Abstract
Multiple genes have been implicated in Parkinson disease pathogenesis, but the relationship between regional expression of these genes and regional dysfunction across the brain is unknown. We address this question by joint analysis of high resolution magnetic resonance imaging data from the Parkinson's Progression Markers Initiative and regional genetic microarray expression data from the Allen Brain Atlas. Regional brain atrophy and genetic expression was co-registered to a common 86 region brain atlas and robust multivariable regression analysis was performed to identify genetic predictors of regional brain atrophy. Top candidate genes from GWAS analysis, as well as genes implicated in trans-synaptic alpha-synuclein transfer and autosomal recessive PD were included in our analysis. We identify three genes with expression patterns that are highly significant predictors of regional brain atrophy. The two most significant predictors are LAG3 and RAB5A, genes implicated in trans-synaptic synuclein transfer. Other well-validated PD-related genes do not have expression patterns that predict regional atrophy, suggesting that they may serve other roles such as disease initiation factors.
Keywords: Parkinson disease, LAG3, Alpha-synuclein, Trans-synaptic spread, Atrophy, Genetic analysis
Highlights
-
•
Joint volumetric and microarray analysis identifies gene expression patterns that predict the PD atrophy pattern.
-
•
The most highly predictive genes, LAG3 and RAB5A, are implicated in trans-synaptic alpha-synuclein transfer.
-
•
The expression patterns of alpha-synuclein and otherPD-related genes do not predict atrophy.
1. Introduction
Parkinson disease (PD) is a common neurodegenerative disease affecting an estimated 630,000 people in the United States (Kowal et al., 2013). Neural circuit dysfunction in PD is pronounced throughout the brain, leading to motor deficits, as well as symptoms that include depression and visuospatial disturbances (Chaudhuri et al., 2006). The advent of functional neurosurgery in PD has led to improved outcomes for some patients with PD (Merola et al., 2014). However, pharmacological therapeutic strategies remain limited to dopamine replacement; an approach introduced more than 50 years ago (Przedborski, 2017). New treatment paradigms are needed, but our understanding of how to develop them is still limited.
One promising approach is to target genes that have been implicated in PD either by large genome wide association studies (GWAS) in humans or by experimental studies in animal models. Variation in multiple genes, including alpha-synuclein (SNCA) and tau (MAPT), increases PD risk in GWAS studies (Nalls et al., 2014). Many additional lines of evidence have established a role for synuclein in PD pathogenesis and progression. In addition to its role as a risk factor in sporadic PD, genetic variants of synuclein cause rare forms of familial PD (Chartier-Harlin et al., 2004). A leading hypothesis postulates that transfer of synuclein between cells explains both the anatomical and temporal progression of PD-related circuit dysfunction and atrophy (Desplats et al., 2009; Luk et al., 2012a, Luk et al., 2012b).
A recent study by Mao et al. (2016) identified four proteins that may mediate trans-synaptic transfer of synuclein in a mouse model of PD. Deletion of one of them, lymphocyte activating gene 3 (LAG3) which directly binds synuclein preformed fibrils, prevents substantia nigra neuron death and corresponding behavioral deficits. Given these findings, we sought to determine whether the regional expression of the genes involved in synuclein transfer in Mao et al., as well as ten top candidate genes (of 24 genes with significant genome-wide association) identified from the PD GWAS meta-analysis by Nalls et al. (2014), and the three genes strongly associated with the typical form of autosomal recessive PD (Bonifati, 2012), can predict the regional pattern of atrophy in PD. These genes do not comprise a complete list of genes implicated in PD, but do represent a set of genes with high a priori probability of being associated with regional PD atrophy.
To investigate the relationship between regional genetic expression and atrophy, we utilized two large well-validated projects with high quality regional imaging and genetic data. We derived regional PD atrophy patterns from the magnetic resonance imaging (MRI) component of the Parkinson's Progression Markers Initiative (PPMI) (Marek et al., 2011) and regional microarray expression data from the Allen Brain Atlas (ABA) (Shen et al., 2012; Goel et al., 2014). As gene expression data is derived from healthy normal subjects, we specifically address whether expression patterns in the normal healthy brain predict PD atrophy. Such expression patterns may represent vulnerability networks or conduits by which PD pathogenesis can occur in the presence of the appropriate initiating factors (Raj et al., 2012; Seeley et al., 2009). Using a similar approach, Romme and colleagues recently showed that expression of schizophrenia-related genes is related to the pattern of cortical disconnectivity (Romme et al., 2017), suggesting that relationships between regional genetic expression and brain atrophy in PD may be detectable with our analysis.
Consistent with our hypothesis, we find that two genes implicated in synuclein transfer, LAG3 and RAB5A, demonstrate expression patterns that are highly predictive of PD regional atrophy. Furthermore, high expressing LAG3 regions in the occipital cortex exhibit markedly more atrophy in patients with hallucinations, suggesting that this gene may be particularly important in mediating visuospatial disturbances that are often present in PD.
2. Materials and methods
2.1. Regional volumetric analysis
Data used in the preparation of this article were obtained from the Parkinson's Progression Markers Initiative (PPMI) database (www.ppmi-info.org/data). Whole brain PPMI MRI data was utilized for PD (n = 149 subjects, n = 378 MR studies) and healthy control (HC, n = 64 subjects, n = 127 MR studies) cohorts. Basic subject meta-data is shown in Supplementary Table 1. Additional details regarding these subjects, including study inclusion and exclusion criteria, are available at the PPMI website (www.ppmi-info.org/data). Sagittal 3D T1-weighted MR images were obtained with MPRAGE or SPGR sequences with the following parameters: slice thickness 1.2 mm, slice gap 0 mm, voxel size 1 mm × 1 mm × 1.2 mm, and matrix 256 × 256 × 170–200. Data were acquired at PPMI centers using scanners from three different manufacturers (GE, Siemens, and Philips). Repetition (TR) and echo (TE) time followed manufacturer recommendations for the relevant sequences. MR images were aligned to an 86 region cortical (Desikan et al., 2006) and subcortical FreeSurfer (Fischl, 2012) atlas as described by Desikan et al. (2006), and regional volumes were computed using FreeSurfer version 5.1. All analysis was performed with this atlas, and other brain parcellation schemes were not used. MRI scans that did not pass FreeSurfer quality control were excluded from analysis. For this purpose initial FreeSurfer segmentation outputs were rated as “Pass”, “Partial” or “Fail” by the operator based on visual inspection in the tkmedit tool, as well as the numerical quality flag returned by the FreeSurfer software. Scans that rated either Partial or Fail were excluded from analysis. Regional volumes were normalized by total intracranial volume. Regional atrophy in PD subjects was quantified by the regional atrophy T-score. For healthy cohort regional volume x and PD cohort regional volume y, with number of samples n and m, and with variances σx2 and σy2, the T-score s is given by the equation . A violin plot of T-scores was computed and displayed using the Matlab routine violin (Holger Hoffman; www.mathworks.com/matlabcentral/fileexchange/45134-violin-plot).
