Significance
Organisms vary in the number of genome copies per cell: ploidy. By altering how DNA is replicated and repaired, ploidy may determine the number and types of mutations that arise, affecting how evolution proceeds. We sequenced the genomes of >200 replicate lines of yeast (Saccharomyces cerevisiae) with one versus two genome copies (haploid versus diploid) after accumulation of thousands of new mutations. Haploids were more susceptible to single-nucleotide mutations, particularly for DNA replicated later in the cell cycle, whereas large changes to genome structure were more common in diploids. Haploid and diploid populations will therefore have access to distinct kinds of genetic variation, contributing to differences in their evolutionary potential.
Keywords: aneuploidy, mitochondria, replication time, RDH54
Abstract
By altering the dynamics of DNA replication and repair, alternative ploidy states may experience different rates and types of new mutations, leading to divergent evolutionary outcomes. We report a direct comparison of the genome-wide spectrum of spontaneous mutations arising in haploids and diploids following a mutation-accumulation experiment in the budding yeast Saccharomyces cerevisiae. Characterizing the number, types, locations, and effects of thousands of mutations revealed that haploids were more prone to single-nucleotide mutations (SNMs) and mitochondrial mutations, while larger structural changes were more common in diploids. Mutations were more likely to be detrimental in diploids, even after accounting for the large impact of structural changes, contrary to the prediction that mutations would have weaker effects, due to masking, in diploids. Haploidy is expected to reduce the opportunity for conservative DNA repair involving homologous chromosomes, increasing the insertion-deletion rate, but we found little support for this idea. Instead, haploids were more susceptible to SNMs in late-replicating genomic regions, resulting in a ploidy difference in the spectrum of substitutions. In diploids, we detect mutation rate variation among chromosomes in association with centromere location, a finding that is supported by published polymorphism data. Diploids are not simply doubled haploids; instead, our results predict that the spectrum of spontaneous mutations will substantially shape the dynamics of genome evolution in haploid and diploid populations.
Mutations play a critical role in evolution and adaptation, but these spontaneous genetic changes are often hazardous, reducing the health of individuals and imposing a genetic load on populations. While individual mutations are chance events, the biological processes that produce or prevent mutation have the potential to vary among genetic contexts, leading to consistent biases in the numbers, locations, and types of genetic changes that occur, affecting how genomes and populations evolve. One dimension of genomic variation that has broad potential to affect the mutation process is ploidy state, the number of homologous chromosome sets per cell. Ploidy varies among cells as a consequence of meiosis and syngamy and can also vary among individuals (e.g., in haplodiploid organisms). Transitions in predominant ploidy level have occurred many times across the tree of life (1, 2), but the evolutionary and ecological drivers and consequences of these changes are still unclear.
All else being equal, the genome-wide mutation rate of diploid cells should be twice that of haploid cells, as they have twice as many nucleotide sites with the potential to mutate. However, ploidy level is likely to have additional effects on the rate and spectrum of mutations. The presence of a homologous chromosome template allows DNA double-strand breaks (DSBs) to be repaired using conservative homology-directed pathways rather than competing error-prone end-joining pathways that generate insertion-deletion (indel) mutations (3). The continual access to homologous template DNA in diploid cells may therefore reduce the rate of indels relative to haploid cells. While diploidy may reduce susceptibility to certain forms of DNA damage, the presence of multiple genome copies may also increase the likelihood of spontaneous structural changes and rearrangements through nonhomologous crossing over (3). There are therefore reasons to expect differences in the mutational properties of alternative ploidy states.
Estimates of genome-wide mutation rates are available from mutation accumulation (MA) experiments with the budding yeast Saccharomyces cerevisiae, which can grow mitotically in either a haploid or diploid state. In these experiments, replicate lines are periodically subjected to single-cell bottlenecks, allowing mutations with mild and moderate effects to accumulate as though they were selectively neutral. Previous studies have examined either haploids or diploids, preventing direct comparison.
Genome-wide patterns of spontaneous mutation have been somewhat better characterized in diploid S. cerevisiae (4, 5) than in haploids, where sampling has been limited (6, 7). At face value, previous estimates suggest that the rate of single-nucleotide mutations (SNMs) per base pair may be greater in haploids than in diploids, and it has been proposed that yeast genomes are more stable in the diploid state (4, 5). However, these different inferred rates could reflect variation among laboratory strains, methodology, or environments. Additionally, in a previous study of haploids, all four lines examined by genome sequencing apparently became diploid spontaneously during the experiment (6), complicating inference of the mutation rate in haploid cells.
To formally investigate the impact of ploidy on the mutation process, we accumulated mutations in isogenic replicate haploid and diploid lines of S. cerevisiae derived from a common haploid ancestor. We maintained 220 lines under relaxed selection by enforcing single-cell bottlenecks every day (∼16 generations) for 100 d, for a total of ∼336,000 generations across the experiment. To address the possible role of homology-directed DSB repair, we deleted the gene RDH54 in the ancestor of half of the lines. There is evidence that this gene is essential for mitotic recombinational repair between homologous chromosomes in diploids but not between sister chromatids in haploids or diploids (8). Genome sequencing of our MA lines revealed over 2,000 mutation events, with clear impacts of ploidy on the rate, spectrum, locations, and fitness consequences of spontaneous mutations but little evidence that homology-directed DSB repair is a major factor in these differences.
Results
Growth Rates and Ploidy.
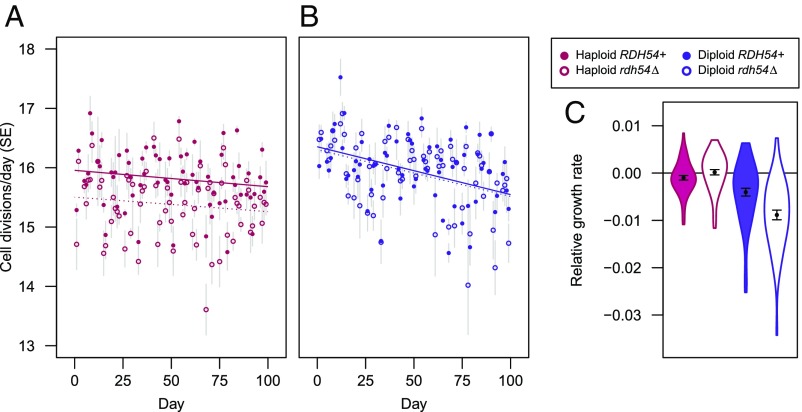
To account for potential differences among treatments in rates of cell division, we measured the number of cells per colony in the MA lines throughout the bottlenecking procedure and used these values to calculate mutation rates per cell division (Table 1). We found small but significant treatment effects on growth rate, with a more rapid decline in diploid lines than in haploid lines over the course of the experiment (Fig. 1 A and B).
Table 1.
Sample sizes
| Haploid | Diploid | |||
| Metric | RDH54+ | rdh54Δ | RDH54+ | rdh54Δ |
| MA lines | 53 | 53 | 63 | 51 |
| Cell divisions per transfer | 15.7 | 15.2 | 15.8 | 15.7 |
| Transfers per line* | 94.9 | 97.5 | 100.0 | 98.9 |
| Median coverage per site per line (range†) | 23 (16, 30) | 22 (10, 35) | 44 (20, 65) | 44 (30, 62) |
| Callable nuclear sites per line‡ | 11,296,337 | 11,255,081 | 23,088,492 | 22,684,822 |
| Callable mt sites per line | 72,036 | 70,426 | 74,868 | 75,028 |
Some lines were reinitialized partway through the procedure, and therefore underwent <100 transfers as unique lines.
Minimum and maximum of median coverage among lines.
Incorporates ploidy, including ancestral trisomy of chromosome XI in some lines.
Fig. 1.
Cell divisions during MA and growth rates following MA. Cell divisions per day (A, haploids; B, diploids) depended on the interaction of ploidy level, RDH54 status, and time (>3,200 colonies measured; linear model, ploidy × RDH54: P < 10−8; ploidy × time: P < 0.01). Solid (dashed) lines show linear regression for RDH54+ (rdh54Δ) treatments. (C) Violin plots of maximum growth rates after 100 transfers relative to ancestral controls, accounting for block effects. Points (error bars) show means (SEs). Steeper declines in cell divisions per day in diploids relative to haploids during MA (A and B) are consistent with relative growth rate estimates following MA (linear mixed model based on 4,000 growth curves: MA × ploidy × RDH54: P < 0.05). Diploid MA lines have significantly reduced growth rates relative to the ancestor (MA: P < 10−6, MA × RDH54: P < 0.05), unlike haploid lines (MA: P = 0.64). Growth-rate data for individual lines are provided in Dataset S2.
