Roots of land plants grow through soil, one of the most microbially rich and diverse environments on earth (1). A long history of work has revealed that plant roots interact with these microbes and help establish microbial communities distinct from the surrounding soil (2). This is partly because roots provide a food source of exuded sugars, metabolites, and dead plant material, which some microbes are better suited to exploit than others (3). It is also partly because the root immune response and secreted antimicrobials provide obstacles to colonization, to which only some microbes are tolerant (4). Thus, roots control microbes not only with “carrots” but also with “sticks.” Because plant genetic variation affects root structure, composition, and exudates, many studies have tried to address the extent of genetic control plants have over their associated root microbiomes (5). These studies have generally shown that plant genotype has an effect on the root microbiome but that this is a small influence relative to environmental effects such as soil composition (4). Much of the work ends here, because the next steps are even more difficult. They involve identifying specific genes whose products interact with root-associated microbes, demonstrating conclusively the relevance of those interactions, and explaining how the genes defined function. The paper in PNAS by Stringlis et al. (6) represents a significant step because it does all these things. The authors use a variety of techniques, from quantifying root exudates to metagenome sequencing in natural soil, to show that an exuded coumarin involved in the plant’s iron starvation response reshapes the microbiome and may also protect plants from pathogenic fungi. Furthermore, this effect is triggered by beneficial bacteria.
Many microbes that are able to colonize roots exert some kind of beneficial effect on the plant, often improving traits of obvious agronomic value such as nutrition, growth, and/or immunity (7). Some beneficial members of the root microbiome can reduce disease caused by foliar pathogens by triggering a defense mechanism known as induced systemic resistance (ISR). The bacterium Pseudomonas simiae WCS417 is probably the most well-characterized microbe capable of eliciting ISR. Although the beneficial effects of disease-suppressive bacteria can, in some instances, be explained by a direct competition for nutrients with pathogens (8, 9), many of them act indirectly by priming plant immunity. A set of important studies published in 1991 demonstrated the ability of rhizobacteria to induce systemic protection (10–12). In particular, van Peer et al. (10) found that the treatment of carnation roots with WCS417 reduced the incidence and severity of infection by the fungal pathogen Fusarium oxysporum in leaves. In their experiment, WCS417 remained spatially separated from the challenging pathogen, implying that a systemic defense mechanism in plants could be triggered by bacteria in the root. Since then, this protective effect (namely ISR) has been observed in multiple plant-pathogen combinations, and a number of studies have provided a wealth of knowledge on how this and other beneficial microbes may promote plant resistance against pathogens (13). However, there is still much to learn.
Large-scale gene expression analyses catalyzed the identification of key players in the establishment of ISR. In the absence of pathogens, leaves of Arabidopsis plants whose roots were treated with WCS417 did not show significant transcriptional alterations. On the other hand, approximately 100 genes were differentially regulated in roots colonized by WCS417 (14). Among these, the gene encoding the root-specific transcription factor MYB72 was induced upon colonization, and subsequently shown to be required for the onset of ISR (15). More recently, the β-glucosidase BGLU42, whose transcript levels are regulated by MYB72, also emerged as a required component of ISR (16). The specific functions of MYB72 and BGLU42 in the context of plant immunity are just starting to emerge. Interestingly, MYB72 is also part of the circuitry that controls the Arabidopsis response to iron starvation (17). When growing in iron-limited soils, plants activate a set of coordinated responses to optimize iron uptake and mitigate nutritional stress (18). Iron deprivation also stimulates the exudation of Fe(III)-chelating coumarins in the rhizosphere to help the mobilization of iron (19). The dual role of MYB72 in the regulation of both ISR and iron starvation responses suggested the existence of functional links between immunity and nutrition, a concept recently demonstrated for phosphate (20).
