Significance
It remains obscure how actin polymerizing and depolymerizing activities cooperate to control diverse actin dynamics. Formins rotate along the long-pitch helix of F-actin during processive actin elongation (helical rotation), which may twist F-actin in the opposite direction of the cofilin-induced twisting. In this study, we show that a mammalian formin mDia1 generates F-actin resistant to cofilin. Tethered F-actin elongating from immobilized mDia1 contained a less twisted portion in EM analysis and exhibited resistance to the severing activity of cofilin. In cells, overexpression of an active mDia1 mutant, which harbors N-terminal regulatory domains, prolonged F-actin lifetime and accelerated dissociation of cofilin. Helical rotation of formins may thus facilitate the formation of stabilized F-actin resistant to actin severing activities of cofilin.
Keywords: actin, formin homology proteins, cofilin, helical rotation, helical structure
Abstract
The complex interplay between actin regulatory proteins facilitates the formation of diverse cellular actin structures. Formin homology proteins (formins) play an essential role in the formation of actin stress fibers and yeast actin cables, to which the major actin depolymerizing factor cofilin barely associates. In vitro, F-actin decorated with cofilin exhibits a marked increase in the filament twist. On the other hand, a mammalian formin mDia1 rotates along the long-pitch actin helix during processive actin elongation (helical rotation). Helical rotation may impose torsional force on F-actin in the opposite direction of the cofilin-induced twisting. Here, we show that helical rotation of mDia1 converts F-actin resistant to cofilin both in vivo and in vitro. F-actin assembled by mDia1 without rotational freedom became more resistant to the severing and binding activities of cofilin than freely rotatable F-actin. Electron micrographic analysis revealed untwisting of the long-pitch helix of F-actin elongating from mDia1 on tethering of both mDia1 and the pointed end side of the filament. In cells, single molecules of mDia1ΔC63, an activated mutant containing N-terminal regulatory domains, showed tethering to cell structures more frequently than autoinhibited wild-type mDia1 and mDia1 devoid of N-terminal domains. Overexpression of mDia1ΔC63 induced the formation of F-actin, which has prolonged lifetime and accelerates dissociation of cofilin. Helical rotation of formins may thus serve as an F-actin stabilizing mechanism by which a barbed end-bound molecule can enhance the stability of a filament over a long range.
Formin homology proteins (formins) play an essential role in the formation of actin stress fibers, yeast actin cables, and contractile rings in cytokinesis (1–5). Formins facilitate actin nucleation and accelerate actin elongation. Formins have two conserved domains, formin homology domain 1 (FH1) and FH2, in the C-terminal half (1). FH2 processively elongates the actin filament (F-actin) by sequentially incorporating actin monomers (G-actins) into the barbed end (6, 7). During processive actin elongation, formins rotate along the long-pitch helix of F-actin (hereinafter referred to as helical rotation) (8).
Helical rotation of a mammalian formin mDia1 is coupled with processive actin elongation (8). When mDia1 and the pointed end side of F-actin are tightly immobilized, mDia1 stops processive actin elongation without forming a bent loop. When mDia1 or a yeast formin Bni1 is weakly immobilized, these formins continue to processively elongate a bent actin filament (8, 9). These observations suggest that processive actin elongation can be limited by the rotational degree of freedom of mDia1 around the filament axis rather than by the force against growth of the filament tip.
The Brownian ratchet theory is believed to explain the relationship between the actin elongation rate and the force against the growing filament tip (10). When actin polymerizes toward a barrier (e.g., a membrane), stochastic intercalation of G-actin into the gap between the barrier and the filament tip generates the force pushing the barrier forward. Similarly, when mDia1 and its processively elongating F-actin are immobilized, mDia1 might work as a barrier against rotational movement around the filament axis, and incorporation of G-actin may impose torsional force on F-actin.
The actin depolymerizing factor cofilin/ADF family plays an essential role in actin recycling. Disruption of the cofilin gene leads to lethal phenotype in Saccharomyces cerevisiae (11). Cofilin localizes to lamellipodia in animal cells (12, 13) and the cortical actin patch in yeast (14, 15) but not to actin stress fibers and yeast actin cables (14, 15). The formation of actin stress fibers and actin cables requires the function of formins (1, 2, 4). The majority of F-actin (85%) in stress fibers slowly disassemble (T1/2 = 311 s), whereas a large portion of F-actin (73%) disassembles fast (T1/2 = 32 s) outside of stress fibers in the lamella region of XTC cells (16). These observations imply that formins might generate F-actin resistant to the activity of cofilin/ADF. Several in vitro studies have shown that cofilin binds cooperatively to F-actin (17–19) and markedly twists the helical structure of F-actin by bridging between two longitudinally associated actin subunits (19). The twisting of F-actin by cofilin weakens the lateral contact between actin subunits in the F-actin (20) and alters the longitudinal contact between actin subunits (21). Cofilin severs F-actin (17, 22, 23) in a nonlinear fashion. It was shown that cofilin severs F-actin at low concentrations more efficiently than at high concentrations (17). In contrast, cofilin severs mDia1-assembled filament in a dose-dependent manner (24). Filament severing by cofilin seems to occur near the boundary between the cofilin-decorated segment and the bare segment (22, 23). Presumably, cofilin-catalyzed actin disassembly involves a filament twisting step.
In this study, we tested whether helical rotation of mDia1 influences F-actin binding and filament severing activities of cofilin, because helical rotation of tethered formins may impose torsional force to twist F-actin in the opposite direction of the cofilin-induced twisting. Our in vitro reconstitution assays revealed that, when both mDia1 and the pointed end side of F-actin were immobilized, F-actin was less twisted and less frequently severed by Xenopus laevis cofilin Xac2 than movable filaments. The binding of cofilin was also reduced compared with spontaneously elongating F-actin. The activity of mDia1 is regulated by the autoinhibitory interaction between the N-terminal half and the C terminus (4). Autoinhibition of mDia1 is released by the binding of small GTPase Rho to its N terminus. Thus, activated mDia1 can be tethered to the cellular structure through the CAAX motif of Rho. Using mDia1-compatible fluorescent actin probes for single-molecule speckle (SiMS) microscopy (16), we found that an active mDia1 mutant tethered to cellular structures through its N-terminal half induces the formation of F-actin with prolonged lifetime and with reduced affinity to cofilin. Thus, torsional force generated by helical rotation of formins stabilizes F-actin against cofilin activities.
Result
Helical Rotation of mDia1 Attenuates F-Actin Severing by Cofilin.
To investigate whether helical rotation of mDia1 prevents filament severing by cofilin, we observed the effect of Xenopus ADF/Cofilin 2 (Xac2) on the F-actin elongating from mDia1 under the condition where the pointed end side of F-actin is occasionally trapped to the glass surface (Fig. 1). We immobilized GST-mDia1ΔN3 (amino acids 543–1,192), which comprises FH1 and FH2 domains, in protein aggregates composed of anti-GST and secondary antibodies (mDia1 aggregates) (7, 8). The rotational freedom of the majority of mDia1 is restricted, whereas a small population of mDia1 may freely rotate in mDia1 aggregates (8). In this study, we used a biotin-conjugated secondary antibody in combination with streptavidin-coated glass coverslips to tether mDia1 aggregates. To incidentally trap the pointed end side of F-actin, we included 3% biotinylated actin in the initial filament nucleation step (8). Biotinylated F-actin was occasionally trapped by the glass surface through the biotin–avidin interaction.
Fig. 1.