2.2. Regional gene expression analysis
17 genes of interest implicated in PD were selected for whole brain expression analysis. The four genes identified as having roles in trans-synaptic synuclein spread in Mao et al. (2016) were analyzed (LAG3, APLP1, NRXN1 and RAB5A). The ten genes with the lowest p-values from the Nalls et al. GWAS meta-analysis for which ABA expression data was available were included: BST1, GBA, GPNMB, HLA-DQB1, LRRK2, MAPT, NUCKS1, SNCA, STK39 and TMEM175. Genetic data for MCC1 and TMEM229B was not available. The three genes strongly associated with typical autosomal recessive PD (Bonifati, 2012) were also included (PINK1, PARK2 and PARK7). Comparison models were created from 51 Alzheimer disease related genes, comprising: ADAM10, ADRA1D, ADRA1A, ADRA2A, ADRA2B, ADRB1, ADRBK2, AP2A2, AP2B1, ALDH1A1, ALDH2, APOE, APP, BDNF, AP2M1, AP2S1, CRYAB, DBH, FKBP5, FOS, GSK3A, GSK3B, DNAJA1, HSPA1A, HSPA1L, HSPA5, HSPB1, JUN, MAOA, MAOB, MAPT, MTF1, PDGFA, PDGFB, PDGFRA, PDGFRB, PRNP, SLC6A2, THY1, ALDH1A2, BAG3, HS3ST2, STUB1, ALDH1L1, AP005668.1, CDC37, BACE1, HSPB8, CDC37L1, PDGFC, and PDGFD (Acosta et al., 2018). For each gene, data was obtained from the publicly available human ABA (Hawrylycz et al., 2012). The ABA includes 926 brain regions, with each region having microarray expression levels from a set of 58,692 probes that correspond to 29,181 distinct genes. The 926 brain regions were mapped to the 86 region Desikan FreeSurfer atlas. As the ABA and Desikan atlas are defined in the same stereotactic space, this was done by collecting all sample locations from ABA that fell within the same Desikan region, using the sample coordinates supplied by the ABA. ABA samples within a 1 voxel margin of gray matter were assigned to the nearest gray matter region. Given the voxel size of 1 mm × 1 mm × 1.2 mm, the maximum distance to a gray matter region of samples assigned in this way is equal to the length of the voxel diagonal, or 1.85 mm. No cortical samples were assigned to subcortical regions. All samples within the same region were then averaged for each gene. Despite differing probe characteristics, a well validated probe weighting procedure has not been identified, and we chose to simply average all probes corresponding to a single gene. White matter tracts were not mapped and were excluded from analysis as done in previous research that successfully linked gene expression to AD vulnerability (Freer et al., 2016). Expression for each gene was averaged for six subject brains. Of these six, four had data from the left hemisphere while the other two had data for both hemispheres. More information on normalization across brain samples can be found at help.brain-map.org/download/attachments/2818165/Normalization_WhitePaper.pdf.
2.3. Univariate correlation analysis
Univariate correlation of regional gene expression with regional atrophy was quantified by the Pearson correlation coefficient. Multiple comparison correction was performed with the Bonferroni method (17 gene comparisons, with threshold p-value = .05/17 = 2.9 × 10−3).
2.4. Robust multivariable regression analysis
Robust multivariable linear regression was performed with the Matlab routine robustfit with atrophy T-score as the dependent variable and average gene expression values (as z-scored expression across all probes and subjects in each brain region) as independent variables. Regression was performed across the entire 86 region atlas. Multiple comparison correction was performed with both Bonferroni method (17 gene comparisons, with threshold p-value = .05/17 = 2.9 × 10−3) and Benjamini-Hochberg method (false discovery rate 0.05). Each method yielded the same significant predictors. Simple multivariable linear regression was performed to estimate overall model R2, F, and p-value. Additional robust regression analysis was performed in the same way for left hemisphere data only.
For comparison to the proposed model, two types of model simulation were performed. Firstly, one million simulated 17 gene models were generated in which the PD-related gene z-scored expression data was randomly permuted across brain regions for each gene. Secondly, one million 17 gene models were derived from a set of 51 Alzheimer related genes. As for the PD-related genes, these AD gene models were normalized by z-scoring expression data for each gene separately. 17 unique genes were randomly selected from the set of 51 genes for each round of simulation. Simple multivariable linear regression was then performed to determine each model R2 and summary statistics were generated describing the distribution of simulated model R2.
2.5. LASSO regression analysis
LASSO regression (Tibshirani, 1996) was performed with the MATLAB routine lasso with 10 fold cross-validation. A simplified model was obtained by minimizing mean squared error by changing the tuning parameter lambda.
2.6. Differential atrophy of highest and lowest expressing regions
Each significant genetic expression predictor in the full regression model was analyzed by computing the average atrophy T-score for the regions ≤10th percentile of average gene expression (for that gene) and ≥90th percentile of average gene expression. The average T-scores for each percentile group were compared using an unpaired t-test.
2.7. Structural covariance analysis
Pairwise Pearson correlation coefficients across all average regional volumes in PD patients were computed. Correlation coefficients were confirmed to be described by a Gaussian distribution. Bonferroni correction was performed across all comparisons (threshold p-value = .05 / ((86 ∗ 85)/2) = 1.37 × 10−5). Correlation coefficients with p-values greater than the threshold p-value were considered to be not statistically significant and were set to 0. The average correlation coefficient across all brain regions was computed (i.e. the mean of the correlation matrix upper triangle, excluding autocorrelation values). The same analysis was performed considering regions with LAG3 expression greater 90th percentile, NUCKS1 expression greater than 90th percentile, and RAB5A regions with expression less than 10th percentile. Gene-specific average correlation coefficients were compared to whole brain average correlation coefficients with an unpaired t-test.
2.8. Behavioral analysis
Movement Disorder Society sponsored Unified Parkinson's Disease Rating Scale (MDS-UPDRS) (Goetz et al., 2008) data was obtained from the PPMI database. The PD cohort was separated into a group with a history of hallucinations and a group without a history of hallucinations. Atrophy T-scores (relative to the HC cohort) for the high expressing LAG3 regions were computed for each of these subgroups. Regional T-scores for subjects with hallucinations were divided by the regional T-scores for subjects without hallucinations to derive the relative T-score change. Average relative T-score changes were calculated for a group comprising the lingual gyrus and cuneus and for another group of other high expressing LAG3 regions excluding these regions. Average relative T-score changes were compared using an unpaired t-test.
3. Results
3.1. Regional atrophy
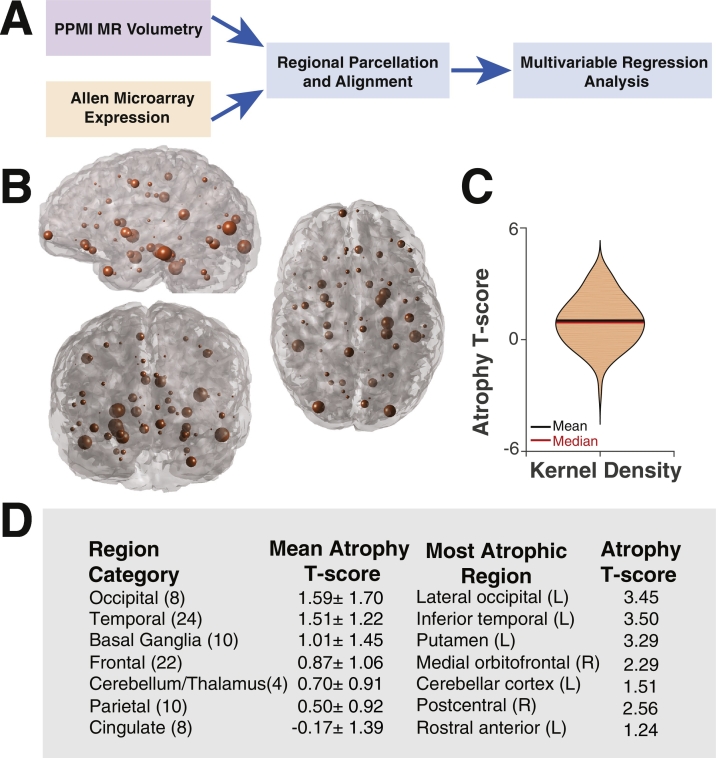
Regional atrophy in PD subjects was computed across an 86 region atlas with the ultimate goal of determining genetic predictors of the overall atrophy pattern (Fig. 1). The occipital lobe was the most atrophic area on average, followed by the temporal lobe and basal ganglia. The parietal and cingulate cortex demonstrated little atrophy.