After 100 bottlenecks, we measured the growth rates of MA lines and ancestral controls in liquid culture, revealing that diploid MA lines, but not haploid MA lines, show reduced growth relative to ancestral controls (Fig. 1C), consistent with the measures of colony growth on plates during MA. All four groups of MA lines (ploidy × RDH54 status) show significant genetic variance for growth rate (all χ2 > 24.8, all P < 10−6), whereas there was no significant genetic variance detected in any group of ancestral control lines (all χ2 < 2.6, all P > 0.11). The finding that diploid but not haploid lines show reduced growth following MA is surprising, as the effects of recessive deleterious mutations would be masked in diploids. We discuss this result in relation to the genomic data in Growth Rates in Relation to Mutations.
Haploid yeast are known to become diploid spontaneously (e.g., through defects in mitosis), and diploids can similarly change ploidy level. Such changes are commonly observed in yeast evolution experiments with large populations, but the spontaneous rate of ploidy change per cell division is unclear. Using flow cytometry, we found that all MA lines retained their original ploidy level at the end of the experiment [Dataset S1; confidence interval (CI) for rate of change from haploid to diploid: 0–2.3 × 10−5 per cell division; results on aneuploidy are discussed below]. This is in contrast to a previous finding (6), where four of four haploid MA lines became diploid over the course of ∼4,800 generations [rate (95% CI): 20.8 (5.7, 53.3) × 10−5]. This difference suggests either that the higher frequency of single-cell bottlenecks in our experiment (daily rather than every 3–4 d) reduced the opportunity for selection favoring diploidy or that yeast strains vary in their propensity to spontaneously change ploidy.
We observed changes in the mating behavior of three MA lines and describe the likely genetic basis for these changes in SI Appendix.
Point Mutations.
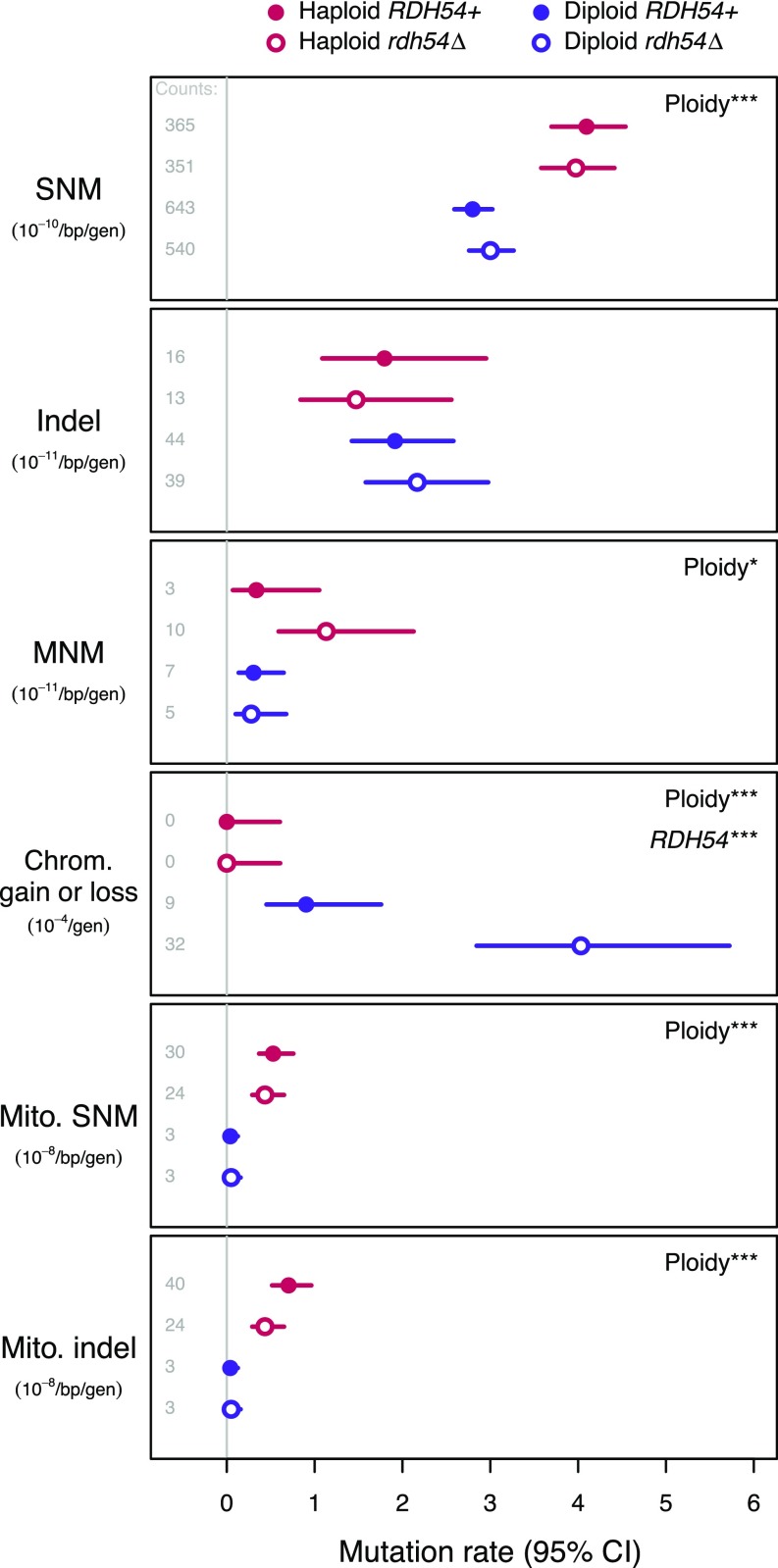
We detected >2,000 point mutations (Dataset S2). Mutation rates, accounting for numbers of cell divisions and callable sites, are shown in Fig. 2. There was no effect of mating type (MAT) on mutation rates within haploids (SI Appendix, Fig. S1), and so we pooled the data for haploid lines of the two mating types (MATa and MATα) throughout our analyses. We detected a significant effect of ploidy level on the rate of SNMs (binomial test: P < 10−11), with no effect of RDH54 status (P > 0.24 in either ploidy level). The per-nucleotide SNM rate was 40% higher in haploids (4.04 × 10−10, 95% CI: 3.75–4.34 × 10−10) than in diploids (2.89 × 10−10, 95% CI: 2.73–3.06 × 10−10). Thus, although diploids have twice as many nucleotide sites as haploids, they incur only 1.43-fold as many SNMs per cell division.
Fig. 2.
Mutation rates in each group of MA lines, with 95% CIs. Panels show SNMs, indels, MNMs, chromosome (Chrom.) gains and losses, mt (Mito.) SNMs, and Mito. indels. Rates represent events per base pair (bp) per generation (gen), except for whole-chromosome gains and losses (events per generation). For mt events, we consider all treatments effectively haploid. The absolute numbers of events observed (“Counts”) are given at the left of each panel; note that detection power differs among groups such that rates are not necessarily proportional to mutation counts. Statistically significant treatment effects are noted at the right of each panel (binomial tests accounting for detection power: *P < 0.05; **P < 0.01; ***P < 0.001). A numeric summary of rates and CIs is provided in Dataset S2.
Because diploidy provides greater access to homologous DSB repair, we expected to see a higher rate of indel mutations in haploids and in diploids lacking the RDH54 gene. However, the indel rate did not depend on ploidy level (binomial test: P = 0.52) or RDH54 status (P > 0.58 in either ploidy level; Fig. 2). In fact, our point estimate of the per-nucleotide rate of indels is 24% greater in diploids (2.03 × 10−11) than in haploids (1.63 × 10−11), such that the indel rate per cell division is 2.48-fold higher in diploids. There was a nonsignificant tendency toward deletions among indel events (62 deletions and 50 insertions; binomial test: P = 0.30).
We considered whether selection during the experiment might have affected rates of MA. Accounting for power to detect mutations in genic and nongenic regions and assuming that genic and nongenic regions are equally likely to mutate, there is no evidence that SNMs were less likely to occur in genes than expected (observed rate: 74.0%, expected rate: 73.9%; binomial test: P = 0.92) or less likely to be nonsynonymous than expected (observed rate: 76.7%, expected rate: 76.1%; binomial test: P = 0.62), with no difference in these rates between ploidy or RDH54 levels. There is therefore no evidence that the accumulation of SNMs was influenced by selection in our experiment.
By contrast, indels were found in genes less often than expected (observed rate: 46.4%, expected rate: 73.9%; binomial test: P < 10−9), as observed in other datasets of this kind (5). Selection acting more effectively against indels in haploids could mask a true difference in indel rate. However, the fraction of indels that accumulated in genes does not differ significantly between ploidy levels (Fisher’s exact test: odds ratio = 1.10, P = 0.83), and there is no indication that nongenic indel rates differed by RDH54 status or ploidy (SI Appendix, Fig. S2). It is therefore not apparent that the pattern of indel accumulation among treatment groups in our experiment was driven by selection against genic indels. The deficit of indels in genes could also stem from differences in sequence complexity between genic and nongenic regions, rather than selection. We observed indels >20-fold more frequently in regions identified by RepeatMasker (9) as simple repeats or a low-complexity sequence (binomial test: P < 10−15), and such regions were >2.8-fold more common outside of genes (odds ratio = 0.34, P < 10−15). Excluding these regions, we still find that indels are less common in genes than expected (binomial test: P < 10−8), but this simple approach is unlikely to account for all repetitive sequences.