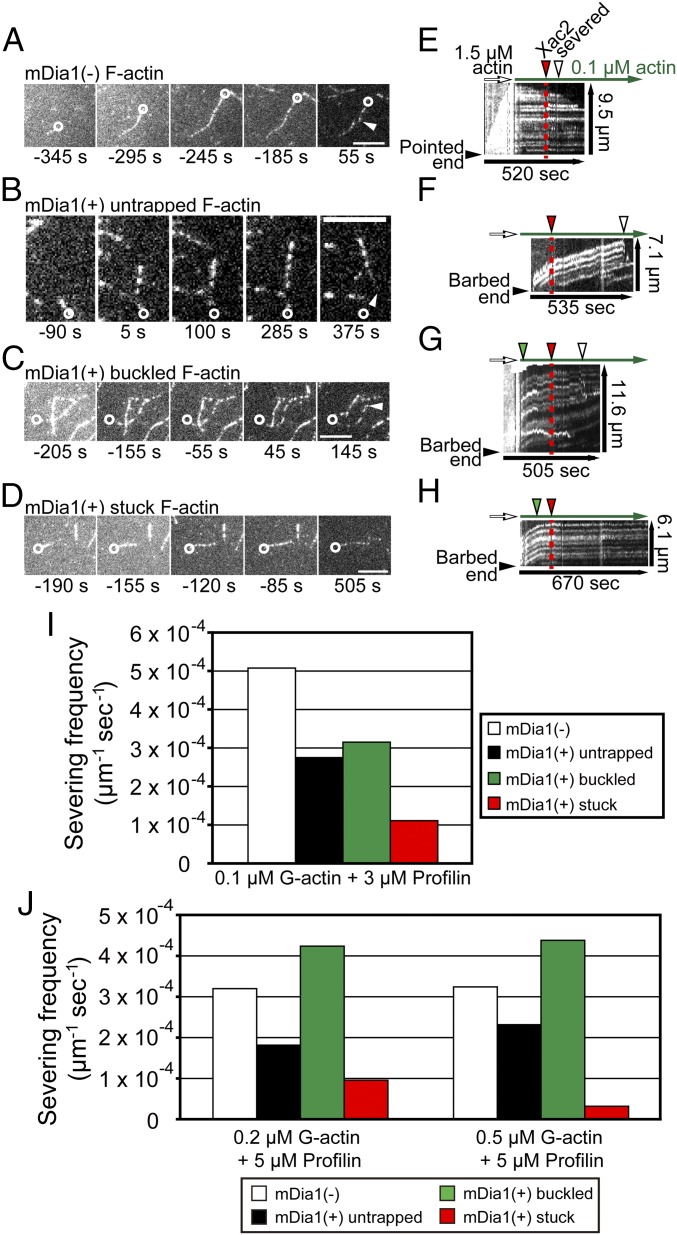
Helical rotation of mDia1 attenuates filament severing by Xac2. (A–H) Representative time-lapse images (A–D) and kymographs (E–H) of mDia1(−) F-actin (A and E), mDia1(+) untrapped F-actin (B and F), mDia1(+) buckled F-actin (C and G), and mDia1(+) stuck F-actin (D and H). These filaments were nucleated for 3 min (white arrows in E–H) and then elongated (green arrows in E–H) in the presence of 0.1 μM DL488-actin and 3 μM profilin. The pointed end sides of buckled and stuck F-actin were trapped during elongation (green arrowheads in G and H). Filament severing was initiated by the addition of 50 nM Xac2 (red arrowheads in E–H), and filaments were severed (white arrowheads in A–C and E–G). In A–D, barbed ends are showed with white circles. Time is after the addition of 50 nM Xac2. (Scale bars: 5 μm.) (I) The severing frequency of four types of F-actin in the presence of 0.1 μM DL488-actin, 3 μM profiling, and 50 nM Xac2. (J) The severing frequency of F-actin induced by 50 nM Xac2 in the presence of 0.2 μM DL488-actin with 5 μM profilin (Left) and 0.5 μM DL488-actin with 5 μM profilin (Right).
Under this condition, we observed four types of F-actin (Fig. 1 A–H; SI Appendix, SI Materials and Methods has details). The first type is the F-actin unbound to mDia1 [mDia1(−) F-actin] (Fig. 1 A and E and Movie S1). mDia1(−) F-actin grows spontaneously at the free barbed end. The second type is the F-actin elongating from immobilized mDia1 aggregates without being captured by the glass surface (untrapped F-actin) (Fig. 1 B and F and Movie S2). The pointed end side of untrapped F-actin freely moves away from the tethered barbed end. The third type is the F-actin, which continues elongation at the tethered barbed end and forms a bent loop after the pointed end side is trapped (buckled F-actin) (Fig. 1 C and G and Movie S3). The fourth type is the F-actin, which stops the filament elongation from immobilized mDia1 without forming a bent loop after the pointed end side is trapped (stuck F-actin) (Fig. 1 D and H and Movie S4).
We previously found that mDia1 nonspecifically adsorbed on the glass surface more frequently forms buckled F-actin than mDia1 trapped by anti-GST and secondary antibodies (8). This altered frequency of stuck and buckled F-actin is attributable to the different rotational degree of freedom of mDia1 arising from the two immobilization methods. In the buckled F-actin, torsional force may be relieved by slippage between mDia1 and the glass surface to continue filament elongation. In stuck F-actin, torsional force presumably accumulates to stop elongation. Furthermore, elongation of stuck F-actin was gradually decelerated after filament was trapped (Fig. 1 G and H and SI Appendix, Fig. S1). This observation implies that mDia1 may act as a rotary Brownian ratchet at the barbed end.
We compared the frequency of Xac2-induced severing between four types of F-actin. We first investigated the severing frequency in the presence of 0.1 μM G-actin and 3 μM profilin. Under this condition, while mDia1(−) F-actin depolymerized at 1.3 ± 1.1 subunits per second at the barbed end (Fig. 1 A and E), untrapped F-actin continued elongation at 6.1 ± 0.9 subunits second−1 (Fig. 1 B and F). The elongation rate of stuck F-actin apparently (>50%) decreased within 50 s after the pointed end side was trapped (Fig. 1 D and H and SI Appendix, Fig. S1), which enabled us to distinguish buckled F-actin (Fig. 1 C and G) from stuck F-actin. The severing frequency with stuck F-actin was the lowest among four types of F-actin (Fig. 1I). Approximately two-thirds of stuck F-actin were not severed within 500 s after the addition of 50 nM Xac2 (SI Appendix, Fig. S2A).
At 0.2 μM G-actin with 5 μM profilin and 0.5 μM G-actin with 5 μM profilin, mDia1 elongated untrapped F-actin at 10.3 ± 1.1 and 19.8 ± 3.7 subunits second−1, respectively. In either case, the severing frequency of stuck F-actin was the lowest among four types of F-actin (Fig. 1J). The larger amount of G-actin was included during the filament elongation step, and the less frequently stuck F-actin was severed (Fig. 1 I and J). Thus, an increase in the probability of adding G-actin into the mDia1-bound barbed end enhances the resistance of stuck F-actin against the filament severing by Xac2.
We also examined the effect of higher concentrations of Xac2. We used carbon-coated glass surface and a modified ratio of mDia1 and antibodies to minimize nonspecific tethering of the side of F-actin and the rotational freedom of mDia1 [SI Appendix, SI Materials and Methods has transmission EM (TEM) observations]. Under this condition, Xac2 promoted F-actin severing in a dose-dependent manner up to 2 μM. The severing rate of stuck F-actin (presence of streptavidin) was lower than that of untrapped F-actin (absence of streptavidin) at 50 and 300 nM Xac2 (SI Appendix, Fig. S3 A, B, and D), whereas 2 μM Xac2 rapidly severed both untrapped and stuck F-actin (SI Appendix, Fig. S3 C and E).
