Abstract
Objective
Currently, there are no reliable biomarkers for predicting therapeutic response in patients with rheumatoid arthritis (RA). The synovium may unlock critical information for determining efficacy as reduction in numbers of sublining synovial macrophages remains the most reproducible biomarker. Thus, a clinically actionable method for collection of synovial tissue, which can be analyzed using high-throughput strategies, must become a reality.
Methods
Rheumatologists at six United States academic sites were trained in minimally invasive ultrasound-guided synovial tissue biopsy. Histology, fluorescence-activated cell sorting and RNA-seq were performed on biopsy synovial tissue from patients with RA and compared with osteoarthritis (OA) samples. An optimized protocol for digesting synovial tissue was developed to generate high quality RNA-seq libraries from isolated macrophage populations. Associations were determined between macrophage transcriptional profiles and clinical parameters of RA patients.
Results
Patients with RA reported minimal adverse effects in response to synovial biopsy. Comparable RNA quality was observed between synovial tissue and isolated macrophages from patients with RA and OA. Whole tissue samples from patients with RA demonstrated a high degree of transcriptional heterogeneity. In contrast, the transcriptional profile of isolated RA synovial macrophages highlighted a subpopulation of patients and identified six novel transcriptional modules that were associated with disease activity and therapy.
Conclusion
Performance of synovial tissue biopsies by rheumatologists in the United States is feasible and generates high-quality samples for research. By utilizing cutting-edge technologies on synovial biopsies with corresponding clinical information, a precision-based medicine approach for patients with RA is attainable.
Introduction
Despite the many therapies for patients with rheumatoid arthritis (RA), there is little information to guide selection of the most effective treatment for an individual patient. Forty-sixty percent of patients with RA respond (defined by ACR50 response criteria) to conventional disease modifying anti-rheumatic drugs (cDMARDs) (1, 2) or cDMARDs plus anti-tumor necrosis factor (TNF) therapy (3–11). Moreover, 20-40% of subjects in clinical trials never demonstrate even a minimal response (ACR20 response criteria) (7–11). Based on a population of over 300 million in the United States, a disease prevalence of 0.6%, and a course of 3-4 months per biologic DMARD therapy, as much as $2.5 billion is wasted annually on inadequate therapy (12, 13). There is a clear need to develop precision-based therapy for patients with RA, whereby clinical information such as novel biomarkers will enhance our ability to predict the therapeutic response and thereby limit ineffective therapy.
Previous studies have linked macrophages to the pathogenesis of RA. Synovial macrophages are highly activated, express elevated levels of toll-like receptor (TLR) 2, 4 and 7 (14, 15) and contribute directly and indirectly to synovial inflammation and destruction of cartilage and bone through the production of degradative enzymes, cytokines, and chemokines. Further, TLR 2, 3 and 7 play essential roles in the development of inflammatory arthritis in mice (16–18). More importantly, macrophages are the central producers of IL-1β, IL-6 and TNFα, which comprise the three essential pro-inflammatory cytokines that contribute to RA pathogenesis. To date, the approved therapeutics including anti-TNFα, anti-IL-6, Jak inhibitors, CTLA4-Ig and anti-CD20 have all been shown to decrease synovial inflammation, bone destruction, and more importantly reduce macrophage numbers in synovial sublining (19–21). Despite the fact that synovial macrophages were discovered more than a half-century ago and are crucial for RA pathogenesis (22), surprisingly very little is known about them.
Biomarkers that indicate sensitivity or resistance to a particular therapy are sorely lacking in RA. For the most part, researchers have utilized peripheral blood, with minimal success, to identify biomarkers for predicting a response to therapy (23). Similarly, the results of genetic approaches have been disappointing (24). More recent studies suggest the synovium, as the target organ in RA, may have greater potential in determining therapeutic response (23). Currently, the most well-known and reproducible biomarker for response to RA therapy is a reduction in the number of sublining synovial macrophages using immunohistochemistry of synovial tissue (25). While arthroscopy has been the most common method to obtain synovial tissue before and after therapy (26–32) and yields substantial amounts of synovial tissue, they are invasive, require surgical suites, and are expensive, thus limiting their usefulness in clinical practice and clinical studies. Synovectomy and joint replacement surgery are other common mechanisms for researchers to obtain synovial tissue, but these patients typically exhibit end-stage disease characteristics and likely do not reflect the overall pathophysiology at the time when therapeutic decisions are made prior to progressive joint damage.