Fig. 1.
Regional brain atrophy pattern in Parkinson disease. (A) Schematic illustrating the overall analysis approach using PPMI MR volumetric data and Allen Brain Atlas genetic expression data. (B) Glass brain representation of regional atrophy in which the radius of the sphere centered on each region is proportional to the regional atrophy T-score (n = 86 regions). (C) Violin plot illustrating the Gaussian distribution of atrophy T-scores. (D) Mean atrophy T-scores (±standard deviation) by brain region category and most atrophic regions within that category. Numbers in parentheses denote the number of brain regions in each category.
3.2. Regional gene expression analysis
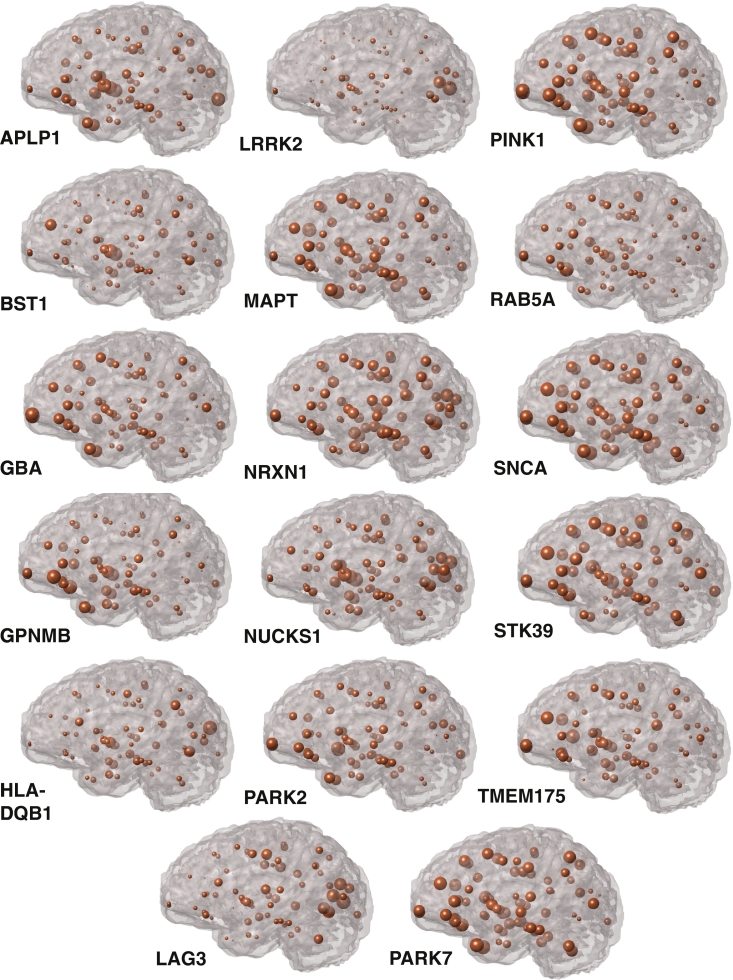
We aligned expression of each of the 17 genes of interest (Table 1) to the same atlas as that used for volumetric analysis. Three categories of genes are included, as shown in Table 1: four spreading synuclein genes from Mao et al. (2016), ten top GWAS hits (meta-analysis p-value ranging from ~10−82 to ~10−13) from Nalls et al. (2014), and the three genes associated with typical autosomal recessive PD (Bonifati, 2012). Regional expression for each gene is shown in Fig. 2. Distinct expression patterns are evident for this set of candidate genes indicating that while these genes may all be involved in PD risk or pathogenesis, there is considerable heterogeneity in gene expression across the brain.
Table 1.
Genes of interest. Symbols, names and analysis categories for each of the 17 genes of interest used in multivariable analysis.
| Symbol | Gene name | Category |
|---|---|---|
| APLP1 | Amyloid beta precursor like protein 1 | Spreading synuclein |
| BST1 | ADP ribosyl cyclase/cyclic ADP-ribose hydrolase 2 | GWAS |
| GBA | beta-Glucocerebrosidase | GWAS |
| GPNMB | Transmembrane glycoprotein NMB | GWAS |
| HLA-DQB1 | Major histocompatibility complex, class II, DQ beta 1 | GWAS |
| LAG3 | Lymphocyte activing gene 3 | Spreading synuclein |
| LRRK2 | Leucine rich repeat kinase 2 | GWAS |
| MAPT | Microtubule associated protein tau | GWAS |
| NRXN1 | Neurexin 1 | Spreading synuclein |
| NUCKS1 | Nuclear case in kinase and cyclin dependent kinase 1 | GWAS |
| PARK2 | Parkin RBR E3 ubiquitin protein ligase | Recessive |
| PARK7 | Parkinsonism associated deglycase | Recessive |
| PINK1 | PTEN induced putative kinase 1 | Recessive |
| RAB5A | Ras-related protein Rab-5a | Spreading synuclein |
| SNCA | Alpha- synuclein | GWAS |
| STK39 | Serine/threonine kinase 39 | GWAS |
| TMEM175 | Transmembrane protein 175 | GWAS |
Fig. 2.
Regional genetic expression profiles. For each of the 17 genes of interest, the regional genetic expression profile is shown with a sphere centered on each brain region with radius proportional to the normalized microarray expression value. Gene expression was normalized by the maximum expression value for that gene to allow for comparison of regional patterns across different genes.
3.3. Prediction of regional atrophy by genetic expression profiles
We next sought to determine whether regional expression of the genes of interest can predict regional atrophy. Univariate correlation between atrophy T-score and normalized gene expression was computed separately for each gene of interest. After correcting for multiple comparisons none of the univariate correlation coefficients was significant (Table 2), possibly suggesting that noise obscures underlying relationships between these variables or that the relationship between atrophy and genetic expression is more complex than can be elucidated by univariate analysis.
Table 2.
Univariate correlation between atrophy and genetic expression. Pearson correlation coefficients were computed separately for atrophy T-score and z-scored regional expression of each gene of interest. After multiple comparison correction, none of the univariate correlation coefficients are significant.