As in previous studies where MA lines have been sequenced, we find evidence for some substitutions occurring in close proximity to one another, which we categorize as multinucleotide mutations (MNMs) (10). As with SNMs, the MNM rate was significantly higher in haploids (0.73 × 10−11) than diploids (0.29 × 10−11; Fig. 2; binomial test: P < 0.05), suggesting that replication errors associated with ploidy generate both SNMs and MNMs. This difference is particularly pronounced in the rdh54Δ haploids (Fig. 2; binomial test of RDH54 effect within haploids: P = 0.056), but the ploidy × RDH54 interaction is nonsignificant (power-adjusted goodness-of-fit test: G = 2.0, P = 0.16).
While asexual diploids are expected to adapt more slowly because beneficial mutations are partially masked, the strength of this effect depends on the rate at which mutations become homozygous (11), due to mitotic crossing over, noncrossover gene conversion, or chromosome loss and duplication (3, 12). We identified 38 homozygous SNMs and one homozygous indel in diploid lines, reflecting loss of heterozygosity (LOH) events. The rate of homozygous mutations did not differ between RDH54 treatments (binomial test: P = 0.63). For six homozygous mutations, another mutation occurred distally on the same chromosome arm in the same MA line, and of these, three were also homozygous, which is a higher rate than expected if all LOH events occurred independently (binomial test: P < 0.001). These pairs of homozygous mutations were separated by 132–451 kb, suggesting that a substantial fraction of LOH events are due to mitotic crossing over or chromosome loss/duplication rather than relatively short gene conversion tracts.
Large-Scale Mutations.
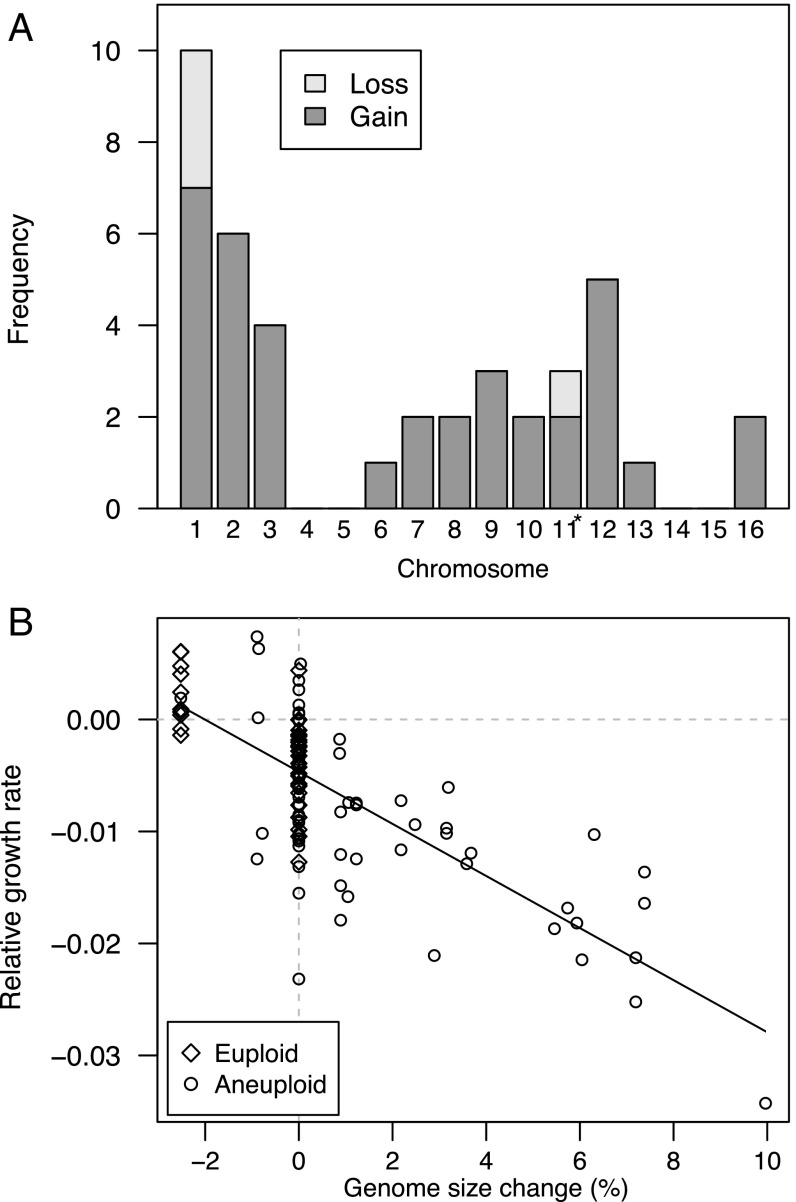
We used sequencing coverage across the genome to detect changes in chromosome copy number (aneuploidy), which result from mitotic nondisjunction. Forty-nine of 63 RDH54+ diploid lines were found to carry three copies of chromosome XI, which we established to be an ancestral polymorphism in that group of lines (Materials and Methods). We also observed 41 de novo chromosome gains and losses, all within the diploid MA lines (Fig. 2). Chromosome gains (35 cases of trisomy and two cases of tetrasomy) were found more often than chromosome losses (four losses, including a loss of a trisomic chromosome XI restoring euploidy; binomial test: P < 10−6; Fig. 3A), as observed in a previous MA experiment (5). While the rarity of chromosome losses relative to gains may reflect stronger selection against monosomy in diploids or a higher chance of reversion to euploidy from monosomy in MA experiments, our observed rates of chromosome loss are generally consistent with previous estimates obtained using other methods (reviewed in ref. 3). The derived versus ancestral allele depths for substitutions conformed well to the expected patterns on aneuploid and nonaneuploid chromosomes (SI Appendix, Fig. S3), confirming that the chromosome XI trisomy occurred before MA and that other aneuploidies occurred uniformly throughout MA.
Fig. 3.
Gains and losses of each chromosome during MA and the fitness consequences of aneuploidy. (A) There was no evidence that the distribution of aneuploidy events among chromosomes depended on RDH54 type (Fisher’s exact test: P = 0.56), but with few events in RDH54+ lines, we have low power to test this hypothesis. Pooling data between RDH54 types, gains were more common than losses (binomial test: P < 10−6) and rates vary among chromosomes (χ2 = 42.1, simulated P < 0.001). Several cases of trisomy for chromosome 11 in RDH54+ diploids were determined to be ancestral and are not scored as gains; in one case, the extra copy was lost, restoring euploidy, and this is scored here as a loss. Aneuploid line identifications are provided in Dataset S2. (B) Genome size relative to controls was negatively correlated with maximum growth rate relative to controls in diploid MA lines (r = −0.74, df = 112, P < 10−15). Diploid RDH54+ controls had trisomy for chromosome 11, so MA lines with only this trisomy were scored as having zero genome size change and MA lines without this trisomy were scored as having a genome size reduction unless other aneuploidy was present. This correlation persists when only aneuploid lines are considered (r = −0.70, df = 78, P < 10−12) or when lines with no change are excluded (r = −0.84, df = 45, P < 10−12).
While the rate of aneuploidy in RDH54+ diploids (0.90 × 10−4) is very similar to previous observations from diploid yeast (1.04 × 10−4) (5), we found that rdh54Δ elevated the rate of aneuploidy fourfold overall and 4.5-fold within diploids (Fig. 2; binomial test: P < 10−4), indicating that RDH54 plays a role in mitotic chromosome segregation in addition to its role in meiosis (8, 13), as suggested previously (14). Given the absence of aneuploidy in haploids, we cannot determine whether the increase in aneuploidy in rdh54Δ lines also applies to haploids or is unique to diploids.
Accounting for the range of haploid aneuploidy rates that is statistically consistent with finding no events and comparing this with the inferred rate in diploids, we find that the rate of aneuploidy is significantly higher in diploids on a per-cell-division basis (Fig. 2; binomial test: P < 10−11). This pattern persists when excluding chromosome losses, which would be lethal in haploids (binomial test: P < 10−10) and when excluding rdh54Δ lines (binomial test: P < 0.05). Accounting for the number of chromosomes per cell that could potentially increase in copy number (twofold higher in diploids), the effect of ploidy on the rate of chromosome gains remains significant overall (binomial test: P < 10−5). Considering only gains in the RDH54+ lines, however, we cannot reject the hypothesis that the opportunity for nondisjunction is simply twofold higher in diploids than in haploids (binomial test: P = 0.20). In any case, our data indicate that diploids experience at least twice the rate of whole-chromosome changes per cell division compared with haploids, in contrast to their lower rate of point mutation (discussed above).