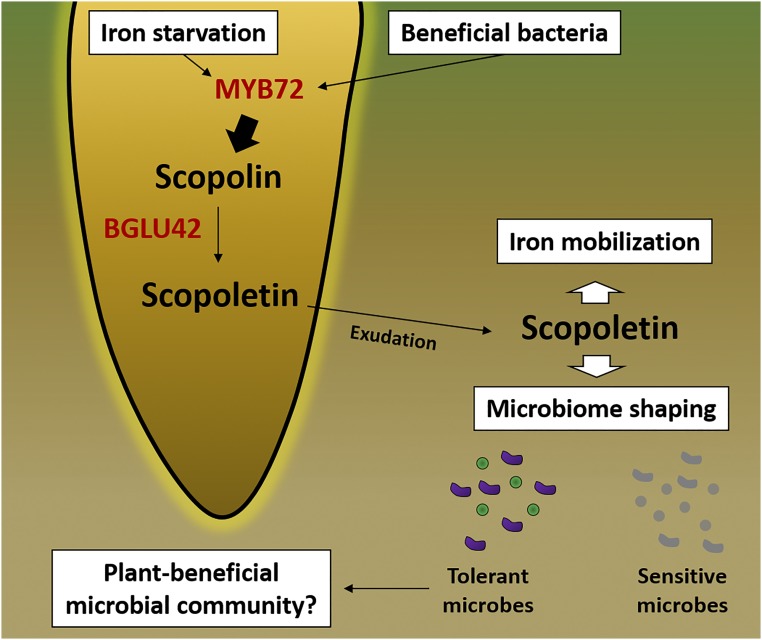
In PNAS, Stringlis et al. (6) show that MYB72 and BGLU42 activities influence the assembly of the Arabidopsis root microbiome by regulating the biosynthesis and exudation of the coumarin scopoletin into the rhizosphere (Fig. 1). Supported by robust genetics, the authors performed a metabolomics survey to identify metabolites produced and secreted by iron-starved roots in a MYB72- and BGLU42-dependent manner. This led to the identification of scopoletin as a major component of the root exudate during iron starvation, which agrees with previous reports and is consistent with the role of coumarins in iron mobilization and uptake (19). As a transcription factor, MYB72 is presumably a major regulator of genes whose products catalyze the biosynthesis of scopoletin (including BGLU42), whereas BGLU42 mediates scopoletin secretion into the rhizosphere by modifying its precursor.
Fig. 1.
Model for the dual action of MYB72 and BGLU42 in iron mobilization and in shaping the root microbiome. The transcription factor MYB72 and the β-glucosidase BGLU42 are both required for the exudation of the coumarin scopoletin by Arabidopsis roots. Biosynthesis and secretion of scopoletin are induced under iron-limiting conditions or by beneficial root-colonizing bacteria such as P. simiae WCS417. Secreted scopoletin is proposed to increase iron mobilization and uptake by the roots. Moreover, scopoletin possess antimicrobial activity, and its accumulation in the rhizosphere may select tolerant microbes with beneficial effects to the plant while suppressing potentially negative microbes, including pathogens. Further research is required to elucidate the significance of scopoletin-selected microbial communities.
A remarkable feature of the paper by Stringlis et al. (6) is the demonstration that root-secreted scopoletin exerts a selective antimicrobial action in the rhizosphere. Rhizosphere metagenome sequencing revealed an altered microbial community surrounding roots of iron-starved plants. A mutant deficient in scopoletin biosynthesis also showed differential abundance of specific microbes. Based on these results, the authors propose that plants can recruit a specific set of coumarin-tolerant microbes during iron starvation by secreting scopoletin. Critically, the authors also show that the beneficial bacteria P. simiae WCS417 and Pseudomonas capeferrum WCS358 are resistant to high doses of scopoletin, whereas the fungal pathogens Fusarium oxysporum f. sp. raphani and Verticillium dahliae JR2 are strongly inhibited by this coumarin. Moreover, WCS417 itself can induce the exudation of scopoletin by the roots. This suggests the exciting possibility that beneficial microbes induce exudation of coumarins into the rhizosphere, thus establishing a favorable niche for its colonization and, in turn, making the plant better adapted against stresses.
The paper by Stringlis et al. (6) offers at once both a broad and detailed view of root microbiome assembly. Not only does it show the effect of a single plant gene and its exuded product on the microbial community but it reveals how beneficial and pathogenic microbes react differently to this challenge, demonstrates how the process is started by a beneficial microbe, and reinforces how plants may repurpose the same stress pathway to effectively respond to abiotic and biotic stress. The paper provides an example of mechanistic microbiome research, revealing how particular genes and particular microbes fit into the complexity of a natural root microbial community. In the future, it will be interesting to explore the limits of the mechanism Stringlis et al. (6) have discovered, particularly whether pathogens have the same forms of coumarin resistance possessed by beneficial bacteria such as WCS417 and, as the authors note, whether coumarin clears the rhizosphere of competing microbes to give coumarin-resistant microbes a competitive advantage. This might open the path for engineering coumarin resistance into desirable rhizosphere microbes to improve their ability to colonize.
