Abstract
Roses hold high cultural and economic importance as ornamentals and for the perfume industry. We report the rose whole genome sequencing and assembly and resequencing of major genotypes that contributed to rose domestication. We generated a homozygous genotype from a heterozygous diploid modern roses progenitor, Rosa chinensis ‘Old Blush’. Using Single Molecule Real-Time sequencing and a meta-assembly approach we obtained one of the most complete plant genomes to date. Diversity analyses highlighted the mosaic origin of ‘La France’, one of the first hybrids combining the growth vigor of European species and recurrent blooming of Chinese species. Genomic segments of Chinese ancestry revealed new candidate genes for recurrent blooming. Reconstructing regulatory and secondary metabolism pathways allowed us to propose a model of interconnected regulation of scent and flower color. This genome provides a foundation for understanding the mechanisms governing rose traits and will accelerate improvement in roses, Rosaceae and ornamentals.
Roses are among the most commonly cultivated ornamental plants worldwide. They have been cultivated by humans since antiquity, e.g. in China. Ornamental features as well as therapeutic and cosmetic values have certainly motivated rose domestication. The genus Rosa contains about 200 species, more than half being polyploid1. Roses have undergone extensive reticulate evolution with interspecific hybridization, introgression and polyploidization. Only 8 to 20 rose species are said to have contributed to the present complex hybrid rose cultivars, namely Rosa x hybrida2. The Chinese rose Rosa chinensis (diploid) was introduced to Europe in the 18th century. It is seen as one of the main species that participated in the subsequent extensive process of hybridization with roses from the European/Mediterranean/Middle-Eastern (mostly tetraploid) sections (Supplementary Notes 1). These crossings gave birth to hybrid tea cultivars which are the parents of the modern roses with their extraordinarily diverse traits3. Among the breeding traits brought by Chinese roses, the capacity of recurrent flowering as well as color and scent signatures are key breeding traits4. Despite progress in the last decade5, the lack of a rose genome sequence has hampered the discovery of the molecular and genetic determinants of these traits and of their breeding history.
Due to natural auto-incompatibility and recent interspecific hybridization, all roses have highly heterozygous genomes6 that are challenging to assemble7 despite their relatively small size (560 Mb)8. So far, attempts to assemble rose genomes with short-reads led to highly fragmented assemblies composed of thousands of scaffolds (83,1399 and 15,938 (this study), respectively). To overcome these bottlenecks towards a genome reference, we obtained a homozygous genome that we sequenced with a long-read sequencing technology. We developed an original in vitro culture protocol combining fine-tuned starvation, cold stress and hormonal treatments to induce R. chinensis ‘Old Blush’ microspores to switch from gametophyte to sporophyte development. This approach allowed microspores to initiate divisions, form homozygous cell clusters, and develop embryogenic callus from which homozygous plantlets could be regenerated (Supplementary Notes 2; Supplementary Fig. 1).
The homozygous rose line was sequenced on the PacBio RS II platform. An 80x sequencing coverage was obtained with 40 single-molecule real-time cells. Preliminary assembly of the rose data with a single assembler generated several hundred of contigs, illustrating the challenge of assembling plant genomes despite long-reads data10,11. A key step in improving the contiguity of the assembly is the detection and the filtering of spurious edges in the graph of overlaps. The assembler CANU implements filter parametrization at the read level, leading to more accurate and contiguous assemblies12. We developed a software called til-r, which implements similar and alternate heuristics to clean the graph of overlaps of the FALCON assembler (Supplementary Fig. 2)13. Then, we used CANU to perform a meta-assembly of six complementary raw assemblies generated by CANU and FALCON/TIL-R (Supplementary Notes 3, see URLs section ). The final assembly was composed of 82 contigs for an N50 of 24Mb, increasing the contiguity metrics of a simple assembly by threefold and demonstrating the power of meta-assembly approaches (Supplementary Fig. 2).
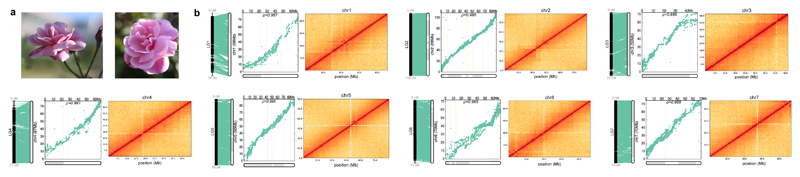
The seven pseudo-chromosomes were built by integrating 86.4% of the 25,695 markers of the K5 rose high-density genetic map14. A large fraction of the assembly (97.7%, 503Mb) was oriented with Pearson's correlation coefficients ranging from 0.986 to 0.996, illustrating the high congruence between sequence and genetic data. The genome structure and quality was confirmed by the mapping of Hi-C chromosomal contact map information data (Figure 1; Supplementary Fig. 3). With very few remaining gaps and high consistency between genetics and sequence data, the rose genome assembly is one of the most contiguous obtained so far for a plant genome.
Figure 1. Chromosome level assembly correlation with genetic map and Hi-C data.
a, Rosa chinensis ‘Old Blush’ mature flowers.
b, Representation of chromosome connections between the physical positions on the reconstructed chromosome and genetic map positions (left panel). Scatter plot with dots representing the physical position on the chromosome (x-axis) versus the map position (y-axis). Rho (ρ) is the Pearson correlation coefficient (middle panel). Hi-C intra-chromosomal contact map for each chromosome (right panel). The intensity of pixels represents the count of Hi-C links between 400kb windows on chromosomes on a logarithmic scale. Darker red color indicates higher contact probability
The rose genome encodes 36,377 inferred protein-coding genes and 3,971 long non-coding RNAs. Annotation assessment with the Plantae BUSCO v2 dataset15 identified 96.5% complete gene models. BUSCO analyses using assembled heterozygous genome of R. chinensis ‘Old Blush’ (Supplementary Notes 4) identified 93.5% complete genes (Supplementary Data 1). Based on transcriptomic data from pooled tissues, 207 miRNA precursors were predicted. Transposable elements (TE) spanned 67.9% of the assembly, 50.6% being LTR retrotransposons (Supplementary Notes 5, Supplementary Fig. 4; Supplementary Table 1). The web portal RchiOBHm-V2 (see URLs section) provides access to the reference genome integrating annotations, polymorphisms, transcriptomic data and the first rose epigenome on rose petals (Supplementary Notes 6).
