13). Among the WCS417-induced genes, the root-specific R2R3-type MYB transcription factor gene MYB72 emerged as a central regulator of the onset of ISR (9, 14). MYB72 is also induced in Arabidopsis roots in response to colonization by the ISR-inducing fungi Trichoderma asperellum T-34 and Trichoderma harzianum T-78 (15). myb72-knockout mutants are unable to mount ISR upon colonization of the roots by WCS417 or T. asperellum T-34 (14, 16), suggesting that MYB72 plays a central role in the regulation of ISR triggered by different root-associated beneficial microbes. Downstream of MYB72 action, the β-glucosidase BGLU42 was identified as an important player in the onset of ISR (12). Constitutive overexpression of BGLU42 resulted in enhanced systemic disease resistance against Botrytis cinerea, Pseudomonas syringae, and Hyaloperonospora arabidopsidis, while the ability of mutant bglu42 to mount WCS417-ISR was blocked.
Besides being essential for the onset of ISR, MYB72 emerged as an integral part of the plant’s adaptive strategy to iron deficiency. Together with its closest paralog, MYB10, MYB72 is essential for plant survival in alkaline soils where iron availability is largely restricted (17). Under iron-limiting conditions, MYB72 is strongly induced in the roots of Arabidopsis as part of a set of coordinated responses that boost iron mobilization and uptake from the soil environment (collectively referred to as the “strategy I iron-deficiency response”) (18, 19). The core of this response consists of rhizosphere acidification by the activity of the H+-ATPase AHA2 (20), which increases the concentration of soluble ferric iron (Fe3+). Fe3+ is subsequently reduced to ferrous iron (Fe2+) by FERRIC REDUCTION OXIDASE2 (FRO2), after which it is transported from the soil environment into root cells via IRON-REGULATED TRANSPORTER1 (IRT1) (21, 22). The basic helix–loop–helix (bHLH) transcription factor FIT (FER-LIKE IRON DEFICIENCY TRANSCRIPTION FACTOR) is a central regulator of the iron-uptake response (19) and also regulates the expression of MYB72 (23).
During iron deprivation, plant roots exude a whole suite of secondary metabolites into the rhizosphere which aid in the mobilization and uptake of iron. These metabolites include phenolics, organic acids, flavins, and flavonoids (24–26). In Arabidopsis roots, MYB72 is required for the biosynthesis of iron-mobilizing fluorescent phenolic compounds, while BGLU42 is important for their subsequent excretion into the rhizosphere (12). These so-called “coumarins” are synthesized in the phenylpropanoid pathway via FERULOYL-COA 6-HYDROXYLASE1 (F6′H1) and are excreted into the rhizosphere by the iron deficiency-regulated ABC transporter PLEIOTROPIC DRUG RESISTANCE9 (PDR9) (24–27). Arabidopsis knockout mutants myb72 and bglu42 are impaired in the excretion of iron-mobilizing coumarins and in the ability to mount rhizobacteria-mediated ISR, suggesting a mechanistic link between the iron-deficiency response and plant immunity (12, 28).
Interplay between plant immunity and adaptive plant responses to nutrient deficiencies is also reported for the phosphate-starvation response (29). Interestingly, phosphate-starved plants release metabolites into the rhizosphere that are also exuded during conditions of iron deficiency (30, 31). Plants responding to changes in their biotic or abiotic environment show changes in root exudation (32, 33), which in turn affect the composition of the root microbiome (1). Likewise, mutations in defense and phosphate-starvation signaling pathways significantly impact the composition of the root-associated microbial community (29, 34, 35). Collectively the picture is emerging that components of the plant immune system and the plant’s adaptive response to nutrient starvation are interlinked in influencing the assembly of the root-associated microbiome, and vice versa (36). However, the molecular mechanisms and the ecological and evolutionary advantages of this apparent relationship are largely unknown.
Because MYB72 is an essential regulator of (i) the onset of ISR by beneficial members of the root microbiome and (ii) the excretion of iron-mobilizing coumarins into the rhizosphere, we set out to investigate the identity of the MYB72-dependent metabolites that are excreted into the rhizosphere and their impact on the root-associated microbiome. We performed metabolite fingerprinting analysis of root extracts and exudates of wild-type Col-0 and mutant myb72 and bglu42 plants. We identified the coumarin scopoletin as a major metabolite that is produced and excreted into the rhizosphere in a MYB72–BGLU42–dependent manner. Scopoletin possesses antimicrobial activity (37) and was previously linked to disease resistance in different plant species (38–40). Metagenome analysis of microbiota associated with roots of Col-0 plants and roots of the scopoletin biosynthesis mutant f6′h1 shows that this antimicrobial and iron-mobilizing coumarin impacts the assembly of the root-associated microbiome. We further show that scopoletin selectively inhibits the growth of two soil-borne fungal pathogens in vitro but has little or no effect on the growth of two beneficial ISR-inducing rhizobacteria. Collectively, our results suggest that the MYB72–BGLU42–mediated production and excretion of scopoletin favors the interaction between plant roots and probiotic members of the root microbiome.
Results
Metabolite Fingerprinting of Root Exudates.
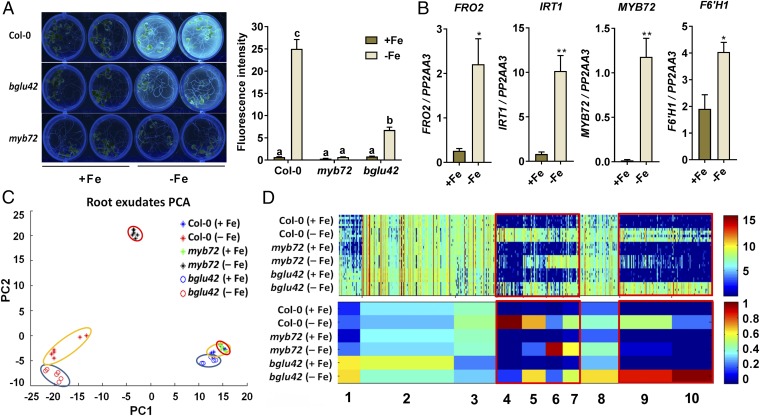
In Arabidopsis roots MYB72 and BGLU42 are both required for the onset of rhizobacteria-mediated ISR. Moreover, MYB72 is required for the biosynthesis of iron-mobilizing fluorescent phenolic compounds, while BGLU42 has a role in their subsequent excretion into the rhizosphere (12). Under iron-starvation conditions, Col-0 roots exuded high quantities of fluorescent phenolic compounds (Fig. 1A) and activated the iron-deficiency marker genes FRO2, IRT1, MYB72, and F6′H1 (Fig. 1B). The mutant myb72 did not exude detectable levels of the fluorescent compounds, and in bglu42 plants their exudation was significantly impaired, confirming previous findings (12). To identify the metabolites that were synthesized and secreted from Arabidopsis roots in a MYB72- and BGLU42-dependent manner, we analyzed the metabolome of root exudates of wild-type Col-0 and mutant myb72 and bglu42 plants. The three genotypes were grown under iron-sufficient and iron-deficient conditions, and the root exudates were analyzed by metabolite fingerprinting using ultra performance liquid chromatography (UPLC)-electrospray ionization (ESI)-TOF-MS (Dataset S1). Filtering the data resulted in a set of 722 metabolite features with a false-discovery rate (FDR) below 0.001. Principal component analysis (PCA) of the profiles of these 722 metabolite features indicates that the exudates of the different genotypes grown under iron-sufficient conditions are similar (Fig. 1C and Fig. S1). In contrast, the root exudates of iron-starved plants were clearly different from those of iron-sufficient plants. The root exudates of iron-starved myb72 plants clearly separated from those of iron-starved Col-0 and bglu42 plants, indicating that the metabolite profiles of myb72 root exudates are markedly different from those of Col-0 and bglu42 plants.
Fig. 1.