In addition, we reproductively observed that the severing frequency of untrapped F-actin was lower than the severing frequency of mDia1(−) F-actin (Fig. 1 I and J). Several studies reported that binding of formins may induce long-range allosteric effects on the F-actin (25–27). However, the severing frequency of buckled F-actin was higher than that of untrapped F-actin (Fig. 1 I and J). This increased severing frequency of buckled F-actin may arise from filament bending. Acanthamoeba coflin severs F-actin frequently at the bent portion (28). Fragmentation of filopodium actin bundles occurs after bending at the transition zone in neuronal growth cones (29, 30). Nevertheless, the severing frequency of stuck F-actin was less than one-half of the severing frequency of untrapped F-actin under all conditions (Fig. 1 I and J). These results indicate that the low severing frequency of stuck F-actin is caused by helical rotation of mDia1 rather than the binding of mDia1 to the barbed end. Helical rotation of mDia1 thus attenuates the F-actin severing by Xac2 when tethered mDia1 polymerizes F-actin that cannot rotate freely.
Helical Rotation of mDia1 Reduces the Binding of Cofilin to F-Actin.
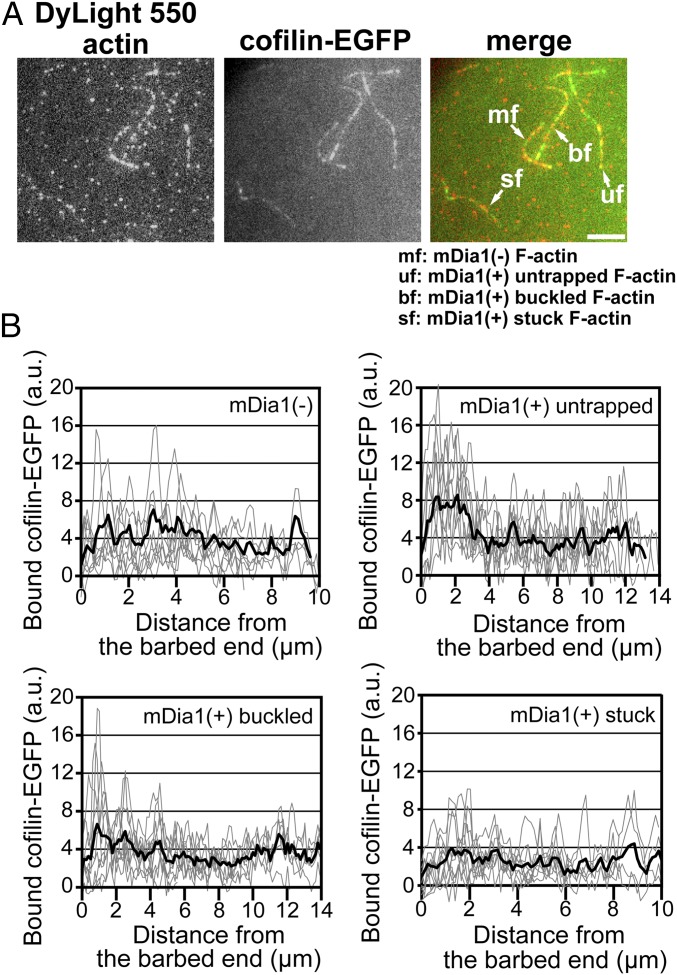
We next investigated the effect of helical rotation of mDia1 on the F-actin binding of cofilin (Fig. 2). To investigate the binding of cofilin while limiting actin depolymerization (21), we observed at pH 6.8. F-actin was elongated from immobilized mDia1 aggregates in the presence of 0.1 μM G-actin and 3 μM profilin using the same procedure as in Fig. 1. We incubated the mixture composed of 1.8 μM recombinant mouse cofilin and 0.2 μM cofilin-EGFP with four types of F-actin at pH 6.8 (Fig. 2A and Movie S5). In yeast, cofilin tagged on its C terminus cannot complement a cofilin null mutation unless overexpressed (31, 32). We verified the binding of fluorescent protein-tagged cofilin with F-actin by using the quenching method (17, 33) of pyrenyl fluorescence on F-actin (SI Appendix, Fig. S4). Cofilin tagged on its C terminus quenched pyrene–F-actin as efficiently as untagged cofilin (SI Appendix, Fig. S4A).
Fig. 2.
Helical rotation of mDia1 reduces binding of cofilin to F-actin. (A) Representative fluorescence images of four types of F-actin bound to cofilin-EGFP 1 min after the addition of 2 μM cofilin-EGFP at pH 6.8. Merged images show DyLight 550-labeled actin (DL550-actin; red) and cofilin-EGFP (green). Note the poor association of cofilin to stuck F-actin. (Scale bars: 5 μm.) (B) Fluorescence intensity of cofilin-EGFP along F-actin 1 min after the addition of 2 μM cofilin-EGFP. Gray lines show the fluorescence intensities of cofilin-EGFP bound to each F-actin [mDia1(−), n = 10; mDia1(+) untrapped, n = 10; mDia1(+) buckled, n = 10; mDia1(+) stuck, n = 13]. Black lines show the average fluorescence intensities of cofilin-EGFP bound to F-actin.
The fluorescence intensity of cofilin-EGFP along the F-actin 1 min after the addition of the cofilin mixture is shown as a function of the distance from the barbed end (Fig. 2B). Cofilin bound evenly along the length of mDia1(−) F-actin and stuck F-actin. The average density of cofilin on stuck F-actin was ∼35% lower than that on mDia1(−) F-actin (Fig. 2). However, cofilin accumulated near the barbed end of mDia1-assembled untrapped F-actin and buckled F-actin without promoting filament severing locally (SI Appendix, Fig. S2B). Thus, the barbed end region of untrapped F-actin seems to favor interaction with cofilin but becomes resistant to its severing activity. In contrast, the barbed end region of stuck F-actin did not show any accumulation of cofilin. Stuck F-actin was less densely labeled with cofilin than those of mDia1(−) F-actin and untrapped F-actin throughout the length of the filament (Fig. 2).
Although we currently do not know how cofilin accumulates near the barbed end of untrapped F-actin, poor association of cofilin with stuck F-actin indicates that helical rotation of mDia1 interferes with the interaction between cofilin and F-actin when helical rotation can impose torsional force on the filament.
The Autoinhibition Defective mDia1 Mutant Containing N-Terminal Domains Attenuates Actin Disassembly in Cells.
The activity of mDia1 is regulated by the autoinhibitory interaction between the Diaphanous inhibitory domain in the N-terminal half and the Diaphanous autoregulatory domain in the C-terminal half. Autoinhibition of mDia1 is released by the binding of Rho GTPases with the N terminus. Our previous study showed that mDia1 lacking C-terminal 63 amino acids no longer retains the autoinhibitory interaction (4).