Comparative genomic investigation allowed us to assess rose paleohistory within the Rosaceae family (Supplementary Notes 7). Conserved gene adjacencies identified an ancestral Rosaceae karyotype (ARK) consisting of 9 protochromosomes with 8,861 protogenes (Supplementary Fig. 5a). Our evolutionary scenario establishes that the ancestral Rosoideae karyotype (ARoK) of strawberry and Rosa genomes, structured into 8 protochromosomes with 13,070 protogenes, was derived from ARK through one ancestral chromosome fission and two fusions. Interestingly, the strawberry genome experienced an extra ancestral chromosome fusion from ARoK to reach its modern genome structure, while the Rosa sp. went through one fission and two fusions, independent from strawberry, to reach its modern genome structure. A phylogeny based on 748 gene sequences showed that Rosa, Fragaria and Rubus diverged within a short timeframe, suggestive of an evolutionary radiation inside the rosoideae subfamily (Supplementary Fig. 5b).
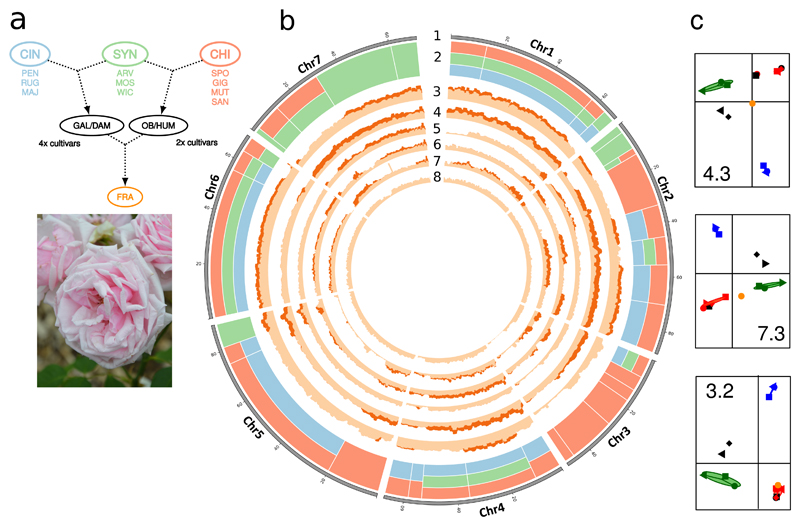
To gain insight into the make-up of modern-day roses, we resequenced representatives of three sections (Synstylae, Chinenses and Cinnamomeae; Supplementary Table 2) that were involved in domestication and breeding that led to rose hybrid cultivar creation (Supplementary notes 1 and 8). We observed discrete levels of variant density along the genomes of hybrid cultivars (Figure 2b), that may reflect different introgression histories. We used the changes in variant density to segment the genome into 35 intervals (2 to 56 Mb) and studied their genetic structure with principal component analyses (Figure 2c, Supplementary Fig. 6). We focused on the modern Rosa x hybrida ‘La France’ (FRA), considered as among the first created hybrids that combine growth vigor traits of European species and recurrent blooming of Chinese species.
Figure 2. Structure of diversity in resequenced genotypes highlights the origin of modern rose cultivars.
a, Genealogy of resequenced genotypes. Sections : CIN = Cinnamoneae ; SYN = Synstylae ; CHI = Chinenses. Genotypes : PEN, R. pendulina ; RUG, R. rugosa ; MAJ, R. majalis ; ARV, R. arvensis ; MOS, R. moschata ; WIC, R. wichurana ; SPO, R. chinensis ‘Spontanea’ ; GIG, R. gigantea ; MUT, R. chinensis ‘Mutabilis’ ; SAN, R. chinensis ‘Sanguinea’ ; GAL, R. gallica ; DAM, R. damascena ; OB, Rosa chinensis ‘Old Blush’ ; HUM, R. chinensis ‘Hume’s Blush’ ; FRA, R. x hybrida ‘La France’ (flower photo).
b, Genetic structure and variant density. 1, circular representation of pseudomolecules. 2, schematic representation of the contribution of Cinnamonea, Synstylae and Chinenses sections to ‘La France’ in 35 chromosomal segments: light red = CHI, light green = SYN, light blue = CIN, multiple bands: mixed origin in the fragment. 3-8, density in heterozygote and homozygote variants (light and dark shades respectively) in 1 Mb sliding windows in ‘La France’, R. gigantea, ‘Hume’s Blush’, ‘Mutabilis’, ‘Sanguinea’, and ‘Old Blush’ heterozygote genotype respectively.
c, Principal component analyses of genetic variation in three illustrative genomic segments. ‘La France’, orange dot; CIN, SYN and CHI in blue, green and red respectively; other cultivars in black. y-axis, 1st component. x-axis, 2nd component. The number indicated in each plot refers to the genomic fragments analyzed (e.g. 4.3 is the third segment of chromosome 4, Supplementary Fig. 6).
Patterns of diversity along the seven chromosomes showed that ‘La France’ genome is a complex mosaic formed by DNA fragments transmitted by the three ancestral pools of diversity represented in the targeted rose sections (Figure 2, Supplementary Notes 8; Supplementary Fig. 6; Supplementary Data 2). For example, chromosome 4 haplotypes are structured by a combination of Cinnamonae, Synstylae and Chinenses genomes, whereas chromosome 7 haplotypes have been transmitted by Synstylae and Chinenses ancestors, without apparent contribution of Cinnamonae.