Metabolite fingerprinting of root exudates of Col-0, myb72, and bglu42 plants grown under iron-sufficient and iron-starvation conditions. (A) Accumulation and secretion of fluorescent phenolic compounds of 20-d-old Col-0, myb72, and bglu42 plants grown in Hoagland medium with (+Fe) or without (−Fe) iron. Visualization of fluorescence was achieved under UV light (365 nm). Fluorescence intensity was quantified in a 96-well microplate reader (excitation: 360 nm; emission: 528 nm) after seedlings were removed from the growth medium. Different letters indicate significant differences (P < 0.05, Tukey’s test, two-way ANOVA). (B) Gene-expression profiles of the iron-deficiency marker genes FRO2, IRT1, MYB72, and F6′H1 in roots of 20-d-old Col-0 plants grown in Hoagland medium with (+Fe) or without (−Fe) iron, quantified by qRT-PCR. Transcript levels were normalized to that of the reference gene PP2AA3 (At1g13320). Data are the means of three biological replicates. Error bars represent the SEM. Asterisks indicate significant differences between treatments: **P < 0.001, *P < 0.05, Student’s t test. (C) PCA plot of root exudates of 20-d-old Col-0 (encircled in yellow), myb72 (encircled in red), and bglu42 (encircled in blue) plants grown in Hoagland medium with (+Fe) or without (−Fe) iron. Data were obtained by metabolite fingerprinting (UPLC-ESI-TOF-MS analysis in positive and negative ionization mode), and the PCA represents the data subset after the filtering. PC1, principal component 1; PC2, principal component 2. (D) 1D-SOM clustering and prototype assignment of 722 high-quality metabolite features (FDR <0.001) derived from the positive as well as the negative ionization mode of the metabolite fingerprinting analysis. The number of features assigned to one prototype determines its width. The raw intensity of individual features (Upper) and prototypes (Lower) mostly affected by iron deficiency are highlighted in the red boxes. Heatmaps correspond to the intensity of each individual feature (Upper) and the average intensity of each cluster/prototype (Lower). The color key shows the range of signal intensities. Three biological replicates per treatment were used for analysis.
To obtain a global overview of the metabolite features and their relative abundances (RAs) in the root exudates, the profiles of the selected 722 metabolite features were clustered by means of one-dimensional self-organizing maps (1D-SOMs) (41). 1D-SOMs organized the intensity profiles in 10 clusters (prototypes) (Fig. 1D). Analogous to the PCA plot, the intensity profiles of the metabolite features of Col-0, myb72, and bglu42 plants grown under iron-sufficient conditions are highly similar. For all three genotypes, the iron-starvation treatment changed the metabolome of root exudates, which resulted mainly in increased signal intensities, most strikingly visible in clusters 4–7 and 9–10. Clusters 1–3 and 8 represent features with rather unaffected intensity profiles or with only a weak accumulation in the bglu42 mutant. Cluster 4 represents features that accumulated highly in exudates of Col-0 plants, while clusters 6 and 10 show features mainly enriched in myb72 or bglu42 exudates, respectively. The metabolite features of clusters 5 and 9 show lower intensities for iron-starved myb72 than for iron-starved Col-0 and bglu42. Collectively, these results indicate that MYB72 activity affects the composition of the root exudate metabolite profile under conditions in which the iron-deficiency response is activated.
Exudation of MYB72- and BGLU42-Dependent Metabolites.
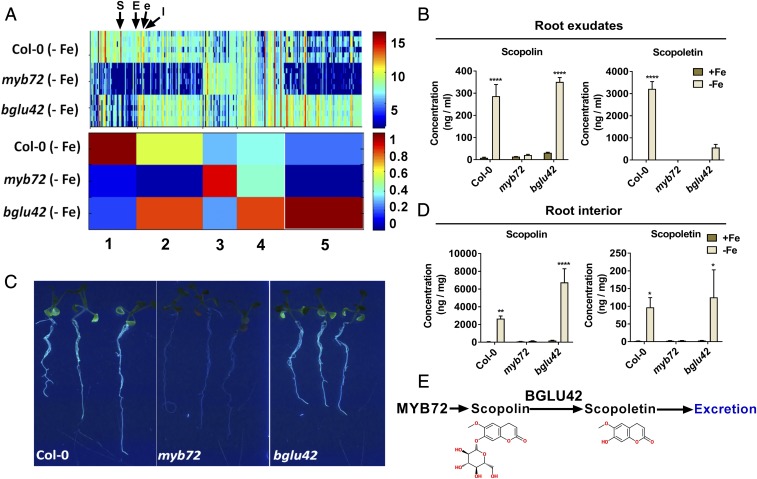
To determine the identity of the MYB72- and BGLU42-dependent metabolites in the root exudates of iron-starved Arabidopsis plants, we generated a data subset of the metabolome data shown in Fig. 1 containing only the metabolite fingerprinting analyses of the root exudates of iron-deprived Col-0, myb72, and bglu42 plants. Filtering the data of this subset resulted in 311 metabolite features with an FDR below 0.001 (Dataset S2). The intensity profiles of these 311 features were clustered by means of 1D-SOMs, resulting in five prototypes (Fig. 2A). Metabolite features in clusters 1, 2, and 5 showed clearly lower intensities in myb72 than in Col-0 exudates, indicating that the abundance of the corresponding metabolites increased in Col-0 root exudates in a MYB72-dependent manner. The metabolite features of prototype 1 showed in addition a depletion in bglu42 exudates, suggesting that the corresponding MYB72-dependent metabolites are excreted in a BGLU42-dependent manner.
Fig. 2.
MYB72- and BGLU42-dependent metabolites in root exudates and roots of Col-0, myb72, and bglu42 plants. (A) 1D-SOM clustering and prototype assignment of 311 high-quality metabolite features (FDR <0.001) in root exudates of iron-starved Col-0, myb72, and bglu42 plants. The number of features assigned to one prototype determines its width. Heatmaps correspond to the intensity of each individual feature (Upper) and the average intensity of each cluster/prototype (Lower). The color key shows the range of signal intensities. Arrows indicate the position of scopoletin (S), esculin (E), esculetin (e), and isofraxidin (I). For details on selected metabolite features, see Fig. S2 and Table S1. (B) HPLC-DAD quantification of scopolin and scopoletin in root exudates of Col-0, myb72, and bglu42 plants grown under iron-sufficient (+Fe) and iron-starved (−Fe) conditions. The data are the means of three replicates of the pooled root exudates of 50–60 plants per replicate. Error bars represent the SEM. Asterisks indicate significant differences between the iron conditions within a genotype: ****P < 0.0001, **P < 0.01, *P < 0.05, two-way ANOVA, Sidak’s test. (C) Photographs of iron-starved Col-0, myb72, and bglu42 plants grown in 12-well plates with liquid Hoagland medium without iron. Visualization of fluorescent phenolic compounds was achieved under UV light (365 nm). (D) HPLC-DAD quantification of scopolin and scopoletin in the roots of Col-0, myb72, and bglu42 plants grown under iron-sufficient and iron-starved conditions. The data are the means of three replicates of the pooled root extracts of 130 plants per replicate. Error bars represent SE. Asterisks indicate significant differences between the iron conditions within a genotype: ****P < 0.0001, **P < 0.01, *P < 0.05, two-way ANOVA, Sidak’s test. (E) Schematic representation of the role of MYB72 in the production of coumarin scopolin and the activity of BGLU42 in the deglycosylation of scopolin and the subsequent production of the aglycone scopoletin before its excretion into the rhizosphere. The presented molecules were created using the website https://www.emolecules.com.
Among the metabolite features that showed depletion in the root exudates of iron-starved myb72 plants, the majority matched to coumarins by a database search on the basis of exact masses comparison. Coumarins play a role in iron mobilization and uptake from alkaline substrates (24–26). The intensity profiles of the coumarins scopolin, scopoletin, esculin, esculetin, and isofraxidin show a strong increase in root exudates of iron-starved Col-0 plants (with scopoletin being the most abundant), while no increase is observed in root exudates of iron-starved myb72 plants (Fig. S2). Most of the features summarized in cluster 1 (Fig. 2A) are related to scopoletin. The diversity of the scopoletin-related features is caused by the intense adduct formation of the highly abundant coumarin scopoletin during ESI. The scopoletin-nonrelated features in cluster 1 showed very low signal intensities and inconclusive database matches. High-resolution tandem mass spectrometry (MS2) analysis confirmed the identity of the detected coumarins (Table S1). Of these excreted MYB72-dependent coumarins, only scopoletin levels were reduced in the root exudates of iron-starved bglu42 plants.
Quantification of scopolin and its aglycone scopoletin by HPLC with diode array UV detection (HPLC-DAD) (Fig. 2B) confirmed their patterns observed in the nontargeted metabolite fingerprinting approach (Fig. S2). Scopoletin accumulated to over 3 µg⋅mL−1 in root exudates of iron-starved Col-0 plants, while it was undetectable or strongly reduced in root exudates of the myb72 and bglu42 plants, respectively. Fig. 2B shows that the glycosylated form of scopoletin, scopolin, also accumulated in a MYB72-dependent manner in the exudates of iron-starved roots, albeit only to about 10% of the concentration of scopoletin (compare the scales of the y axes). However, the bglu42 mutation had no effect on the iron-starvation–induced levels of scopolin. Collectively, these results show that scopoletin is the major MYB72- and BGLU42-dependent metabolite that is secreted by roots of iron-starved Arabidopsis plants. Besides coumarins, the coumarin precursor phenylalanine and the citric acid cycle components citrate, malate, and succinate also increased in abundance in root exudates of iron-starved Col-0 plants (Fig. S2 and Table S1), confirming previous findings (24). However, their excretion was not dependent on MYB72 and BGLU42.