| Gene | Correlation coefficient | p-Value |
|---|---|---|
| APLP1 | 0.12 | .27 |
| BST1 | 0.03 | .81 |
| GBA | 0.01 | .95 |
| GPNMB | 0.07 | .51 |
| HLA-DQB1 | 0.003 | .98 |
| LAG3 | 0.13 | .24 |
| LRRK2 | −0.05 | .66 |
| MAPT | −0.03 | .78 |
| NRXN1 | 0.02 | .89 |
| NUCKS1 | 0.12 | .28 |
| PARK2 | 0.27 | .01 |
| PARK7 | 0.02 | .87 |
| PINK1 | −0.02 | .84 |
| RAB5A | −0.20 | .06 |
| SNCA | 0.02 | .82 |
| STK39 | −0.08 | .48 |
| TMEM175 | −0.003 | .98 |
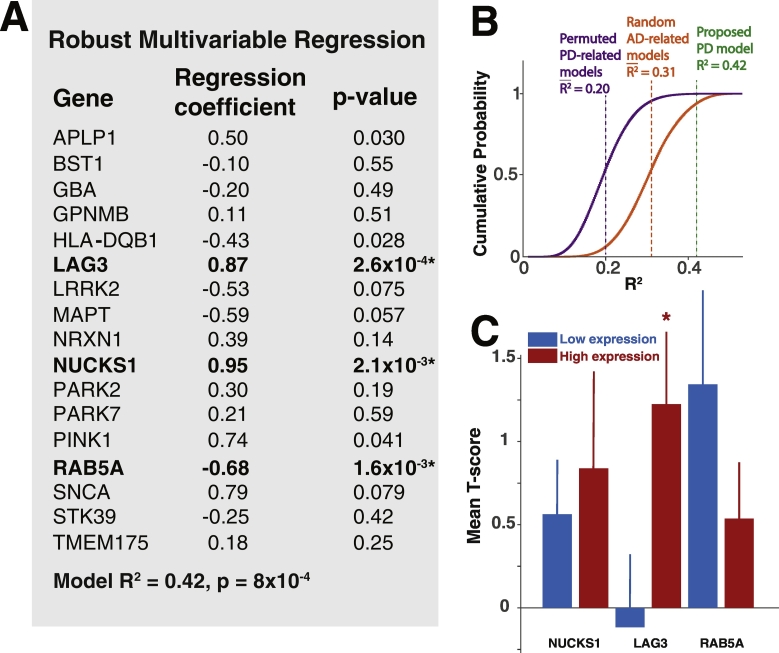
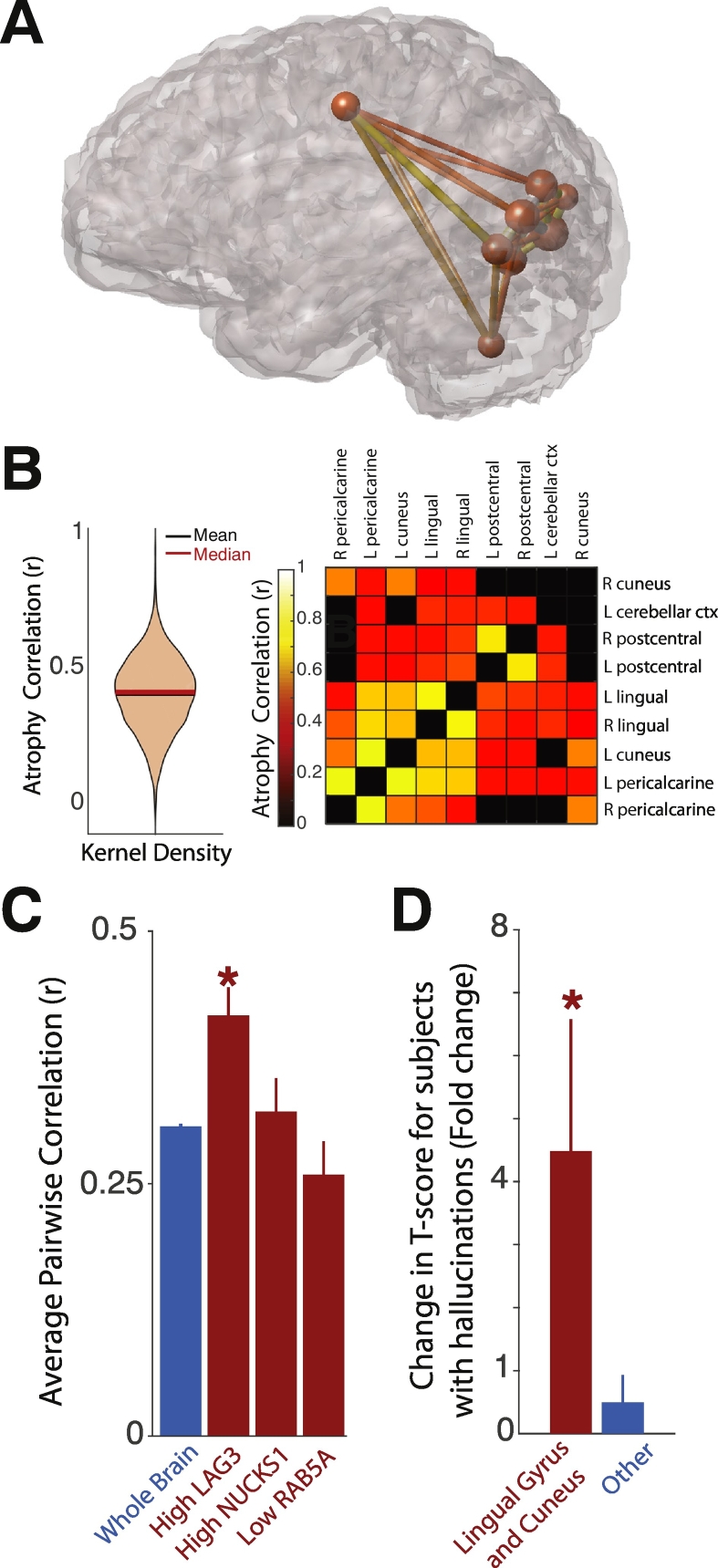
To determine whether this is the case, we performed a robust multivariable linear regression analysis with atrophy T-score as the dependent variable and the normalized (z-scored) expression of each of the genes of interest as independent variables. Robust multivariable methods are particularly well suited to detecting underlying relationships between variables in complex datasets by minimizing the effects of outlier bias (Huber, 2004). Using this approach, we find that LAG3, RAB5A and NUCKS1 are each highly significant predictors of atrophy T-score (Fig. 3A) even after correcting for multiple comparisons. LAG3 and NUCKS1 expression are positively correlated with atrophy, while RAB5A expression is negatively correlated with atrophy. The overall fit of the regression model was estimated with simple linear regression, yielding R2 = 0.42 and p = 8 × 10−4, indicating that regional gene expression explains a considerable amount of variance in the regional atrophy pattern.
Fig. 3.
Genetic predictors of regional atrophy. (A) Regression coefficients and p-values for each gene from a robust multivariable regression model with regional T-score as the dependent variable and regional z-scored expression values for each gene as independent variables. Genes that are bolded remain significant predictors after multiple comparison correction with either Bonferroni or Benjamin-Hochberg methods. (B) Cumulative probability plots of R2 from simulated regression models with randomly permuted PD-related genetic expression values across brain regions (purple, n = 106 simulated models), and random AD-related genes (orange, n = 106 simulated models), using regional atrophy as the dependent variable. Mean simulated R2, and R2 from the proposed PD-related gene model, are denoted. (C) Mean T-scores (±SEM) for the brain regions ≤10th percentile (blue) (n = 9 regions) or ≥90th percentile (red) (n = 9 regions) of gene expression for each of the significant predictors in (A). LAG3 high-expressing and low-expressing regions demonstrate significant differences in mean atrophy T-score (NUCKS1, p = .69; LAG3, p = .045; RAB5A, p = .25).
To determine the predictive power of other possible genetic models, we first simulated one million 17 gene models by randomly permuting genetic expression values across brain regions for each gene in our proposed model. These simulated models therefore address whether the spatial distribution of gene expression in the proposed model better predicts the atrophy pattern than other possible distributions. Simulated model R2 was estimated with simple multivariable linear regression. The simulated model R2 is normally distributed with mean of 0.2 and standard deviation of 0.061. As an additional control, we compared our proposed model to one million simulated 17 gene models derived from a set of 51 Alzheimer disease related genes (Acosta et al., 2018, in press). As most of the AD-related genes have not been implicated in PD pathology, the fit of resultant models therefore estimates the predictive power of mostly spurious associations. The AD gene simulated model R2 is normally distributed with mean of 0.31 and standard deviation of 0.070. The higher mean R2 for these models may be related to roles for some of the AD-related genes in trans-synaptic transmission and protein aggregation, which may be relevant to both AD and PD. The proposed PD gene model outperforms nearly all simulated models, as shown in Fig. 3B.