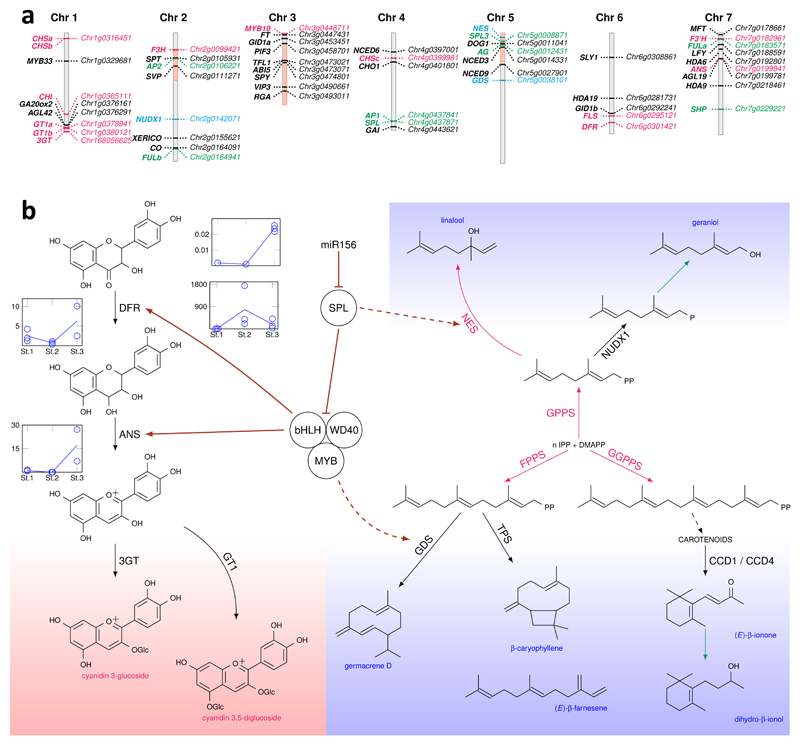
We took advantage of the transmission of genomic bits of Chinenses hybrids to ‘La France’ to identify new candidate genes involved in recurrent blooming. The insertion of a transposable element in TFL1 (RoKSN), a repressor of floral transition responsive to activation by gibberellic acid (GA), is considered a major determinant of recurrent blooming16. We identified that this transposable element was transmitted to ‘La France’ by R. chinensis cultivars and thus may participate to its recurrent blooming. A recent segregation analyses of a R. chinensis ‘Old Blush’ x R. wichurana backcross progeny, showed that recurrent blooming likely involves at least a second independent locus17. This second locus may have been transmitted to ‘La France’ only by R. chinensis, and thus could lie on chromosomal segments such as those originating from the Chinenses section, i.e. segments 2.4 and 5.1 (Figure 2). On these segments, we identified the putative homologues of the transcription factor SPT (segment 2.4, Figure 3a), known to control flowering in Arabidopsis18,19 and of DOG1 (segment 5.1, Figure 3a), known to modify flowering by acting on miR15620. These genes are thus other promising determinant candidates associated with recurrent blooming in roses.
Figure 3. Inter-regulatory connections between color biosynthesis and some scent pathways.
a, Schematic representation of the rose chromosomes together with the position of candidate genes for anthocyanin pigments and volatile molecules biosynthesis and for flowering. Chromosome segments 2.4, 3.2-3.6 and 5.1 originating only from R. chinensis are indicated in light red. Anthocyanin synthesis genes are indicated in red; terpene biosynthesis genes in blue; flowering time genes in black; and development genes in green.
b, Schematic representation of interconnections between color (pink background) and scent (blue background) pathways. Gene expression data show the anti-correlation between miR156 and SPL9 genes during petal development. RT-qPCR was performed on petals harvested at three successive stages: Non-colored petals early during development (St1); Petals at onset of anthocyanin synthesis (St2); Fully colored petals (St3).
Black arrows: biosynthetic steps reported in the rose. Red arrows: biosynthetic steps reported in other species, but not in the rose. Green arrows: putative steps with unknown enzymes. Dashed black arrow: Several enzymatic steps. Maroon arrows: Gene regulation reported in A. thaliana, but not in the rose. Dashed maroon arrow: putative gene regulations. IPP: isopentenyl diphosphate, DMAPP: dimethylallyl diphosphate, DFR: dihydroflavonol-4-reductase, ANS: anthocyanidin synthase, 3GT: anthocyanidin 3-O-glucosyltransferase, GT1: anthocyanidin 3,5-diglucosyltransferase, GPPS: geranyl diphosphate synthase, FPPS: farnesyl diphosphate synthase, GGPPS: geranylgeranyl diphosphate synthase, GDS: germacrene D synthase, TPS: terpene synthase, NES: linalool/nerolidol synthase, CCD1/4 : carotenoid cleavage dioxygenases 1/4, NUDX1: nudix hydrolase1.
Roses exhibit a huge diversity of flower fragrance and color for which biochemical and regulatory determinants are only partially elucidated (Supplementary Notes 9; Supplementary Fig. 7). Data mining of the rose genome combined with in-depth biochemical and molecular analyses of volatile organic compounds (VOCs) permitted identification of at least 22 biosynthetic steps in the terpenes pathway that have not been characterized in the rose, two among which have never been characterized in other species (Supplementary Notes 9; Supplementary Fig. 7).
To study the relationships between color and scent pathways, we performed biochemical and molecular analyses on cyanidin, whose glucosylated derivatives represent more than 99% of total anthocyanin pigments21, and on germacrene D, a VOC produced in petal cells of R. chinensis ‘Old Blush’ (Supplementary Data 3). Our analyses suggest a coordinated biosynthesis of these two compounds achieved through the miR156-SPL9 regulatory module. In Arabidopsis, SPL9 is considered as a repressor of anthocyanin synthesis in cells of aging plants22. miR156 negatively regulates SPL9 in cells of young plants which enables the formation of a MYB-bHLH-WD40 protein complex that activates anthocyanin production22. Analysis of this module in petals of ‘Old Blush’ showed that the expression of SPL9 peaks before maximum expression of ANTHOCYANIDIN SYNTHASE (ANS) expression (Supplementary Fig. 8). In fully colored petals, we observed induced expression of miR156 which correlated with downregulation of SPL9 expression and upregulation of ANS expression (Figure 3b, Supplementary Fig. 8; Supplementary Fig. 9). The maximum expression of GDS, which encodes the enzyme catalyzing germacrene D synthesis, also correlates with miR156 and ANS activation and with the down-regulation of SPL9 (Figure 3; Supplementary Fig. 8). This observation, together with the previous demonstration that ANS and GDS can be activated in rose petals by expression of the Arabidopsis AtPAP1 MYB transcription factor23, suggests that anthocyanin and germacrene D biosynthesis could be coupled by the miR156-SPL9 regulatory module, possibly acting on a MYB-bHLH-WD40 complex. Although PAP1 is not expressed in ‘Old Blush’ petals, we found that the expression pattern of RhMYB10, previously described as a regulator of anthocyanin biosynthetic pathway in Rosaceae24, is compatible with a role in cyanidin and germacrene D synthesis co-activation in petal epidermal cells (Supplementary Fig. 8).
The biosynthesis of terpenes, major scent compounds in roses, has been shown to involve TERPENE SYNTHASES (TPS), such as NEROLIDOL SYNTHASE (NES)25. Search for TPS in the rose genome revealed a cluster of NES genes on chromosome 5 that has a counterpart in Fragaria26. These genes were not significantly expressed in rose petals (Supplementary Data 4). In Arabidopsis, some TPS are activated by SPL927. In rose petals, the downregulation of SPL9 through activation of miR156 (Figure 3b; Supplementary Fig. 8) might explain the absence of expression of NES genes and likely why they do not participate in the production of some terpenes in rose flowers. Our data provide hints about why alternative routes to produce terpenes, such as the one involving NUDX128, have been employed in rose flowers.