Role of BGLU42 in the Excretion of Scopoletin.
BGLU42 encodes a β-glucosidase that belongs to glycoside hydrolase (GH) family 1, and it was previously shown that the GH family 1 members BGLU21, BGLU22 and BGLU23 can hydrolyze scopolin, resulting in the production of scopoletin (42). Glycosylation and deglycosylation can change phenylpropanoid solubility, stability, and toxic potential and can influence their compartmentalization and biological activity (43). While myb72 plants are blocked in the ability to produce fluorescent phenolic compounds in their roots, iron-starved bglu42 roots still accumulate significant amounts (Fig. 2C). Hence, we hypothesized that if BGLU42 activity facilitates the excretion of scopoletin, then iron-starved bglu42 mutant plants should still accumulate the glycosylated form scopolin within their roots. To test this, we analyzed the metabolite profiles of the polar and nonpolar extractions of the roots of Col-0, myb72, and bglu42 plants grown under iron-sufficient and iron-starved conditions by UPLC-ESI-TOF-MS (Datasets S3 and S4). Similar to the observations for the root exudates, the root metabolome showed different metabolite profiles in terms of iron treatment and plant genotype (Figs. S1 and S3 A and B). Metabolite features related to coumarins were abundantly present in the polar extracts of the root, with scopolin, scopoletin, and esculin showing highly abundant signal intensities (Fig. S3C and Table S2). Scopolin, scopoletin, and esculin were produced within the roots in a MYB72-dependent manner, and BGLU42 did not negatively impact their abundance in the roots (Fig. 2D and Fig. S3C). Scopolin even increased to higher levels in the roots of iron-starved bglu42 plants than in the roots of Col-0 plants, supporting the notion that BGLU42-mediated deglycosylation of scopolin is impaired in this mutant. As opposed to root exudates of iron-starved Col-0 plants, where scopoletin levels were about 10-fold higher than those of scopolin (Fig. 2B), within the roots of iron-starved Col-0 plants the scopoletin levels were over 25-fold lower than those of scopolin (Fig. 2D). Collectively, these results indicate that roots of iron-starved Col-0 plants produce scopolin and scopoletin in a MYB72-dependent manner and that BGLU42 activity is important for the deglycosylation of scopolin and the subsequent excretion of the aglycone scopoletin into the rhizosphere (Fig. 2E).
In addition to the polar fraction, we also analyzed the nonpolar fraction of the root extracts (Fig. S3B and Dataset S4). Metabolite fingerprinting yielded a rich source of metabolite features with either MYB72- or BGLU42-dependent patterns. However, because none were impaired in both the myb72 and the bglu42 mutant background, they are not described further in this study.
Effect of Scopoletin on Rhizosphere Microbiome Assembly.
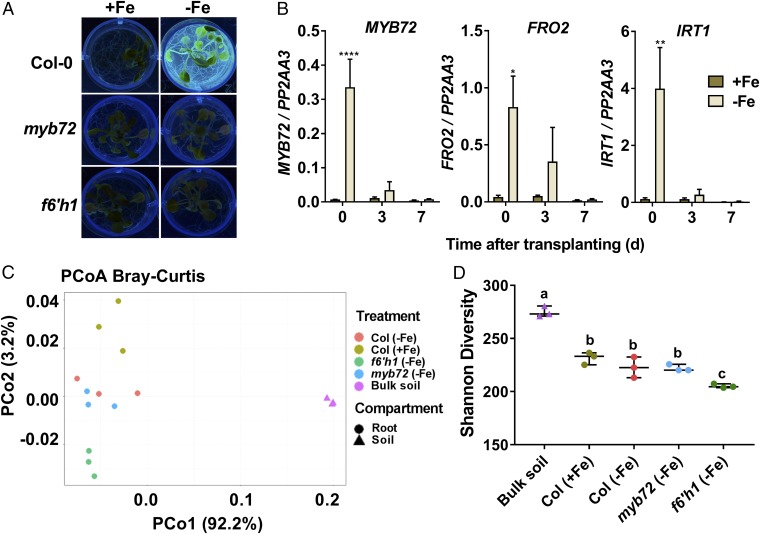
Previously, coumarin derivatives were shown to possess antimicrobial activity (37). Because beneficial ISR-inducing members of the root microbiome induce MYB72 and coumarin biosynthesis genes upon colonization of the roots (12), we hypothesized that coumarins may play a role in shaping the microbial community in the rhizosphere. To investigate the effect of MYB72-dependent coumarins, in particular scopoletin, on the composition of the root-associated microbial community, we analyzed the root-associated microbiomes of Col-0 and myb72 and the scopoletin biosynthesis mutant f6′h1 (26, 44). Arabidopsis seedlings were grown in vitro under iron-sufficient or iron-starved conditions to verify their coumarin-exudation status at the start of the experiment. As expected, iron-starved Col-0 seedlings abundantly exuded fluorescent phenolic compounds, while myb72 and f6′h1 seedlings did not (Fig. 3A). Subsequently, the plants were transplanted into natural Reijerscamp soil (45) that was limed to maintain iron limitation and differential coumarin excretion patterns. At the moment of transplantation, MYB72, FRO2, and IRT1 were strongly induced in the roots of iron-starved Col-0 seedlings (Fig. 3B). Their induced expression leveled off over time but was still detectable at day 3 after transplantation. In this experimental setup, differences in iron availability of the Col-0 seedlings after transplantation to the natural Reijerscamp soil had no major effect on the expression of the root immunity marker gene CYP71A12, the glucosinolate biosynthesis genes CYP79B2 and CYP79F2, and the pathogenesis-related protein gene PR3 (Fig. S4). Hence, potential differences in microbiome composition are not likely the result of differences in basal root defenses. Next, we analyzed the microbiota associated with the roots of iron-starved Col-0, myb72, and f6′h1 plants that were grown in limed soil for 3 d. As a control, we characterized the root microbiota of Col-0 plants that were pregrown under iron-sufficient conditions. Day 3 after transplantation was chosen because enhanced expression levels of MYB72, FRO2, and IRT1 were still detectable at that time point (Fig. 3B), and phenolics excreted by the roots can reside in the soil for several days (46). To analyze the effect of MYB72 and F6′H1 activity on the composition of the root microbiome, genomic DNA of the root-associated microbiota was subjected to shotgun metagenome sequencing. Per sample, Illumina NextSeq. 500 sequencing yielded between 52.4 and 108.7 million paired-end reads with a length of 150 bp (Table S3). For the analysis of the metagenomes, we followed a classification-first approach using Kaiju, a program that estimates the sequence similarity of metagenomic reads with reference prokaryotic and eukaryotic microbial protein databases (47). For the root samples, Kaiju classified 28–32% of the reads at the genus level to Arabidopsis and 21–24% of the reads to microbiota in the reference database (Table S3). For the bulk soil samples, a very small number of reads compared with the total sample reads were classified to Arabidopsis (sample B1: 10,995, 0.03%; sample B2: 3,534, 0.01%; sample B3: 2,802, 0.01%), while 37% matched with microbiota in the reference database. For all samples, 45–60% of the reads could not be classified at the genus level.
Fig. 3.
Metagenome analysis of bulk soil and root-associated microbiomes of Col-0, myb72, and f6′h1 plants. (A) Excretion of fluorescent phenolic compounds by 26-d-old Col-0, myb72, and f6′h1 plants grown in Hoagland medium with (+Fe) or without (−Fe) iron. Visualization of fluorescence was achieved under UV light (365 nm). (B) Gene-expression profiles of MYB72 and the iron-deficiency marker genes FRO2 and IRT1 in roots of Col-0 plants pregrown for 14 d in Hoagland medium with (+Fe) or without (−Fe) iron and transplanted on day 0 to limed Reijerscamp soil. Gene expression was quantified by qRT-PCR. Transcript levels were normalized to that of reference gene PP2AA3 (At1g13320). Data are means of three biological replicates. Error bars represent the SEM. Asterisks indicate significant differences between treatments: ****P < 0.0001, **P < 0.01, *P < 0.05, two-way ANOVA, Sidak’s test. (C) PCoA using Bray–Curtis metrics displays the dissimilarity of microbial communities in soil (triangles) and root samples (circles) and of Col-0 plants pregrown under iron-sufficient (+Fe) conditions and Col-0, myb72, and f6′h1 plants pregrown under iron-starved conditions (−Fe). (D) Shannon diversity (effective number of species) displaying the within-sample diversity of different treatments. Horizontal bars correspond to the median and interquartile range of values. Different letters indicate significant differences: P < 0.05, one-way ANOVA, Tukey’s test.