Because all of the six Allen Brain subjects had data from the left hemisphere, and two of the six had data from both hemispheres, we repeated our robust regression analysis using only left hemisphere data. Despite the loss of statistical power from eliminating half of the regions in the analysis, we obtained similar results with LAG3 remaining a highly significant predictor (Supplementary Table 2). NUCKS1 and RAB5A had two of the lowest p-values in this analysis (p = .033 and p = .088), although neither was significant after multiple comparison correction. Because only two subjects had right hemisphere expression data we were unable to analyze the right hemisphere exclusively, although comparison of the two hemispheres may reveal additional laterality-related differences that are functionally relevant.
3.4. Differential atrophy of highest and lowest expressing LAG3 regions
We further analyzed the expression of significant predictors from the full model by comparing the average atrophy T-score for the highest (≥90th percentile) and lowest expressing (≤10th percentile) brain regions for each of the significant predictor genes. Atrophy was significantly greater in high expressing LAG3 regions as compared with low expressing regions (Fig. 3C). Differences in both NUCKS1 and RAB5A atrophy were not statistically significant, suggesting that while their expression profiles predict atrophy, there is less difference in atrophy at the extremes of expression values.
3.5. Genetic predictor selection by LASSO
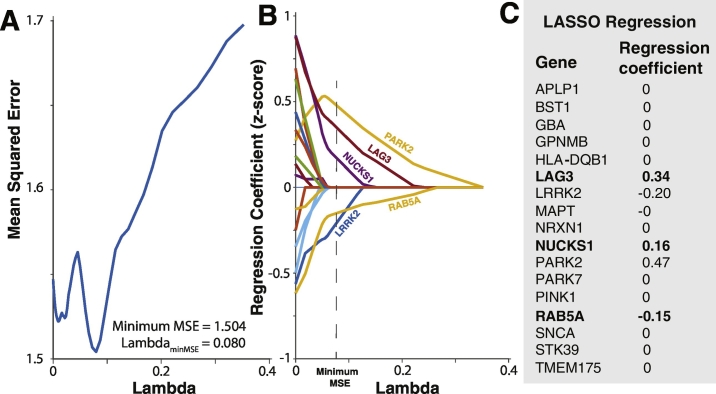
To determine whether a simpler model could be rigorously derived from our full multivariable model, we employed least absolute shrinkage selection operator (LASSO) (Tibshirani, 1996) regression. By changing the tuning parameter lambda, LASSO regression minimizes model mean squared error (MSE) and in the process reduces non- or weakly-predictive independent variable regression coefficient to zero. The most highly predictive variables with non-zero coefficients remain in the model. We hypothesized that this technique should retain the three previously identified highly significant predictors LAG3, RAB5A and NUCKS1 in the simplified model. Indeed, the model with the lowest MSE included only five predictor variables: LAG3, RAB5A, NUCKS1, LRRK2 and PARK2 (Fig. 4). Although most variables were eliminated, this model is unlikely to result from overfitting as 10-fold cross-validation was utilized to minimize this possibility. Although LRRK2 and PARK2 were not significant predictors in the full model, these results suggest that they may play a role in regional atrophy, possibly below the resolution of technique in this study. Importantly, these results are consistent with the full robust multivariable regression analysis, in that a small number of independent variables predict regional atrophy.
Fig. 4.
LASSO regression analysis. (A) Model mean squared error (MSE) as a function of the tuning parameter lambda. MSE is minimized with lambda = 0.080. (B) Gene regression coefficients as a function of lambda. As lambda increases to the value which minimizes model MSE all regression coefficients decay to zero, with the exception of five remaining predictor variables: LAG3, LRRK2, NUCKS1, PARK2, and RAB5A. (C) Gene regression coefficients for the model with minimum MSE. The three significant predictors from the robust regression analysis are shown in bold.
3.6. Correlated atrophy of high expressing LAG3 regions
Because there was markedly more atrophy in high expressing LAG3 regions, we further assessed these regions for the presence of correlated atrophy. We hypothesized that the presence of correlated atrophy may be due to either the presence of high levels of LAG3, elevated interregional connectivity promoting synuclein transfer, or both. To answer this question, we computed the average pairwise correlation coefficient for high expressing (≥90th percentile) LAG3 regions, and for the entire brain. Pairwise correlation coefficients measure the strength of correlated volume changes between brain regions across PD subjects. Thus, two brain regions that frequently atrophy together across PD subjects will exhibit a high correlation coefficient. We employed multiple comparison correction to decrease detection of false positive correlations and set correlation coefficients below the significance threshold to zero. The remaining non-zero correlation coefficients therefore represent significant correlated atrophy between two brain regions. The high expressing LAG3 regions exhibited significantly higher average atrophy correlation than that observed across the entire brain (Fig. 5A–C), suggesting that high levels of LAG3 or differential white matter connectivity may drive coordinated atrophy of these regions. In contrast, neither NUCKS1 high-expressing nor RAB5A low-expressing regions demonstrated correlated atrophy different from that of the whole brain (Fig. 5C).
Fig. 5.
Atrophy of high expressing LAG3 regions. (A) Glass brain representation of the brain regions in the 90th percentile of LAG3 expression (n = 9 regions). The sphere radius centered on each brain region is proportional to microarray expression. Pipes connecting the brain regions are color-coded by the pairwise atrophy correlation between those brain regions using the same color scale as in (B). (B) (Left) violin plot of all atrophy pairwise correlation coefficients across the entire brain, demonstrating a Gaussian distribution. (Right) heat map illustrating pairwise atrophy correlation for LAG3 high expressing brain regions show in (A). (C) Quantification of average pairwise atrophy correlation for the high expressing LAG3 and NUCKS1 regions, and low expressing RAB5A regions, versus the whole brain. LAG3 average r is significantly greater than that for whole brain (p = 1.8 × 10−4), while NUCKS1 and RAB5A average r are not (p = .66 and 0.14, respectively). (D) Change in average T-score for subjects with hallucinations versus those without hallucinations. A group comprising the lingual gyrus and cuneus (left) demonstrates a significant difference in T-score across subject groups (p = .014), while a group comprising the other high expressing LAG3 regions (right) does not (p = .84).
3.7. Hallucinations related to selective atrophy of LAG3 high expressing lingual gyrus and cuneus
As the set of high expressing LAG3 regions is enriched in occipital cortical regions, we next sought to determine whether visuospatial processing may be affected by atrophy of these regions. The lingual gyrus and cuneus, in particular, have been shown to be involved in the pathogenesis of visual hallucinations (Goldman et al., 2014). To this end, we analyzed MDS-UPDRS data in the same cohort of PD patients. We hypothesized that the lingual gyrus and cuneus would exhibit more atrophy in subjects reporting hallucinations than in those that have not experienced hallucinations. We combined data from these two brain regions and computed the change in T-score across subject groups. As predicted, the average atrophy T-score for these brain regions is approximately 4-fold greater in subjects with hallucinations (Fig. 5D). In contrast, there was no difference in average atrophy for the other high expressing LAG3 regions across these subject subgroups. These findings support the hypothesis that dysfunction of the lingual gyrus and cuneus is related to visuospatial disturbance.