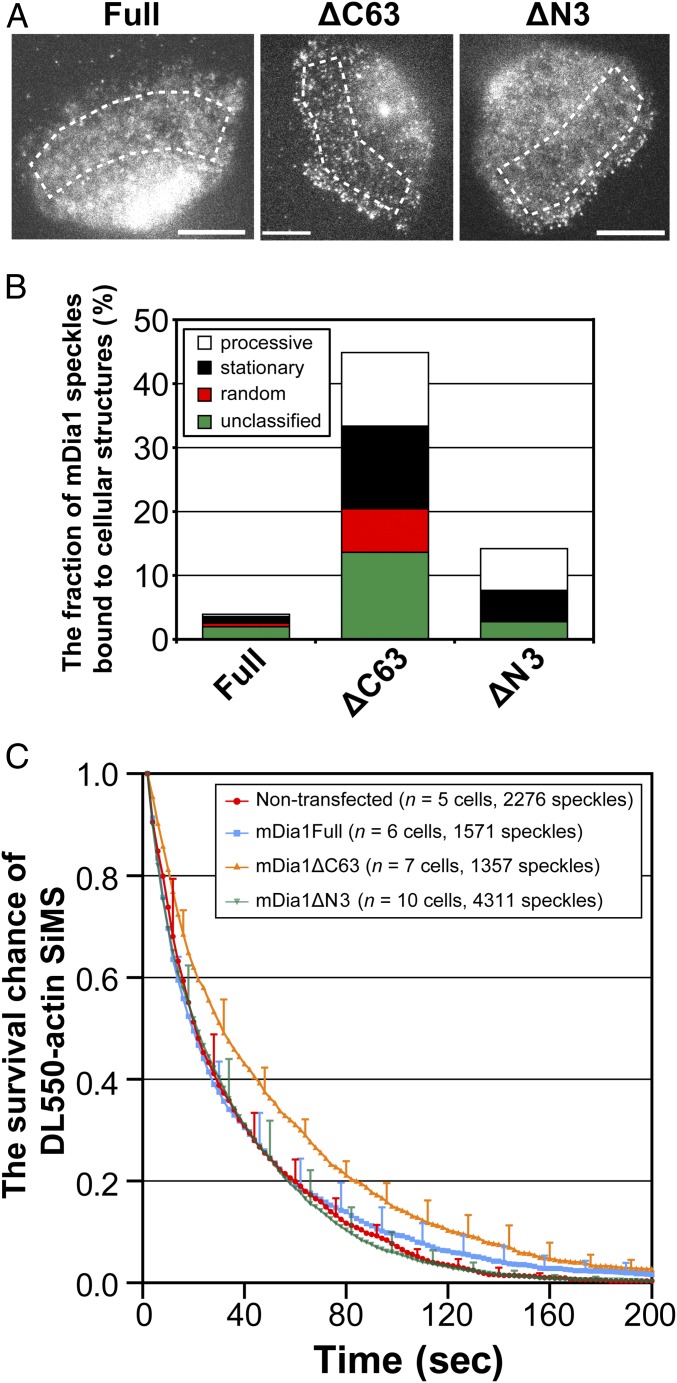
We first compared the fraction of cell structure-associated mDia1 molecules between the wild type and mutants using SiMS microscopy (34). When fluorescently labeled proteins are expressed at a very low density, individual fluorescent proteins bound to cellular structures are observed as a discrete spot. The fluorescence intensity of SiMS of EGFP-mDia1 was comparable with that of two EGFP molecules as mDia1 forms a dimer. The fraction of structure-bound molecules was calculated by dividing the number of the observed SiMS by the total intensity of EGFP fluorescence in the observed region. Comparison was made between wild-type mDia1 (mDia1Full; amino acids 1–1,255), the autoinhibition-defective mDia1 mutant (mDia1ΔC63; amino acids 1–1,192), and the mDia1 FH1-FH2 mutant (mDia1ΔN3; amino acids 543–1,192) (Fig. 3 A and B).
Fig. 3.
The autoinhibition defective mDia1 mutant containing N-terminal domains attenuates actin disassembly in cells. (A) Representative images of EGFP-tagged mDia1 and its mutants expressed in XTC cells at a low density. Under this condition, individual EGFP-mDia1 molecules bound to cellular structures were observed as a discrete spot. Dotted lines indicate the area for the measurement in B. (Scale bars: 10 μm.) (B) The fraction of speckles of mDia1 and its mutants bound to cellular structures. mDia1 speckles were classified according to the criteria in our previous report (36). Processive is the fraction showing directional motion over five consecutive images (white), stationary is the fraction that stops the motion presumably trapped by rigid cell structures (black), random is the fraction showing slow random diffusing motion (red), and unclassified is the fraction that did not fall into three categories within the observation time (green). (C) The cumulative survival chance of DL550-actin SiMS incorporated into F-actin in nontransfected XTC cells and in cells overexpressing mDia1 and its mutants. Error bars show SD.
As reported previously (7, 35, 36), only a small fraction of mDia1Full appeared as a speckle in cells (Fig. 3A and Movie S6). The fraction of structure-associated mDia1Full was 3.9% (Fig. 3B), suggesting that the majority of expressed mDia1Full does not bind cellular structures. However, the fraction of structure-associated mDia1ΔC63 was 44.8%, which is 12-fold higher than that of mDia1Full. The fraction of cell structure-associated mDia1ΔN3 was 14.2%, which was substantially lower than that of EGFP-mDia1ΔC63. These results indicate that a large fraction of mDia1ΔC63 is tethered to cell structures through its N-terminal half.
We further classified the cell structure-associated speckles of mDia1 and its mutants into four categories, which are “processive,” “stationary,” “random,” and “unclassified” (Fig. 3B), according to the criteria used in our previous study (36). The fraction of stationary mDia1ΔC63 speckles (12.9%) was higher than those of mDia1Full (1.0%) and mDia1ΔN3 (4.8%). The fraction of random mDia1ΔC63 speckles (6.8%) was also higher than those of mDia1Full (0.58%) and mDia1ΔN3 (0.10%). The fraction of random speckles of all three mDia1 constructs exhibited motion typical of plasma membrane-associating molecules. These results indicate that, after autoinhibition is relieved, mDia1 is frequently tethered to either the cytoskeleton or the plasma membrane through its N-terminal half. With regard to the frequency of processive actin polymerization, the processive fraction of mDia1Full (0.32%) was much lower than those of mDia1ΔC63 (11.5%) and mDia1ΔN3 (6.5%). In addition, we frequently observed that the behavior of processively moving mDia1ΔC63 and mDia1ΔN3 switched to either stationary or random motion. Based on these observations, we speculate that a certain population of stationary and random fractions of mDia1ΔC63 and mDia1ΔN3 is attached to the actin barbed end and able to elongate F-actin until actin elongation is physically constrained.
Next, we examined F-actin lifetime in cells overexpressing mDia1 and its mutants. The lifetime of F-actin was measured by the speckle lifetime method (37) with improved fluorescent actin probes. Chen and Pollard (31) reported that GFP-tagged actin is eliminated from the contractile ring formed by the fission yeast formin Cdc12p (38). In accordance with this finding, processive movement of mDia1ΔN3 frequently exhibits arrest in cells expressing a high level of mRFP1-actin (16). Thus, the fluorescent protein-tagged actin may not reliably report the turnover of formin-assembled actin filaments.
To solve this issue, we recently developed improved SiMS microscopy, which uses an actin probe, DyLight 550-labeled actin (DL550-actin) (16). DL550-actin incorporates into the mDia1-bound barbed end at a similar efficiency (77%) to the free barbed end. Additionally, DL550-actin shows excellent photostability and brightness, allowing simultaneous lifetime measurement of actin SiMS of short and long lifetimes. We introduced DL550-actin into XTC cells overexpressing mDia1 or its mutants using electroporation, which enables us to label all of the cells at a similar low density with DL550-actin molecules (Movie S7).
Fig. 3C shows the cumulative survival chance of DL550-actin after assembly into the cellular F-actin, which was calculated from the lifetime distribution of single-molecule DL550-actin speckles (SI Appendix, Fig. S5). The cumulative survival chance of actin SiMS was markedly higher in cells overexpressing mDia1ΔC63 than in nontransfected cells (P < 0.0001, log-rank test). In contrast, the cumulative survival chance in cells overexpressing mDia1Full and mDia1ΔN3 was similar to that of nontransfected cells (P = 0.0441 for mDia1Full and P = 0.2351 for mDia1ΔN3, log-rank test). These results indicate that, on relief of autoinhibition, mDia1 containing intact N-terminal and FH1-FH2 domains may promote the formation of stabilized F-actin in cells.
