Significance
Obesity has reached pandemic levels, prompting the need for novel therapeutics. The immune system has been suggested to be critically linked to metabolic health, leading to the prospect of immune-directed therapies. We report that obese fat tissue contains multiple distinct populations of macrophages with unique tissue distributions, transcriptomes, chromatin landscapes, and functions. These results provide a higher resolution of the cellular and functional heterogeneity within adipose macrophages and provide a framework within which to develop new immune-directed therapies for the treatment of obesity and related inflammatory comorbidities.
Keywords: macrophage, adipose tissue, exosome, obesity, inflammation
Abstract
Obesity is characterized by an accumulation of macrophages in adipose, some of which form distinct crown-like structures (CLS) around fat cells. While multiple discrete adipose tissue macrophage (ATM) subsets are thought to exist, their respective effects on adipose tissue, and the transcriptional mechanisms that underlie the functional differences between ATM subsets, are not well understood. We report that obese fat tissue of mice and humans contain multiple distinct populations of ATMs with unique tissue distributions, transcriptomes, chromatin landscapes, and functions. Mouse Ly6c ATMs reside outside of CLS and are adipogenic, while CD9 ATMs reside within CLS, are lipid-laden, and are proinflammatory. Adoptive transfer of Ly6c ATMs into lean mice activates gene programs typical of normal adipocyte physiology. By contrast, adoptive transfer of CD9 ATMs drives gene expression that is characteristic of obesity. Importantly, human adipose tissue contains similar ATM populations, including lipid-laden CD9 ATMs that increase with body mass. These results provide a higher resolution of the cellular and functional heterogeneity within ATMs and provide a framework within which to develop new immune-directed therapies for the treatment of obesity and related sequela.
There is currently a worldwide pandemic of obesity that threatens population health by predisposing to diabetes, cardiovascular disease, and cancer (1). Our limited understanding of the mechanisms that direct adipose homeostasis in response to high fat diet (HFD) exposure has hindered the development of new therapies. There is a growing body of literature to suggest that the immune system is a critical regulator of normal adipose physiology and the response to fatty foods (2). This observation has led to the prospect of immune-directed therapies for the treatment of obesity and related inflammatory sequela.
Macrophages are the predominant immune cell in adipose tissue, and they accumulate in obese mice and humans (3, 4). The increased number of adipose tissue macrophages (ATMs) in obese adipose tissue is in part due to recruitment of peripheral cells (5–7), but local proliferation also plays a role as a subset of ATMs express Ki67 and markers of S phase (8–11). Proliferation seems to occur predominantly at sites known as crown-like structures (CLSs), characterized by collections of ATMs that form around dead adipocytes (12). There is evidence that accumulation of ATMs contributes to obesity-induced insulin resistance (13, 14). For example, deletion of chemokine (C-C motif) ligand 2 (CCL2) reduces ATM accumulation and improves insulin sensitivity in some but not all cases (5, 6, 15, 16), while overexpression of CCL2 promotes insulin resistance (7). Moreover, chemical or genetic disruption of macrophages or their functions improves insulin sensitivity (17–21).
Despite these advances, the identity of the macrophages that accumulate in obese adipose tissue is uncertain. Soon after the initial description of ATM infiltration in obesity, it was suggested that this was associated with a switch from an antiinflammatory (M2) phenotype to an inflammatory (M1) activation state (22, 23). However, an alternative view is that ATMs accumulate in obesity as a homeostatic mechanism that evolved to protect the metabolic health of the organism. Indeed, recent studies suggest that obesity-associated ATMs can adopt a metabolic activation state (24, 25) that facilitates clearance of dead adipocytes (26). It has also been shown that more than one population of ATMs exist (27), although the exact number and functions of ATM subsets in obese adipose, and the transcriptional mechanisms that define their unique activation states, are not known.