The probability of aneuploidy was not uniform across chromosomes (Fig. 3A), and there was a negative but nonsignificant correlation between the rate of gain or loss and chromosome length (all lines: r = −0.41, P = 0.11; RDH54+ lines: r = −0.36, P = 0.17; rdh54Δ lines: r = −0.41, P = 0.11), supporting the view that chromosome size is not the sole determinant of aneuploidy rates (3, 5). Variation in aneuploidy rates among chromosomes in our experiment was correlated with aneuploidy rates observed previously in diploids exhibiting chromosome instability (r = 0.57, P < 0.05) (15). We found that growth rates were highly correlated with genome size among aneuploid lines (Fig. 3B).
In addition to aneuploidy, coverage profiles revealed three cases of large segmental duplications, all heterozygous in diploids (SI Appendix, Fig. S4), ranging in size from 17 kb to 211 kb. The approximate breakpoints of all three duplications are closer to known Ty elements (mean distance = 700 bp, range: 100–1,662 bp) than expected by chance (simulation of 104 sets of six breakpoints; mean distance to Ty ≤ 700; P < 10−4), supporting the view that Ty elements often anchor large duplications in yeast (16–19). Our estimate of the large duplication rate in diploids is 1.68 (95% CI: 0.34, 5.24) × 10−5 per cell division, consistent with a previous observation of diploid MA lines (∼1.00 × 10−5) (5).
The presence of mutations could increase the subsequent mutation rate by directly altering DNA repair genes, by increasing genome instability (3, 7), or if genetic quality affects DNA repair (20). However, we found no evidence for an effect of aneuploidy on the point mutation rate and no evidence for additional variation in the number of mutations per line beyond that expected under a Poisson distribution (i.e., no evidence of overdispersion) (SI Appendix).
Mitochondrial Mutations.
The cellular environment, replication mechanisms and nucleotide composition of mitochondrial (mt) genomes are distinct from those of nuclear genomes, but it is not clear how nuclear genome ploidy might interact with mt mutation. Unexpectedly, we found that the rate of mt mutation was an order of magnitude higher in haploids (SNM: 4.82 × 10−9, indel: 5.71 × 10−9) than in diploids (SNM: 4.47 × 10−10, indel: 4.47 × 10−10; Fig. 2 and SI Appendix, Table S1). This pattern remained when we relaxed our filtering criteria to detect putative heteroplasmic mutations, which are not fixed within an MA line (SI Appendix). Mt mutation rate estimates based on fewer events from a previous study of four haploid MA lines (which became diploid during MA) are even higher than the haploid rates we observed (SNM: 12.2 × 10−9, indel: 10.4 × 10−9) (6). As in this previous study, we found that the haploid substitution rate was higher in the mt genome than in the nuclear genome (11.9-fold difference, binomial test: P < 10−15), but this was not the case for diploids (1.6-fold difference, binomial test: P = 0.30).
As with nuclear point mutations, there was no evidence that mt SNMs occurred in genes less often than expected (observed proportion: 0.27, expected: 0.35; binomial test: P = 0.23), but mt indels were less likely to accumulate in genes than expected (observed: 0.19; binomial test: P < 0.01). The fraction of events in genes did not differ between ploidy levels for either SNMs (G = 0.32, P = 0.57) or indels (G = 0.07, P = 0.79), and so there is no indication that the elevated rate of mt mutations in haploids relative to diploids is the result of more effective selection against such events in diploids. Small insertions and deletions were about equally prevalent among mt events (37 deletions, 33 insertions; binomial test: P = 0.72).
Mt deletions or other mutations can sometimes result in a “petite” phenotype due to defects in respiratory function. Although we attempted to prevent the accumulation of such variants, two haploid lines had a petite phenotype by the end of the experiment. An examination of sequencing coverage in these lines indicated large deletions in COX1 with very similar breakpoints (deleted locations: ∼16,438–24,230 and 16,435–24,592). Mt deletions with these approximate breakpoints are known to occur frequently, possibly due to autocatalytic activity (21). Additionally, two haploid lines showed large mt deletions in nongenic regions based on coverage (deleted locations: ∼4,824–5,751 and 80,559–83,041). An additional haploid line with a missense mutation in COX1 also showed poor respiration. Thus, in the mt genome, both point mutations and structural changes were observed more often in haploids, unlike in the nuclear genome, where structural changes were observed more often in diploids.
Spectrum of Nucleotide Changes.
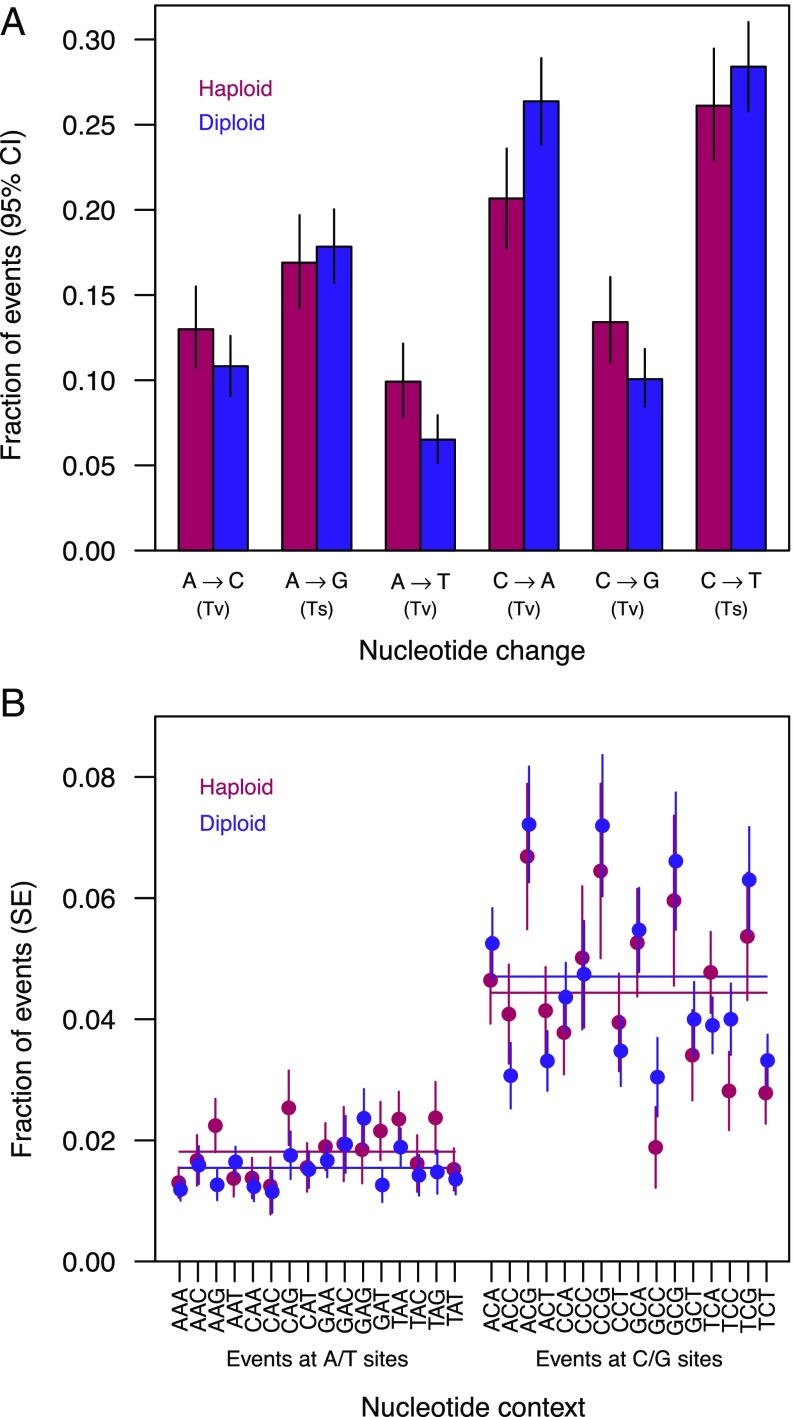
Haploids and diploids differed in the spectrum of SNMs (Fig. 4A). In particular, A-to-T and C-to-G changes were more common among haploid SNMs than among diploid SNMs, whereas the reverse was true for C-to-A changes. Overall, mutations at A/T sites made up a larger fraction of SNMs in haploids than in diploids (Fig. 4B), although all six types of substitution occurred more frequently in haploids than in diploids (SI Appendix, Fig. S5). Such mutational biases may contribute to different substitution spectra in adapting haploid and diploid populations (22).
Fig. 4.