Cells overexpressing mDia1ΔC63 contained condensed actin fibers, whereas cells overexpressing mDia1ΔN3 contained massive thin actin fibers (SI Appendix, Fig. S6). We previously reported that mDia1ΔN3 reverses ROCK-induced heavily condensed F-actin into morphologically normal actin stress fibers (4), although the mechanism of this antagonism between mDia1 and ROCK remains unclear. Nevertheless, mDia1 tethered to cellular structures through its N terminus seems to contribute to the formation of condensed F-actin bundles better than mDia1FH1-FH2 alone, implying that interaction with Rho may render mDia1 more favorable for the formation of actin stress fibers.
mDia1ΔC63 Accelerates Dissociation of Cofilin from F-Actin in Cells.
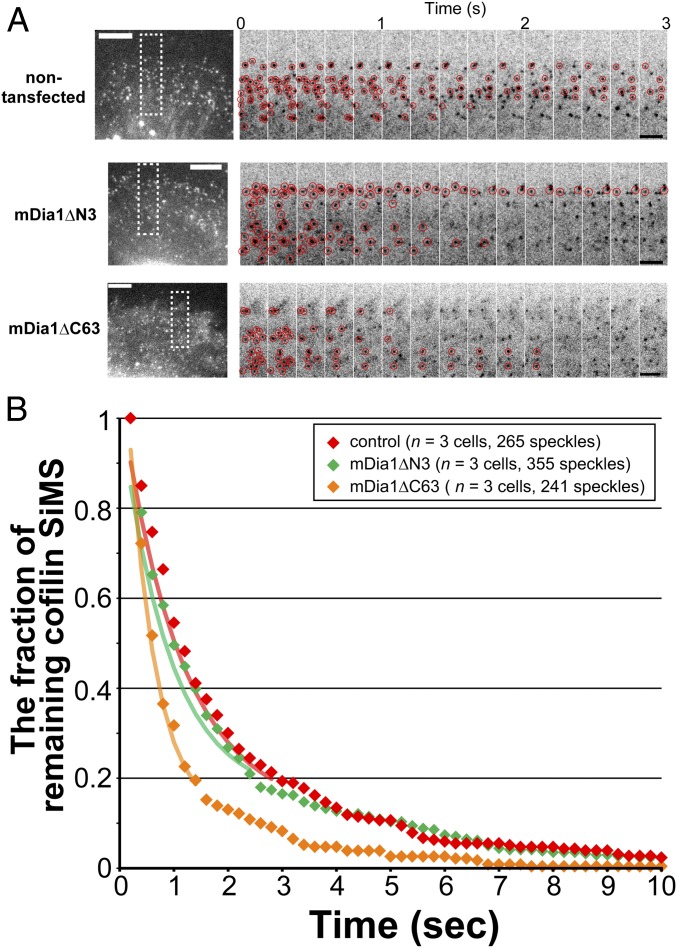
We next investigated the dissociation kinetics of cofilin from F-actin in cells overexpressing mDia1ΔN3 and mDia1ΔC63. After cells started expression of these mDia1 mutants, recombinant cofilin-EGFP was introduced by electroporation, which allows constant labeling of cells (16). We analyzed kinetics of cofilin-EGFP SiMS within 15 μm from the leading edge (Fig. 4A). We carried out regression analysis (37) by measuring the surviving fraction of preexisting cofilin-EGFP SiMS (Fig. 4B). Dissociation of cofilin-EGFP in cells overexpressing mDia1ΔC63 was significantly faster than nontransfected cells (Fig. 4B) (P < 0.0001, log-rank test). In contrast, dissociation of cofilin-EGFP in cells overexpressing mDia1ΔN3 was similar to that of nontransfected cells (Fig. 4B) (P = 0.303, log-rank test). The initial decay rates between the remaining fractions 1.0 and 0.2 were 1.96 s−1 in cells overexpressing mDia1ΔC63, 0.88 s−1 in control cells, and 1.09 s−1 in mDia1ΔN3-expressing cells.
Fig. 4.
The autoinhibition-defective mDia1 mutant containing N-terminal domains accelerates dissociation of cofilin from cellular F-actin. (A) Representative time-lapse images of EGFP-cofilin SiMS in XTC cells. Images indicated by rectangles are paneled. Red circles indicate cofilin-EGFP SiMS remaining from the first frame. (Scale bars: Left, 10 μm; Right, 5 μm.) (B) The fraction of cofilin-EGFP SiMS persisting from the initial image frame is plotted after correction for the photobleaching rate of cofilin-EGFP (T1/2 = 14.84 s). The decay rates between 1.0 and 0.2 were obtained by fitting the data with single exponentials (continuous lines).
In addition, we investigated the distribution of cofilin-EGFP in cells overexpressing mDia1 mutants (SI Appendix, Fig. S7). Cofilin-EGFP in nontransfected cells showed similar distribution to endogenous Xac2 in XTC cells (13). Cells overexpressing mDia1 mutants contained abundant F-actin throughout the cytoplasm (SI Appendix, Fig. S7 A and C). Although cofilin-EGFP in nontransfected cells was enriched in lamellipodia, cofilin-EGFP in cells overexpressing both mDia1ΔN3 and mDia1ΔC63 accumulated only in a narrow region near the cell edge, despite the wide distribution of abundant F-actin throughout the cytoplasm (SI Appendix, Fig. S7 B and D).
Thus, while both mDia1ΔN3 and mDia1ΔC63 can prevent the formation of the cofilin-rich lamellipodial actin network, only mDia1ΔC63 promotes dissociation of cofilin from cellular F-actin. Together with the effects on actin SiMS (Fig. 3), tethered and activated mDia1 may enhance the stability of F-actin by preventing stable association of cofilin with F-actin.
mDia1 Untwists the F-Actin Long-Pitch Helical Structure.
We next investigated using TEM whether helical rotation of mDia1 may alter the structure of F-actin. We established the sample preparation method by which F-actin elongating from immobilized mDia1 aggregates can be trapped on the carbon-coated EM grid. We noted that, without methylcellulose, it was difficult to keep the elongating F-actin near the grid surface and that methylcellulose interferes with EM observation by forming inhomogeneous background. We, therefore, replaced the solution with a methylcellulose-free solution containing the same concentration of G-actin after trapping the pointed end side and incubated for another 6 min. We also modified the ratio between mDia1 and antibodies to lessen the rotational freedom of mDia1. Under this condition, when the pointed end side was trapped on the carbon surface by streptavidin, most of F-actin ceased elongation (Movie S8).