4. Discussion
In this study we used the PPMI and ABA databases to characterize the regional PD atrophy pattern with high spatial resolution, and to identify genes with expression patterns that are significantly predictive of regional atrophy. Similar to other groups, we found that there is atrophy of both cortical and subcortical brain regions (Tinaz et al., 2011; Rosenberg-Katz et al., 2013; Weintraub et al., 2012). The basal ganglia are notably atrophic, the degree of which may correlate with the degree of motor impairment (Rosenberg-Katz et al., 2013). However, we found that the occipital and temporal lobes demonstrate even more atrophy. The prominence of posterior cortical atrophy has been described in prior studies, such as by Tinaz et al. in which these areas were shown to exhibit disproportionate cortical thinning (Tinaz et al., 2011). These findings reinforce the importance of non-motor symptoms that often reduce quality of life for PD patients, such as the cognitive and memory deficits that correlate with temporal atrophy (Weintraub et al., 2012); and the visuospatial deficits that correlate with occipital atrophy (Goldman et al., 2014).
We found that there are multiple genes in the normal brain with expression patterns that are highly correlated with the PD regional atrophy pattern. A certain number of genes in the human genome are expected to have regional expression profiles similar to that of the PD atrophy pattern purely by chance. Our approach to identifying genes with expression causally related to PD atrophy was designed to be highly stringent to minimize the likelihood of identifying these false positive genes. To that end, we analyzed only genes with high a priori probabilities of being involved in PD pathogenesis. We used robust regression analysis to minimize the influence of outlier data, stringent multiple comparison correction, LASSO feature selection, and large scale model simulation, to identify our small subset of candidate genes comprising LAG3, NUCKS1, and RAB5A. Because these gene expression patterns exist in the normal healthy brain, it is possible that PD utilizes these normal patterns of expression as conduits for spatial propagation after disease initiation has occurred.
The power of combining imaging data with ABA genetic expression data has also been recently demonstrated by other groups. For example, Rittman et al. compared genetic expression with regional brain connectivity in PD and found that synuclein expression did not correlate with loss of functional connectivity (Rittman et al., 2016). In an imaging-genetic expression study of schizophrenia, Romme et al. showed that particular classes of schizophrenia-related genes had expression profiles that correlated with brain disconnectivity (Romme et al., 2017). This group used a slightly different analysis approach, in which multiple genes within a certain gene class, such as synaptic function, were separately correlated with the brain disconnectivity pattern. Using this approach, particular gene classes were shown to be more predictive of brain disconnectivity than others, providing important mechanistic insight into schizophrenia pathogenesis. Along similar lines, Powell et al. recently showed that the DISC1 genetic expression profile is associated with age-related changes in white matter disconnectivity in schizophrenia (Powell et al., 2017). Other studies have found that genetic expression poorly predicts brain pathology. For example, the Alzheimer disease atrophy pattern is poorly explained by single AD-related genetic expression profiles, despite accurate prediction of spreading dysfunction by trans-synaptic mechanisms (Acosta et al., 2018, in press).
One of the atrophy-predictive genes in our study, NUCKS1, is a highly significant predictor of PD risk in GWAS studies. NUCKS1, a gene involved in DNA damage response (Parplys et al., 2015), is positively correlated with atrophy. Little is known about the function of NUCKS1. However, its role in DNA damage response suggests that it could be involved in cell-autonomous pathology that contributes to dysfunction when regional accumulation of synuclein mediated damage occurs. The two other genes with expression correlated with regional atrophy, LAG3 and RAB5A, are genes that are implicated in trans-synaptic synuclein transfer. LAG3 expression is positively correlated with atrophy, while RAB5A expression is negatively correlated with atrophy. Elevated LAG3 levels may contribute to regional dysfunction and atrophy by promoting binding and endocytosis of extracellular synuclein preformed fibrils (Mao et al., 2016). Increasing the intracellular concentration of synuclein has deleterious effects such as interference with mitochondrial fusion (Kamp et al., 2010) and reduction of neurotransmitter release by inhibition of synaptic vesicle clustering (Nemani et al., 2010). In contrast, RAB5A may act as a protective factor, possibly by sequestering synuclein or promoting its disposal within the cell. The role of RAB5A as an important mediator of endocytic pathways (Numrich and Ungermann, 2014) positions it to potentially control the transport of synuclein inside the cell.
Other well validated PD risk factors such as synuclein itself, tau, GBA and the PARK genes (Gan-Or et al., 2015) do not have expression patterns similar to that of the PD atrophy pattern, suggesting that these genes may serve as initiation factors rather than determinants of disease propagation. Particularly intriguing is the absence of a correlation of SNCA expression with the regional atrophy pattern. There is ample evidence that increasing gene dosage of SNCA by duplication or triplication causes familial cases of Parkinson disease, possibly by increasing the prevalence of pathological aggregates of synuclein (Devine et al., 2011). However, our results suggest that normal expression patterns of SNCA do not predict the ultimate regional pattern of degeneration. These results are concordant with another recent study in which synuclein expression did not correlate with loss of functional connectivity in PD (Rittman et al., 2016). One hypothesis that reconciles these findings is that increasing synuclein expression may initiate disease in one or several brain regions, while additional pathways, potentially including LAG3 and RAB5A, control regional propagation. Similar arguments may apply to other risk factors such as GBA, tau and PARK genes.
The identification of LAG3 and RAB5A as the genes with expression most highly associated with regional atrophy strongly indicates that PD atrophy patterns are mainly dependent on cell non-autonomous function, rather than impairment of cell-autonomous function. This provides indirect support to the emerging concept - based on histopathological studies - that most proteinopathies, including PD, Alzheimer disease and other dementias, spread in stereotypical patterns from one brain region to another (Braak et al., 2003). Multiple studies suggest that misfolded synuclein can spread via a trans-neuronal “prion-like” mechanism, as can other proteins such as tau, beta-amyloid, TDP-43 and Huntingtin, which are implicated in Alzheimer disease, frontotemporal dementia and Huntington disease, respectively (Frost and Diamond, 2010; Angot et al., 2010; Polymenidou and Cleveland, 2012; Jucker and Walker, 2011). Misfolded proteins trigger misfolding of same-species proteins by a corruptive templating process, which in turn is thought to cascade along neuronal pathways (Frost et al., 2009a, Frost et al., 2009b).
Transmission of misfolded synuclein is thought to follow network projections, thus suggesting that connectivity is a determinant of susceptibility. Our group was one of the first to demonstrate that a connectivity-based model of spread, encoded in a graph-theoretic network-diffusion model, was able to capture the brain-wide pattern of trans-neuronal spread in Alzheimer disease and frontotemporal dementia (Raj et al., 2012). Similar conclusions were also made in a study using functional connectivity networks (Zhou et al., 2012). Given our current findings, in which the most highly implicated genes are involved in pathology transfer between cells, it is likely that a similar non-cell autonomous process is involved in PD pathophysiology.
The occipital cortex is selectively enriched in high expressing LAG3 regions, which may be particularly relevant to the pathogenesis of the visuospatial deficits that are prominent features of PD in some patients (Goldman et al., 2014). Indeed, we found that patients reporting hallucinations demonstrated markedly more atrophy of the cuneus and lingual gyrus, two higher order visual processing centers. These findings also provide support to the hypothesis that the LAG3 pathway mediating synuclein transfer may be closely related to PD pathology.
Because the relevant gene expression patterns are present in the normal brain, it may be possible to target LAG3 and RAB5A in a pre-symptomatic or early stage of the disease. In this context, decreasing LAG3 function, or increasing RAB5A function, may be therapeutically beneficial. Multiple LAG3 antagonists are currently in clinical trials, primarily as adjuncts to other forms of cancer immunotherapy (Andrews et al., 2017). One of these trials is designed to assess responses of glioblastoma to anti-LAG3 monoclonal antibodies (clinicaltrials.gov, trial ID NCT02658981), suggesting that anti-LAG3 mAb may be able to cross the blood-brain barrier. If these agents prove safe in initial studies, it may be warranted to attempt a small clinical trial in PD patients. We predict that inhibition of LAG3 function may impede the spread of synuclein and possibly slow the progression of disease.