Spectrum of nuclear SNMs. (A) Fraction of SNMs of each type, including complementary changes. Ts, transition; Tv, transversion. RDH54 status did not significantly affect the SNM spectrum (G = 4.5, P = 0.48), but the SNM spectrum differed between ploidy levels (G = 19.8, P < 0.01), particularly among transversion mutations (G = 17.9, P < 0.001). By contrast, the spectrum of transition mutations and the overall transition-to-transversion ratio did not vary significantly by ploidy (G = 0.04, P = 0.84 and odds ratio = 0.88, P = 0.18, respectively). (B) Fraction of SNMs in each 3-bp context, including the complementary context, centered on the focal site, accounting for the frequency of each context in the genome. While the context of SNMs did not differ significantly between haploids and diploids when considering all contexts (G = 21.5, P = 0.90), the fraction of mutations at A/T versus C/G sites differed between ploidy levels (odds ratio = 1.22, P < 0.05; horizontal lines show means across A/T or C/G sites for each ploidy level). The overall rate of SNMs depended on the 3-bp context at G/C sites (G = 81.9, P < 10−10) but not at A/T sites (G = 20.9, P = 0.14). The mutation rate in XCG contexts (where X is any base) was elevated relative to the rate for C/G sites in other contexts (binomial test: P < 10−11).
The transition/transversion ratio did not differ significantly between ploidy levels (haploid = 0.75, diploid = 0.86; odds ratio = 0.88; P = 0.18), and the overall ratio [0.82 (95% CI: 0.75–0.90)] fell between previous estimates for initially haploid MA lines (0.62) (6) and diploid MA lines (0.95) (5).
SNM rates were affected by adjacent nucleotide context at C/G sites but not at A/T sites, with no differences in context effects between ploidy levels (Fig. 4B). In our experiment, both haploids and diploids show an elevated substitution rate at CpG sites compared with C/G sites in other contexts (binomial test: P < 10−11; Fig. 4B), with an approximately twofold elevation in the rate of C-to-T mutations in this context (binomial test: P < 10−14). Although S. cerevisiae shows little evidence for cytosine methylation (but see ref. 23), which can increase the frequency of C-to-T mutations in CpG contexts (24), such an increase has been observed in both diploid S. cerevisiae (5) and haploid Schizosaccharomyces pombe (25, 26).
Genomic Locations of Mutations.
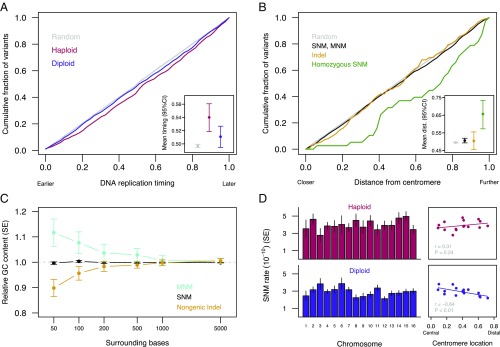
Previous studies have found evidence for increased mutation in late-replicating genomic regions in haploids (27) and nonsignificant trends in diploids (5). Here, we use published information on replication across the yeast genome (28) to compare the effect of replication timing between ploidy levels. Replication timing across the genome is very similar between ploidy levels (28), but we find that SNMs were more likely to occur in later-replicating regions in haploids, whereas there was no effect of replication timing on the rate of SNMs in diploids (Fig. 5A). This effect of replication timing in haploids at least partly explains the overall difference in SNM rates between ploidy levels: We found that haploid and diploid SNM rates were similar within early-replicating genomic regions but diverged in later-replicating regions (SI Appendix, Fig. S6).
Fig. 5.
Genomic context of nuclear mutations. (A) DNA replication timing (data from ref. 28) of mutated sites differed significantly between haploids (red) and diploids (blue) (Wilcoxon test: P < 0.05). Haploid mutations arose in later-replicating positions relative to random callable sites (Wilcoxon test: P < 10−4), whereas diploid mutations did not differ significantly from the random expectation (Wilcoxon test: P = 0.09). (Inset) Mean replication timing for each group (also SI Appendix, Fig. S6). (B) Mutations versus distance from the centromere, as a fraction of maximum possible distance. All event types except indels occurred further from the centromere than random callable sites (Wilcoxon test, SNM and MNM: P < 0.05, Wilcoxon test, homozygous: P < 0.001), and homozygous diploid SNMs are further from the centromere than heterozygous SNMs (Wilcoxon test: P < 0.001). (Inset) Mean distance from the centromere for each event type. (C) GC content surrounding mutations relative to GC content surrounding equivalent random sites (A/T or C/G sites for substitutions, genic or nongenic sites for all mutation types). We consider only nongenic indels here because of possible selection against genic indels. MNM values represent the mean for the component sites. MNMs were associated with higher GC content within 50 bp (bootstrap P < 0.05), whereas nongenic indels were associated with lower GC content within 50 bp (bootstrap P < 0.01). (D) Variation in SNM rate among chromosomes (Left; diploids: G = 23.7, P = 0.07; haploids: G = 12.0, P = 0.68) was correlated with the relative distance between the centromere and the chromosome midpoint in diploids (Right), and this correlation differs significantly between ploidy levels (bootstrap P < 0.01). Centromere location was calculated as (Lq − Lp)/(Lq + Lp), where Lq is the length of the long arm and Lp is the length of the short arm. A summary of variants by chromosome is provided in Dataset S2.
We found that substitutions, but not indels, occurred further from the centromere than expected by chance (Fig. 5B), but neither ploidy nor RDH54 status affected the average distance of mutations from the centromere (linear models: all |t| < 0.99, all P > 0.32). Homozygous SNMs were especially likely to occur far from the centromere (Fig. 5B), which is expected if LOH is caused by mitotic crossing over, although noncrossover gene conversion has also been found to be more frequent further from the centromere (20). Homozygous variants occurred nonrandomly among chromosomes (χ2 = 114, P < 10−15), and the distribution differs from that of heterozygous variants (χ2 = 143, P < 10−15). This pattern is driven almost entirely by an excess of homozygous variants on chromosome XII (21 of the 39 events, all in different lines; binomial test: P < 10−12), all distal to the rDNA tandem repeat region (SI Appendix, Fig. S7), which is known to exhibit a high LOH rate (29, 30). Excluding events on chromosome XII, homozygous variants still occur further from the centromere than expected by chance (Wilcoxon and Kolmogorov–Smirnov tests: P < 0.01).
Previous studies have found evidence that mutation rates are correlated with the guanine-cytosine (GC) content of surrounding regions (20, 31). While we find no correlation between SNM rate and GC content (in accordance with ref. 5), we find that indels are associated with low GC content (in accordance with ref. 20), whereas MNMs are associated with high GC content (Fig. 5C). These associations diminish rapidly with distance from the focal site.
While the number of SNMs on a given chromosome was highly correlated with callable chromosome length (haploids: r = 0.96, P < 10−8; diploids: r = 0.96, P < 10−8), there was some evidence for residual variation (Fig. 5D), and we found that diploid SNM rates were significantly lower on chromosomes where the centromere is relatively distal, whereas this correlation was absent, and possibly reversed, in haploids (Fig. 5D). We reasoned that if this unexpected result reflects genuine mutation rate variation among chromosomes, the pattern might also be evident in the locations of polymorphisms. We found that polymorphic sites in wild and domesticated yeast isolates (32) coincided with SNM sites more often than expected by chance (odds ratio = 1.52, P < 0.05) and that polymorphism rates by chromosome were more strongly correlated with our diploid SNM rates (ρ = 0.79, 95% CI: 0.45–0.95, P < 0.001; SI Appendix, Fig. S8) than our haploid SNM rates (ρ = −0.05, 95% CI: −0.50 to 0.41, P = 0.87), as expected if polymorphism reflects mutation rates and given that S. cerevisiae is generally diploid. As with SNMs in diploids, polymorphism rates tend to be negatively correlated with centromere location (ρ = −0.47, P = 0.066).
Examining the spatial pattern of SNMs within chromosomes, we found that the extent of spatial correlation between ploidy levels was itself negatively correlated with centromere location (r = −0.70, P < 0.01; SI Appendix, Fig. S9), which is unlikely to occur by chance (5,000 simulated datasets; P < 0.01). This suggests that the mutation rate in diploids is reduced in particular regions of chromosomes with distal centromeres. Additional possible explanatory factors are addressed in SI Appendix. The relative position of centromeres will affect the physical location of chromosome arms within the nucleus, as well as interactions between homologous loci (33–35), which might differentially affect haploid and diploid genome stability.
Growth Rates in Relation to Mutations.
In our study, MA led to reduced growth in diploids but not haploids (Fig. 1C). This difference was driven, at least in part, by a particular class of mutations, aneuploidy events, that arose in diploid but not haploid lines (Fig. 3B). Accounting for these large-effect variants, including the presence of chromosome XI trisomy in the ancestral diploid RDH54+ control, we modeled the effects of genome size and number of nonsynonymous nuclear and mt point mutations using mixed models (SI Appendix, Fig. S10, and details are provided in SI Appendix). We confirmed a highly significant effect of genome size change (χ2 = 291.37, P < 10−15) and detected an interaction between ploidy and point mutations (χ2 = 5.91, P < 0.05). Separate models for haploids and diploids showed a negative but nonsignificant effect of point mutations in haploids (χ2 = 0.32, P = 0.57) and a significant negative effect in diploids (χ2 = 19.56, P < 10−5).