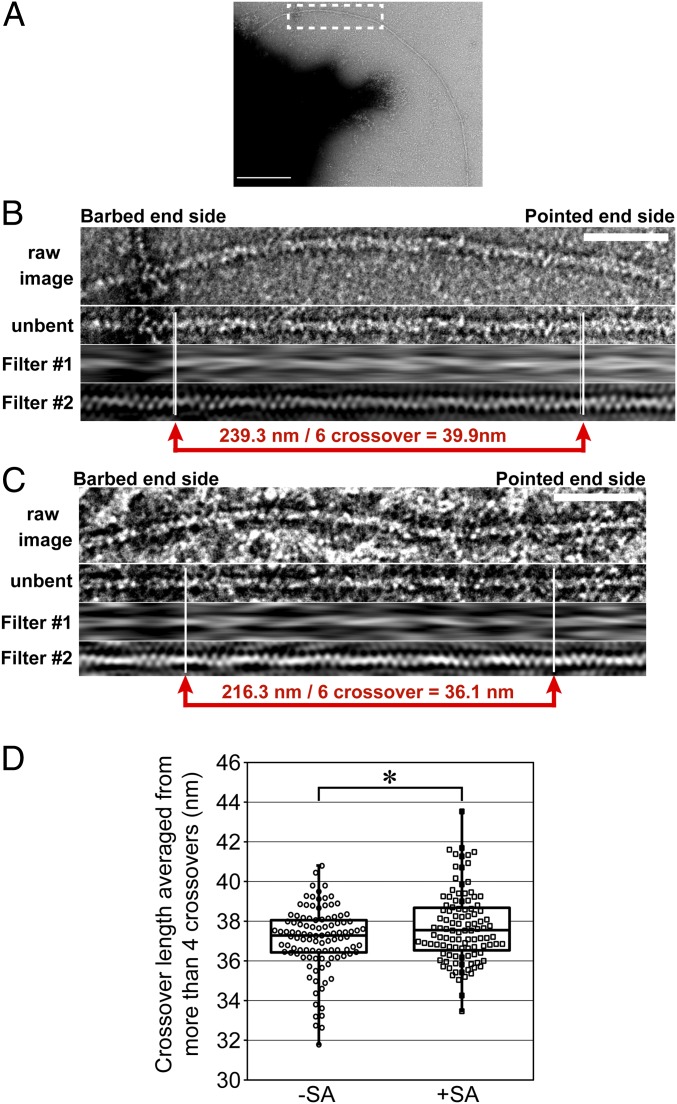
We compared the helical structure of F-actin elongating from immobilized mDia1 in the absence and presence of streptavidin (Fig. 5). To determine the cross-over position in F-actin, we applied digital processing to raw micrographs of F-actin (Fig. 5 A–C). For the digital processing, we created a bandpass filter (SI Appendix, Fig. S8). The bandpass filter enables identification of the cross-over position of the double-strand F-actin structure (2 in Fig. 5 B and C and SI Appendix, SI Materials and Methods). We measured the average cross-over length between two precisely identifiable cross-over positions flanking 4–10 cross-overs. The average cross-over length in the presence of streptavidin, 37.7 ± 3.2 nm, was longer than that in the absence of streptavidin, 37.1 ± 2.7 nm (Fig. 5D). The fractions more than 39.5 nm, the upper limit of the 95% confidence interval in the absence of streptavidin, were 13.1 and 4.7% in the presence and absence of streptavidin, respectively. Under this condition, helical rotation increased the length of actin cross-overs by only up to 10%. This change is less marked than 75% shortening of cross-over length in cofilin-decorated F-actin (19). Nevertheless, this subtle change can interfere with cofilin-induced severing (Fig. 1). These results indicate that helical rotation of mDia1 twists the helical structure of F-actin in the opposite direction of the cofilin-induced twist when mDia1 and the pointed end side are immobilized.
Fig. 5.
Helical rotation of mDia1 untwists the helical structure of F-actin when mDia1 and the pointed end side are immobilized. (A) Representative micrograph of a negatively stained F-actin elongated from an mDia1 aggregate in the presence of streptavidin. The black area on the left side of the image is an mDia1 aggregate. The rectangle indicates the area for digital processing in B. (Scale bar: 200 nm.) (B) Digital processing of electron micrographs. An extracted F-actin image was straightened (unbent) and then filtered in the Fourier space (1 and 2). The 1 image shows the filament after masking out the spatial frequencies >(30.0 nm)−1 in the Fourier space according to the method by Bremer et al. (57). The 2 image shows the filament filtered by a bandpass filter (SI Appendix, Fig. S8). In the 2 image, white lines indicate the position of cross-over. (Scale bar: 50 nm.) (C) Digital processing of electron micrographs of F-actin elongated from immobilized mDia1 aggregates in the absence of streptavidin. The procedure of digital processing is the same as B. (Scale bar: 50 nm.) (D) Box and whisker plot of cross-over length averaged from F-actin containing more than four consecutive cross-overs in the absence of streptavidin (−SA; n = 106) and the presence of streptavidin (+SA; n = 107). *Statistical significance (P < 0.01).
Discussion
In vitro, when F-actin elongating from immobilized mDia1 is tethered on the glass surface, helical rotation of mDia1 enhances resistance of the F-actin against the severing activity of cofilin. Helical rotation also abrogates the binding of cofilin with F-actin. Importantly, resistance of stuck F-actin against cofilin-induced severing is enhanced with an increase in the concentration of G-actin. TEM analysis shows that helical rotation of mDia1 twists F-actin in the opposite direction of cofilin-induced twist. Because elongation of stuck F-actin is gradually decelerated after the filament is trapped, mDia1 and its bound actin barbed end likely act as a rotary ratchet and generate torsional force as postulated in the Brownian ratchet theory. Our results show that the fate of F-actin is determined by the opposite actions of formins and cofilin on the twist of F-actin.
In cells, mDia1ΔC63, an active mutant that contains N-terminal domains, induces the formation of F-actin with prolonged lifetime. Furthermore, overexpression of mDia1ΔC63 accelerates dissociation of cofilin from F-actin. These effects are not induced by mDia1FH1-FH2 alone. F-actins are highly cross-linked with each other in cells as evidenced by the constant relative position of DL-actin SiMS along the retrograde actin flow (Movie S7) (16). Taken together with our in vitro results, these findings suggest that helical rotation of tethered mDia1 imposes torsional force on F-actin, which accelerates dissociation of cofilin, thereby inhibiting filament disassembly. This mechanism may perhaps contribute to the formation of actin stress fibers downstream of Rho signaling.
Cyclic cell stretch (39) and extracellular ligands (40), such as PDGF and lysophosphatidic acid, induce the remodeling and the formation of actin stress fibers through Rho signaling. ROCK, another Rho effector, enhances phosphorylation of myosin light chain (MLC) (41, 42). Phosphorylation of MLC evokes condensation of actin fibers (43) by enhancing actomyosin contractility (44). ROCK also phosphorylates LIM kinase, which inhibits actin disassembly by cofilin. Overexpression of LIMK1 induces the formation of stress fibers in a manner dependent on ROCK (45). Before this study, only ROCK had been implicated in inhibition of cofilin downstream of Rho signaling.
The N-terminal sequences of mDia bind to Rho and scaffold proteins, such as anillin (46) and liprin-α (47), and these proteins regulate subcellular localization of mDia. For example, Rho and anillin recruit mDia2 to the cleavage furrow during cytokinesis. This translocation of mDia2 is essential for the induction of actin-based contractile rings (46). Liprin-α negatively regulates the formation of actin stress fibers by displacing mDia1 from the plasma membrane (47). Thus, localization of mDia to the cell surface structures through its N-terminal sequence plays a pivotal role in the formation of Rho-induced actin structures. In cells, F-actin induced by mDia1ΔC63 showed higher resistance against filament disassembly than that induced by mDia1ΔN3, which does not contain the N-terminal sequences. Fractions of stationary and random speckles of mDia1ΔC63 were 13 and 6.8%, which are 2.7- and 67-fold higher than those of mDia1ΔN3, respectively. These results suggest that tethering mDia1 to cellular structures through its N-terminal sequences may contribute to the efficient formation of actin stress fibers.
In vitro, we found stabilization of F-actin against cofilin by helical rotation of mDia1. Suarez et al. (23) found that the region near the barbed end is not decorated with cofilin in polymerizing F-actin, which persists ∼90 s after assembly at the barbed end. Under faster elongation conditions, we also noticed that the barbed end region of mDia1ΔN3-assembled F-actin was poorly decorated by cofilin. Cofilin binding had a delay time of ∼36 s (SI Appendix, Fig. S9). In the data in Fig. 2B, only a narrow region (<0.73 μm on average) near the barbed end of untrapped F-actin is predicted to be devoid of cofilin, which was not clear at the optical resolution. The other three types of F-actin had the cofilin-free region less than 0.2 μm. Since the average length of untrapped F-actin at the time of severing was 7.8 μm, only a limited portion (9.4%) of untrapped F-actin could have been protected by this mechanism. The observed difference in the severing frequency is not due to the mechanism proposed by Suarez et al. (23).