Genus-level classification using publicly available taxonomy databases as in ref. 48 allowed us to track shifts in higher taxonomic ranking between the root-associated microbiota and those assembled in the unplanted soil (Fig. S5). Taxonomic classification at the genus level resulted in the assignment of reads to 4,046 genera belonging to the domains of Bacteria, Eukaryota, and Archaea (Dataset S5). Bacteria were the most dominant domain in terms of RA, with Eukaryota being the second and Archaea representing only a small fraction of the microbial communities (Fig. S5A). Bacteria displayed a significant decrease in the root samples of all genotypes compared with bulk soil. In contrast, Eukaryota showed a significant increase in the root samples, while the RA of Archaea remained unaffected in both compartments (Fig. S5A). At the phylum level, we focused on phyla with an RA of over 0.5% (Fig. S5B). Proteobacteria were the most abundant (around 50% RA) with Actinobacteria, Firmicutes, and Bacteroidetes being overrepresented as well but with lower RA. The phyla Acidobacteria, Actinobacteria, Bacteroidetes, and Firmicutes showed statistically significant changes in RA between soil and all root/rhizosphere samples. For the phyla Chlorophyta, Mucoromycota, Planctomycetes, and Proteobacteria only a subset of the genotype–treatment combinations showed significant changes in the RA in root/rhizosphere versus bulk soil samples, while the phyla Verrucomicrobia, Cyanobacteria, Chloroflexi, Basidiomycota, and Ascomycota remained unaffected (Fig. S5B).
To gain further insight into the effect of plant genotype and treatment on root microbial diversity, we performed β- and α-diversity analyses. In the principal coordinate analysis (PCoA) of Bray–Curtis similarities (β-diversity), the microbial diversity of bulk soil clearly separates from those of the different plant genotypes (Fig. 3C), indicating a significant effect of the plant on root microbiome assembly. Along the second principal coordinate, the root-associated microbial communities of iron-sufficient (+Fe) and iron-starved (−Fe) Col-0 plants differentiate (Fig. 3C), suggesting that root exudates produced during iron starvation affected microbiome assembly. The microbial communities of iron-starved Col-0 and myb72 plants both displayed intermediate separation from that of iron-sufficient Col-0 plants, while the microbial community on the scopoletin biosynthesis mutant f6′h1 diverged most distinctly. To pinpoint genotype-mediated differences within the root-associated microbiota, the microbial diversity within each sample (α-diversity) was calculated as Shannon diversity (Fig. 3D). This calculation showed that microbial communities in unplanted bulk soil are significantly more diverse and complex than the root-associated microbial communities and that the Shannon diversity for the f6′h1 root-associated microbiota was significantly lower than that for the other genotypes. The observation that the scopoletin-biosynthesis mutant f6′h1 clearly separates from the other genotypes in both between- and within-samples diversity estimations suggests that F6′H1 activity affects root microbiome assembly.
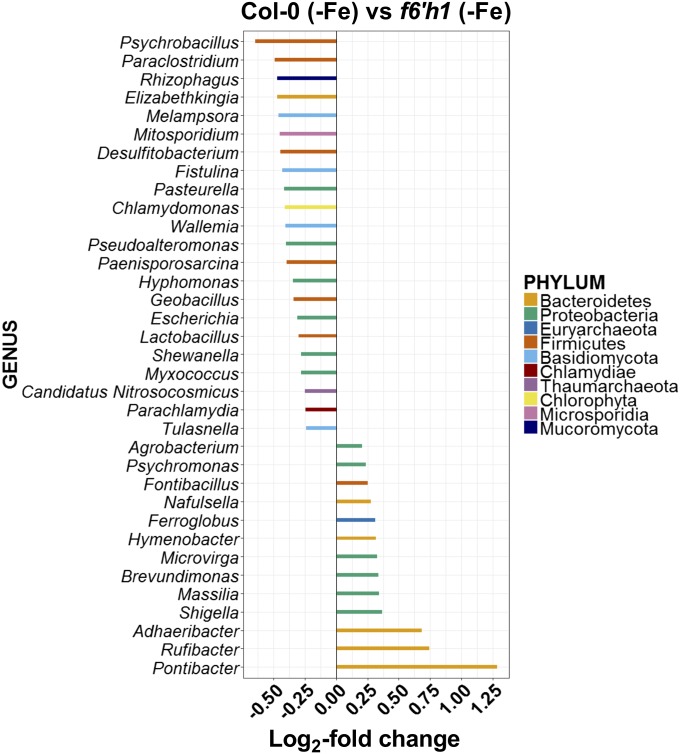
To dissect the plant genotype-mediated differences in root microbiome structure on lower taxonomic ranking, we performed pairwise comparisons using DESeq2. Pairwise comparison between the root-associated microbiota of iron-starved and iron-sufficient Col-0 plants revealed 21 genera with differential abundance, predominantly from the Proteobacteria and Firmicutes phyla (Fig. S6A). Psychrobacillus, Stenotrophomonas, Paeinisporosarcina, and Dyella were the genera with the highest differential abundance in the root microbiomes of iron-starved Col-0 plants, indicating that these genera grow better in the rhizosphere of iron-starved Col-0 plants than in that of iron-sufficient Col-0 plants. Conversely, Adhaeribacter, Niastella, and Hymenobacter were most highly enriched on roots of iron-sufficient Col-0 plants, indicating that they grow better in the rhizosphere of iron-sufficient Col-0 plants than in that of iron-starved Col-0 plants. Comparison between the root microbiomes of iron-starved Col-0 and myb72 plants yielded only three genera above the limit of statistical significance (Fig. S6B). However, these genera were also detected in the comparison between iron-starved Col-0 and f6′h1 plants, which revealed the largest number (35) of genera with differential abundance (Fig. 4). The 22 genera significantly enriched on Col-0 over f6′h1 roots (growth positively affected by F6′H1/scopoletin activity) represent seven phyla, with Psychrobacillus and Paraclostridium from the Firmicutes phylum and Rhizophagus, a genus of mycorrhizal fungi from the phylum Mucoromycota, being among the most strongly stimulated genera. The 13 genera significantly enriched on f6′h1 over Col-0 roots (growth negatively affected by F6′H1/scopoletin activity) represent four phyla, with Pontibacter, Rufibacter, and Adhaeribacter, all belonging to the Hymenobacteraceae family, being among the most strongly affected genera. Interestingly, Adhaeribacter and Hymenobacter were both significantly less abundant on the roots of iron-starved Col-0 plants than on the roots of iron-starved f6′h1 and iron-sufficient Col-0 plants, suggesting that these genera are particularly sensitive to F6′H1-dependent coumarins that are secreted under conditions of iron starvation. Together, these results demonstrate that the F6′H1-dependent root exudation patterns, with the coumarin scopoletin as a major compound, can positively and negatively influence the abundance of specific genera in the root-associated microbiome of Arabidopsis, thereby impacting the assembly of the root microbiome.
Fig. 4.
Differential abundance of microbial genera on Arabidopsis roots with different scopoletin exudation patterns. Differentially abundant bacterial and fungal genera in root samples of Col-0 (−Fe) and f6′h1 (−Fe) plants as determined using DESeq2. Comparisons were performed at the genus level using an FDR <0.05 to select for significance. In all graphs, negative log2 fold-change values relate to genera that are significantly enriched in iron-starved Col-0 (−Fe) root samples in comparison with the contrasting genotype/treatment combination. Different bar colors represent different phyla as shown in the key.
Differential Antimicrobial Effect of Scopoletin.