Building on prior work, we hypothesized that the dissimilar properties attributed to ATMs in different studies could be due to functional heterogeneity. To address this hypothesis in a comprehensive and unbiased manner, we examined macrophages from adipose tissue of obese mice by flow cytometry and single-cell transcriptomics. This led to the detection of multiple distinct ATM populations with unique morphology, tissue localization, and transcriptomes. Ly6c ATMs reside outside of CLSs and are adipogenic, while CD9 ATMs reside within CLSs, are lipid-laden, and are proinflammatory. We detect similar ATM subsets in human adipose tissue, including a CD9+, lipid-laden group, which is directly associated with obesity. Thus, functional heterogeneity is a conserved feature of ATM biology between mice and humans and directs inflammatory as well as physiologic changes during obesity. This observation has implications for the development of new, immune-directed therapies for the treatment of obesity and related inflammatory sequela.
Results
CD9 and Ly6c Define Unique Populations of ATMs in Obese Adipose.
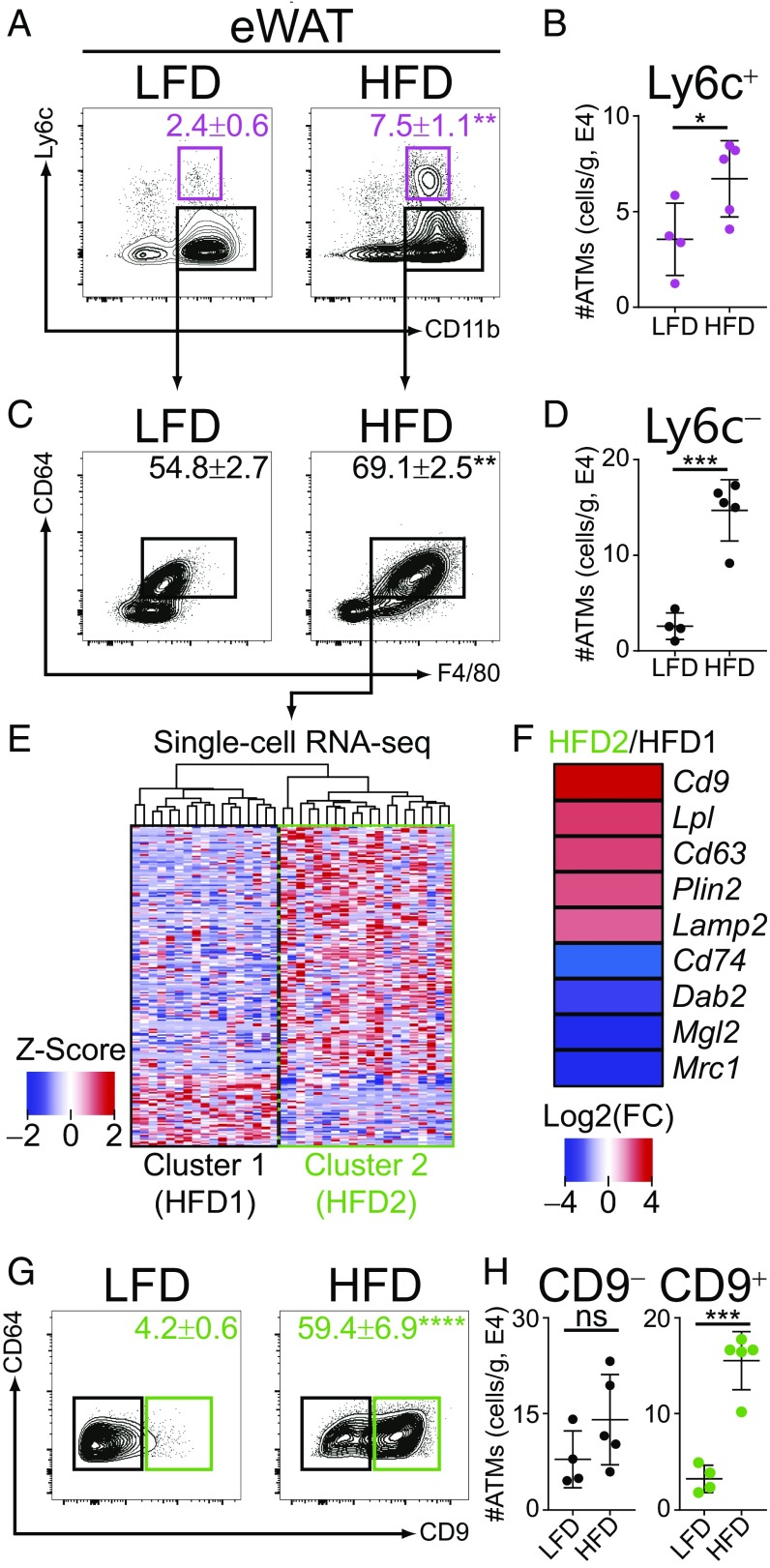
To evaluate the number and functions of ATM populations in adipose in a comprehensive and unbiased manner, cohorts of B6 mice were subjected to HFD or control low-fat diet (LFD) for 12 wk, resulting in a time-dependent weight gain (SI Appendix, Fig. S1A). We used flow cytometry to characterize ATM populations in epididymal white adipose tissue (eWAT), based on established cell markers and conventions (28). A population of CD11b+ Ly6c+ cells, likely representing monocyte-derived cells, accumulated with obesity (Fig. 1 A and B) (29). There was also an increase in CD11b+ Ly6c− cells that coexpressed the macrophage markers F4/80 and CD64 (Fig. 1 C and D), therefore fitting the classic description of ATMs that accumulate with obesity (3).
Fig. 1.
CD9 and Ly6c define unique populations of ATMs in obese adipose. (A) Flow-cytometric analysis of ATMs from eWAT of mice fed LFD or HFD for 12 wk. Gated on CD45+, CD3−, CD4−, CD8−, CD19−, Ly6g−, and SiglecF−. Numbers indicate percentage of parent gate with mean ± SEM displayed (n = 4–5 per group, statistical comparison by Student’s t test). (B) Number of CD11b+, Ly6c+ (Ly6c+) ATMs in eWAT of LFD or HFD mice. (C) Flow-cytometric analysis of CD11b+, Ly6c− ATMs from eWAT of LFD or HFD mice. (D) Number of CD11b+, Ly6c−, CD64+, F4/80+ (Ly6c−) ATMs in eWAT of LFD or HFD mice. (E) Heatmap of differentially expressed genes in CD11b+, Ly6c−, CD64+, F4/80+ ATMs of HFD mice (n = 37 cells). (F) Examples of differentially expressed genes in HFD2 compared with HFD1 ATMs, calculated as log2 fold change (FC)(HFD2/HFD1). (G) Flow-cytometric analysis of CD11b+, Ly6c−, CD64+, F4/80+ ATMs from eWAT of LFD or HFD mice. (H) Number of CD11b+, Ly6c−, CD64+, F4/80+, CD9− (CD9−) or CD11b+, Ly6c−, CD64+, F4/80+, CD9+ (CD9+) ATMs in eWAT of LFD or HFD mice. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant. Representative of three or more independent experiments.
To test for heterogeneity among CD11b+ Ly6c− ATMs, cells were pooled from eWAT of five mice, sort-purified, and single-cell RNA-sequencing (RNA-seq) was performed. Two transcriptionally distinct populations of Ly6c− ATMs were observed upon unbiased hierarchical clustering (Fig. 1E, HFD1 vs. HFD2). Examination of the most differentially expressed genes between these groups revealed that the HFD2 group of ATMs expressed high levels of the tetraspanin family member Cd9 (Fig. 1F) (30), as well as a number of genes related to lipid metabolism (Lpl, Plin2) and intracellular vesicle function (Cd63, Lamp2), consistent with known ATM characteristics (24, 25).
Subsequent flow-cytometric analysis revealed that two populations of Ly6c− ATMs could be identified based on their differential surface expression of CD9 (Fig. 1G). CD9+ ATMs accumulated with HFD, while CD9− ATMs did not (Fig. 1 G and H), indicating that CD9+ ATMs are the predominant Ly6c− ATM type that increases with obesity. While Ly6c and CD9 clearly distinguished the three ATM populations in question, expression of other canonical macrophage markers was more similar among the various ATM groups (SI Appendix, Fig. S1B). Together, these results indicate that three populations of ATMs exist in adipose and identify CD9+ and Ly6c+ ATMs (hereafter referred to as CD9 and Ly6c, respectively) as the predominant populations associated with obesity.
CD9 and Ly6c ATMs Are Bone Marrow-Derived.
Accumulation of ATMs in obese adipose is thought to be due in part to recruitment of bone marrow-derived monocytes (5–7). To determine whether CD9 and Ly6c ATMs are hematopoietically derived, we created bone marrow chimeras by irradiating B6 CD45.1+ recipients and reconstituting their hematopoietic compartment with bone marrow of B6 CD45.2+ donors (SI Appendix, Fig. S2A). After reconstitution, CD45.1+ recipients were fed HFD for 12 wk, and systemic and adipose chimerism was determined. Recipients showed a high degree of systemic chimerism as 94% of splenic lymphocytes expressed the donor congenic marker CD45.2 (SI Appendix, Fig. S2B). A similar proportion of CD9 and Ly6c ATMs expressed the donor congenic marker CD45.2 (SI Appendix, Fig. S2C), indicating that both CD9 and Ly6c ATMs are hematopoietically derived.
CD9 ATMs Localize to CLSs, Are Filled with Lipids, and Secrete Exosomes.