In addition, cofilin accumulates in the portion ∼2 μm apart from the barbed end of untrapped F-actin. The rate of γ-phosphate (Pi) release from formin-bound F-actin is 0.0047 s−1, which is as slow as from native F-actin (48). Thus, the observed accumulation of cofilin does not arise from acceleration of Pi release. We postulate two possible mechanisms.
First, increased helical structural variability of F-actin might enhance the association of cofilin. In accordance with this possibility, F-actin untwisted by mDia1 exhibits a wider cross-over length distribution than pointed end-free F-actin in TEM analysis (Fig. 5D). mDia1 has been reported to increase flexibility of F-actin (25–27). However, these studies were performed using high concentrations of mDia1, which could lead to massive formation of short actin fragments. These studies might have detected the property of short actin segments near mDia1.
Second, cofilin might bind effectively during the transition from the FH2-bound actin structure to the typical F-actin structure. In the cocrystal with the Bni1p FH2 domain, actin dimers are arranged in the rotational symmetry of order 2 (49). However, F-actin elongating from mDia1 forms the typical F-actin helical repeat away from the barbed end (8, 50). In addition, while two major domains of actin subunit flatten during the G- to F-actin transition (51, 52), they rotate in the opposite direction on binding of cofilin (21). Structural transitions in F-actin might render accumulation of cofilin, which we speculate might be enhanced near the formin-bound barbed end. ATP–G-actin but not ADP–G-actin is capable of accelerating actin elongation by formins (8). It is also tempting to speculate that ATP hydrolysis might contribute to such structural transitions.
In summary, mDia1 assembles F-actin resistant to cofilin through helical rotation in vivo and in vitro. Our results provide additional support for the idea that twisting of F-actin plays an important role in cofilin-induced filament severing. Helical rotation-induced actin stabilization may play a pivotal role in the formation of high-order actin structure, such as actin stress fibers and actin cables. After bundled, mDia1-assembled F-actin may become resistant to the severing activity of ADF/coflin (24). Tropomyosin may also contribute to the actin fiber formation through increased affinity with formin-assembled F-actin (53). Our findings provide an important insight into our understanding of how the formation of diverse actin-based structures of different stabilities and dynamics is facilitated in cells.
Materials and Methods
Detailed experimental procedures used in this study are described in SI Appendix, SI Materials and Methods.
Measurement of the Rate of Cofilin-Induced Filament Severing with Native and mDia1-Assembled F-Actin.
Preparation of streptavidin-coated glass coverslip and immobilization of GST-tagged mDia1FH1-FH2 to the flow cell were performed according to the previous method (8, 54). For actin nucleation, immobilized mDia1 was incubated with 1.5 μM DyLight488-labeled actin (DL488-actin, 3.1% labeled) and 45 nM biotinylated actin in the Basic buffer (50 mM KCl, 10 mM imidazole-HCl, pH 7.0, 1 mM MgCl2, 1 mM EGTA, 50 μM CaCl2, 0.5% methylcellulose, 100 μg/mL glucose oxydase, 20 μg/mL catalase, 4.5 mg/mL glucose, 2 mM ATP). After 3 min, the flow cell was perfused with the Severing buffer containing 0.1, 0.2, or 0.5 μM DL488-actin and 3 or 5 μM profilin. The Severing buffer is the same as the Basic buffer, except that 10 mM imidazole-HCl (pH 7.0) is replaced with 10 mM Tris⋅HCl (pH 8.0). After 3 min, 50 nM Xac2 in the Severing buffer containing the same concentrations of DL488-actin and profilin was perfused to the flow cell to initiate filament severing. The severing frequency was calculated by the initial and final lengths of each filament and its severing-free duration after the addition of Xac2. DL488-actin images were acquired using an Olympus total internal reflection fluorescence (TIRF) microscope equipped with CoolSNAP HQ (Roper Scientific) at 5-s intervals. Images were analyzed using ImageJ and the JFilament plug-in (55).
In Vitro Binding of Cofilin with Native and mDia1-Assembled F-Actin.
Actin was nucleated by immobilized mDia1 with 1.5 μM DyLight550-labled actin (DL550-actin, 2.7% labeled) and 45 nM biotinylated actin in the Basic buffer. After 3 min, the flow cell was perfused with 0.1 μM DL550-actin and 3 μM profilin in the Binding buffer. Binding buffer is the same as the Basic buffer, except that 10 mM imidazole-HCl (pH 7.0) is replaced with 10 mM Piperazine-1,4-bis(2-ethanesulfonic acid)-KOH (pH 6.8). After 3 min, 2 μM mouse cofilin (10% cofilin-EGFP) in the Binding buffer containing the same concentrations of DL550-actin and profilin was perfused to the flow cell. Images of DL550-actin and cofilin-EGFP were acquired using TIRF microscopy at 5-s intervals.
Analysis of the Fraction of mDia1 and Its Mutants Bound to Cellular Structures.
SiMS imaging of mDia1 and its mutants was performed as described (7, 34, 36, 37). We calculated the fraction of cell structure-bound mDia1 by dividing the number of mDia1 speckles with the number of expressed EGFP-mDia1 in the measurement area. The number of expressed mDia1 molecules was calculated by dividing the total fluorescence intensity of EGFP in the measurement area by the average fluorescence intensity of single-molecule EGFP-mDia1 dimers that stuck on the glass surface outside of the cell area. We classified speckles of mDia1 and its mutants bound to the cellular structure according to the criteria in our previous report (36).
Lifetime Measurement of F-Actin in XTC Cells.
We introduced DL550-actin into XTC cells by electroporation using the Neon Transfection System (Invitrogen) (16). DL550-actin was excited by a mercury lamp through the 0.75% neural density (ND) filter and acquired at 2-s intervals. Lifetime of incorporated DL550-actin was measured by SiMS imaging as described (16, 34, 37). Images were analyzed using ImageJ and the Speckle Tracker J plug-in (56). DL550-actin fluorescence was reduced by only 7% at 200 s under continuous mercury excitation of the whole-cell area through the 0.75% ND filter. Therefore, the data shown for lifetime distribution of DL550-actin are without normalization for photobleaching. We calculated the cumulative survival chance of DL550-actin incorporated in the cellular F-actin (Fig. 3C) from the lifetime distribution data (SI Appendix, Fig. S5) using
where S(i) is the cumulative survival chance of DL550-actin at the ith frame, i and j are the frame numbers after the appearance of DL550-actin speckles, nj is the number of speckles with lifetime of j frames, and N is the total number of measured speckles.
Regression Analysis of Cofilin-EGFP SiMS in Cells Overexpressing mDia1.
To observe cofilin-EGFP in control and mDia1-expressing cells (Fig. 4), we introduced recombinant cofilin-EGFP using electroporation after transfection. We first introduced the plasmid encoding mCherry-mDia1ΔN3 or mCherry-mDia1ΔC63 using polyethylenimin (34). Several days later, cells were collected and suspended at a density of 1.86 × 107 cells per 1 mL with buffer R containing 0.5 μM cofilin-EGFP. Electroporation was carried out using the Neon Transfection System (Invitrogen) (16). After electroporation, cells were washed with serum-free 70% Leibovitz’s L15 medium and spread on poly-l-lysine–coated glass coverslips. We carried out regression analysis on the surviving fraction of preexisting cofilin SiMS as described (37).
TEM Observation and Digital Processing of Micrograph.