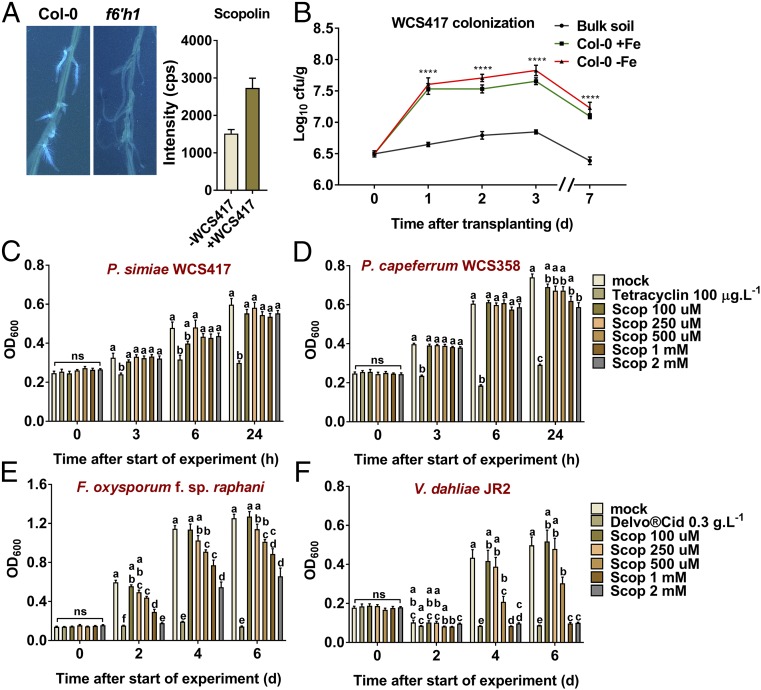
MYB72 and BGLU42 are both important for the onset of rhizobacteria-mediated ISR and the excretion of iron-mobilizing coumarins into the rhizosphere (12, 23). Metabolite fingerprinting using UPLC-ESI-TOF-MS confirmed that at 48 h after colonization by P. simiae WCS417, Col-0 roots already show increased accumulation of the coumarins scopolin, scopoletin, and esculin and the tricarboxylic acid (TCA) cycle intermediates citrate, malate, and succinate (Fig. 5A, Fig. S7, and Dataset S6). Production of coumarins in response to colonization of Arabidopsis roots by ISR-inducing P. simiae WCS417 bacteria occurs in a F6′H1-dependent manner (Fig. 5A), which raises the question how and to what extent such plant-beneficial rhizobacteria benefit from the excretion of antimicrobial compounds into the rhizosphere. To assess the effect of coumarin production on root colonization by WCS417, we monitored the growth of WCS417 on roots of iron-starved and iron-sufficient Col-0 plants over a time period of 7 d after the plants were transplanted into limed Reijerscamp soil (Fig. 5B; same experimental setup as used for the microbiome analysis). Within 1 d, the density of WCS417 bacteria had increased by 10-fold in the rhizosphere of Col-0 plants in comparison with the density in unplanted bulk soil. This increase in WCS417 density was maintained for the 7-d period of monitoring. No difference was observed in the level of colonization on iron-starved and iron-sufficient Col-0 roots, suggesting that WCS417 is insensitive to possible antimicrobial effects of root exudates that are excreted in the rhizosphere during conditions of iron starvation.
Fig. 5.
Effect of scopoletin on P. simiae WCS417 colonization and on the growth of selected soil-inhabiting microbes. (A, Left) Visualization of fluorescent phenolic compounds produced by roots of iron-sufficient Col-0 and scopoletin biosynthesis mutant f6′h1 plants in response to colonization by P. simiae WCS417. Visualization of fluorescent phenolic compounds was achieved under UV light (365 nm). Photographs were taken from roots of 20-d-old in vitro-grown Arabidopsis plants 7 d after colonization by the rhizobacteria. (Right) Bars show signal intensity in counts per second of the coumarin scopolin (detected as [M+H]+) in Col-0 roots 2 d after colonization by P. simiae WCS417. Shown data are the means (± SEM) of three biological replicates (see also Fig. S5). (B) Number of P. simiae WCS417 bacteria recovered from rhizospheres of Col-0 plants grown in limed Reijerscamp soil that was amended with 105 cfu⋅g−1 WCS417 bacteria. Root colonization was determined at the indicated days after the seedlings were transplanted from iron-sufficient (+Fe) or iron-starved (−Fe) Hoagland growth medium into the WCS417-amended Reijerscamp soil. Values for each time point were calculated from five rhizosphere or bulk soil samples. Asterisks indicate significant differences between bulk soil and colonized plants: ****P < 0.0001, two-way ANOVA, Tukey’s test. (C–F) Graphs showing growth (OD600) of P. simiae WCS417 (C), P. capeferrum WCS358 (D), F. oxysporum f.sp. raphani (E), and V. dahliae JR2 (F) in medium containing the indicated concentrations of scopoletin (Scop). Tetracycline and Delvocid were used as positive controls for bacteria and fungi, respectively. Growth measurements were performed over a period of 24 h (bacteria) or 6 d (fungi). The data shown are the means of 8–10 replicates. Error bars represent the SE of the mean. Different letters represent significant differences between treatments: P < 0.05, two-way ANOVA, Tukey’s test.
To confirm that P. simiae WCS417 is insensitive to coumarins, we tested the effect of scopoletin on the growth of WCS417 in vitro, using the antibiotic tetracycline as a positive control (Fig. 5C) (40). While tetracycline prevented the growth of WCS417, scopoletin concentrations of up to 2 mM had no effect on the growth of this PGPR. Additionally, we tested the effect of scopoletin on another well-characterized ISR- and MYB72-inducing PGPR, Pseudomonas capeferrum WCS358 (7, 23). WCS358 was also highly insensitive to scopoletin (Fig. 5D). We then reasoned that the excretion of coumarins would aid the rhizobacteria that induce their production in selectively outcompeting other microbes in the rhizosphere that compete for the same niche. To provide proof of concept for this hypothesis, we tested the effect of scopoletin on the growth of two soil-borne pathogens of Arabidopsis, Fusarium oxysporum f. sp. raphani and Verticillium dahliae JR2 (Fig. 5 E and F) (49, 50). Scopoletin concentrations of 500 µM and higher significantly inhibited the growth of both fungi in vitro and inhibited the growth of V. dahliae to the same extent as the fungicide Delvocid. Since we detected scopoletin concentrations of over 3 µg⋅mL−1 (∼15 µM) in the growth medium of iron-starved Col-0 plants (Fig. 2B), it is likely that in the narrow region of soil directly surrounding the roots much higher concentrations of scopoletin can occur. Together, these results indicate that scopoletin has a differential antimicrobial effect to which the tested soil-borne fungal pathogens F. oxysporum f. sp. raphani and V. dahliae JR2 are sensitive but the ISR- and MYB72-inducing PGPR P. simiae WCS417 and P. capeferrum WCS358 are not.
To further investigate the activity of scopoletin on fungal physiology and growth, we assessed the effect of 500 µM scopoletin on F. oxysporum f. sp. raphani radial growth on potato dextrose agar (PDA) plates. Scopoletin reduced the radial growth of the fungus and inhibited the formation of pigment in the mycelial mat (Fig. S8A). Metabolites involved in fungal pigmentation can be associated with protection against abiotic stresses or competition with other microbes (51, 52), indicating that scopoletin potentially exerts its effect on multiple determinants of fungal performance. To study the effect of scopoletin in root exudates on F. oxysporum f. sp. raphani hyphal growth, we performed a chemotropism assay developed by Turrà et al. (53). We used root exudates of iron-starved Col-0 and scopoletin biosynthesis-mutant f6′h1 plants, increasing concentrations of scopoletin, and pectin as a positive control. Fig. S8B shows that fungal hyphae from germinating conidia are chemotropically attracted toward pectin, confirming previous findings (53). Conversely, scopoletin deterred the fungal hyphae, which preferentially grew away from this antimicrobial coumarin. Root exudates of iron-starved Col-0 plants, which exude large amounts of scopoletin (Figs. 1 and 2), similarly deterred the fungal hyphae, while root exudates of iron-starved f6′h1 plants significantly attracted the fungal hyphae. Together, these results indicate that scopoletin in root exudates can have a multifaceted negative effect on the performance of scopoletin-sensitive microbes in the rhizosphere.
Discussion
MYB72 and BGLU42 Are Required for the Production and Secretion of Scopoletin.