Here, we propose that the miR156-SPL9 regulatory hub orchestrates the coordination of production of both colored anthocyanins and certain terpenes, by permitting the complexation of pre-existing MYB-bHLH-WD40 proteins to modulate different components of both pathways (Figure 3). Therefore, anthocyanin synthesis in rose flowers may be linked to the production of some volatile compounds, providing a regulatory reason for the evolution of non-standard terpene biosynthesis pathways. Moreover, this co-regulation may hamper combining pigmentation and specific scents in rose hybrids.
The very high-quality rose genome sequence reported in this study combined with an expert annotation of the main pathways of interest for the rose (Supplementary Notes 9-13; Supplementary Figs. 7 to 23; Supplementary Table 3; Supplementary Data 5 to 10), give unprecedented insights into the genome dynamics of this woody ornamental, and offers a basis to disentangle seemingly mandatory trait associations or exclusions. Furthermore, access to candidate genes, such as the ones involved in abscisic acid synthesis and signaling, paves the way for improving rose quality with better water use efficiency, and increased vase-life. Breeding for other characteristics such as increased resistance to pathogens should also benefit from these data and may lead to reduced use of pesticides.
Online Methods
Production of homozygous rose line derived from heterozygous Rosa chinensis ‘Old Blush’
Flower buds were harvested from R. chinensis ‘Old Blush’ plants when most microspores were at the mid-late uninucleate/early bicellular development stages (Supplementary Fig. 1). Microspores were aseptically isolated from anthers, suspended in starvation medium, and pretreated at 4°C in darkness for 21 days. About 160,000 microspores were suspended in AT12 medium corresponding to AT3 medium29 supplemented with 4.5 µM 2,4-D and 0.44 µM BAP, pH 5.8, and then incubated at 25°C in the dark. Developing micro-calli (ca. 0.5 mm diameter) were observed after about 11 weeks and then subcultured individually in the same conditions (Supplementary notes 2). Developed calli were then plated onto solid MS salts medium complemented with B5 vitamins, 30 g/L sucrose, 2.5 mM MES, 4.5 µM 2,4D, 0.44 µM BAP and 6.5 g/L VitroAgar, (Kalys Biotechnologie, Saint Ismier, France) pH 5.8. A callus that displayed somatic embryos (designated RcHzRDP12; Supplementary Fig. 1g) was selected. Homozygosity status and ploidy level of this callus were confirmed, respectively, by DNA genotyping and by fluorescence-activated cell sorting (FACS) analysis as previously described30.
Samples preparation and sequencing
High quality nuclear DNA was prepared from RcHzRDP12 homozygous callus propagated on callus maintenance medium (Supplementary Notes 2) mainly as previously described31 with the following modifications. Ten % fresh weight of PVP40 was added to callus cells upon grinding in liquid nitrogen. Purified nuclei pellets were processed with Qiagen DNeasy Plant kit (Qiagen, MD, USA). DNA integrity was checked via gel electrophoresis (0.7% agarose) and total DNA was quantified by fluorometry using Picogreen® (Applied Biosystems/Life Technologies, Carlsbad CA, USA.
To sequence R. chinensis ‘Old Blush’ genome, we used in vitro cultured plants obtained through adventitious shoot organogenesis from Type 1 somatic embryo (RcOBType1), as described32. Axenic in vitro R. chinensis ‘Old Blush’ plantlets, were ground in liquid nitrogen and nuclei were purified as previously described31. Nuclei pellets were then processed with Qiagen DNeasy Plant kit (Qiagen, MD, USA), according to the protocol provided by the supplier.
High quality DNA was extracted from leaf samples of Rosa species and cultivars grown at ENS-Lyon, at Lyon botanical garden, in the rose garden “O. Masquelier, La Bonne Maison, Lyon-France” or in the rose garden “Jardin Expérimental de Colmar, France” (Supplementary Notes 8).
DNA integrity was inspected by gel electrophoresis (0.7% agarose) and then quantified by fluorometry using Picogreen® (Applied Biosystems/Life Technologies, Carlsbad CA, USA).
Paired-end sequencing DNA libraries were constructed using Illumina’s TruSeq DNA LT kit following the manufacturer recommendations (Supplementary Tables 4 and 5). The distributions of DNA fragment lengths in the libraries were checked with Agilent BioAnalyzer High Sensitivity DNA chip assays. Whole genome sequencing of R. chinensis ‘Old Blush’ was performed on Illumina HiSeq 2000. Sequences from paired-end and mate-pair reads of the multiple libraries were assembled using the ALLPathsLG software33 (Supplementary Table 6).
Three-dimensional proximity information obtained by chromosome conformation capture sequencing (Hi-C)
Leaf tissues were fixed in 1% (v/v) formaldehyde and were then used for the preparation of 2 independent in situ Hi-C libraries. Nuclei extraction, nuclei permeabilization, chromatin digestion and proximity ligation treatments were performed essentially as previously described34. DpnII was used as restriction enzyme. The recovery of Hi-C DNA and subsequent DNA manipulations were performed as previously described35. Libraries were sequenced on an Illumina NextSeq instrument with 2 x 75 bp reads. Hi-C libraries were independently analyzed with HiC-Pro pipeline (default parameters and LIGATION_SITE=GATCGATC36). Valid ligation products from each library were merged together for the interaction matrix construction. The genome was divided into bins of equal size and number of contacts observed between each pair of reported bins. Finally contact maps were plotted with HiCPlotter software37.
Genome assembly
A software program called til-r was developed to implement heuristics aiming at filtering the graph of overlap generated by FALCON (Supplementary Notes 3). A meta-assembly combining two CANU and four FALCON assemblies was generated by CANU 1.4 (Supplementary Fig. 2; Supplementary Notes 3).
Pseudomolecules building
Pseudomolecules were built by anchoring the 82 contigs to the K5 SNP genetic linkage map14 using the ALLMAPS software38. Four chimeric breakpoints were identified and corrected by identifying the primary contigs in which the problematic regions were not merged. Three chimeric breakpoints were absent in CANU assemblies and the fourth was absent in all primary assemblies. Finally, ALLMAPS was applied on the corrected meta-assembly enabling the building of seven pseudomolecules corresponding to rose haploid chromosome number by anchoring and orienting 97.7% of the contigs (503Mb) based on 86.4% of the genetic markers. The final assembly consists of seven pseudo-chromosomes, the mitochondrial and chloroplast genomes plus 46 unanchored contigs spanning 11.2 Mb (Supplementary Fig. 2a).