EM sample preparation is described in SI Appendix, SI Materials and Methods. EM was performed using a Hitachi H-7650 electron microscope operated at an accelerating voltage of 80 kV. Micrographs were recorded under low-dose condition at 50,000× nominal magnification on an AMT XR-41C CCD camera system (Advanced Microscopy Techniques). Acquired images were cropped, straightened, and then low pass-filtered at >(30.0 nm)−1 in the Fourier space (1 in Fig. 5) according to the method by Bremer et al. (57). Separately, straightened filaments were filtered by a bandpass filter (2), which passes regions around typical three-layer lines on the F-actin diffraction pattern (SI Appendix, Fig. S8). Using images of 2, we determined the position of cross-over (SI Appendix, SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank Haruyasu Kohda and Keiko Okamoto-Furuta (Division of Electron Microscopic Study, Center for Anatomical Studies, Kyoto University Graduate School of Medicine) for skillful assistance in EM. This work was supported by Japan Society for the Promotion of Science KAKENHI Grant 24770175 (to H.M.), Japan Science and Technology Agency CREST Grant JPMJCR15G5 (to N.W.), and the Uehara Memorial Foundation (N.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803415115/-/DCSupplemental.
References
- 1.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 2.Evangelista M, Zigmond S, Boone C. Formins: Signaling effectors for assembly and polarization of actin filaments. J Cell Sci. 2003;116:2603–2611. doi: 10.1242/jcs.00611. [DOI] [PubMed] [Google Scholar]
- 3.Watanabe S, et al. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watanabe N, Kato T, Fujita A, Ishizaki T, Narumiya S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat Cell Biol. 1999;1:136–143. doi: 10.1038/11056. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe N. Inside view of cell locomotion through single-molecule: Fast F-/G-actin cycle and G-actin regulation of polymer restoration. Proc Jpn Acad Ser B Phys Biol Sci. 2010;86:62–83. doi: 10.2183/pjab.86.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42:486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- 7.Higashida C, et al. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303:2007–2010. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- 8.Mizuno H, et al. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011;331:80–83. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- 9.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peskin CS, Odell GM, Oster GF. Cellular motions and thermal fluctuations: The Brownian ratchet. Biophys J. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bamburg JR. Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 12.Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuji T, Miyoshi T, Higashida C, Narumiya S, Watanabe N. An order of magnitude faster AIP1-associated actin disruption than nucleation by the Arp2/3 complex in lamellipodia. PLoS One. 2009;4:e4921. doi: 10.1371/journal.pone.0004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moon AL, Janmey PA, Louie KA, Drubin DG. Cofilin is an essential component of the yeast cortical cytoskeleton. J Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulholland J, et al. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamashiro S, et al. New single-molecule speckle microscopy reveals modification of the retrograde actin flow by focal adhesions at nanometer scales. Mol Biol Cell. 2014;25:1010–1024. doi: 10.1091/mbc.E13-03-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Hawkins M, Pope B, Maciver SK, Weeds AG. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 1993;32:9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- 19.McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGough A, Chiu W. ADF/cofilin weakens lateral contacts in the actin filament. J Mol Biol. 1999;291:513–519. doi: 10.1006/jmbi.1999.2968. [DOI] [PubMed] [Google Scholar]
- 21.Galkin VE, et al. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci USA. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCullough BR, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J. 2011;101:151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suarez C, et al. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michelot A, et al. Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr Biol. 2007;17:825–833. doi: 10.1016/j.cub.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 25.Kupi T, Gróf P, Nyitrai M, Belágyi J. The uncoupling of the effects of formins on the local and global dynamics of actin filaments. Biophys J. 2009;96:2901–2911. doi: 10.1016/j.bpj.2008.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papp G, et al. Conformational changes in actin filaments induced by formin binding to the barbed end. Biophys J. 2006;91:2564–2572. doi: 10.1529/biophysj.106.087775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bugyi B, et al. Formins regulate actin filament flexibility through long range allosteric interactions. J Biol Chem. 2006;281:10727–10736. doi: 10.1074/jbc.M510252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- 30.Welnhofer EA, Zhao L, Cohan CS. Calcium influx alters actin bundle dynamics and retrograde flow in Helisoma growth cones. J Neurosci. 1999;19:7971–7982. doi: 10.1523/JNEUROSCI.19-18-07971.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Q, Pollard TD. Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. J Cell Biol. 2011;195:485–498. doi: 10.1083/jcb.201103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okreglak V, Drubin DG. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J Cell Biol. 2007;178:1251–1264. doi: 10.1083/jcb.200703092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De La Cruz EM. Cofilin binding to muscle and non-muscle actin filaments: Isoform-dependent cooperative interactions. J Mol Biol. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe N. Fluorescence single-molecule imaging of actin turnover and regulatory mechanisms. Methods Enzymol. 2012;505:219–232. doi: 10.1016/B978-0-12-388448-0.00020-6. [DOI] [PubMed] [Google Scholar]
- 35.Higashida C, et al. F- and G-actin homeostasis regulates mechanosensitive actin nucleation by formins. Nat Cell Biol. 2013;15:395–405. doi: 10.1038/ncb2693. [DOI] [PubMed] [Google Scholar]
- 36.Higashida C, et al. G-actin regulates rapid induction of actin nucleation by mDia1 to restore cellular actin polymers. J Cell Sci. 2008;121:3403–3412. doi: 10.1242/jcs.030940. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- 38.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 39.Kaunas R, Nguyen P, Usami S, Chien S. Cooperative effects of Rho and mechanical stretch on stress fiber organization. Proc Natl Acad Sci USA. 2005;102:15895–15900. doi: 10.1073/pnas.0506041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 41.Kimura K, et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 42.Kureishi Y, et al. Rho-associated kinase directly induces smooth muscle contraction through myosin light chain phosphorylation. J Biol Chem. 1997;272:12257–12260. doi: 10.1074/jbc.272.19.12257. [DOI] [PubMed] [Google Scholar]
- 43.Ishizaki T, et al. p160ROCK, a Rho-associated coiled-coil forming protein kinase, works downstream of Rho and induces focal adhesions. FEBS Lett. 1997;404:118–124. doi: 10.1016/s0014-5793(97)00107-5. [DOI] [PubMed] [Google Scholar]
- 44.Uehata M, et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 45.Maekawa M, et al. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe S, et al. Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol Biol Cell. 2010;21:3193–3204. doi: 10.1091/mbc.E10-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakamoto S, et al. Liprin-α controls stress fiber formation by binding to mDia and regulating its membrane localization. J Cell Sci. 2012;125:108–120. doi: 10.1242/jcs.087411. [DOI] [PubMed] [Google Scholar]
- 48.Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19, and correction (2008) 18:233. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 50.Mizuno H, Watanabe N. mDia1 and formins: Screw cap of the actin filament. Biophysics (Nagoya-shi) 2012;8:95–102. doi: 10.2142/biophysics.8.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 52.Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 53.Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20:2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizuno H, Watanabe N. Rotational movement of formins evaluated by using single-molecule fluorescence polarization. Methods Enzymol. 2014;540:73–94. doi: 10.1016/B978-0-12-397924-7.00005-4. [DOI] [PubMed] [Google Scholar]
- 55.Smith MB, et al. Segmentation and tracking of cytoskeletal filaments using open active contours. Cytoskeleton (Hoboken) 2010;67:693–705. doi: 10.1002/cm.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith MB, et al. Interactive, computer-assisted tracking of speckle trajectories in fluorescence microscopy: Application to actin polymerization and membrane fusion. Biophys J. 2011;101:1794–1804. doi: 10.1016/j.bpj.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bremer A, et al. The structural basis for the intrinsic disorder of the actin filament: The “lateral slipping” model. J Cell Biol. 1991;115:689–703. doi: 10.1083/jcb.115.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.