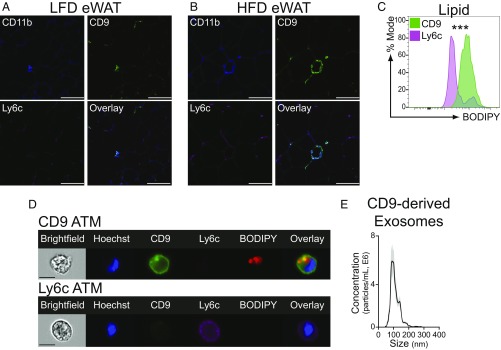
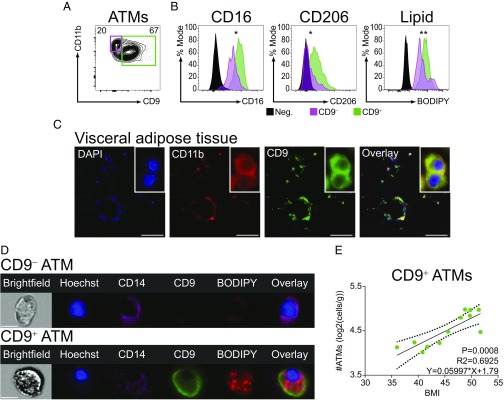
It is known that obesity is associated with ATM accumulation around dead adipocytes, although the identity of these CLS-associated ATMs is uncertain (24, 25). Therefore, we examined the tissue localization of CD9 and Ly6c ATMs via immunohistochemistry. Those CD9 and Ly6c ATMs that were present in lean eWAT were equally dispersed among adipocytes (Fig. 2A). However, the distribution of ATMs in eWAT changed upon exposure of mice to HFD diet, with CD9 ATMs predominating in CLSs (Fig. 2B) (12). It has been proposed that ATMs can exist in a metabolic activation state that is characterized by intracellular lipid accumulation and lysosomal-dependent lipid metabolism (24–26). We therefore examined intracellular lipid levels in CD9 and Ly6c ATMs using flow cytometry, and the hydrophobic lipid dye BODIPY. Compared with Ly6c ATMs, CD9 ATMs contained high levels of intracellular lipid (Fig. 2C) within vacuolar structures, which likely represent lysosomes based on prior studies (Fig. 2D) (25). As CD9 is relevant to exosome function (30), we examined whether CD9 ATMs secrete exosomes. CD9 ATMs secreted exosome-sized vesicles (Fig. 2E) as measured by nanoparticle detector analysis, consistent with recent reports of a role for these structures in mediating ATM functions (31). Together, these findings indicate that CD9 and Ly6c ATMs display distinct cellular characteristics and tissue locations and identify CD9 ATMs as the predominant CLS-associated ATM subtype.
Fig. 2.
CD9 ATMs localize to CLSs and are filled with lipids. (A and B) Immunohistochemistry of eWAT from LFD (A) or HFD (B) mice. Sections were stained for CD11b, CD9, and Ly6c. (C) Comparison of intracellular lipid levels in HFD eWAT CD9 or Ly6c ATMs by mean fluorescence intensity (normalized to mode, n = 5 per group, statistical comparison by Student’s t test, ***P < 0.001). (D) Representative imaging cytometry of HFD eWAT CD9 or Ly6c ATMs. (Scale bars: A and B, 100 μm; D, 10 μm.) (E) Nanoparticle detector analysis of CD9-derived exosomes secreted into culture media (SEM of technical replicates in gray). Representative of two or more independent experiments.
CD9 ATMs Express Proinflammatory Transcriptomes.
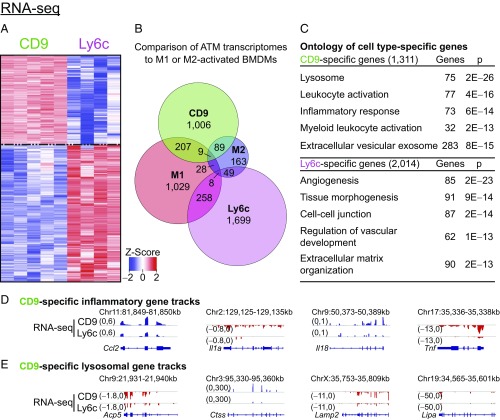
Having identified subgroups of ATMs with distinct tissue localizations, we sought to interrogate their function in adipose. CD9 and Ly6c ATMs were sort-purified and bulk RNA-seq analysis was performed, which confirmed the identity of the HFD2 ATM cluster identified with single-cell RNA-seq as CD9 cells, and revealed 1,311 CD9-specific genes and 2,014 Ly6c-specific genes (Fig. 3A). Notably, neither obesity-associated ATM subtype shared predominant features of classical M1 or M2 activation by comparative transcriptome analysis (Fig. 3B and SI Appendix, Fig. S3 A and B). Rather, unbiased clustering and gene ontology analysis indicated that genes more highly expressed in CD9 ATMs were enriched for lysosomal pathways and proinflammatory mediators (Fig. 3C and SI Appendix, Fig. S3C). Among the notable genes more highly expressed in CD9 ATMs were Ccl2, Il1a, Il18, and Tnf (Fig. 3D) as well as Acp5, Ctss, Lamp2, and Lipa (Fig. 3E), suggesting they share features of both inflammatory and metabolic activation (25). Conversely, Ly6c ATMs expressed factors that support vascular development and organization (Fig. 3C). These results indicate that CD9 and Ly6c ATMs are activating distinct transcriptional networks, likely mediated by cell type-specific regulatory elements (32).
Fig. 3.
CD9 ATMs express proinflammatory transcriptomes, while Ly6c ATMs are homeostatic. (A) Heatmap of differentially expressed genes between CD9 and Ly6c ATMs (fold change > 2, false discovery rate < 0.01, n = 4–5 per group). Genes ordered by hierarchical clustering. (B) Venn diagram of genes characteristic of CD9, Ly6c, M1, or M2 transcriptomes. M1- and M2-specific genes were determined by comparing LPS or IL-4 treated bone marrow-derived macrophages (BMDM) to control-treated cells, as detailed in SI Appendix, Fig. S3. (C) Ontology of genes more highly expressed in CD9 or Ly6c ATMs. Most significant, nonredundant biologic process, molecular function, or cellular component terms with gene number and adjusted P value are shown. (D) Representative CD9-specific inflammatory gene tracks. (E) Representative CD9-specific lysosomal gene tracks. Representative of one or more independent experiments.
CD9 ATMs Have an Inflammatory Chromatin Landscape.
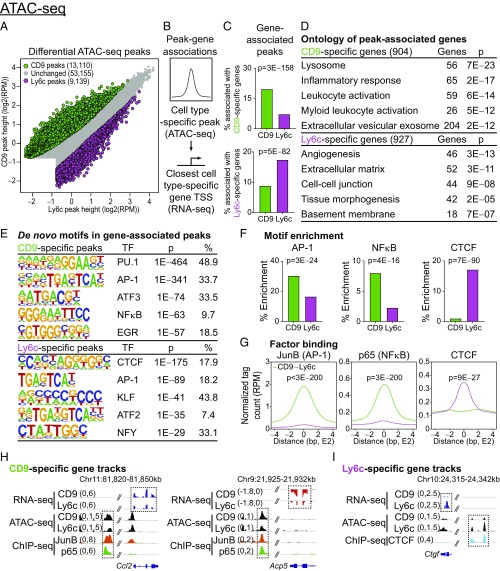
To investigate cell type-specific chromatin regulatory elements and transcription factors driving the functionally divergent transcriptomes of CD9 or Ly6c ATMs, we performed Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq). Differential peak analysis revealed 13,110 CD9-specific and 9,139 Ly6c-specific ATAC-seq peaks (Fig. 4A). To enhance the specificity of our analysis for unique gene regulation programs, we next associated cell type-specific peaks with the closest corresponding cell type-specific gene (Fig. 4B). CD9 or Ly6c-specific peaks were enriched nearby corresponding CD9 or Ly6c-specific genes, respectively, suggesting that these regions of open chromatin are functioning as enhancers (Fig. 4C). Interrogation of cell type-specific genes associated with respective cell type-specific peaks by gene ontology analysis confirmed the proinflammatory versus tissue-homeostatic gene programs of CD9 and Ly6c ATMs, respectively (Fig. 4D).