Acknowledgments
D.S.L. was supported by the Human Frontier Science Program (Grant LT000565/2015). P.J.P.L.T. was supported by the Pew Latin American Fellows Program in the Biomedical Sciences (Grant 00026198) and by National Science Foundation Interdisciplinary Research and Education (INSPIRE) Grant IOS-1343020 to Jeffery L. Dangl (University of North Carolina at Chapel Hill).
Footnotes
The authors declare no conflict of interest.
See companion article on page E5213.
References
- 1.Howe AC, et al. Tackling soil diversity with the assembly of large, complex metagenomes. Proc Natl Acad Sci USA. 2014;111:4904–4909. doi: 10.1073/pnas.1402564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sánchez-Cañizares C, Jorrín B, Poole PS, Tkacz A. Understanding the holobiont: The interdependence of plants and their microbiome. Curr Opin Microbiol. 2017;38:188–196. doi: 10.1016/j.mib.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Zhalnina K, et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol. 2018;3:470–480. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 4.Doornbos RF, van Loon LC, Bakker PA. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron Sustain Dev. 2012;32:227–243. [Google Scholar]
- 5.Fitzpatrick CR, et al. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc Natl Acad Sci USA. 2018;115:E1157–E1165. doi: 10.1073/pnas.1717617115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stringlis IA, et al. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci USA. 2018;115:E5213–E5222. doi: 10.1073/pnas.1722335115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 8.Raaijmakers JM, et al. Dose-response relationships in biological control of fusarium wilt of radish by Pseudomonas spp. Phytopathology. 1995;85:1075–1080. [Google Scholar]
- 9.Lemanceau P, Bakker PA, De Kogel WJ, Alabouvette C, Schippers B. Effect of pseudobactin 358 production by Pseudomonas putida WCS358 on suppression of fusarium wilt of carnations by nonpathogenic Fusarium oxysporum Fo47. Appl Environ Microbiol. 1992;58:2978–2982. doi: 10.1128/aem.58.9.2978-2982.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Peer R, Niemann G, Schippers B. Induced resistance and phytoalexin accumulation in biological control of Fusarium wilt of carnation by Pseudomonas sp. strain WCS 417 r. Phytopathology. 1991;81:728–734. [Google Scholar]
- 11.Alström S. Induction of disease resistance in common bean susceptible to halo blight bacterial pathogen after seed bacterization with rhizosphere pseudomonads. J Gen Appl Microbiol. 1991;37:495–501. [Google Scholar]
- 12.Wei G, Kloepper JW, Tuzun S. Induction of systemic resistance of cucumber to Colletotrichum orbiculare by select strains of plant growth-promoting rhizobacteria. Phytopathology. 1991;81:1508–1512. [Google Scholar]
- 13.Pieterse CM, et al. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 14.Verhagen BW, et al. The transcriptome of rhizobacteria-induced systemic resistance in arabidopsis. Mol Plant Microbe Interact. 2004;17:895–908. doi: 10.1094/MPMI.2004.17.8.895. [DOI] [PubMed] [Google Scholar]
- 15.Van der Ent S, et al. MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 2008;146:1293–1304. doi: 10.1104/pp.107.113829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zamioudis C, Hanson J, Pieterse CM. β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 2014;204:368–379. doi: 10.1111/nph.12980. [DOI] [PubMed] [Google Scholar]
- 17.Palmer CM, Hindt MN, Schmidt H, Clemens S, Guerinot ML. MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet. 2013;9:e1003953. doi: 10.1371/journal.pgen.1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer CM, Guerinot ML. Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat Chem Biol. 2009;5:333–340. doi: 10.1038/nchembio.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmid NB, et al. Feruloyl-CoA 6′-Hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol. 2014;164:160–172. doi: 10.1104/pp.113.228544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castrillo G, et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]