The genome was first polished by quiver39 using stringent alignment cutoffs (--minLength 3000 --maxHits 1). Then, a run of pilon40 (version 1.21, --mindepth 30 --fix bases) using homozygous ‘Old Blush’ Illumina paired-end reads edited 7,444 SNPs, 107,249 small insertions and 33 small deletions. The final genome assembly is composed of 515,588,973 nucleotides including the 3,300 “N” for the 33 gaps of which seven represent centromeres. Biological centromeres were located by identifying tandem repeats using the TRF software41, selecting patterns of an over-represented length in the genome, assembling them in contigs and visually inspecting their distribution along the pseudomolecules (Supplementary Notes 3).
Localization of putative crossing-overs and segmental conservation between genotypes
Identification of putative loci of crossing-overs was performed by mapping Illumina reads from the heterozygous genome (5 distinct libraries) on the constructed pseudo-chromosomes using the BWA software42 and by counting pairs in which only one read had a match, in 10kb long windows. We observed 50 windows with over-represented one-end mapped pairs in at least two libraries and kept them as candidate crossing-over loci (Supplementary Fig. 12, yellow frame). To confirm them, when possible, we used the sequence conservation with genotypes related to the inferred parents of ‘Old Blush’ (Supplementary Fig. 12, red plots; Supplementary Notes 4.2).
Annotation of protein-coding genes and lncRNAs
Gene models were predicted using a fully automated and parallelized pipeline egn-ep (see URLs section) that carries out probabilistic sequence models training, genome masking, transcript and protein alignments computation and integrative gene modeling by the EuGene software43 (release 4.2a). The configuration of the egn-ep pipeline is detailed in Supplementary Notes 5. The inferred mRNAs were assessed by BUSCO v215 which found 1,389 complete, 23 fragmented and 28 missing gene models (96.5%, 1.6% and 1.9% respectively). 36,377 genes were retained after the removal of annotated repeated elements (see below). Correspondence between gene models in homozygous and heterozygous annotations were established by best reciprocal hits (Supplementary Table 7; Supplementary Data 1).
Functional annotation of protein-coding genes
The protocol described by Schläpfer et al44 was used to annotate enzymes and build the metabolic network. Two cut-offs were modified to increase stringency, BLAST e-value cutoff lowered to 10-5 and pathway-prediction-score set to 0.3 in pathway-tools. Nineteen pathways considered as false positives were removed. A MetExplore instance45 gives access to the network (see URLs section).
Protein coding genes were annotated by integrating five sources depending on their expected accuracy. Priorities were successively given to i) a search of reciprocal best hits with the 218 Rosaceae proteins tagged as "reviewed" in the UniProt database (90% span, 80% identity)46, ii) the description of the 8,512 previously annotated enzymes, iii) transcription factors and kinases identified (2,414 and 1,885 respectively) by ITAK47, iv) the 3,954 transcription factors identified by PlantTFCat48, v) the InterPro analysis matching 31,853 proteins49. Finally, the annotations were tested and edited when needed to follow consistency rules defined by GenBank (see URLs section).
De novo transposable element and repeat annotation
The pseudo-chromosomes were deconstructed into “virtual” contigs by removing stretches >11 undefined bases (Ns) to exclude gaps. We generated 2,742 « virtual » contigs with a N50 of 22 Mb for a total length of 515 Mb. TEdenovo pipeline50,51 from the REPET package v2.5 (see URLs section) was used to detect transposable elements (TEs) in these contigs and to build a consensus sequence for each TE family using a minimum of 5 sequences per group. A library of 28,545 consensus, classified according to structural and functional features (similarities with characterized TEs from RepBase database v21.0152 and domains from Pfam27.0), was generated. After removing redundancy and filtering consensus classified as satellites (labelled SSR) and unclassified consensus constructed with <10 copies in the genome, a library of 8,226 consensus was used to annotate TE copies in the whole homozygote genome using the TEannot pipeline with default parameters53. To refine TE annotation, consensus showing no full-length fragments (i.e. fragment covering more than 95% of the consensus sequence) in the genome were filtered out and a subset of 3,933 consensus was used to run a second TEannot iteration. After a step of manual curation to re-classify some consensus, final annotation files were renamed with this new classification and this library was used to annotate the heterozygote genome (15,938 scaffolds for a total length without Ns of 746 Mb) with the TEannot pipeline. Consensus classified as potential host genes (PHG) harboring Pfam domains, were manually curated and removed from the TE set (453 consensus).
Annotation of miRNA precursors and mature miRNAs
To identify R. chinensis miRNA genes an RNA library was constructed using mixed RNAs from pooled organs. After adapter cleaning and removal of rRNA/tRNA related sequences, we identified 38 million putative small RNAs displaying a size distribution ranging between 20 and 25nt, with two peaks at 21 nt (17 million) and 24 nt (11.8 million). Genome wide annotation of miRNA precursors was performed with an updated version of the pipeline described by Formey et al54, modified to integrate stringent criteria proposed by miRBase (e.g., expression of both 5p and 3p matures55. A total of 207 miRNA precursor loci were predicted to correspond to 636 expressed mature precursors (328 5p and 308 3p). miRNA targets were predicted using miRanda v3.0 (see URLs section). Known mature miRNAs not found by the automatic and stringent process were annotated using blastn.
Genetic structure and genome segmentation
Illumina data mapping and SNP calling was performed as described in Supplementary Notes 8. The number of homozygote and heterozygote variants by sliding windows of 1 Mb was computed for each genotype using functions of the bedtools suite (bedtools makewindows, bedtools intersect, bedtools groupby)56 on genic SNPs. To compute the density of variants per window, the number of variants was divided by the number of informative sites (mapping coverage between 5 and 60 for the fourteen resequenced species and between 50 and 300 for the heterozygote Old Blush genotype). We use the term variants in tetraploid species to refer both to allelic differences and differences between homeologues (i.e., between genes of different sub-genomes). Due to vegetative multiplication of rose cultivars, limited recombination has occurred after hybridization, and introgressed fragments should be of large size. If the genomes or sub-genomes involved in hybridization events have a different distance with the reference genome, genomic regions with different introgression histories should display different levels of variant density in resequenced hybrid cultivars. We used the changes in variant density in the genotypes FRA, GIG, HUM, MUT and SAN to segment the genome into 35 intervals (ranging from 2 to 56 Mb). The genomic boundaries were defined as the start of the windows corresponding to the inflexion points in density files. For each of the thirty-five genome segments, the genetic structure was inferred on bi-allelic SNPs with no missing data and not overlapping with repeated elements. Principal component analyses57 were performed with the glPCA function of the adegenet package (version 2.0.1) 58. Axes 1 and 2 of the PCA explained a significant proportion of the variance (29.29 to 40.53% and 12.07 to 19.89% respectively). Therefore, we present only the analyses of these two axes.