The rhizosphere microbiome is highly diverse, and its interplay with plant roots has a marked influence on plant fitness (2). Specific root microbiome members can trigger ISR through the activity of the root-specific transcription factor MYB72 and the MYB72-regulated β-glucosidase BGLU42 (6, 14). MYB72 and BGLU42 also have a role in the production and excretion of fluorescent phenolic compounds when plants grow under conditions of iron starvation (12). We used this knowledge to identify the metabolites that are produced and excreted by Arabidopsis roots in a MYB72- and BGLU42-dependent manner and investigated their possible role in plant–microbiome interactions. Untargeted UPLC-ESI-TOF-MS metabolomics of root exudates and root extracts revealed coumarins as a major group of compounds whose production relies on MYB72 activity (Fig. 2 and Figs. S2 and S3). Coumarins, such as scopolin, scopoletin, esculin, esculetin, and isofraxidin, are produced via the phenylpropanoid pathway (54) and accumulate abundantly in roots and exudates of iron-starved plants (24–26) where they play a role in the mobilization and uptake of iron (26, 27). Mutant bglu42 plants produced wild-type levels of coumarins but were specifically impaired in the excretion of scopoletin, the most abundant coumarin in the root exudates (Fig. 2 and Figs. S2 and S3). However, the glycosylated form of scopoletin, scopolin, accumulated to high levels in iron-starved bglu42 roots, which suggests that BGLU42 functions in the hydrolysis of scopolin and that this activity is required for the excretion of scopoletin into the rhizosphere. This is in line with previous findings in which the Arabidopsis β-glucosidases BGLU21, BGLU22, and BGLU23 were shown to specifically hydrolyze scopolin into scopoletin in vitro (42). β-Glucosidases can hydrolyze glycosidic bonds either between two carbohydrates or between a carbohydrate and a noncarbohydrate (55), often resulting in the release of bioactive derivatives (55, 56). The activity of BGLU42 seems to be specific for scopolin/scopoletin, since other coumarin pairs such as esculin/esculetin were not affected by bglu42 (Figs. S2 and S3). We thus conclude that BGLU42 activity in iron-starved Arabidopsis roots plays an important role in the processing of scopolin to scopoletin, resulting in the excretion of scopoletin into the rhizosphere.
Scopoletin Affects Root Microbiome Assembly.
Scopoletin possesses antimicrobial activity (37). Hence, its excretion likely influences the composition of the microbial community in the rhizosphere. In our search for scopoletin-mediated effects on root microbiome assembly, shotgun sequencing of the microbial communities in bulk soil and on roots of Col-0, myb72, and f6′h1 plants confirmed a number of concepts that have emerged in the field of root microbiome research. First, bulk soil and root-associated microbial communities were largely different (Fig. 3), resembling the rhizosphere effect (57). Second, the RA of Bacteria in the rhizosphere was reduced in comparison with bulk soil, while that of Eukaryota was higher, supporting previous findings in crop species (Fig. S5A) (58). Third, at the phylum level, the RA of Proteobacteria, Firmicutes, and Bacteroidetes was generally increased in root rhizosphere samples compared with bulk soil, whereas the RA of Actinobacteria and Acidobacteria was decreased (Fig. S5B). Such shifts in phylum-level abundance were also observed in other rhizosphere microbiome studies of Arabidopsis, rice, and barley plants that were grown for prolonged periods in the soil (35, 59–61). Moreover, the α-diversity estimation with Shannon’s diversity (Fig. 3D) showed a diversity gradient from higher to lower between soil and rhizosphere samples, suggesting that microbial selection is occurring in the rhizosphere (59, 61, 62). In this study, we analyzed the structure of the root-associated microbial communities that were formed within 3 d after transplantation into the soil. The fact that general concepts of microbiome assembly were already detectable within this short time frame highlights the speed with which root-associated microbial communities are established in response to chemical and structural cues in the rhizosphere.
In comparison with the large differences between root-associated and bulk soil microbial communities, the plant genotype-mediated differences in root microbiome composition were more subtle (Fig. 3C), which is in line with previous findings (29, 35, 63). In comparison with the microbiomes assembled on Col-0 roots, those assembled on myb72 roots showed fewer differences than those assembled on f6′h1 roots, possibly because in iron-starved soil MYB72 functions redundantly with MYB10 (17). The microbiomes assembled on the roots of Col-0 and f6′h1 plants displayed the largest differences, indicating that excretion of scopoletin affects the composition of the root microbiome. Genus-level analysis showed that the abundance of genera such as Stenotrophomonas, Lactobacillus, Psychrobacillus, Elizabethkingia, Chlamydomonas, and Geobacillus was enhanced on scopoletin-producing roots (Fig. 4). Interestingly, these genera were previously found to be able to hydrolyze different coumarins (64–68). Among the most strongly promoted genera on coumarin-excreting roots are genera that promote plant growth (Psychrobacillus, Variovorax), facilitate metal uptake in plants (Psychrobacillus), have a role in nitrogen cycling in soil (Candidatus Nitrosocosmicus), or possess antimicrobial potential (Pseudoalteromonas) (69–72). Also, the genera Rhizophagus and Tulasnella, both of which are mycorrhizal fungi, performed better in coumarin-containing rhizospheres, which is in line with the observation that phenolic compounds stimulate the growth and root colonization of arbuscular mycorrhizal fungi (73).
Among the genera that performed better on coumarin-nonproducing roots (Col-0/+Fe and/or f6′h1/−Fe plants) was the genus Nafulsella, which was previously found to be incapable of hydrolyzing the coumarin esculin (74). Moreover, the genera Adhaeribacter, Hymenobacter, and Pontibacter, all belonging to the Hymenobacteraceae family, were enriched in coumarin-nonproducing rhizospheres compared with coumarin-producing ones (Fig. 4 and Fig. S6). Genera of this family can grow in poor substrates and can form strong biofilms, and their motility is not based on flagellar movement (gliding motility) (75–77). However, the extent to which these characteristics are related to the effect of coumarins on their performance remains elusive.
Differential Antimicrobial Activity of Scopoletin: Selecting Friends from Foes?
Exudation of coumarins is a response that is shared by the iron- and the phosphate-starvation response of plants (28, 30, 31). In analogy to our findings with iron uptake-deficient mutants, Castrillo et al. (29) observed that phosphate uptake-deficient and phosphate-hyperaccumulating Arabidopsis genotypes assembled significantly different root-associated microbiomes than wild-type plants. Additionally, amendment of root exudates rich in phenolic compounds into the soil showed more pronounced effects on the microbial community than other classes of compounds (78). Hence, phenolic compounds in root exudates can be potent modulators of root microbiome assembly. This raises the question how ISR-inducing and plant growth-promoting microbes in the rhizosphere benefit from the induction of the MYB72- and BGLU42-dependent exudation of scopoletin in the rhizosphere. Scopoletin and other coumarins play a role in the mobilization and uptake of Fe3+ from the soil environment (26, 27) and may thereby contribute to the mutually beneficial growth-promoting effect that such PGPRs have on the plant. Besides its role in the iron-uptake response, scopoletin possesses antimicrobial activities. In Arabidopsis and tobacco, scopoletin has been shown to accumulate around infection sites, where it inhibited the growth of the foliar pathogens B. cinerea, Alternaria alternata, and P. syringae (38, 40, 79). Other studies have shown that coumarins can negatively affect biofilm formation and the virulence of pathogenic bacteria in different systems (80, 81). In our experimental setup, scopoletin had no growth-inhibiting effect on the ISR-inducing PGPR P. simiae WCS417, either in vivo on Arabidopsis roots or in vitro, and mildly reduced growth of ISR-inducer P. capeferrum WCS358 only at high concentrations (Fig. 5). By contrast, scopoletin strongly inhibited the growth of the soil-borne fungal pathogens F. oxysporum and V. dahliae (Fig. 5). We found that, in addition to reducing the growth of these fungi, scopoletin deters the germination of F. oxysporum spores (Fig. S8B) and inhibits pigmentation of F. oxysporum mycelium (Fig. S8A). Such mycelial pigments can have protective activities against environmental stresses (52, 82). Hence, the inhibition of their formation by scopoletin could render this fungus less viable in the rhizosphere.
Collectively, our microbiome and targeted microbial growth analyses show that some soil-borne microbes can be sensitive to the antimicrobial activity of scopoletin, while others are tolerant. Beneficial ISR-inducing rhizobacteria, such as P. simiae WCS417 and P. capeferrum WCS358, induce MYB72 and BGLU42, which leads to the onset of ISR and the excretion of scopoletin in the rhizosphere. Like P. simiae WCS417 (Fig. 5A), the ISR inducers Pseudomonas fluorescens SS101 and Paenibacillus polymyxa BFKC01 (83, 84) also stimulate the production of coumarins upon colonization of Arabidopsis roots. It is tempting to speculate that in these mutualistic plant–microbe interactions, the rhizobacteria benefit because scopoletin outcompetes scopoletin-sensitive microbes in the same root niche, while the plant benefits because the population of plant-beneficial microbes increases while scopoletin-sensitive soil-borne pathogens are suppressed.
Materials and Methods
Cultivation of Plants and Microbes.