Despite identifying several gene predictors of spatial atrophy, there are several limitations to our approach which we hope to address with future work. The first is that while we study a subset of genes with high a priori probability of being involved in regional atrophy patterning, there are genes which may be involved which are not included in our analysis. Although there are methodological issues with including larger number of genes in our analysis (e.g. multiple comparisons and increased false negative detection), there are probably other genes excluded from our analysis which play a functional role in the spatial propagation of disease. As we learn more about the spread of PD pathology throughout the brain, we hope to further leverage our combined imaging-genetic approach to identify additional genes important in this process. For instance, analysis of additional endocytosis-related genes which may regulate trans-synaptic alpha-synuclein transfer, and genes controlling alpha-synuclein aggregation may be particularly promising.
Another limitation of our approach is related to the specificity of the ABA genetic expression data. Although there are multiple expression probes for each gene of interest, the specificity of some of these probes has been questioned. For example, one of the four MAPT probes has been reported to have only 88% homology to the MAPT gene (Rittman et al., 2016). We did not attempt to correct for probe non-specificity, and for completeness have included all available ABA probes for all genes. Further investigation of appropriateness criteria for probe inclusion, exclusion and differential weighting for studies using ABA data is warranted.
The third important limitation of our study is that imaging and genetic data are derived from different subject populations. While we are able to use high quality imaging data for PD subjects, no data similar to the ABA exists for this population. The incredibly resource intensive creation of the ABA from a small number of healthy subjects underscores how difficult it currently remains to derive similar data from subjects within specific disease populations. For that reason, we are limited to comparing genetic expression from healthy subjects and atrophy data from PD subjects. It is possible that genetic expression patterns are altered in important ways in PD subjects, but we cannot answer that question with our current resources. Based on our results, we hypothesize that PD pathogenesis exploits pre-existing genetic expression networks present in healthy people, but further investigation is required in this area. Identification of such disease-enabling networks in the healthy brain may be extremely important, as these networks may be amenable to prophylactic or therapeutic manipulation in pre-disease or early disease states.
Acknowledgments
Acknowledgments
PPMI – a public-private partnership – is funded by the Michael J. Fox Foundation for Parkinson's Research funding partners Abbvie, Avid Radiopharmaceuticals, Biogen Idec, BioLegend, Bristol-Myers Squibb, Eli Lilly and Company, F. Hoffmann-La Roche Ltd., GE Healthcare, Genentech, GlaxoSmithKline, Lundbeck Foundation, Merck, MesoScale Discovery, Piramal, Pfizer, Sanofi Genzyme, Servier, Takeda, Teva, and UCB.
We gratefully acknowledge funding from NIH grant R01NS092802 (AR).
Author contributions
BF and AR designed the study, analyzed data and wrote the manuscript. DA and YZ performed genetic expression analysis. SP aided in volumetric analysis and developed data visualization tools. The authors have no financial disclosures.
Appendix A.
Supplementary Table 1.
Subject characteristics. Mean (± standard deviation) age and cognitive assessment (MOCA) for healthy control and PD subjects. Mean Hoehn & Yahr stage for PD subjects.
| Healthy Control | Parkinson Disease | |
|---|---|---|
| Age (years) | 59.9 ± 10.7 | 60.9 ± 9.7 |
| MOCA | 27.5 ± 2.0 | 26.9 ± 3.0 |
| Hoehn & Yahr | 1.8 ± 0.6 |
Supplementary Table 2.
Left hemisphere robust multivariable regression. Using only left hemisphere data, LAG3 expression remains a highly significant predictor of atrophy.
| Gene | Regression coefficient | p-value |
|---|---|---|
| APLP1 | 0.17 | 0.59 |
| BST1 | -0.36 | 0.13 |
| GBA | 0.86 | 0.064 |
| GPNMB | 0.14 | 0.59 |
| HLA-DQB1 | 0.15 | 0.68 |
| LAG3 | 1.18 | 5.2x10-4⁎ |
| LRRK2 | -0.92 | 0.062 |
| MAPT | -0.34 | 0.54 |
| NRXN1 | -0.16 | 0.62 |
| NUCKS1 | 0.98 | 0.033 |
| PARK2 | 0.60 | 0.10 |
| PARK7 | -0.71 | 0.21 |
| PINK1 | 0.14 | 0.83 |
| RAB5A | -0.47 | 0.088 |
| SNCA | 0.63 | 0.30 |
| STK39 | -0.10 | 0.84 |
| TMEM175 | 0.14 | 0.53 |
Model R2 = 0.69, p = 3.6x10-3.
References
- Acosta D., Powell F., Zhao Y., Raj A. Regional vulnerability in Alzheimer's: The role of cell-autonomous and transneuronal processes. Alzheimers Dement. 2018 doi: 10.1016/j.jalz.2017.11.014. (Jan 4), pii: S1552-5260(17)33857-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews L.P., Marciscano A.E., Drake C.G., Vignali D.A.A. LAG3 (CD223) as a cancer immunotherapy target. Immunol. Rev. 2017;276(1):80–96. doi: 10.1111/imr.12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angot E., J a Steiner, Hansen C., Li J.-Y., Brundin P. Are synucleinopathies prion-like disorders? Lancet Neurol. 2010;9(11):1128–1138. doi: 10.1016/S1474-4422(10)70213-1. [DOI] [PubMed] [Google Scholar]
- Bonifati V. Autosomal recessive parkinsonism. Parkinsonism Relat. Disord. 2012;18:S4–S6. doi: 10.1016/S1353-8020(11)70004-9. [DOI] [PubMed] [Google Scholar]
- Braak H., Del Tredici K., Rüb U., de Vos R.A.I., Jansen Steur E.N.H., Braak E. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol. Aging. 2003;24(2):197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- Chartier-Harlin M.C., Kachergus J., Roumier C. alpha-Synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364(9440):1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- Chaudhuri K.R., Healy D.G., Schapira A.H. Non-motor symptoms of Parkinson's disease: diagnosis and management. Lancet Neurol. 2006;5(3):235–245. doi: 10.1016/S1474-4422(06)70373-8. [DOI] [PubMed] [Google Scholar]
- Desikan R.S., Ségonne F., Fischl B. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Desplats P., Lee H.-J., Bae E.-J. Inclusion formation and neuronal cell death through neuron-to-neuron transmission of -synuclein. Proc. Natl. Acad. Sci. 2009;106(31):13010–13015. doi: 10.1073/pnas.0903691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine M.J., Gwinn K., Singleton A., Hardy J. Parkinson's disease and α-synuclein expression. Mov. Disord. 2011;26(12):2160–2168. doi: 10.1002/mds.23948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B. FreeSurfer. NeuroImage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freer R., Sormanni P., Vecchi G., Ciryam P., Dobson C.M., Vendruscolo M. A protein homeostasis signature in healthy brains recapitulates tissue vulnerability to Alzheimer's disease. Sci Adv. 2016;2(8) doi: 10.1126/sciadv.1600947. eCollection 2016 Aug. PMID:27532054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B., Diamond M.I. Prion-like mechanisms in neurodegenerative diseases. Nat. Rev. Neurosci. 2010;11(3):155–159. doi: 10.1038/nrn2786. (Accessed July 21, 2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B., Ollesch J., Wille H., Diamond M.I. Conformational diversity of wild-type Tau fibrils specified by templated conformation change. J. Biol. Chem. 2009;284(6):3546–3551. doi: 10.1074/jbc.M805627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B., Jacks R.L., Diamond M.I. Propagation of tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 2009;284(19):12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan-Or Z., Amshalom I., Kilarski L.L. Differential effects of severe vs mild GBA mutations on Parkinson disease. Neurology. 2015;84(9):880–887. doi: 10.1212/WNL.0000000000001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel P., Kuceyeski A., Locastro E., Raj A. Spatial patterns of genome-wide expression profiles reflect anatomic and fiber connectivity architecture of healthy human brain. Hum. Brain Mapp. 2014;35(8):4204–4218. doi: 10.1002/hbm.22471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz C.G., Tilley B.C., Shaftman S.R. Movement disorder society-sponsored revision of the unified Parkinson's disease rating scale (MDS-UPDRS): scale presentation and clinimetric testing results. Mov. Disord. 2008;23(15):2129–2170. doi: 10.1002/mds.22340. [DOI] [PubMed] [Google Scholar]