Ultrasound technology has significantly advanced and is widely used by rheumatologists as a mechanism for determining the degree of synovitis and inflammation, for detecting erosions, and for identifying sites for therapeutic injection (33). Over the past decade, ultrasound has been used to facilitate collection of synovial tissue (29). Minimally invasive ultrasound-guided synovial tissue biopsies have been performed for research purposes throughout Europe and the standardization for these procedures has been fully evaluated (23, 29, 34–46). However, there are currently no published studies from the United States describing the utilization of ultrasound-guided synovial tissue biopsies for research. The potential reasons that this technique has not been commonly adopted for research in the United States include a lack of training, differences in the medical system and patient populations between Europe and the United States, and a lack of “buy-in” from rheumatologists who would recommend the procedure to their patients.
We assembled a consortium of established academic rheumatology groups in the United States including the University of Alabama at Birmingham (UAB), Columbia University, Mayo Clinic, Washington University, University of Michigan, and Northwestern University to form the RhEumatoid Arthritis SynOvial tissue Network (REASON). Our consortium was trained in minimally invasive ultrasound-guided synovial biopsy techniques in the UK and has since performed over 41 biopsies in the United States on RA patients with active disease. RNA was extracted from whole synovial tissue and from FACS-sorted synovial macrophages for RNA-seq analysis. The transcriptional profiles of isolated macrophages were used to distinguish between RA patient groups and identify modules of co-regulated genes that were associated with clinical disease and medication. We believe that these studies demonstrate the utility of isolating individual populations of cells within the synovium to understand pathobiology of disease and to establish a precision-medicine-based approach for RA patients.
Materials and Methods
Patients
Adult patients with RA, defined by either the 1987 ACR criteria or 2010 ACR/EULAR criteria, were selected as candidates for ultrasound-guided synovial biopsy based on the presence of palpable synovitis documented by clinical examination (47–49). To increase uniformity of the collected tissue, only wrists were sampled in this study. Candidate joints were scanned with standard two-dimensional B-mode ultrasound with and without Doppler (SonoSite M-MSK with 15-6 MHz linear probe, FujiFilm SonoSite), and were included in the study if they had a gray scale synovitis score of ≥2 on a 4-point scale (35). Exclusion criteria included uncontrolled comorbid diseases, therapeutic anticoagulation (low-dose aspirin and non-steroidal anti-inflammatories were allowed), use of systemic steroids in excess of prednisone 10mg daily or equivalent, administration of intramuscular steroids within the previous 4 weeks or intra-articular steroids into the target joint within the past 8 weeks, chronic or recurrent infection, intolerance to lidocaine or chlorhexidine, inability to communicate effectively in English, and membership in a vulnerable population (prisoners, pregnant women, etc.).
Training for Ultrasound-Guided Synovial Biopsies
The REASON consortium was created to adopt the ultrasound-guided synovial biopsies for research purposes in the United States. Rheumatologists from REASON who were experienced in ultrasonography traveled to the UK for a two-day training session with Drs. Andrew Filer, Christopher Buckley, Stephen Kelly, and Costantino Pitzalis on ultrasound-guided synovial biopsies. This session included observation of ultrasound-guided synovial biopsies followed by a practice session using cadavers. Following the training session, the rheumatologists from REASON attended two additional practice sessions using cadavers at Northwestern University for refresher training.