Fig. 4.
CD9 ATMs have an inflammatory chromatin landscape driven by activating transcription factors. (A) Differential analysis of ATAC-seq peaks from CD9 or Ly6c ATMs of HFD-fed B6 mice [n = 3 per arm; fold change > 1.5, false discovery rate < 0.05, and >0.5 reads assigned per million mapped reads (RPM)]. (B) Diagram describing association between cell type-specific peaks and closest cell type-specific gene [within 50 kb of the transcription start site (TSS)]. (C) Percent of ATAC-seq peaks associated with CD9- or Ly6c-specific genes (P values determined by Fisher’s exact test). (D) Gene ontology for cell type-specific genes, associated with corresponding cell-type specific peaks. Most significant, nonredundant biologic process, molecular function, or cellular component terms with gene number and adjusted P value are shown. (E) De novo motif search within ATAC-seq peaks associated with a corresponding cell type-specific gene [consensus motif, transcription factor (TF), P value, and percentage of targets shown]. (F) Percent enrichment of AP-1, NF-κB, and CTCF motifs within CD9- or Ly6c-specific peaks (P values determined y Fisher’s exact test). (G) Occupancy of JunB (AP-1 subunit) and CTCF at CD9- or Ly6c-specific regions of open chromatin (P values determined by Wilcoxon test). (H and I) Representative RNA-seq, ATAC-seq, and ChIP-seq browser tracks displaying CD9- (H) or Ly6c-specific (I) gene loci. Legacy ChIP-seq tracks for JunB (AP-1) or p65 (NF-κB) in lipopolysaccharide-treated BMDM or CTCF in untreated BMDM are displayed. Representative of two or more independent experiments.
We next performed de novo motif search within peaks associated with cell type-specific genes to elucidate the transcription factors (TFs) responsible for driving CD9 and Ly6c-specific gene programs. Areas of open chromatin shared by both ATM subsets were enriched for the macrophage lineage-determining factor PU.1 motif (SI Appendix, Fig. S4A). However, the chromatin architecture of CD9 and Ly6c ATMs was otherwise distinct, with CD9-specific peaks enriched for proinflammatory TF motifs including AP-1 and NF-κB, and Ly6c-specific peaks enriched for the architectural factor CTCF (Fig. 4 E and F) (33). Integrative analysis of macrophage cistromes in the public domain revealed that binding of AP-1 subunit JunB, and NFκB subunit p65, were enriched at CD9-specific peaks, while CTCF binding was enriched at Ly6c-specific peaks (Fig. 4G), indicating a high degree of correlation between TF binding sites and ATM-specific enhancers across the genome. This correlation was confirmed at the individual gene level (Fig. 4 H and I and SI Appendix, Fig. S4 B and C). Together, these results indicate that CD9 and Ly6c ATMs have distinct transcriptomes driven by unique chromatin landscapes activated by distinct combinations of TFs, with CD9 ATMs manifesting primarily proinflammatory characteristics.
CD9 ATMs Confer an Inflammatory Response to Lean Adipose Tissue.
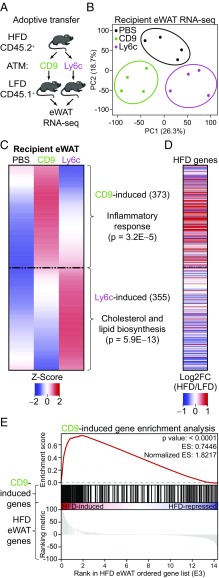
To better understand the functions of CD9 and Ly6c ATMs in adipose, we next utilized adoptive cell transfer. CD9 and Ly6c ATMs were sort-purified from eWAT of obese, CD45.2+ B6 donor mice and transferred into the peritoneal cavity of lean, CD45.1+ B6 recipients (Fig. 5A). Transferred ATMs were identifiable in the eWAT of CD45.1+ recipients, as confirmed via expression of CD9 or Ly6c, and the congenic marker CD45.2 (SI Appendix, Fig. S5A). Bulk RNA-seq of recipient eWAT was performed, and transcriptional changes were compared with PBS-injected controls. Principal component analysis revealed distinct clustering for each of the experimental arms (Fig. 5B), indicating that CD9 and Ly6c ATMs induce robust and unique changes in the adipose transcriptome. This conclusion was supported by collective examination of differentially expressed genes. In hierarchical clustering (SI Appendix, Fig. S5B), we identified three distinct groups of genes, two of which represented genes uniquely induced by either CD9 or Ly6c ATM transfer (Fig. 5C). Genes induced by CD9 ATM transfer were proinflammatory in nature, while those induced by Ly6c ATM transfer were characteristic of normal adipocyte physiology. To determine whether CD9 ATMs were the origin of the proinflammatory gene signature observed in obese adipose tissue, we compared genes induced by CD9 ATM transfer identified from the clustering analysis to the transcriptional changes induced by HFD exposure (34). Remarkably, CD9 ATMs induced a gene expression profile in lean adipose tissue that was highly similar to that of adipose tissue from obese mice (Fig. 5 D and E). Together, these results indicate that CD9 and Ly6c ATMs exert different effects on adipose tissue and that CD9 ATMs are predominantly responsible for the inflammatory changes observed with obesity.
Fig. 5.
CD9 ATMs confer an inflammatory response to lean adipose tissue. (A) Experimental design of adoptive transfer. (B) PCA plot of transfer groups (n = 4 per group). (C) Hierarchical clustering heatmap and ontology of genes differentially regulated in eWAT after CD9 or Ly6c ATM transfer, compared with PBS injection [fold change (FC) > 2 and false discovery rate < 0.05]. Average of four replicates displayed. Most significant, nonredundant biologic process terms with gene number and adjusted P value are shown. (D) Heatmap of genes expressed in eWAT after 12 wk of HFD [displayed as log2FC(HFD/LFD)]. (E) Gene set enrichment analysis of CD9-induced genes within the HFD eWAT transcriptome. Representative of two or more independent experiments.
CD9 ATMs Are Present in Human Adipose and Correlate with Body Mass Index.
Accumulation of ATMs in adipose is a feature of obesity in humans (3, 24). To determine whether similarities exist between mouse and human ATMs, visceral adipose tissue (VAT) samples were obtained from obese patients undergoing bariatric surgery (Table 1). ATMs were readily identified by flow cytometry in VATs of obese individuals (SI Appendix, Fig. S6A). As Ly6c is not present in humans, we utilized several common human myeloid surface markers to identify and characterize ATMs including CD11b and CD14, which are expressed on monocyte-derived cells, and CD16 and CD206, which are more characteristic of tissue-resident cells (24, 28, 35). We found that CD9 defined two populations of human ATMs (Fig. 6A). Compared with CD9− ATMs, CD9+ATMs of obese patients expressed higher levels of CD16 and CD206 and had higher intracellular lipid content (Fig. 6B). To further assess for features of ATM biology that are conserved between mice and humans, we examined tissue localization of ATMs in obese adipose tissue by immunohistochemistry. Consistent with prior studies, VAT of obese individuals displayed abundant CLSs (3). Notably, CD9+ ATMs localized prominently to CLSs (Fig. 6C). Like their murine counterparts, human CD9+ ATMs contained lipid in lysosome-like structures, while CD9− ATMs did not (Fig. 6D). Remarkably, the number of CD9+ ATMs in VAT was positively correlated with body mass index (BMI) (Fig. 6E), whereas CD9− ATMs were not (SI Appendix, Fig. S6B), suggesting a similar pathophysiologic role to that of murine CD9 ATMs. Thus, similar to mice, CD9 defines a subset of ATMs in humans that are lipid-laden, predominate in CLSs, and positively correlate with body mass.