- Goldman J.G., Stebbins G.T., Dinh V. Visuoperceptive region atrophy independent of cognitive status in patients with Parkinson's disease with hallucinations. Brain. 2014;137(3):849–859. doi: 10.1093/brain/awt360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L., Abajian C., Beckmann C.F., Bernard A., Bertagnolli D., Boe A.F., Cartagena P.M., Chakravarty M.M., Chapin M., Chong J., Dalley R.A., David Daly B., Dang C., Datta S., Dee N., Dolbeare T.A., Faber V., Feng D., Fowler D.R., Goldy J., Gregor B.W., Haradon Z., Haynor D.R., Hohmann J.G., Horvath S., Howard R.E., Jeromin A., Jochim J.M., Kinnunen M., Lau C., Lazarz E.T., Lee C., Lemon T.A., Li L., Li Y., Morris J.A., Overly C.C., Parker P.D., Parry S.E., Reding M., Royall J.J., Schulkin J., Sequeira P.A., Slaughterbeck C.R., Smith S.C., Sodt A.J., Sunkin S.M., Swanson B.E., Vawter M.P., Williams D., Wohnoutka P., Zielke H.R., Geschwind D.H., Hof P.R., Smith S.M., Koch C., Grant S.G.N., Jones AR. An anatomically comprehensive atlas of the adult human brain transcriptome. 2012;489(7416):391–399. doi: 10.1038/nature11405. (Sep 20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber P.J. Robust statistics. Statistics (Ber) 2004;60(1986):1–11. [Google Scholar]
- Jucker M., Walker L.C. Pathogenic protein seeding in Alzheimer disease and other neurodegenerative disorders. Ann. Neurol. 2011;70(4):532–540. doi: 10.1002/ana.22615. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3203752&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp F., Exner N., Lutz A.K. Inhibition of mitochondrial fusion by α-synuclein is rescued by PINK1, Parkin and DJ-1. EMBO J. 2010;29(20):3571–3589. doi: 10.1038/emboj.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowal S.L., Dall T.M., Chakrabarti R., Storm M.V., Jain A. The current and projected economic burden of Parkinson's disease in the United States. Mov. Disord. 2013;28(3):311–318. doi: 10.1002/mds.25292. [DOI] [PubMed] [Google Scholar]
- Luk K.C., Kehm V., Carroll J. Pathological-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science. 2012;338(6109):949–953. doi: 10.1126/science.1227157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk K.C., Kehm V.M., Zhang B., O'Brien P., Trojanowski J.Q., Lee V.M.Y. Intracerebral inoculation of pathological α-synuclein initiates a rapidly progressive neurodegenerative α-synucleinopathy in mice. J. Exp. Med. 2012;209(5):975–986. doi: 10.1084/jem.20112457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Ou M.T., Karuppagounder S.S. Pathological-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science. 2016;353(6307) doi: 10.1126/science.aah3374. (aah3374-aah3374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek K., Jennings D., Lasch S. The Parkinson progression marker initiative (PPMI) Prog. Neurobiol. 2011;95(4):629–635. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merola A., Rizzi L., Zibetti M. Medical therapy and subthalamic deep brain stimulation in advanced Parkinson's disease: a different long-term outcome? J. Neurol. Neurosurg. Psychiatry. 2014;85(5):552–559. doi: 10.1136/jnnp-2013-305271. [DOI] [PubMed] [Google Scholar]
- Nalls M.A., Pankratz N., Lill C.M. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet. 2014;46(9):989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemani V.M., Lu W., Berge V. Increased expression of α-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron. 2010;65(1):66–79. doi: 10.1016/j.neuron.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numrich J., Ungermann C. Endocytic Rabs in membrane trafficking and signaling. Biol. Chem. 2014;395(3):327–333. doi: 10.1515/hsz-2013-0258. [DOI] [PubMed] [Google Scholar]
- Parplys A.C., Zhao W., Sharma N. NUCKS1 is a novel RAD51AP1 paralog important for homologous recombination and genome stability. Nucleic Acids Res. 2015;43(20):9817–9834. doi: 10.1093/nar/gkv859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M., Cleveland D.W. Prion-like spread of protein aggregates in neurodegeneration. J. Exp. Med. 2012;209(5):889–893. doi: 10.1084/jem.20120741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell F.J., LoCastro E., Acosta D.M. Age-related changes in topological degradation of white matter networks and gene expression in chronic schizophrenia. Brain Connect. 2017;7(9) doi: 10.1089/brain.2017.0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przedborski S. The two-century journey of Parkinson disease research. Nat. Rev. Neurosci. 2017;18(4):251–259. doi: 10.1038/nrn.2017.25. [DOI] [PubMed] [Google Scholar]
- Raj A., Kuceyeski A., Weiner M. A network diffusion model of disease progression in dementia. Neuron. 2012;73(6):1204–1215. doi: 10.1016/j.neuron.2011.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittman T., Rubinov M., Vértes P.E. Regional expression of the MAPT gene is associated with loss of hubs in brain networks and cognitive impairment in Parkinson disease and progressive supranuclear palsy. Neurobiol. Aging. 2016;48:153–160. doi: 10.1016/j.neurobiolaging.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romme I.A.C., de Reus M.A., Ophoff R.A., Kahn R.S., van den Heuvel M.P. Connectome disconnectivity and cortical gene expression in patients with schizophrenia. Biol. Psychiatry. 2017;81(6):495–502. doi: 10.1016/j.biopsych.2016.07.012. [DOI] [PubMed] [Google Scholar]
- Rosenberg-Katz K., Herman T., Jacob Y., Giladi N., Hendler T., Hausdorff J.M. Gray matter atrophy distinguishes between Parkinson disease motor subtypes. Neurology. 2013;80(16):1476–1484. doi: 10.1212/WNL.0b013e31828cfaa4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley W.W., Crawford R.K., Zhou J., Miller B.L., Greicius M.D. Neurodegenerative diseases target large-scale human brain networks. Neuron. 2009;62(1):42–52. doi: 10.1016/j.neuron.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen E.H., Overly C.C., Jones A.R. The Allen Human Brain Atlas. Comprehensive gene expression mapping of the human brain. Trends Neurosci. 2012;35(12):711–714. doi: 10.1016/j.tins.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Tibshirani R. Regression selection and shrinkage via the lasso. J. R. Stat. Soc. B. 1996;58(1):267–288. [Google Scholar]
- Tinaz S., Courtney M.G., Stern C.E. Focal cortical and subcortical atrophy in early Parkinson's disease. Mov. Disord. 2011;26(3):436–441. doi: 10.1002/mds.23453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub D., Dietz N., Duda J.E. Alzheimer's disease pattern of brain atrophy predicts cognitive decline in Parkinson's disease. Brain. 2012;135(1):170–180. doi: 10.1093/brain/awr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Gennatas E.D., Kramer J.H., Miller B.L., Seeley W.W. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73(6):1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]