Ultrasound-Guided Synovial Biopsy Procedure
Procedures were performed either in exam rooms in outpatient rheumatology clinics or in designated research space. After re-confirming the presence of synovitis by sonographic criteria as above using customary non-sterile techniques, patients were dressed in a lab cover-up or exam gown, surgical mask, and surgical hair net. Rheumatologists performing the biopsy were in surgical scrubs with cap, mask, sterile surgical gown, and sterile gloves. The ultrasound probe was placed in a sterile cover. The subject’s hand and arm were scrubbed with chlorhexidine from fingertips to mid-forearm, and the subject then placed the hand palm down onto a surgical wrist support in a prepared sterile field. The arm was draped in a sterile manner with a fenestrated sheet centered on the wrist, and sterile ultrasound gel was applied. Ultrasound scanning over the dorsal aspect of the wrist joint in both the longitudinal and transverse planes was then used to locate the region of greatest synovitis in the wrist, usually immediately dorsal/superficial to either the proximal or the distal row of carpal bones. A wheal of lidocaine was used to anesthetize the skin at the ulnar aspect of the wrist. While monitoring the procedure under ultrasound in real time with the probe in the transverse position, an initial ultrasound-guided lidocaine pass into the target joint was made with a 25-gauge by 1.5-inch needle. A second ultrasound-guided lidocaine pass was made into the same wound and needle track with an 18-gauge by 1.5-inch needle to ensure anesthesia and a clear path for the biopsy device. Care was taken to avoid neurovascular structures and tendons, especially the extensor digiti minimi. An appropriately-sized Quick Core Biopsy needle (Cook Medical, Bloomington, IN) was then selected based on the target wrist (usually 18-gauge by 9cm, with 10mm throw) and introduced into the same needle track.
Using continuous real-time ultrasound imaging, the jaw of the device was positioned within the synovium [defined by OMERACT (50) as intra-articular, hypoechoic, non-displaceable, poorly-compressible tissue] at the point of its greatest abundance, usually between the common extensor tendon bundle and the underlying carpal row. The device was triggered and removed. The sample was removed from the device by scraping with a sterile 21-gauge needle and placed into PBS (Thermo Fisher). The process was repeated to obtain a total of 12 samples. Not all biopsy passes yielded tissue; most procedures resulted in 1-3 “empty passes”, and thus most subjects underwent a total of 13-15 passes to obtain 12 samples of synovium. All samples were taken through the same skin wound but with varying positions of the jaw of the device within the region of most abundant synovitis, with the intent of sampling the entire chosen region uniformly. Variations in position of the device also included variations in the radial orientation of the jaw with respect to the axis of the device. For example, some samples were taken from 12 o’clock, closer to the extensor tendon bundle, and some from 6 o’clock, closer to the carpal row. At the completion of the procedure the biopsy site was washed, and after confirming hemostasis, an adhesive bandage was applied to the single puncture wound. No subject required a suture. Patients were instructed in routine after-care and encouraged to use over-the-counter acetaminophen for any discomfort, with permission to escalate to over-the-counter NSAIDs if needed and to contact the research team by telephone to report any adverse events, including delayed healing or pain not controlled with over-the-counter agents as above. Most procedures lasted a total of 45-60 minutes from the non-sterile scout imaging to the subject departing the procedure room. Subjects did not require conscious sedation and were sent home immediately with no post-procedure observation/recovery period and no activity restrictions.
Tolerability
Tolerability of the procedure was assessed by questionnaires administered before and after the procedure as previously described (36). Patients were asked to rate pain, swelling and stiffness on a 10-point visual analog scale (VAS). After the procedure, patients were also asked by questionnaire to rate their discomfort during the procedure (none, mild, moderate, severe) and to rate their likelihood (very likely, somewhat likely, unsure, somewhat unlikely, very unlikely) to agree to undergo another procedure.
Tissue Preparation and Flow Cytometry
From the 12 pieces of synovial tissue obtained by needle biopsies, four were selected at random and placed into 10% neutral buffered formalin (NBF) for histology; four more randomly-selected pieces were placed into RNALater (Ambion) for whole tissue processing; and the remaining four were placed into PBS for tissue digestion. Osteoarthritis (OA) synovial tissue was received from the National Disease Research Interchange (NDRI) that was shipped in DMEM and antibiotics overnight on wet ice. Only soft tissues containing meniscus and synovium were processed. OA synovial tissue was processed identically to tissue from patients with RA.