Rose and Rosaceae paleogenomics
Two parameters were defined as previously described59 to increase the stringency and significance of BLAST sequence alignment by parsing BLAST results and rebuilding HSPs (High Scoring Pairs) or pairwise sequence alignments to identify accurate paralogous and orthologous relationships between Rosa (7 chromosomes, 49,767 genes), apricot (8 chromosomes, 31390 genes), peach (8 chromosomes, 27,864 genes), apple (17 chromosomes, 63,514 genes), pear (17 chromosomes, 42,812 genes) and strawberry (7 chromosomes, 32,831 genes). From the previous orthologous and paralogous relationships, ancestral karyotypes were reconstructed as defined in Salse (2016)59 where the ancestral genome is a ‘median’ or ‘intermediate’ genome consisting of a clean reference gene order common to the extant species investigated.
Biochemical analyses of scent composition in roses
Volatile compounds were extracted from petals and stamens of the different rose genotypes with hexane, mainly as previously described28 (Supplementary Notes 9). Camphor was used as an internal standard to estimate compound quantities. Hexane sample fraction was analyzed with a gas chromatograph coupled to an electron ionization mass spectrometer detector (EI-MS; Agilent 6850) detector operated under an ion source temperature of 230°C, a trap emission current of 35 µA and a 70 eV ionization energy. All experiments were performed at least twice. Chromatograph data were analyzed using Agilent Data Analysis software and the volatile substances were identified by screening the WILEY 275, NIST 08 and CNRS libraries to compare MS spectra. The Kovats retention indexes (KI) of each substance were calculated using data of the injection of a homologous set of n-alkane (C8-C20) according to the Kovats formula60. Mass spectra similarities combined with KI were then used for compound identification. Concentrations were calculated by comparison of the camphor area as the internal standard.
ChIP-seq assay
Petal were collected from R. chinensis ‘Old Blush’ and fixed in 1% (v/v) formaldehyde. ChIP assays were performed using anti-H3K9ac (Millipore, ref. 07-352) or anti-H3K27me3 (Millipore, ref. 07-449) antibodies according to a procedure adapted from Veluchamy et al61. Library quality was assessed with Agilent 2100 Bioanalyzer (Agilent) and the libraries were subjected to high-throughput sequencing on Illumina NextSeq 500. After trimming, reads were aligned onto R. chinensis genome with bowtie262 and a maximum mismatch of 1 bp and unique mapping reported. To determine the target regions of H3K9ac ChIP-seq, the Model-based Analysis of ChIP-seq (MACS2)63 was used. Detection of H3K27me3 modification regions was performed using SICER64. HOMER65 was used to annotate H3K9ac peaks with nearby genes if peaks were located into -2k to +1kb window around the gene TSS. For H3K27me3 peaks, bedtools intersect 56 was used and only genes that are overlapped with this specific modification were kept. Clustering of H3K9ac and H3K27me3 peaks was performed using SeqMINER66. Rstudio, Circos67 and NGSplot68 were used for graphic representation of histone modifications.
RNA preparation and qPCR analyses
Total RNA and small RNA were prepared from petals at the three following developmental stages: Non-colored petals early during development (Closed bud; Stage 1); Petals at onset of anthocyanin synthesis (Closed bud; Stage 2); Fully colored petals with maximum anthocyanin content (Bud opening; Stage 3). Total RNA was prepared as previously described69. One microgram of RNA was used in reverse transcription assay and qPCR as previously described70 using gene specific primers (Supplementary Notes 10; Supplementary Tables 8 and 9). Small RNAs were extracted using Macherey-Nagel NucleoSpin® miRNA. Contaminating DNA was removed using the Ambion® DNA-free kit (Cambridgeshire, UK). RNA concentration was measured on a NanoDrop ND-1000 Micro-Volume (NanoDrop Technologies) before and after DNase treatment. Small RNA quantification was performed using stem-loop RT-PCR as previously described71. Reverse transcription was performed with RevertAid kit (Thermo Fisher Scientific). Primers specific to 5.8S rRNA or stem-loop RT-primer for miR156 (Supplementary Notes 10; Supplementary Table 8) were used. 5.8S rRNA and miR156 expression were quantified on QuantStudio™ 6 Flex Real-Time PCR 384 (Applied Biosystems) using Fast SYBR® Green Master Mix kit (Roche Diagnostic) and specific primers (Supplementary Notes 10). Data were collected for three independent biological replicates.
URLs
Genome browser and genomic resources, https://lipm-browsers.toulouse.inra.fr/pub/RchiOBHm-V2/. MetExplore, https://metexplore.toulouse.inra.fr/metexplore2/?idBioSource=5104. EuGene plant pipeline, http://eugene.toulouse.inra.fr/Downloads/egnep-Linux-x86_64.1.4.tar.gz. tbl2asn2, https://www.ncbi.nlm.nih.gov/genbank/tbl2asn2/. REPET, https://urgi.versailles.inra.fr/Tools/REPET. miRanda, http://www.microrna.org. til-r, http://lipm-bioinfo.toulouse.inra.fr/download/til-r/
Supplementary Material
Acknowledgements
We thank J. Thomas, T. Goujon and C. Bendahmane for critical reading of the manuscript. We thank Alain Meilland for helpful discussion. We thank Alexis Lacroix, Patrice Bolland and Justin Berger, the “Lyon Botanical Garden-France” and the Rose Garden “O. Masquelier, La Bonne Maison, Lyon, France” for providing plant material. We thank Loïs Taulelle and Emmanuel Quemener for computing resources.
We thank the Genotoul bioinformatics platform Toulouse Midi-Pyrenées for providing help and computing resources. We gratefully acknowledge support from the Pôle Scientifique de Modélisation Numérique of the ENS de Lyon for the computing resources. We thank the epigenomic platform of the IPS2-University Paris-Sud-Orsay France. We thank the platforms AniRA-Cytometry and ‘Analyse génétiques et cellulaire’ of the IFR BioScience Lyon (UMS3444/US8) for HRM and FACS experiments. PacBio sequencing was supported by the GET-PACBIO program (Programme operationnel FEDER-FSE MIDI-PYRENEES ET GARONNE 2014-2020).