These results, as well as other modeling approaches (SI Appendix), indicate that the accumulated point mutations had stronger negative effects on diploids than haploids, controlling for the effects of aneuploidy. This unexpected pattern could arise if the accumulation of deleterious alleles was opposed by selection during MA in haploids but not in diploids, although we did not find evidence of selection, at least among the SNMs, when examining the fraction of genic or nonsynonymous events. Furthermore, the effective population sizes of our lines should permit the accumulation of mutations with effects less than ∼12%, which is more extreme than the largest fitness decline we observed (∼3%), suggesting that the vast majority of mutations had weak effects and would accumulate as though they were neutral. Even if selection had no influence on which mutations accumulated, it is possible that the types or locations of point mutations that accumulated in diploids had more severe fitness effects on average. Alternatively, it is possible that beneficial mutations account for the difference in average fitness between haploid and diploid MA lines, as the initial strain was not given the opportunity to adapt to the specific conditions of the growth rate assay. The haploid MA lines exhibited significant genetic variance in growth rate but no significant change in average growth rate, consistent with the presence of beneficial mutations (36–38); beneficial mutations could occur at both ploidy levels but be partially recessive in diploids, which would account for the lower average growth rate of diploids. More work is needed to test these and other possible explanations, and to relate the molecular changes caused by spontaneous mutation to their effects on fitness (e.g., ref. 39).
Discussion
Despite intense study of DNA replication and repair in yeast, the effect that ploidy has on the rate and spectrum of spontaneous, unselected mutations has not been investigated on a genome-wide scale before this study. Populations of haploid and diploid yeast frequently show distinct evolutionary behavior in experimental systems (17, 18, 40–42), but it is unclear what role ploidy-specific mutational variation plays in these differences. We find evidence that multiple dimensions of the mutation spectrum depend on ploidy state in an otherwise isogenic background. In particular, diploidy confers greater replication fidelity with respect to single-nucleotide changes and mt mutations, but diploids are also more prone to large-scale mutations with deleterious fitness effects.
Models of life cycle evolution assume that twice as many mutations occur in diploid genomes than in haploid genomes (e.g., refs. 43, 44, as well as many others), but our findings indicate that this classic and seemingly universal assumption does not hold. Rather, the genomic diploid/haploid mutation rate ratio is only ∼1.4 in the wild-type genetic background we studied. Considering the “mutation load” (the reduction in fitness caused by deleterious mutations in a constant environment), our results, if confirmed in other organisms, imply that diploid life cycles would be favored across a broader range of parameters than suggested by previous theory (as reviewed in ref. 2). On the other hand, diploids would also have less access to beneficial substitutions, reducing their ability to proliferate in a changing environment even more than predicted by previous theory (45).
The higher mutation rate per base pair we observed in haploids appears to be linked to replication timing and could reflect a haploid-specific mechanistic constraint on replication fidelity. It is also possible that our lines carry haploid-specific mutator alleles, which might be subject to weak purifying selection, given their indirect effects on fitness (46) and the apparent rarity of haploid cells in natural populations of this species (47). Comparing mutation rates in haploid and diploid forms of predominantly haploid organisms (e.g., using nonsporulating S. pombe) would be an important next step in differentiating these hypotheses.
While we accounted for rates of cell division and applied similar mutation-calling criteria in haploids and diploids, it is worth considering whether other differences between ploidy levels could have biased our mutation rate comparisons. To examine whether our coverage criteria might differentially affect haploid and diploid mutation rate estimates, we reestimated SNM and indel rates using progressively more stringent cutoffs and found no indication that mutation-calling criteria affected our results (SI Appendix, Fig. S11).
Selection might have also influenced our results. In particular, recessive lethal mutations can accumulate in diploids but not in haploids, which could lead us to underestimate the haploid mutation rate. This would not explain the higher SNM rate we observed in haploids, but it could affect our comparisons of indel rates. Using the viability of spores from diploid MA lines, Zhu et al. (5) estimated the rate of recessive lethal mutations as 3.2 × 10−5 per diploid genome per generation, involving SNMs and indels. Applying half of this rate to haploids, we would have missed only approximately five mutations among all of the haploid lines due to recessive lethality (a bias of <1%). Similarly, aneuploidy might not be observed in haploid lines if such changes are highly deleterious. However, aneuploidy for 10 different chromosomes has been observed in haploid MA lines with mutator genotypes (7), and doubling times are similar to the wild type for at least some disomic haploids (48), suggesting that selection does not fully explain the lack of aneuploidy events in haploids.
More generally, mutations might accumulate at a lower rate in haploids than diploids because they are fully exposed to selection and not masked in heterozygous form. However, we found no evidence for selection on SNMs, and a consistent pattern of apparent selection across ploidy levels based on genic versus nongenic indel rates (SI Appendix, Fig. S2). While the dearth of genic indels suggests purifying selection, mutations may not always arise at the same rate in genic and nongenic regions (49), complicating this interpretation.
We predicted that the rate of indels might be greater in haploids and rdh54Δ diploids owing to their reduced opportunity for homologous DNA repair, which did not prove to be the case (Fig. 2). We have lower statistical power with respect to indels, which are rare relative to SNMs. However, given the mean experiment-wide indel rate observed, we would detect a twofold higher indel rate in haploids ∼89% of the time at the α = 0.05 level. Similarly, given the mean diploid indel rate, we would detect a twofold higher indel rate in rdh54Δ lines versus RDH54+ lines ∼85% of the time.
The lack of a difference in indel accumulation between ploidy levels or any interaction with RDH54 status suggests that most small indels are the result of processes other than DSB repair, such as replication slippage (50), or that most DSBs are repaired when sister chromatids are available (3, 51). However, our results point to another possible effect of ploidy on DNA repair: The finding that replication timing affects mutation rates more strongly in haploid than diploid genomes suggests that the activity of error-prone DNA repair processes in late-replicating regions [specifically translesion synthesis (27)] is more important in haploids, which may explain why an effect of replication timing is detected in some studies [e.g., in haploids (27)] but not others [e.g., in diploids (5)]. Replication timing may also explain the difference we observed in the spectrum of SNMs between ploidy levels, as this difference increased with replication timing (SI Appendix, Fig. S12), with late-replicating regions more prone to A-to-T and C-to-G transversions in haploids. Rather than increasing the rate of indels, the most obvious effect of rdh54Δ was to increase the rate of aneuploidy, suggesting that this gene plays a role in mitotic segregation.
LOH in diploids will convert some heterozygous mutant sites into sites homozygous for the ancestral allele, causing us to underestimate diploid point mutation rates. Using maximum likelihood to jointly estimate the rate of LOH and the true mutation rate (SI Appendix), we find that the rate of LOH (95% CI) is 7.92 (5.69, 10.65) × 10−5 per base pair per generation and that the observed diploid mutation rates are not substantially downwardly biased by LOH [3% bias; corrected diploid rates: SNMs: 2.97 (2.81, 3.15) × 10−10, indels: 2.09 (1.67, 2.57) × 10−11]. These relative rates of mutation and LOH predict that few heterozygous sites will exist in equilibrium asexual populations (SI Appendix).
Haploid and diploid yeast differ in development, gene expression, and physiology. Some of these differences are due to ploidy per se, including differences in cell size and expression profiles (52), while others are due to differences in mating competency as determined by the MAT genotype (53). In our study, we chose to examine diploids with the standard MATa/MATα genotype. Thus, results that we ascribe to ploidy differences may also stem from differences in MAT genotype. Although we did not detect an effect of MATa versus MATα in haploids, it would be valuable to examine the mutational profiles of MATa/MATa and MATα/MATα diploids to clarify the role of MAT heterozygosity versus ploidy per se, especially given that MAT is known to affect repair pathway regulation (3) and competitive fitness (54), although at least some differences in DNA repair appear to be independent of MAT genotype (55). Similarly, haploids experienced less fitness decline than diploids after having accumulated mutations in our experiment, but additional data are needed to determine whether this fitness difference stems from cell size and content effects of ploidy per se, MAT composition, or explanations involving beneficial mutations and dominance.
Previous studies have reported faster rates of adaptation in haploid yeast compared with either MATa/MATα diploids (30, 42) or MATa/MATa diploids (41). Explanations have focused on the greater efficacy of selection in haploids than in diploids because the benefits of mutations are partially masked in diploids, assuming all else is equal. Our observation that haploids experience a higher SNP mutation rate, however, indicates that not all else is equal, and previous observations of more rapid haploid adaptation could, in part, reflect a higher mutation rate.