The length of digestion (30 to 60 min.) and intensity of mechanical disaggregation pre- and post-incubation were varied to optimize macrophage isolation. Mechanical disaggregation was performed on a GentleMACS dissociator (Miltenyi Biotec). The pre-set gentleMACS programs m_lung_01 and m_brain_01 were used to test moderate and aggressive mechanical disaggregation, respectively. Sorted macrophages used for analysis were processed by infusing tissue with a digestion buffer [RPMI 1640 (Sigma), Liberase TL (Roche, 0.1mg/mL) and DNase (Roche, 0.1mg/mL)] and minced with scissors. Tissue suspensions were transferred to C-tubes (Miltenyi Biotec) and incubated for one hour at 37°C with aggressive disaggregation pre- and post- incubation. The digestion reaction was quenched with MACS buffer (Miltenyi Biotec) and the tissue suspension was filtered over a 40-micron filter. Red blood cells were lysed (BD Pharm Lyse) and then washed twice with HBSS (Thermo Fisher). Cells were counted (Invitrogen Countess) and stained with a viability dye (Supplemental Table 1; 0.5 μL/mL, 15 min., 25°C, dark). Cells were then washed twice with MACS buffer, incubated with Fc block (BD Biosciences; 6μL/60μL total volume, max 5×106 cells, 4°C, 15 min., dark), stained with antibody cocktail (Supplemental Table 1; 4°C, 30 min., dark), washed twice and re-suspended in MACS buffer and kept on ice until sorting. Synovial macrophages (CD45+, CD11b+, HLA-DR+, CD15−, CD1c−, CD206+) were sorted on a BD FACSAria SORP instrument (BD Bioscience) at the Northwestern University Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Core Facility. Cells from RA synovial biopsies were sorted directly into 100μL PicoPure RNA extraction buffer (Arcturus Bioscience, Inc.). Cells from OA tissues were sorted (more than 10,000 cells) into cold MACS buffer (Miltenyi Biotec), immediately centrifuged at 4°C, supernatant removed, and resuspended in 100μL PicoPure RNA extraction buffer. All cells were stored at −80°C until RNA was extracted.
Histopathology and Immunohistochemistry
The 4 biopsy pieces fixed in 10% neutral buffered formalin for histology were stored overnight and submitted to the Pathology Core Facility at Northwestern University. Paraffin-embedded tissue sections were stained with hematoxylin and eosin (H&E), and for CD45 or CD68. Slide images were taken at 40× and 100× magnification using an Olympus BX41 microscope and Olympus DP21 camera. Since not all samples using this procedure demonstrate synovial lining, other characteristics including CD45 and CD68 staining were included to provide a semi-quantitative or -qualitative analysis of inflammation in the biopsied tissue. RA synovial biopsy sections stained for Hematoxylin and Eosin (H&E) or with antibodies to either CD45 or CD68 antigens were scored for percent synovium of all four pieces of tissues. The CD45 and CD68 score was based on a 0-4 modified scoring system which described the percent of CD45 or CD68 positivity in identified synovium (35). All scoring was performed by an experienced rheumatologist blinded to the identity of the samples.
Preparation of RNA Library
RNA was isolated from whole synovial tissue by homogenizing tissue with 3.0 mm high impact zirconium beads and a Bead Blaster 24 microtube homogenizer (Benchmark Scientific). RNA was extracted from the cell homogenate using a QIAGEN Plus Mini kit. RNA from sorted macrophages was extracted using a PicoPure RNA isolation kit according to manufacturer’s instructions (Arcturus Bioscience, Inc.). RNA quality and quantity were measured using a High Sensitivity RNA ScreenTape System (Agilent Technologies). Whole synovial tissue RNA-seq libraries were prepared from 90ng of total RNA using the NEBNext Ultra Kit with polyA-enrichment (NEB). RNA-seq libraries from sorted macrophage populations were prepared using a SMART-Seq v4 Ultra Low Input RNA Kit (Clontech Laboratories, Inc.) followed by Nextera XT protocol (Illumina). RNA-seq libraries were sequenced on an NextSeq 500 instrument (Illumina Inc.) with ~5-10×106 aligned reads per sample. A commercially available universal human RNA reference (uhRNA) was prepared along with the synovial RNA to represent background RNA expression.