Table 1.
Demographic and clinical characteristics of the patient cohort
| Patient | Sex | Ethnicity | BMI | T2DM | A1C | T. Chol. | LDL | HDL | Tri. |
| 1 | Female | Black, non-Hispanic | 44 | Yes | 9.4 | 144 | 82 | 41 | 103 |
| 2 | Female | Black, non-Hispanic | 51 | Pre | 6.2 | 162 | 94 | 54 | 69 |
| 3 | Female | Black, non-Hispanic | 36 | Yes | 7.0 | 145 | 59 | 50 | 178 |
| 4 | Male | Black, non-Hispanic | 48 | Yes | 6.8 | 134 | 62 | 28 | 220 |
| 5 | Female | White, non-Hispanic | 50 | Pre | 5.7 | 127 | 57 | 49 | 103 |
| 6 | Female | White, non-Hispanic | 52 | Yes | 8.9 | 166 | 70 | 46 | 250 |
| 7 | Male | White, Hispanic | 50 | Yes | 6.7 | 178 | 111 | 36 | 156 |
| 8 | Female | White, non-Hispanic | 41 | Yes | 8.7 | 215 | 139 | 41 | 174 |
| 9 | Female | Black, non-Hispanic | 49 | No | 4.8 | 141 | 83 | 43 | 74 |
| 10 | Female | White, non-Hispanic | 42 | No | 5.7 | 226 | 132 | 68 | 128 |
| 11 | Male | White, Hispanic | 46 | Pre | 5.8 | 171 | 109 | 50 | 60 |
| 12 | Female | Black, non-Hispanic | 39 | Yes | 9.6 | 178 | 90 | 67 | 106 |
T2DM, type 2 diabetes mellitus; T. Chol., total cholesterol; Tri., triglycerides.
Fig. 6.
CD9 ATMs are present in CLSs of human adipose and correlate with BMI. (A) Flow-cytometric analysis of VAT ATMs (gating strategy in SI Appendix, Fig. S6). (B) Surface marker expression and intracellular lipid levels of VAT ATMs (normalized to percentage mode of MFI; Neg., Lymphocyte gate; statistical comparison between CD9− and CD9+ ATMs by ratio paired Student’s t test, *P < 0.05, **P < 0.01; n = 8–12). (C) Immunohistochemistry of VAT from obese individuals. Sections stained for DAPI, CD11b, and CD9. (D) Representative imaging flow cytometry of VAT CD9− or CD9+ ATMs. (Scale bars: C, 50 μm; D, 10 μm.) (E) Correlation between CD9+ ATM number and BMI (analysis by linear regression, n = 12).
Discussion
The accumulation of macrophages in adipose was initially thought to accompany a pathogenic transition from an antiinflammatory (M2) to a proinflammatory (M1) activation state. However, this model is evolving as a result of the identification of a distinct, metabolic activation state for obesity-associated ATMs (24–26, 36). Here, we identify three discrete populations of ATMs in mice, two of which (CD9 and Ly6c) are characteristic of obesity. Neither obesity-associated ATM subtype shares predominant features of classical M1 or M2 activation by comprehensive transcriptome analysis, consistent with an evolving view that the M1/M2 paradigm is not directly applicable to in vivo ATM populations (24–26).
The three ATM populations identified here express similar levels of several canonical macrophage surface markers including F4/80 and CD11b. As such, prior studies that utilized one or both of these markers for identification or isolation of ATMs likely studied average characteristics of some or all of these populations. Additionally, ATMs have been previously subdivided based on expression of CD11c, although the functional significance of this distinction is still under investigation (22, 27). It is difficult to integrate the identities of previously identified CD11c-low and CD11c-high populations with those identified here, as the three ATM populations (CD9+, CD9−, and Ly6c+) express CD11c at different levels. However, based on other cellular characteristics (namely the presence of intracellular lipid and characteristics of metabolic activation), we suspect that CD9+ ATMs are most similar to the CD11c-high population identified in prior studies (25, 27).
As prior studies of ATMs were limited by utilization of similarly expressed canonical macrophage markers, localization of distinct ATM subsets within adipose tissue has not been previously possible. Through the utilization of CD9 and Ly6c, we show that these two populations have distinct tissue localization, with CD9 ATMs predominating in CLSs. CD9 ATMs were also found to contain high amounts of intracellular lipid in lysosome-like structures, and express genes related to lysosomal-dependent lipid metabolism, consistent with features noted in bulk analyses of obese ATMs (24–26). This observation could indicate a role for these cells in homeostatic handling of lipid. Further, CD9 ATMs secrete exosome-size vesicles, implicating CD9 ATMs as one potential source of these important signaling structures that may influence mammalian metabolism (31). Importantly, adoptive transfer of CD9 ATMs revealed that they are sufficient to promulgate obesity-associated adipose inflammation.
In contrast to CD9 ATMs, Ly6c ATMs are uniformly distributed in obese adipose and do not contain large amounts of intracellular lipid. The fact that they express high levels of Ly6c suggest that they are monocyte-derived cells and may be recent immigrants to the adipose (29). Rather than genes related to metabolic activation, Ly6c ATMs express genes related to angiogenesis and tissue organization. This observation provides additional insights into the role of angiogenesis in adipose homeostasis and insulin resistance (37, 38). Furthermore, Ly6c ATMs appear to support normal adipose physiology upon adoptive transfer by inducing genes related to cholesterol and lipid biosynthesis. This observation is consistent with prior studies that have reported an angiogenic and adipogenic role for ATMs (39, 40) and suggests a higher degree of complexity dictating the role of the immune system adipose homeostasis (41). In sum, these findings reveal distinct obesity-associated ATM subtypes with potentially synergistic functions whereby metabolically activated CD9 ATMs respond to tissue injury, express immune modulatory factors including CCL2 (5–7), and facilitate the recruitment of tissue-regulatory Ly6c ATMs (SI Appendix, Fig. S7).
Given the unique activation states of CD9 and Ly6c ATMs, we took an in-depth systems approach to understanding the regulation of their transcriptional programs. Open chromatin of both CD9 and Ly6c ATMs is enriched for the macrophage lineage-defining PU.1 motif. Remarkably however, the chromatin landscapes of these two populations are otherwise quite distinct. CD9 ATM chromatin is enriched for the proinflammatory AP-1 and NF-κB motifs, consistent with known roles for these TFs in ATM activation and survival (42, 43), and the proposed role of CD9 ATMs in mediating the inflammatory changes observed with obesity. Conversely, the motif most enriched in open Ly6c chromatin is for the genome-organizing factor CTCF (33). Classically thought of as a genomic insulator, CTCF is known to play critical roles in development and gene regulation by spatially orienting cis-acting regulatory elements to specific promoters within topologically associating domains (44). Consistent with this role, CTCF binding has been shown to be dynamically controlled during normal development, as well as pathologic states, by various mechanisms including DNA methylation, which inhibits its binding. It has also been specifically implicated in regulation of expression of proangiogenic growth factors (45, 46). It is therefore plausible that CTCF acts to dynamically define Ly6c ATM transcriptional programs and functional responses, which are tailored to the cell’s microenvironment.