This work was supported by funds from the French National Institute of Agronomic Research (INRA), by the program Fonds Recherche of Ecole Normale Supérieure-Lyon-France to M.Bend and O.R., by the Genoscope to P.W., by French National Research Agency programs DODO (ANR-16CE20-0024-03 to M.Bend. and M.Va.), and AUXIFLO (ANR-12-BSV6-0005 to T.V), and by the European Research Council (ERC-SEXYPARTH) and by the Labex Saclay Plant Sciences -SPS (ANR-10-LABX-0040-SPS) to A.Be.
Footnotes
Life Sciences Reporting Summary. Further information on experimental design is available in the Life Sciences Reporting Summary.
Code availability. Source code (in C) and linux binaries of the til-r software are available (see URLs section) under the GPL license.
Data availability.
Rosa chinensis ‘Old Blush’ homozygous genome has been deposited at DDBJ/ENA/GenBank under the accession PDCK00000000. PacBio raw data under the study accession SRP119907. Rosa chinensis ‘Old Blush’ heterozygous genome has been deposited under the accession PRJEB24406.
Resequencing sequence reads have been deposited in the database as study SRP119986.
Hi-C data were deposited under SRA numbers SRR6189546 and SRR6189547 and ChIPseq data under SRA numbers SRR6167310, SRR6167311, SRR6167312 and SRR6167313 and under GEO number GSE109433.
Author contributions.
O.R., C.B. performed DNA extraction. P.Ve., P.Vi. produced the rose homozygous line. C.L-R., M.Bend. PacBio sequencing data production. J.Sz. performed flux cytometry experiments. P.H. provided Rosa material. M.Ve, D.L., M.Benh., M.P. performed epigenome analysis. O.R., X-P.F., S-H.Y., A.Du., M.L-B., M.Bend. contributed DNA/RNA sample collection and data production. J.J., M.Ve. M.L., L.F, O.R. performed RNAseq sequencing and analyses of gene expression. M.Benh., M.Ve., C.L. A.Bo., A.Be. contributed chromosome conformation capture Hi-C. M.Benh., M.Ve. integrated assembly and genetic map to build pseudo-chromosomes. J.G. developed bioinformatics tools and assembled the PacBio homozygous genome. M.Benh., M.Ve. validated the assembly with HiC and genetic data. P.W., A.Lem. performed Illumina sequencing. A.Bo., A.Be, contributed resequencing sequencing data. A.C, J-MA., A.M., K.L., P.W. assembled the rose heterozygous Illumina sequencing data. N.C., H.Q., J.J. conducted Repetitive DNA analysis. J.G., N.C., J.J. annotated protein coding genes, Transposable Elements and miRNA. J.G., H.B., J.J, L.C. performed bioinformatics analyses. S.C. built the Rosa web portal. J.Sa., C.P. conducted paleo-evolution analyses. J.J., O.R. conducted miRNA analyses. A.La., T.V. J.J. performed integrated analyses on auxin genes. SM, J-C.C., S.B. performed GC-MS analyses of scent compounds. SM, J-C.C., S.B., O.R., J.J. performed integrated analyses on scent genes. O.R., L.P., F.P., L.F., M.Ve. performed integrated analyses on flowering genes. M.Va. performed integrated analyses on MADS transcription factor genes. O.R., L.F., J.J., L.P., J.S., V.B. performed integrated analyses on color genes. M.L-B., B.G., Y.L. performed integrated analyses on meiosis genes. H.B., O.R. J.G., J.J., F.P performed diversity analysis. M.Bend., J.G. coordinated the rose genome consortium. M.Bend., O.R., H.B., J.G. wrote the manuscript.
Competing interests.
The authors declare no competing financial or non-financial interests.
References
- 1.Fougere-Danezan M, Joly S, Bruneau A, Gao XF, Zhang LB. Phylogeny and biogeography of wild roses with specific attention to polyploids. Ann Bot. 2015;115:275–91. doi: 10.1093/aob/mcu245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Vries DP, Dubois L. Rose breeding : past, present, prospects. Acta Horticulturae. 1996;424:241–248. [Google Scholar]
- 3.Martin M, Piola F, Chessel D, Jay M, Heizmann P. The domestication process of the Modern Rose: genetic structure and allelic composition of the rose complex. Theoretical-and-Applied-Genetics. 2001;102:398–404. [Google Scholar]
- 4.Hurst CC. Notes on the orig in and evolution of our garden roses. Journal of the Royal Horticultural Society. 1941;66:73–82. [Google Scholar]
- 5.Bendahmane M, Dubois A, Raymond O, Bris ML. Genetics and genomics of flower initiation and development in roses. J Exp Bot. 2013;64:847–57. doi: 10.1093/jxb/ers387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esselink GD, Smulders MJ, Vosman B. Identification of cut rose (Rosa hybrida) and rootstock varieties using robust sequence tagged microsatellite site markers. Theor Appl Genet. 2003;106:277–86. doi: 10.1007/s00122-002-1122-y. [DOI] [PubMed] [Google Scholar]
- 7.Zharkikh A, et al. Sequencing and assembly of highly heterozygous genome of Vitis vinifera L. cv Pinot Noir: problems and solutions. J Biotechnol. 2008;136:38–43. doi: 10.1016/j.jbiotec.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Yokoya K, Roberts AV, Mottley J, Lewis R, Brandham PE. Nuclear DNA Amounts in Roses. Annals of Botany. 2000;85:557–561. [Google Scholar]
- 9.Nakamura N, et al. Genome structure of Rosa multiflora, a wild ancestor of cultivated roses. DNA Res. 2017 doi: 10.1093/dnares/dsx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badouin H, et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature. 2017;546:148–152. doi: 10.1038/nature22380. [DOI] [PubMed] [Google Scholar]
- 11.VanBuren R, et al. Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature. 2015;527:508–11. doi: 10.1038/nature15714. [DOI] [PubMed] [Google Scholar]
- 12.Koren S, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin CS, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourke PM, et al. Partial preferential chromosome pairing is genotype dependent in tetraploid rose. Plant J. 2017;90:330–343. doi: 10.1111/tpj.13496. [DOI] [PubMed] [Google Scholar]
- 15.Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 16.Iwata H, et al. The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 2012;69:116–25. doi: 10.1111/j.1365-313X.2011.04776.x. [DOI] [PubMed] [Google Scholar]
- 17.Li S, et al. Inheritance of Perpetual Blooming in Rosa chinensis ‘Old Blush’. Horticultural Plant Journal. 2015;1:108–112. [Google Scholar]
- 18.Mouradov A, Cremer F, Coupland G. Control of flowering time: interacting pathways as a basis for diversity. Plant Cell. 2002;14(Suppl):S111–30. doi: 10.1105/tpc.001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaistij FE, et al. Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc Natl Acad Sci U S A. 2013;110:10866–71. doi: 10.1073/pnas.1301647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huo H, Wei S, Bradford KJ. DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc Natl Acad Sci U S A. 2016;113:E2199–206. doi: 10.1073/pnas.1600558113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han Y, et al. Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis. Sci Rep. 2017;7:43382. doi: 10.1038/srep43382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–22. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zvi MM, et al. PAP1 transcription factor enhances production of phenylpropanoid and terpenoid scent compounds in rose flowers. New Phytol. 2012;195:335–45. doi: 10.1111/j.1469-8137.2012.04161.x. [DOI] [PubMed] [Google Scholar]