RNA-seq Analysis
RNA-seq data were de-multiplexed using bcl2fastq, and RNA-seq reads were aligned to the human reference genome (NCBI, hg19) using TopHat2 (version 2.17.1.14) aligners (51). Gene coverage of samples was calculated using the RseQC package (52). Normalized gene counts were calculated using HTSeq (53). For our analysis, we focused on genes in which at least two samples had Fragments Per Kilobase per Million (log2(FPKM)) expression > 3. For visualization, GENE-E (https://software.broadinstitute.org/GENE-E/) was used to generate Pearson pairwise correlation matrices and to perform K-means and hierarchical clustering. Differential gene expression between RA and OA in the whole tissue dataset was determined using the edgeR Bioconductor packages parameters as described (adjusted p-value<0.01) (54, 55). Gene Ontology (GO) associations were determined by GOrilla (56). To account for the increased noise of the low input macrophage-specific RNA-seq, we focused our analysis in Figure 5 on genes in which at least two samples had log2(FPKM+1) expression over 5 and raised all lower values to 5. We defined differentially expressed genes across patients with RA as those with an adjusted range (log2-fold change between the second highest and second lowest samples) > 1. We removed genes driven by one outlier sample (defined as genes where the difference between the Max–2nd highest > distance between 2nd highest-2nd lowest) leaving 553 genes for the analysis. Modules were identified by clustering genes using K-means clustering and calculating the pairwise Pearson correlation between each gene. The enrichment/depletion of modules within each patient was determined by a Kolmogorov-Smirnov (KS) test between the expression level of genes in a given module compared to all 553 genes (P-value<0.05).
Figure 5.
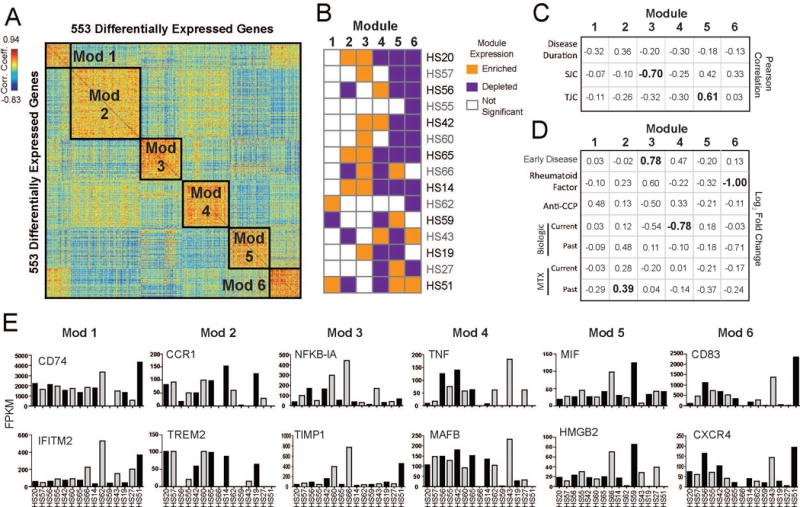
A. Pairwise Pearson correlations between 553 differentially expressed genes of their expression across sorted macrophages from RA patients. Genes are clustered using k-means to identify modules of co-regulated genes (Modules 1-6). B. Kolmogorov-Smirnov test to determine if expression of module genes is enriched (orange) or depleted (purple) in each patient (p-value<0.05). C. Table of association displaying the Pearson correlation between the median expression of gene modules in patients and the given clinical parameters (continuous variables) were calculated. Correlation coefficients with a p-value <0.05 are shown in bold. D. Table of association displaying the average fold-change (Log2FC) between median expression of gene modules in patients that were positive vs. negative for the given clinical parameters (categorical variables). Current medication treatment compares patients who were currently on the medication against those who were not on the medication at the time of biopsy. Past medication treatment compares patients who had stopped medication against those who were never on a given medication. Significant comparisons (p-value<0.05, t-test) are shown in bold. MTX=methotrexate. E. Bar graph showing the expression levels (FPKM) across patients with RA of individual genes selected across the 6 Modules.