Finally, we find that CD9 expression distinguishes two populations of ATMs in human adipose. Human CD9 ATMs share many features of CD9 ATMs in mice, including being lipid-laden and localizing to CLSs. Notably, human CD9+ ATMs express higher levels of CD16 and CD206 compared with CD9− ATMs suggesting that, akin to our model in mice, CD9− ATMs are recent tissue immigrants and CD9+ ATMs have a tissue-resident phenotype. Perhaps most dramatically, human CD9+ ATMs positively correlate with body mass. Together, these findings indicate that cellular and functional heterogeneity is a conserved feature of ATM biology and identify CLS-associated CD9 ATMs as the predominant ATM group responsible for the inflammatory signature of obese adipose tissue.
Our study is limited by the examination of a relatively small number of mouse ATMs by single-cell RNA-seq. However, the fact that clear transcriptional heterogeneity was detected with a limited number of cells strengthens the reliability of our conclusion that expression of CD9 distinguishes the two predominant Ly6c− ATM groups in murine adipose. Further, our subsequent flow-cytometric identification and use of bulk RNA-seq and ATAC-seq analyses ensure that our genomic studies are of adequate depth and breadth to provide a truly comprehensive analysis of the transcriptional networks active in each ATM subtype. Nevertheless, it is possible that additional transcriptional and cellular heterogeneity exists within ATMs and will be revealed with the analysis of more cell numbers. Additionally, in our bone marrow transplant studies, we utilized total body irradiation that may damage tissue-resident immune cells (10). It is therefore possible that bone marrow-derived macrophages can reconstitute local ATMs after total body irradiation, but ATMs may also be derived locally as a result of proliferation or other resident progenitor during normal physiology (8).
Together, our results reveal a heightened degree of complexity within ATMs of mice and humans and provide a framework for understanding the inflammatory and homoeostatic roles of these important cells. Notable similarities were observed between our animal models and human subjects, indicating that this paradigm is applicable to our understanding of human physiology and pathophysiology. The functional heterogeneity described herein should be closely considered in the development of novel, immune-directed therapeutics for the treatment or prevention of obesity, as well as related inflammatory sequela, as approaches that target ATMs en masse may have seemingly paradoxical and undesirable consequences.
Materials and Methods
Statistics.
Data are presented as mean ± SEM unless otherwise stated. GraphPad Prism software was used for graphing and statistical analysis. All statistical tests are fully described in the figure legends. For comparison between groups, a Student’s t test, ratio-paired Student’s t test, Fisher’s exact test, binomial test, or Wilcoxon test were used. For determining associations between a scalar-dependent variable and an independent variable, linear regression was used.
Animal Studies.
All experiments used male age-matched C57BL/6J mice housed at four or five animals per cage. HFD studies were conducted by feeding mice a purified-ingredient diet composed of 60:20:20 kcal percentage of fat:carbohydrate:protein (Research Diets, D12492) or control diet (Research Diets, D12450B) from 6 to 18 wk of age.
Primary Adipose Stromal-Fraction Isolation.
eWAT depots were isolated from mice and VAT were obtained from obese individuals. Adipose samples were placed in 1 mL of DMEM, and individual depots were finely minced under sterile conditions with a razorblade before digestion in 5 mL of DMEM with 6 mg/1 mL collagenase D (Roche) or 3 mg/1 mL collagenase IV (Gibco) at 37 °C in a shaking incubator for 30 min. Cells were pelleted at 700 × g and adipocyte fraction removed. After resuspension of the stromal fraction in 5 mL of FACS buffer (1× PBS, 2% calf serum, 1 mM EDTA, 0.1% sodium azide) for flow-cytometric analysis, or MACS buffer (1× PBS, 2% calf serum, 25 mM Hepes, 2 mM EDTA) for cell sorting, cells were purified through a 100-μm filter (Falcon), pelleted at 700 × g, and resuspended in 1 mL of RBC lysis buffer (0.15 M NH4Cl, 0.01 M NaHCO3, 0.0013 M EDTA) for 5 min at room temperature. Cells were washed again in FACS or MACS buffer before staining for flow cytometry or sorting, respectively.
Flow Cytometry and Cell Sorting.
Murine single-cell suspensions were Fc-receptor blocked with CD16/32 (2.4G2, 1:50, BD) and Rat IgG (1:50, Fisher Scientific) before staining with anti-mouse fluorochrome-conjugated monoclonal antibodies (BD Bioscience, BioLegend, eBioscience; clone and concentration shown) specific for CD3ε (145-2C11, 1:500), CD4 (GK1.5, 1:500), CD8 (53-6.7, 1:500), CD9 (MZ3, 1:500), CD11b (M1/70, 1:500), CD11c (N418, 1:500), CD19 (6D5, 1:500), CD45.1 (A20, 1:100), CD45.2 (104, 1:100), CD64 (X54-5/7.1, 1:500), CD68 (FA-11, 1:500), F4/80 (BM8, 1:250), Ly6c (HK1.4, 1:500), Ly6g (1A8, 1:500), or SiglecF (E50-2440, 1:500), or BODIPY FL (0.5 μg/mL, Thermo Fisher Scientific).
Human single-cell suspensions were treated with human Fc-receptor block (BD Pharmingen) before staining with anti-human fluorochrome-conjugated monoclonal antibodies (BioLegend; clone and concentration shown) specific for CD1c (L161, 1:500), CD3ε (HIT3a, 1:500), CD9 (HI9a, 1:500), CD11b (M1/70, 1:500), CD11c (3.9, 1:500), CD14 (M5E2, 1:500), CD15 (W6D3, 1:500), CD16 (3G8, 1:500), CD20 (HIB19, 1:500), CD45 (HI30, 1:500), or CD206 (15-2, 1:500). Cells were also stained with DAPI (2 ng/mL), Hoechst (10 µM, Thermo Fisher Scientific), or BODIPY FL (0.5 μg/mL, Thermo Fisher Scientific). Compensation was performed with cells or OneComp eBeads beads (Thermo Fisher Scientific). Cells were analyzed or sorted with a BD LSR II, Amnis ImageStream, or BD FACS Aria II with 100-μm nozzle and running DiVa software (BD Bioscience) and analyzed with FlowJo software (version 10.4; Tree Star).
Histology.
Adipose tissue was fixed in 4% paraformaldehyde/1× PBS overnight at 4 °C and dehydrated through sequential ethanol washes. Tissue was embedded in paraffin before sectioning and staining with antibodies against CD11b (Abcam ab133357), Ly6c (Abcam ab15627), or CD9 (Abcam ab92726) with species-specific secondary antibodies conjugated to Alexa 488, TSA tetramethylrhodamine, or streptavidin-635. Sections were visualized and photographed with a Leica confocal fluorescence microscope.