We observed an order of magnitude higher rate of MA in mt sequences within haploid cells than diploid cells. The number of mt genomes differs from that of nuclear genomes, as multiple mt genomes are transmitted to daughter cells (56). However, mt segregation restores homoplasmy in under ∼20 cell divisions following a mating event between cells with different mt genotypes (56, 57), suggesting that heteroplasmy should have been transient in our 1,600-generation MA experiment. Furthermore, relaxing our filters to include heteroplasmic mt variants had little effect on our results (SI Appendix).
We also found that nuclear coverage relative to mt coverage was consistent across ploidy levels (t = 1.62, df = 218, P = 0.11), suggesting that our diploid lines have about twice the mtDNA of haploids [qualitatively similar results are reported elsewhere (58)]. Assuming sequencing and mapping are equally efficient for nuclear and mtDNA, we estimate that there are three to four sequenced mtDNA genomes per haploid cell, fewer than most previous estimates, although there is substantial variation among strains and conditions (e.g., refs. 56–58). Furthermore, we also see substantial variation in coverage across the mt genome, so coverage metrics may poorly estimate the number of mt genomes per cell.
While we cannot exclude the possibility that a higher effective mt population size in diploids permitted more effective selection against new mutations, most of the mt mutations we observed were nongenic and presumably had minimal fitness effects. Instead, the factors causing a higher nuclear mutation rate in haploids may have had an even greater impact on mt sequences, or haploids may incur a higher level of mtDNA damage. A study using a reporter gene approach found that mt microsatellites were 100-fold more stable in diploids than in haploids (59), attributable to nuclear ploidy per se rather than MAT-specific gene expression or mt copy number (60). The effect of ploidy on mt genome maintenance clearly deserves further study.
In an experimental evolution context, haploid populations of S. cerevisiae are sometimes found to attain a diploid state (61–63), but it is unclear to what extent this is due to a high rate of diploidization or a large selective advantage of diploidy. We observed no changes in ploidy, indicating that spontaneous diploidization does not occur at a high rate in the strain and experimental conditions we used. Assuming a rate of diploidization equal to our upper 95% confidence limit, a haploid population would incur >130 nonsynonymous point mutations for every ploidy change event. However, our data also suggest that shifts to diploidy will increase the rate of large structural mutations, consistent with analyses of adaptive genome evolution in haploids and diploids, where diploids are more likely to incur beneficial amplifications or deletions of chromosome segments (17, 18, 63). Polyploid yeast seem to adapt even more rapidly than diploids for the same reason (64). Ploidy shifts, which often seem to lack immediate benefits in yeast (61, 65), may therefore be favored in some environments due to their effects on the mutation spectrum, with our results suggesting that haploids have more access to single-nucleotide changes, while diploids experience more structural variation.
We find that the mutation rate and spectrum differ dramatically between the haploid and diploid forms of a common genetic background. Our results contribute to the growing understanding that the mutation process can vary in response to genetic and genomic context, impacting the ability of organisms to withstand the load of deleterious mutations and to adapt to a changing world.
Materials and Methods
Details on growth rate analyses, phenotyping, bioinformatics, and flow cytometry are provided in SI Appendix.
Isogenic haploid and diploid strains were generated starting with a single haploid cell, using single-colony bottlenecks throughout the procedure. The strain SEY6211 (MATa, ho, leu2-3 112, ura3-52, his3-Δ200, trp1-Δ901, ade2-101, suc2-Δ9) was obtained from the American Type Culture Collection and induced to switch mating type using a standard plasmid transformation protocol. RDH54 was deleted (positions 383,065–386,180 on chromosome II), replaced with KanMX in cells of each mating type via transformation, and confirmed by Sanger sequencing. MATa and MATα haploids were mated to generate diploids with and without the RDH54 deletion. These strains were frozen as ancestral controls and plated to single colonies to begin the bottlenecking procedure.
MA was conducted on solid yeast-extract-peptone-dextrose media, supplemented with 40 mg/L adenine sulfate, on 6-cm-diameter plates. Plates were incubated at 30 °C, and plates from the previous day were stored at 4 °C as backup. Petite mt mutations result in very small 24-h colonies, and we used the backup plate if all colonies on a plate were petite, if individual colonies could not be distinguished, or if colonies were absent. Backup plates were required in 0.4% of transfers, with no significant differences among treatments.
Early in the experiment, 13 putatively haploid MATα RDH54+ lines were found to be diploid due to mating immediately following the mating type switch during strain construction, leading to a mixed colony of MATα and diploid cells; we continued to propagate these diploids and added 15 MATα RDH54+ lines by subdividing five existing lines confirmed to be haploid. We later found that the diploid RDH54+ lines generated by controlled crossing, but not those obtained by unintentional mating, were trisomic for chromosome XI; furthermore, the ancestral diploid RDH54+ strain (but not rdh54Δ) used in phenotype assays carries this trisomy (confirmed by Illumina sequencing).
Concurrent with bottleneck 100, each ancestral control genotype was thawed and plated in the same fashion as the MA lines. The next day, a single colony of each MA line and three replicate colonies from each control plate were transferred to 3 mL of liquid media, grown for 3 d at 30 °C, and frozen in 15% glycerol. These frozen cultures were subsequently used for DNA extraction and sequencing, flow cytometry, and phenotyping.
DNA was extracted from 10-mL saturated cultures using a standard phenol/chloroform method and quantified with fluorometry. DNA (0.4 ng) was used to construct libraries using the Illumina Nextera XT kit and protocol. Libraries were pooled such that diploid samples had twice the concentration of haploid samples (to give equivalent coverage per chromosome) and sequenced in a single Illumina NextSeq lane with paired-end 150-bp reads (average coverage per line before screening: haploid 24.5×, diploid 47.6×). Sequence data are available from the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP139886).
Reads were aligned with Burrows–Wheeler aligner mem (66), and mutations in nonrepetitive regions were called using GATK HaplotypeCaller (67), following recommended practices. We screened sites based on multiple metrics and calculated mutation rates accounting for the number of callable sites in each line.
Supplementary Material
Acknowledgments
We thank A. Kuzmin for assistance with sequencing and transformations; A. Gerstein, M. Dunham, and M. Whitlock for helpful discussions; A. Johnson for flow cytometry support; C. Landry for providing plasmids; and anonymous reviewers for constructive comments on a previous version of the manuscript. This work was supported by a Banting Postdoctoral Fellowship (to N.P.S.) and Natural Sciences and Engineering Research Council Discovery Grant RGPIN-2016-03711 (to S.P.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the National Center for Biotechnology Information Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra/SRP139886 (accession no. SRP139886).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801040115/-/DCSupplemental.
References
- 1.Otto SP, Gerstein AC. The evolution of haploidy and diploidy. Curr Biol. 2008;18:R1121–R1124. doi: 10.1016/j.cub.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 2.Mable BK, Otto SP. The evolution of life cycles with haploid and diploid phases. BioEssays. 1998;20:453–462. [Google Scholar]
- 3.Skoneczna A, Kaniak A, Skoneczny M. Genetic instability in budding and fission yeast-sources and mechanisms. FEMS Microbiol Rev. 2015;39:917–967. doi: 10.1093/femsre/fuv028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishant KT, et al. The baker’s yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLoS Genet. 2010;6:e1001109-15. doi: 10.1371/journal.pgen.1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu YO, Siegal ML, Hall DW, Petrov DA. Precise estimates of mutation rate and spectrum in yeast. Proc Natl Acad Sci USA. 2014;111:E2310–E2318. doi: 10.1073/pnas.1323011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lynch M, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci USA. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serero A, Jubin C, Loeillet S, Legoix-Né P, Nicolas AG. Mutational landscape of yeast mutator strains. Proc Natl Acad Sci USA. 2014;111:1897–1902. doi: 10.1073/pnas.1314423111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein HL. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smit A, Hubley R, Green P. 2015 RepeatMasker Open-4.0. Available at www.repeatmasker.org/. Accessed March 7, 2018.