A detailed description of the growth conditions of wild-type and mutant A. thaliana plants for the metabolite fingerprinting and root microbiome analyses and of the cultivation conditions of P. simiae WCS417, P. capeferrum WCS358, F. oxysporum, and V. dahliae in the different assays used in this study are provided in SI Materials and Methods.
Metabolite Fingerprinting Analysis.
The protocol used for the UPLC-ESI-TOF-MS metabolite fingerprinting analysis of the root extracts and root exudates is described in detail in SI Materials and Methods.
Root Microbiome Analysis.
The experimental setup used to study the root microbiomes and the metagenome analysis pipeline is described in detail in SI Materials and Methods.
Quantification, Visualization, and Determination of Biological Effects of Coumarins.
Quantification and visualization of fluorescent coumarins in root extracts and root exudates and assessment of the effect of scopoletin on the performance of P. simiae WCS417, P. capeferrum WCS358, F. oxysporum, and V. dahliae are described in detail in SI Materials and Methods.
All other methods are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank David Turrà for his suggestions on the chemotropism assay, Bart Thomma for providing Verticillium dahliae JR2, Jürgen Zeier for f6′h1 seeds, Richard Hickman for discussions on the statistical analysis of microbiome data, and Sabine Freitag and Hans Van Pelt for technical assistance. This work was supported by European Research Council Advanced Grant 269072 (to C.M.J.P.), a China Scholarship Council fellowship (to K.Y.), Research Foundation Flanders postdoctoral fellowship 12B8116N (to R.d.J.), and Deutsche Forschungsgemeinschaft Grant ZUK 45/2010 (to I.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this article have been deposited in the National Center for Biotechnology Information Short Read Archive BioProject database (BioProject ID: PRJNA435676).
See Commentary on page 5629.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722335115/-/DCSupplemental.
References
- 1.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 2.Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Mendes R, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 4.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 5.Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P. Interplay between innate immunity and the plant microbiota. Annu Rev Phytopathol. 2017;55:565–589. doi: 10.1146/annurev-phyto-080516-035623. [DOI] [PubMed] [Google Scholar]
- 6.Pieterse CMJ, et al. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 7.Berendsen RL, et al. Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genomics. 2015;16:539. doi: 10.1186/s12864-015-1632-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pieterse CMJ, et al. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 1998;10:1571–1580. doi: 10.1105/tpc.10.9.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verhagen BW, et al. The transcriptome of rhizobacteria-induced systemic resistance in arabidopsis. Mol Plant Microbe Interact. 2004;17:895–908. doi: 10.1094/MPMI.2004.17.8.895. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Medina A, et al. Recognizing plant defense priming. Trends Plant Sci. 2016;21:818–822. doi: 10.1016/j.tplants.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 11.Pozo MJ, Van Der Ent S, Van Loon LC, Pieterse CMJ. Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol. 2008;180:511–523. doi: 10.1111/j.1469-8137.2008.02578.x. [DOI] [PubMed] [Google Scholar]
- 12.Zamioudis C, Hanson J, Pieterse CMJ. β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 2014;204:368–379. doi: 10.1111/nph.12980. [DOI] [PubMed] [Google Scholar]
- 13.Stringlis IA, et al. Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 2018;93:166–180. doi: 10.1111/tpj.13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van der Ent S, et al. MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 2008;146:1293–1304. doi: 10.1104/pp.107.113829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez-Medina A, Van Wees SCM, Pieterse CMJ. Airborne signals from Trichoderma fungi stimulate iron uptake responses in roots resulting in priming of jasmonic acid-dependent defences in shoots of Arabidopsis thaliana and Solanum lycopersicum. Plant Cell Environ. 2017;40:2691–2705. doi: 10.1111/pce.13016. [DOI] [PubMed] [Google Scholar]
- 16.Segarra G, Van der Ent S, Trillas I, Pieterse CMJ. MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol (Stuttg) 2009;11:90–96. doi: 10.1111/j.1438-8677.2008.00162.x. [DOI] [PubMed] [Google Scholar]
- 17.Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML. MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet. 2013;9:e1003953. doi: 10.1371/journal.pgen.1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi T, Nishizawa NK. Iron uptake, translocation, and regulation in higher plants. Annu Rev Plant Biol. 2012;63:131–152. doi: 10.1146/annurev-arplant-042811-105522. [DOI] [PubMed] [Google Scholar]
- 19.Colangelo EP, Guerinot ML. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell. 2004;16:3400–3412. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santi S, Schmidt W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009;183:1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x. [DOI] [PubMed] [Google Scholar]
- 21.Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson NJ, Procter CM, Connolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- 23.Zamioudis C, et al. Rhizobacterial volatiles and photosynthesis-related signals coordinate MYB72 expression in Arabidopsis roots during onset of induced systemic resistance and iron-deficiency responses. Plant J. 2015;84:309–322. doi: 10.1111/tpj.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fourcroy P, et al. Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 2014;201:155–167. doi: 10.1111/nph.12471. [DOI] [PubMed] [Google Scholar]
- 25.Rodríguez-Celma J, et al. Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol. 2013;162:1473–1485. doi: 10.1104/pp.113.220426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmid NB, et al. Feruloyl-CoA 6′-Hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol. 2014;164:160–172. doi: 10.1104/pp.113.228544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fourcroy P, Tissot N, Gaymard F, Briat JF, Dubos C. Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high-affinity root Fe(2+) transport system. Mol Plant. 2016;9:485–488. doi: 10.1016/j.molp.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Verbon EH, et al. Iron and immunity. Annu Rev Phytopathol. 2017;55:355–375. doi: 10.1146/annurev-phyto-080516-035537. [DOI] [PubMed] [Google Scholar]
- 29.Castrillo G, et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai HH, Schmidt W. One way. Or another? Iron uptake in plants. New Phytol. 2017;214:500–505. doi: 10.1111/nph.14477. [DOI] [PubMed] [Google Scholar]
- 31.Ziegler J, et al. Non-targeted profiling of semi-polar metabolites in Arabidopsis root exudates uncovers a role for coumarin secretion and lignification during the local response to phosphate limitation. J Exp Bot. 2016;67:1421–1432. doi: 10.1093/jxb/erv539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carvalhais LC, et al. Activation of the jasmonic acid plant defence pathway alters the composition of rhizosphere bacterial communities. PLoS One. 2013;8:e56457. doi: 10.1371/journal.pone.0056457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dakora FD, Phillips DA. Root exudates as mediators of mineral acquisition in low-nutrient environments. Plant Soil. 2002;245:35–47. [Google Scholar]
- 34.Carvalhais LC, et al. Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol Plant Microbe Interact. 2015;28:1049–1058. doi: 10.1094/MPMI-01-15-0016-R. [DOI] [PubMed] [Google Scholar]
- 35.Lebeis SL, et al. PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 36.Bakker PAHM, Pieterse CMJ, de Jonge R, Berendsen RL. The soil-borne legacy. Cell. 2018;172:1178–1180. doi: 10.1016/j.cell.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 37.Gnonlonfin GJB, Sanni A, Brimer L. Review scopoletin–A coumarin phytoalexin with medicinal properties. Crit Rev Plant Sci. 2012;31:47–56. [Google Scholar]
- 38.El Oirdi M, Trapani A, Bouarab K. The nature of tobacco resistance against Botrytis cinerea depends on the infection structures of the pathogen. Environ Microbiol. 2010;12:239–253. doi: 10.1111/j.1462-2920.2009.02063.x. [DOI] [PubMed] [Google Scholar]
- 39.Kim YH, et al. Scopoletin production related to induced resistance of tobacco plants against Tobacco mosaic virus. Plant Pathol J. 2000;16:264–268. [Google Scholar]