Nanoparticle Detector Analysis.
CD9 ATMs were sort-purified from HFD-fed B6 mice and cultured at a density of 100,000 cells per mL in serum-free media (Gibco) for 48 h, after which culture supernatant was removed and filtered through a 0.8-μm filter (Millipore). Exosomes were isolated using the ExoQuick-TC PLUS kit (Systems Biosciences) and analyzed with a NanoSight NS300 nanoparticle detector (Malvern Panalytical) in the light scatter mode for quantitation and size distribution.
Bone Marrow-Derived Macrophage Generation and Cell Culture.
Bone marrow cells were isolated under sterile conditions and cultured in the presence of mCSF at a density of 5 × 106 cells per 10 mL of complete media for 6 d. On day six of culture, cells were replated at a density of 3 × 106 cells and cultured for 24 h in the presence of BSA (control), LPS (5 ng/mL), or IL-4 (10 ng/mL).
RNA Isolation and Single-Cell and Bulk RNA-Seq Library Preparation.
For single-cell RNA-seq, libraries were prepared from sort-purified cell populations pooled from five HFD-fed B6 mice via a C1 platform (Fluidigm) and barcoded via Nextera XT DNA Library Preparation Kit (Illumina). For bulk RNA-seq of sort-purified cell populations, libraries were prepared using the SMARTer Stranded RNA-seq kit (Clontech) and TruSeq RNA library prep kit (Illumina). For bulk RNA-seq of adipose tissue, total RNA was isolated from snap-frozen tissue samples. Tissues were mechanically homogenized in 500 μL of TRIzol (Life Technologies) in a TissueLyser (Qiagen) for 4 min, at a frequency of 22 s−1. After homogenization, samples were centrifuged at 4 °C and homogenates transferred to a fresh tube for RNA extraction with chloroform. Aqueous chloroform fractions were mixed with equal volumes of 70% ethanol/30% water and RNA further purified with RNeasy Mini spin columns (Qiagen) and oncolumn DNase-digestion (Qiagen). One microgram of purified DNase-treated total RNA from biological replicates was processed with a Ribo-Zero Magnetic rRNA removal kit (Epicentre, MRZH11124). Bulk adipose RNA libraries were prepared using a TruSeq Stranded Total RNA Library Prep Gold (Illumina, 200020599) and standard Illumina protocol. RNA-seq libraries were sequenced at single or paired-end, 75- to 100-bp read length on an Illumina HiSeq 2000.
ATAC-Seq Library Preparation.
ATAC-seq was performed using a modified protocol based on the technique published by Buenrostro et al (47). Briefly, sort-purified cell populations were washed in PBS and lysed with lysis buffer (1 M Tris·Cl, pH 7.4, 5 M NaCl, 1 M MgCl2; 10% Nonidet P-40 in nuclease-free H2O) and gentle agitation before treatment with Tn5 transposase. DNA was purified (Qiagen MinElute Reaction Cleanup Kit) and amplified with NEBNext High-Fidelity 2× PCR Master Mix (New England Biolabs) using barcoded primers. DNA library was purified with AMPure XP beads, and library quality was assayed via Qubit and Agilent High Sensitivity DNA Bioanalysis chip. Libraries were sequenced via 40-bp paired-end sequencing using the Illumina NextSeq 500 platform.
Single-Cell RNA-Seq Analysis.
Sequencing reads were aligned to UCSC mm9 genome using default options in STAR aligner (48). After filtering chrM reads, poor quality libraries were eliminated (<1 million aligned reads or <30% of reads aligned to exonic regions). Gene expression levels were quantified and normalized using cuffquant and cuffnorm (49). First, genes expressed >1 fragments per kilobase of transcript per million mapped reads (FPKM) in at least 10 cells were selected for downstream analysis. Two distinct subpopulations of ATMs in eWAT of HFD mice were identified by hierarchical clustering of cells. Differentially expressed genes between the two subpopulations were defined by Wilcoxon test as having a P value <0.05, and >1 FPKM in more than 30% of cells per subpopulation. Hierarchical clustering of genes was performed using Spearman correlation coefficient as a similarity measure under the Ward’s criterion.
Bulk RNA-Seq and ATAC-Seq Analyses.
Bulk RNA-seq and ATAC-seq reads were aligned to UCSC mm9 genome using STAR aligner and analyzed as described in the SI Appendix, SI Materials and Methods.
Bone Marrow Chimera Generation and Adoptive Transfer.
For bone marrow chimera generation, lean CD45.1+ recipients were treated with 12 Gy of total body irradiation, split into two 6-Gy treatments with a 12-h interval between doses. Recipients were injected i.v. with 5 million Thy1.2-T cell depleted bone marrow cells from lean CD45.2+ donors. Mice were allowed to reconstitute after transplant for 6 wk, at which time mice were placed on HFD for 12 wk before sacrifice for subsequent analyses. For adoptive transfer, 75,000–125,000 cells were sort-purified from obese, CD45.2+ donors and transferred via i.p. injection three times over the course of 1 wk into lean, CD45.1+ recipients. Recipient mice were allowed to rest 24–48 h before sacrifice for subsequent analyses.
Study Approval and Data Availability.
Animal experiments were performed in accordance with ethical regulations and protocols approved by the Institutional Animal Care and Use Committee of the Perelman School of Medicine at the University of Pennsylvania and the Children’s Hospital of Philadelphia. VAT samples were obtained from obese individuals with participant informed consent obtained after the nature and possible consequences of the studies were explained under protocols approved by the Institutional Review Boards of the Perelman School of Medicine at the University of Pennsylvania and the Children’s Hospital of Philadelphia. Genomic data are available in the Gene Expression Omnibus (GSE113595).
Supplementary Material
Acknowledgments
We thank Marine Adlanmerini and the other members of the M.A.L. laboratory for helpful discussions and comments and Erika Briggs for assistance with animal husbandry. We acknowledge the Functional Genomics Core and Human Metabolic Tissue Bank of the Penn Diabetes Research Center through Grant P30 DK19525, as well as the Penn Flow-Cytometry and Cell Sorting Core, Histology and Gene Expression Core, and the Cell and Developmental Biology Microscopy Core for assistance with these studies. This work was supported by NIH Grants DK49780 (to M.A.L.), T32 HD043021 (to D.A.H.), F30 DK112507 (to Y.H.K.), K08 NHLBI 132099 (to P.V.); pilot Grant P30 MH097488 from the Penn Mental Health AIDS Research Center (to M.A.L. and D.A.H.); the JPB Foundation (to M.A.L.); a Children’s Hospital of Philadelphia Senior Fellow K-readiness grant (to D.A.H.); and American Heart Association Postdoctoral Fellowship 16POST29680002 (to V.L.N.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE113595).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802611115/-/DCSupplemental.