- 10.Schrider DR, Hourmozdi JN, Hahn MW. Pervasive multinucleotide mutational events in eukaryotes. Curr Biol. 2011;21:1051–1054. doi: 10.1016/j.cub.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandegar MA, Otto SP. Mitotic recombination counteracts the benefits of genetic segregation. Proc Biol Sci. 2007;274:1301–1307. doi: 10.1098/rspb.2007.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Symington LS, Rothstein R, Lisby M. Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics. 2014;198:795–835. doi: 10.1534/genetics.114.166140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shinohara M, et al. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tosato V, Sidari S, Bruschi CV. Bridge-induced chromosome translocation in yeast relies upon a Rad54/Rdh54-dependent, Pol32-independent pathway. PLoS One. 2013;8:e60926-17. doi: 10.1371/journal.pone.0060926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCulley JL, Petes TD. Chromosome rearrangements and aneuploidy in yeast strains lacking both Tel1p and Mec1p reflect deficiencies in two different mechanisms. Proc Natl Acad Sci USA. 2010;107:11465–11470. doi: 10.1073/pnas.1006281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dujon B. Yeast evolutionary genomics. Nat Rev Genet. 2010;11:512–524. doi: 10.1038/nrg2811. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, et al. Gene copy-number variation in haploid and diploid strains of the yeast Saccharomyces cerevisiae. Genetics. 2013;193:785–801. doi: 10.1534/genetics.112.146522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gresham D, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303-19. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Argueso JL, et al. Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci USA. 2008;105:11845–11850. doi: 10.1073/pnas.0804529105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp NP, Agrawal AF. Low genetic quality alters key dimensions of the mutational spectrum. PLoS Biol. 2016;14:e1002419-18. doi: 10.1371/journal.pbio.1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiller GF. Frequent site-specific mit- deletions at cryptic exon-intron junctions in the COX1 gene of yeast mtDNA. Curr Genet. 1994;26:542–545. doi: 10.1007/BF00309947. [DOI] [PubMed] [Google Scholar]
- 22.Anderson JB, Sirjusingh C, Ricker N. Haploidy, diploidy and evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics. 2004;168:1915–1923. doi: 10.1534/genetics.104.033266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Y, Gao X-D, Wang Y, Yuan B-F, Feng Y-Q. Widespread existence of cytosine methylation in yeast DNA measured by gas chromatography/mass spectrometry. Anal Chem. 2012;84:7249–7255. doi: 10.1021/ac301727c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Capuano F, Mülleder M, Kok R, Blom HJ, Ralser M. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal Chem. 2014;86:3697–3702. doi: 10.1021/ac500447w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farlow A, et al. The spontaneous mutation rate in the fission yeast Schizosaccharomyces pombe. Genetics. 2015;201:737–744. doi: 10.1534/genetics.115.177329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behringer MG, Hall DW. Genome-wide estimates of mutation rates and spectrum in Schizosaccharomyces pombe indicate CpG sites are highly mutagenic despite the absence of DNA methylation. G3 (Bethesda) 2016;6:149–160. doi: 10.1534/g3.115.022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lang GI, Murray AW. Mutation rates across budding yeast chromosome VI are correlated with replication timing. Genome Biol Evol. 2011;3:799–811. doi: 10.1093/gbe/evr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller CA, et al. The dynamics of genome replication using deep sequencing. Nucleic Acids Res. 2014;42:e3. doi: 10.1093/nar/gkt878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magwene PM, et al. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2011;108:1987–1992. doi: 10.1073/pnas.1012544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marad DA, Buskirk SW, Lang GI. Altered access to beneficial mutations slows adaptation and biases fixed mutations in diploids. Nat Ecol Evol. 2018;2:882–889. doi: 10.1038/s41559-018-0503-9. [DOI] [PubMed] [Google Scholar]
- 31.Ness RW, Morgan AD, Vasanthakrishnan RB, Colegrave N, Keightley PD. Extensive de novo mutation rate variation between individuals and across the genome of Chlamydomonas reinhardtii. Genome Res. 2015;25:1739–1749. doi: 10.1101/gr.191494.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parada L, Misteli T. Chromosome positioning in the interphase nucleus. Trends Cell Biol. 2002;12:425–432. doi: 10.1016/s0962-8924(02)02351-6. [DOI] [PubMed] [Google Scholar]
- 34.Therizols P, Duong T, Dujon B, Zimmer C, Fabre E. Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres. Proc Natl Acad Sci USA. 2010;107:2025–2030. doi: 10.1073/pnas.0914187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, et al. The dynamic three-dimensional organization of the diploid yeast genome. eLife. 2017;6:e23623. doi: 10.7554/eLife.23623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall DW, Joseph SB. A high frequency of beneficial mutations across multiple fitness components in Saccharomyces cerevisiae. Genetics. 2010;185:1397–1409. doi: 10.1534/genetics.110.118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joseph SB, Hall DW. Spontaneous mutations in diploid Saccharomyces cerevisiae: More beneficial than expected. Genetics. 2004;168:1817–1825. doi: 10.1534/genetics.104.033761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerstein AC. Mutational effects depend on ploidy level: All else is not equal. Biol Lett. 2013;9:20120614. doi: 10.1098/rsbl.2012.0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraemer SA, Böndel KB, Ness RW, Keightley PD, Colegrave N. Fitness change in relation to mutation number in spontaneous mutation accumulation lines of Chlamydomonas reinhardtii. Evolution. 2017;71:2918–2929. doi: 10.1111/evo.13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerstein AC, Otto SP. Ploidy and the causes of genomic evolution. J Hered. 2009;100:571–581. doi: 10.1093/jhered/esp057. [DOI] [PubMed] [Google Scholar]
- 41.Gerstein AC, Cleathero LA, Mandegar MA, Otto SP. Haploids adapt faster than diploids across a range of environments. J Evol Biol. 2011;24:531–540. doi: 10.1111/j.1420-9101.2010.02188.x. [DOI] [PubMed] [Google Scholar]
- 42.Zeyl C, Vanderford T, Carter M. An evolutionary advantage of haploidy in large yeast populations. Science. 2003;299:555–558. doi: 10.1126/science.1078417. [DOI] [PubMed] [Google Scholar]
- 43.Crow JF, Kimura M. Evolution in sexual and asexual populations. Am Nat. 1965;99:439–450. [Google Scholar]
- 44.Kondrashov AS, Crow JF. Haploidy or diploidy: Which is better? Nature. 1991;351:314–315. doi: 10.1038/351314a0. [DOI] [PubMed] [Google Scholar]
- 45.Orr HA, Otto SP. Does diploidy increase the rate of adaptation? Genetics. 1994;136:1475–1480. doi: 10.1093/genetics/136.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ezov TK, et al. Molecular-genetic biodiversity in a natural population of the yeast Saccharomyces cerevisiae from “Evolution Canyon”: Microsatellite polymorphism, ploidy and controversial sexual status. Genetics. 2006;174:1455–1468. doi: 10.1534/genetics.106.062745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 49.Frigola J, et al. Reduced mutation rate in exons due to differential mismatch repair. Nat Genet. 2017;49:1684–1692. doi: 10.1038/ng.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fortune JM, et al. Saccharomyces cerevisiae DNA polymerase δ: High fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 51.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. Ploidy regulation of gene expression. Science. 1999;285:251–254. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- 53.de Godoy LMF, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 54.Selk E, Wills C. Mismatch repair and the accumulation of deleterious mutations influence the competitive advantage of MAT (mating type) heterozygosity in the yeast Saccharomyces cerevisiae. Genet Res. 1998;71:1–10. doi: 10.1017/s001667239700308x. [DOI] [PubMed] [Google Scholar]
- 55.Li XC, Tye BK. Ploidy dictates repair pathway choice under DNA replication stress. Genetics. 2011;187:1031–1040. doi: 10.1534/genetics.110.125450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Birky CW, Strausberg RL, Forster JL, Perlman PS. Vegetative segregation of mitochondria in yeast: Estimating parameters using a random model. Mol Gen Genet. 1978;158:251–261. [Google Scholar]
- 57.Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- 58.Grimes GW, Mahler HR, Perlman RS. Nuclear gene dosage effects on mitochondrial mass and DNA. J Cell Biol. 1974;61:565–574. doi: 10.1083/jcb.61.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sia EA, et al. Analysis of microsatellite mutations in the mitochondrial DNA of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:250–255. doi: 10.1073/pnas.97.1.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sia RAL, Urbonas BL, Sia EA. Effects of ploidy, growth conditions and the mitochondrial nucleoid-associated protein Ilv5p on the rate of mutation of mitochondrial DNA in Saccharomyces cerevisiae. Curr Genet. 2003;44:26–37. doi: 10.1007/s00294-003-0420-5. [DOI] [PubMed] [Google Scholar]
- 61.McDonald MJ, Hsieh Y-Y, Yu Y-H, Chang S-L, Leu J-Y. The evolution of low mutation rates in experimental mutator populations of Saccharomyces cerevisiae. Curr Biol. 2012;22:1235–1240. doi: 10.1016/j.cub.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 62.Gerstein AC, Chun HJ, Grant A, Otto SP. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2006;2:e145. doi: 10.1371/journal.pgen.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fisher KJ, Buskirk SW, Vignogna RC, Marad DA, Lang GI. 2018. Cause and consequences of genome duplication in haploid yeast populations. bioRxiv:247320. [DOI] [PMC free article] [PubMed]
- 64.Selmecki AM, et al. Polyploidy can drive rapid adaptation in yeast. Nature. 2015;519:349–352. doi: 10.1038/nature14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerstein AC, Otto SP. Cryptic fitness advantage: Diploids invade haploid populations despite lacking any apparent advantage as measured by standard fitness assays. PLoS One. 2011;6:e26599-13. doi: 10.1371/journal.pone.0026599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.