- 40.Sun H, et al. Scopoletin is a phytoalexin against Alternaria alternata in wild tobacco dependent on jasmonate signalling. J Exp Bot. 2014;65:4305–4315. doi: 10.1093/jxb/eru203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaever A, et al. MarVis-pathway: Integrative and exploratory pathway analysis of non-targeted metabolomics data. Metabolomics. 2015;11:764–777. doi: 10.1007/s11306-014-0734-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahn YO, et al. Scopolin-hydrolyzing beta-glucosidases in roots of Arabidopsis. Plant Cell Physiol. 2010;51:132–143. doi: 10.1093/pcp/pcp174. [DOI] [PubMed] [Google Scholar]
- 43.Le Roy J, Huss B, Creach A, Hawkins S, Neutelings G. Glycosylation is a major regulator of phenylpropanoid availability and biological activity in plants. Front Plant Sci. 2016;7:735. doi: 10.3389/fpls.2016.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kai K, et al. Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J. 2008;55:989–999. doi: 10.1111/j.1365-313X.2008.03568.x. [DOI] [PubMed] [Google Scholar]
- 45.Berendsen RL, et al. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018 doi: 10.1038/s41396-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mimmo T, et al. Rhizospheric organic compounds in the soil-microorganism-plant system: Their role in iron availability. Eur J Soil Sci. 2014;65:629–642. [Google Scholar]
- 47.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016;7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Federhen S. The NCBI Taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fradin EF, et al. Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 2009;150:320–332. doi: 10.1104/pp.109.136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pieterse CMJ, van Wees SCM, Hoffland E, van Pelt JA, van Loon LC. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell. 1996;8:1225–1237. doi: 10.1105/tpc.8.8.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Medentsev AG, Arinbasarova AIu, Akimenko VK. Biosynthesis of naphthoquinone pigments by fungi of the genus Fusarium. Prikl Biokhim Mikrobiol. 2005;41:573–577. [PubMed] [Google Scholar]
- 52.Son SW, et al. Bikaverin and fusaric acid from Fusarium oxysporum show antioomycete activity against Phytophthora infestans. J Appl Microbiol. 2008;104:692–698. doi: 10.1111/j.1365-2672.2007.03581.x. [DOI] [PubMed] [Google Scholar]
- 53.Turrà D, El Ghalid M, Rossi F, Di Pietro A. Fungal pathogen uses sex pheromone receptor for chemotropic sensing of host plant signals. Nature. 2015;527:521–524. doi: 10.1038/nature15516. [DOI] [PubMed] [Google Scholar]
- 54.Liu J, Osbourn A, Ma P. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant. 2015;8:689–708. doi: 10.1016/j.molp.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 55.Morant AV, et al. beta-Glucosidases as detonators of plant chemical defense. Phytochemistry. 2008;69:1795–1813. doi: 10.1016/j.phytochem.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 56.Jones P, Vogt T. Glycosyltransferases in secondary plant metabolism: Tranquilizers and stimulant controllers. Planta. 2001;213:164–174. doi: 10.1007/s004250000492. [DOI] [PubMed] [Google Scholar]
- 57.Bakker PAHM, Berendsen RL, Doornbos RF, Wintermans PC, Pieterse CMJ. The rhizosphere revisited: Root microbiomics. Front Plant Sci. 2013;4:165. doi: 10.3389/fpls.2013.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turner TR, et al. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 2013;7:2248–2258. doi: 10.1038/ismej.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bulgarelli D, et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17:392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edwards J, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA. 2015;112:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zgadzaj R, et al. Root nodule symbiosis in Lotus japonicus drives the establishment of distinctive rhizosphere, root, and nodule bacterial communities. Proc Natl Acad Sci USA. 2016;113:E7996–E8005. doi: 10.1073/pnas.1616564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, et al. Huanglongbing impairs the rhizosphere-to-rhizoplane enrichment process of the citrus root-associated microbiome. Microbiome. 2017;5:97. doi: 10.1186/s40168-017-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aliotta G, De Feo V, Pinto G, Pollio A. In vitro inhibition of algal growth by Ruta graveolens L. extracts: Biological and chemical aspects. Plant Biosyst. 1999;133:185–191. [Google Scholar]
- 65.Kim KK, Kim MK, Lim JH, Park HY, Lee ST. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. nov. Int J Syst Evol Microbiol. 2005;55:1287–1293. doi: 10.1099/ijs.0.63541-0. [DOI] [PubMed] [Google Scholar]
- 66.Krishnamurthi S, Ruckmani A, Pukall R, Chakrabarti T. Psychrobacillus gen. nov. and proposal for reclassification of Bacillus insolitus Larkin & Stokes, 1967, B. psychrotolerans Abd-El Rahman et al., 2002 and B. psychrodurans Abd-El Rahman et al., 2002 as Psychrobacillus insolitus comb. nov., Psychrobacillus psychrotolerans comb. nov. and Psychrobacillus psychrodurans comb. nov. Syst Appl Microbiol. 2010;33:367–373. doi: 10.1016/j.syapm.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 67.Nazina TN, et al. Taxonomic study of aerobic thermophilic bacilli: Descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. th. Int J Syst Evol Microbiol. 2001;51:433–446. doi: 10.1099/00207713-51-2-433. [DOI] [PubMed] [Google Scholar]
- 68.Guan S, et al. Aflatoxin B(1) degradation by Stenotrophomonas maltophilia and other microbes selected using coumarin medium. Int J Mol Sci. 2008;9:1489–1503. doi: 10.3390/ijms9081489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Nys R, Steinberg PD. Linking marine biology and biotechnology. Curr Opin Biotechnol. 2002;13:244–248. doi: 10.1016/s0958-1669(02)00311-7. [DOI] [PubMed] [Google Scholar]
- 70.Han JI, et al. Complete genome sequence of the metabolically versatile plant growth-promoting endophyte Variovorax paradoxus S110. J Bacteriol. 2011;193:1183–1190. doi: 10.1128/JB.00925-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pérez Rodríguez N, et al. The role of bacterial consortium and organic amendment in Cu and Fe isotope fractionation in plants on a polluted mine site. Environ Sci Pollut Res Int. 2014;21:6836–6844. doi: 10.1007/s11356-013-2156-1. [DOI] [PubMed] [Google Scholar]
- 72.Sauder LA, et al. Cultivation and characterization of Candidatus Nitrosocosmicus exaquare, an ammonia-oxidizing archaeon from a municipal wastewater treatment system. ISME J. 2017;11:1142–1157. doi: 10.1038/ismej.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fries LLM, Pacovsky RS, Safir GR, Siqueira JO. Plant growth and arbuscular mycorrhizal fungal colonization affected by exogenously applied phenolic compounds. J Chem Ecol. 1997;23:1755–1767. [Google Scholar]
- 74.Zhang L, Shen X, Liu Y, Li S. Nafulsella turpanensis gen. nov., sp. nov., a member of the phylum Bacteroidetes isolated from soil. Int J Syst Evol Microbiol. 2013;63:1639–1645. doi: 10.1099/ijs.0.044651-0. [DOI] [PubMed] [Google Scholar]
- 75.Dastager SG, Deepa CK, Pandey A. Plant growth promoting potential of Pontibacter niistensis in cowpea (Vigna unguiculata (L.) Walp.) Appl Soil Ecol. 2011;49:250–255. [Google Scholar]
- 76.McBride MJ, Liu W, Lu X, Zhu Y, Zhang W. The Prokaryotes. Springer; Berlin: 2014. The family Cytophagaceae; pp. 577–593. [Google Scholar]
- 77.Srinivasan S, et al. Complete genome sequence of Rufibacter sp. DG31D, a bacterium resistant to gamma and UV radiation toxicity. Mol Cell Toxicol. 2015;11:415–421. [Google Scholar]
- 78.Badri DV, Chaparro JM, Zhang R, Shen Q, Vivanco JM. Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J Biol Chem. 2013;288:4502–4512. doi: 10.1074/jbc.M112.433300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simon C, et al. The differential spatial distribution of secondary metabolites in Arabidopsis leaves reacting hypersensitively to Pseudomonas syringae pv. tomato is dependent on the oxidative burst. J Exp Bot. 2010;61:3355–3370. doi: 10.1093/jxb/erq157. [DOI] [PubMed] [Google Scholar]
- 80.Gutiérrez-Barranquero JA, Reen FJ, McCarthy RR, O’Gara F. Deciphering the role of coumarin as a novel quorum sensing inhibitor suppressing virulence phenotypes in bacterial pathogens. Appl Microbiol Biotechnol. 2015;99:3303–3316. doi: 10.1007/s00253-015-6436-1. [DOI] [PubMed] [Google Scholar]
- 81.Lee JH, et al. Coumarins reduce biofilm formation and the virulence of Escherichia coli O157:H7. Phytomedicine. 2014;21:1037–1042. doi: 10.1016/j.phymed.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 82.Limón MC, Rodríguez-Ortiz R, Avalos J. Bikaverin production and applications. Appl Microbiol Biotechnol. 2010;87:21–29. doi: 10.1007/s00253-010-2551-1. [DOI] [PubMed] [Google Scholar]
- 83.van de Mortel JE, et al. Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol. 2012;160:2173–2188. doi: 10.1104/pp.112.207324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou C, et al. Paenibacillus polymyxa BFKC01 enhances plant iron absorption via improved root systems and activated iron acquisition mechanisms. Plant Physiol Biochem. 2016;105:162–173. doi: 10.1016/j.plaphy.2016.04.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.