References
- 1.Heymsfield SB, Wadden TA. Mechanisms, pathophysiology, and management of obesity. N Engl J Med. 2017;376:254–266. doi: 10.1056/NEJMra1514009. [DOI] [PubMed] [Google Scholar]
- 2.Ferrante AW., Jr Macrophages, fat, and the emergence of immunometabolism. J Clin Invest. 2013;123:4992–4993. doi: 10.1172/JCI73658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weisberg SP, et al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112:1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu H, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanda H, et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest. 2006;116:1494–1505. doi: 10.1172/JCI26498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weisberg SP, et al. CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J Clin Invest. 2006;116:115–124. doi: 10.1172/JCI24335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamei N, et al. Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J Biol Chem. 2006;281:26602–26614. doi: 10.1074/jbc.M601284200. [DOI] [PubMed] [Google Scholar]
- 8.Amano SU, et al. Local proliferation of macrophages contributes to obesity-associated adipose tissue inflammation. Cell Metab. 2014;19:162–171. doi: 10.1016/j.cmet.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haase J, et al. Local proliferation of macrophages in adipose tissue during obesity-induced inflammation. Diabetologia. 2014;57:562–571. doi: 10.1007/s00125-013-3139-y. [DOI] [PubMed] [Google Scholar]
- 10.Zheng C, et al. Local proliferation initiates macrophage accumulation in adipose tissue during obesity. Cell Death Dis. 2016;7:e2167. doi: 10.1038/cddis.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zamarron BF, et al. Macrophage proliferation sustains adipose tissue inflammation in formerly obese mice. Diabetes. 2017;66:392–406. doi: 10.2337/db16-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cinti S, et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res. 2005;46:2347–2355. doi: 10.1194/jlr.M500294-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Hardy OT, et al. Body mass index-independent inflammation in omental adipose tissue associated with insulin resistance in morbid obesity. Surg Obes Relat Dis. 2011;7:60–67. doi: 10.1016/j.soard.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odegaard JI, Chawla A. Type 2 responses at the interface between immunity and fat metabolism. Curr Opin Immunol. 2015;36:67–72. doi: 10.1016/j.coi.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang H, et al. Synergistic modulation of inflammatory but not metabolic effects of high-fat feeding by CCR2 and CX3CR1. Obesity (Silver Spring) 2017;25:1410–1420. doi: 10.1002/oby.21900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolus WR, Gutierrez DA, Kennedy AJ, Anderson-Baucum EK, Hasty AH. CCR2 deficiency leads to increased eosinophils, alternative macrophage activation, and type 2 cytokine expression in adipose tissue. J Leukoc Biol. 2015;98:467–477. doi: 10.1189/jlb.3HI0115-018R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patsouris D, et al. Ablation of CD11c-positive cells normalizes insulin sensitivity in obese insulin resistant animals. Cell Metab. 2008;8:301–309. doi: 10.1016/j.cmet.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nawaz A, et al. CD206+ M2-like macrophages regulate systemic glucose metabolism by inhibiting proliferation of adipocyte progenitors. Nat Commun. 2017;8:286. doi: 10.1038/s41467-017-00231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng B, et al. Clodronate liposomes improve metabolic profile and reduce visceral adipose macrophage content in diet-induced obese mice. PLoS One. 2011;6:e24358. doi: 10.1371/journal.pone.0024358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho KW, et al. An MHC II-dependent activation loop between adipose tissue macrophages and CD4+ T cells controls obesity-induced inflammation. Cell Rep. 2014;9:605–617. doi: 10.1016/j.celrep.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris DL, et al. CD40 promotes MHC class II expression on adipose tissue macrophages and regulates adipose tissue CD4+ T cells with obesity. J Leukoc Biol. 2016;99:1107–1119. doi: 10.1189/jlb.3A0115-009R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu D, et al. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 2011;332:243–247. doi: 10.1126/science.1201475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kratz M, et al. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 2014;20:614–625. doi: 10.1016/j.cmet.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu X, et al. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coats BR, et al. Metabolically activated adipose tissue macrophages perform detrimental and beneficial functions during diet-induced obesity. Cell Rep. 2017;20:3149–3161. doi: 10.1016/j.celrep.2017.08.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li P, et al. Functional heterogeneity of CD11c-positive adipose tissue macrophages in diet-induced obese mice. J Biol Chem. 2010;285:15333–15345. doi: 10.1074/jbc.M110.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guilliams M, et al. Dendritic cells, monocytes and macrophages: A unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14:571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deiuliis JA, et al. CXCR3 modulates obesity-induced visceral adipose inflammation and systemic insulin resistance. Obesity (Silver Spring) 2014;22:1264–1274. doi: 10.1002/oby.20642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andreu Z, Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ying W, et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171:372–384.e12. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 32.Gosselin D, Glass CK. Epigenomics of macrophages. Immunol Rev. 2014;262:96–112. doi: 10.1111/imr.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolic T, et al. The DNA-binding factor Ctcf critically controls gene expression in macrophages. Cell Mol Immunol. 2014;11:58–70. doi: 10.1038/cmi.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soccio RE, et al. Targeting PPARγ in the epigenome rescues genetic metabolic defects in mice. J Clin Invest. 2017;127:1451–1462. doi: 10.1172/JCI91211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Italiani P, Boraschi D. From monocytes to M1/M2 macrophages: Phenotypical vs. functional differentiation. Front Immunol. 2014;5:514. doi: 10.3389/fimmu.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phan AT, Goldrath AW, Glass CK. Metabolic and epigenetic coordination of T cell and macrophage immunity. Immunity. 2017;46:714–729. doi: 10.1016/j.immuni.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gealekman O, et al. Control of adipose tissue expandability in response to high fat diet by the insulin-like growth factor-binding protein-4. J Biol Chem. 2014;289:18327–18338. doi: 10.1074/jbc.M113.545798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corvera S, Gealekman O. Adipose tissue angiogenesis: Impact on obesity and type-2 diabetes. Biochim Biophys Acta. 2014;1842:463–472. doi: 10.1016/j.bbadis.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YH, Petkova AP, Granneman JG. Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell Metab. 2013;18:355–367. doi: 10.1016/j.cmet.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho CH, et al. Angiogenic role of LYVE-1-positive macrophages in adipose tissue. Circ Res. 2007;100:e47–e57. doi: 10.1161/01.RES.0000259564.92792.93. [DOI] [PubMed] [Google Scholar]
- 41.Bolus WR, et al. Elevating adipose eosinophils in obese mice to physiologically normal levels does not rescue metabolic impairments. Mol Metab. 2017;8:86–95. doi: 10.1016/j.molmet.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill AA, et al. Activation of NF-κB drives the enhanced survival of adipose tissue macrophages in an obesogenic environment. Mol Metab. 2015;4:665–677. doi: 10.1016/j.molmet.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanssen LLP, et al. Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat Cell Biol. 2017;19:952–961. doi: 10.1038/ncb3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu J, Tang M. CTCF-dependent chromatin insulator as a built-in attenuator of angiogenesis. Transcription. 2012;3:73–77. doi: 10.4161/trns.19634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang M, et al. Restraint of angiogenesis by zinc finger transcription factor CTCF-dependent chromatin insulation. Proc Natl Acad Sci USA. 2011;108:15231–15236. doi: 10.1073/pnas.1104662108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murray PJ. Macrophage polarization. Annu Rev Physiol. 2017;79:541–566. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.