- 24.Lin-Wang K, et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010;10:50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aharoni A, et al. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell. 2004;16:3110–31. doi: 10.1105/tpc.104.023895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shulaev V, et al. The genome of woodland strawberry (Fragaria vesca) Nat Genet. 2011;43:109–16. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu ZX, et al. Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and Patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Mol Plant. 2015;8:98–110. doi: 10.1016/j.molp.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Magnard JL, et al. PLANT VOLATILES. Biosynthesis of monoterpene scent compounds in roses. Science. 2015;349:81–3. doi: 10.1126/science.aab0696. [DOI] [PubMed] [Google Scholar]
- 29.Touraev A, Heberle-Bors E. Microspore embryogenesis and in vitro pollen maturation in tobacco. Methods Mol Biol. 1999;111:281–91. doi: 10.1385/1-59259-583-9:281. [DOI] [PubMed] [Google Scholar]
- 30.Brioudes F, Thierry AM, Chambrier P, Mollereau B, Bendahmane M. Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc Natl Acad Sci U S A. 2010;107:16384–9. doi: 10.1073/pnas.1007926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrier G, et al. An efficient and rapid protocol for plant nuclear DNA preparation suitable for next generation sequencing methods. Am J Bot. 2011;98:e13–5. doi: 10.3732/ajb.1000371. [DOI] [PubMed] [Google Scholar]
- 32.Vergne P, et al. Somatic embryogenesis and transformation of the diploid rose Rosa chinensis cv ‘Old Blush'. Plant Cell Tissue and Organ Culture. 2010;100:73–81. [Google Scholar]
- 33.Gnerre S, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 2011;108:1513–8. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu W, et al. Altered chromatin compaction and histone methylation drive non-additive gene expression in an interspecific Arabidopsis hybrid. Genome Biol. 2017;18:157. doi: 10.1186/s13059-017-1281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C, et al. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 2015;25:246–56. doi: 10.1101/gr.170332.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Servant N, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akdemir KC, Chin L. HiCPlotter integrates genomic data with interaction matrices. Genome Biol. 2015;16:198. doi: 10.1186/s13059-015-0767-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang H, et al. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 2015;16:3. doi: 10.1186/s13059-014-0573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10:563–9. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 40.Walker BJ, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–80. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Foissac S, G J, Rombauts S, Mathe C, Amselem J, Sterck L, Van de Peer Y, Rouze P, Schiex T. Genome annotation in plants and fungi: EuGene as a model platform. Current Bioinformatics. 2008;3:87–97. [Google Scholar]
- 44.Schlapfer P, et al. Genome-Wide Prediction of Metabolic Enzymes, Pathways, and Gene Clusters in Plants. Plant physiology. 2017;173:2041–2059. doi: 10.1104/pp.16.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cottret L, et al. MetExplore: a web server to link metabolomic experiments and genome-scale metabolic networks. Nucleic Acids Research. 2010;38:W132–nW137. doi: 10.1093/nar/gkq312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.UniProt: the universal protein knowledgebase. Nucleic Acids Research. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Y, et al. iTAK: A Program for Genome-wide Prediction and Classification of Plant Transcription Factors, Transcriptional Regulators, and Protein Kinases. Molecular Plant. 2016;9:1667–1670. doi: 10.1016/j.molp.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 48.Dai X, Sinharoy S, Udvardi M, Zhao PX. PlantTFcat: an online plant transcription factor and transcriptional regulator categorization and analysis tool. BMC bioinformatics. 2013;14:321. doi: 10.1186/1471-2105-14-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finn RD, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic acids research. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flutre T, Duprat E, Feuillet C, Quesneville H. Considering transposable element diversification in de novo annotation approaches. PLoS One. 2011;6:e16526. doi: 10.1371/journal.pone.0016526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoede C, et al. PASTEC: an automatic transposable element classification tool. PLoS One. 2014;9:e91929. doi: 10.1371/journal.pone.0091929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–7. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 53.Quesneville H, et al. Combined evidence annotation of transposable elements in genome sequences. PLoS Comput Biol. 2005;1:166–75. doi: 10.1371/journal.pcbi.0010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Formey D, et al. The small RNA diversity from Medicago truncatula roots under biotic interactions evidences the environmental plasticity of the miRNAome. Genome biology. 2014;15:457. doi: 10.1186/s13059-014-0457-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearson K. LIII. On lines and planes of closest fit to systems of points in space. The London, Edinburgh, and Dublin Philosophical Magazine and Journal of Science. 1901;2:559–572. [Google Scholar]
- 58.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–5. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 59.Salse J. Ancestors of modern plant crops. Curr Opin Plant Biol. 2016;30:134–42. doi: 10.1016/j.pbi.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 60.Adams RP. Identification of Essential Oil Components By Gas Chromatography/Mass Spectrometry. 4th Edition. Allured Publishing Corporation; Carol Stream, Illinois, USA: 2007. [Google Scholar]
- 61.Veluchamy A, et al. LHP1 Regulates H3K27me3 Spreading and Shapes the Three-Dimensional Conformation of the Arabidopsis Genome. PLoS One. 2016;11:e0158936. doi: 10.1371/journal.pone.0158936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–8. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ye T, et al. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39:e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dubois A, et al. Tinkering with the C-function: a molecular frame for the selection of double flowers in cultivated roses. PLoS One. 2010;5:e9288. doi: 10.1371/journal.pone.0009288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dubois A, et al. Genomic approach to study floral development genes in Rosa sp. PLoS One. 2011;6:e28455. doi: 10.1371/journal.pone.0028455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marcial-Quino J, et al. Stem-Loop RT-qPCR as an Efficient Tool for the Detection and Quantification of Small RNAs in Giardia lamblia. Genes (Basel) 2016;7 doi: 10.3390/genes7120131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.