Statistical Analysis
Association of RNA-seq expression patterns in patients was determined with the clinical parameters including disease duration, swollen joint count (SJC), tender joint count (TJC), early disease duration (defined as patients with disease < 2 years), rheumatoid factor (RF), Anti-cyclic citrullinated peptide (anti-CCP) antibodies and patient treatment history with biologics and methotrexate (Table 1). Disease duration, SJC and TJC were recorded as continuous variables. Early disease duration, rheumatoid factor, anti-CCP antibodies and patient medication history were recorded as categorical data of either positive or negative. Patient treatment and clinical data association with groups 1 and 2 in Figure 4A were determined using Fisher’s exact test for categorical data and students’ t-test for continuous data. Pearson correlation was used to determine association between patient’s median gene expression for each module and patient information with continuous variables. The log2-fold change of the average median module expression between patients with a positive compared to negative clinical response/treatment regimen was used to calculate association to patient information with categorical variables (Supplemental Table 2). Significant changes in gene expression were determined using student’s t-test.
Table 1.
Patient and Sample Information. Anti-CCP-cyclic citrullinated peptide; ESR-Erythrocyte Sedimentation Rate; DAS-Disease Activity Score; CDAI-Clinical Disease Activity Index; RAPID-Routine Assessment of Patient Index Data; PGA-Physician Global Assessment; HAQ-Health Assessment Questionnaire; TJC-tender joint count; SJC-swollen joint count. Treatment is divided by medication type and whether the patient is currently taking or has previously taken the medication. Standard deviation or percent of population is reported for patient information and treatment. Histological Scoring of Synovial Tissue. All slides were scored by same physician blinded to the origin of the samples. The amount of synovial tissue in each biopsy was estimated by structure, lining and leukocyte content. The number of CD45- and CD68-positive cells were scored on modified scale from 0-4. SEM is reported for Histology and RNA-seq library stats.
|
Patient Information
| ||||
| Age (years) | 57.5 | ± | 11.4 | |
| Gender (female) | 29 | (71 %) | ||
| Gender (Male) | 12 | (29%) | ||
| Disease duration, years n=40 | 9.3 | ± | 8.4 | |
| Early disease (<2 years), n= 40 | 9 | (23%) | ||
| Positive Rheumatoid Factor (RF) n=40 | 23 | (58%) | ||
| Positive Anti-CCP n=36 | 19 | (53%) | ||
| ESR (mm/hour) n=14 | 33.5 | ± | 21.7 | |
| DAS28 n=17 | 4.7 | ± | 1.3 | |
| CDAI n=20 | 21.5 | ± | 10.6 | |
| RAPID3 n=25 | 11.4 | ± | 7.4 | |
| PGA n=24 | 4.7 | ± | 3.1 | |
| HAQ n=18 | 0.7 | ± | 0.5 | |
| TJC n=31 | 5.8 | ± | 6.0 | |
| SJC n=31 | 7.1 | ± | 4.9 | |
|
Treatment (n=31) | ||||
| no Treatment (naïve) | 3 | (10%) | ||
| Methotrexate, current | 15 | (48%) | ||
| Methotrexate, past | 9 | (29%) | ||
| TNF-inhibitor, current | 12 | (38%) | ||
| TNF-inhibitor, past | 17 | (55%) | ||
| IL-6 inhibitor, current | 2 | (6%) | ||
| IL-6 inhibitor, past | 2 | (6%) | ||
| JAK-inhibitor, current | 3 | (10%) | ||
| JAK-inhibitor, past | 5 | (16%) | ||
| Other treatment, current | 11 | (35%) | ||
| Other treatment, past | 12 | (39%) | ||
| Prednisone, <5 mg, current | 1 | (3%) | ||
| Prednisone, 5-10 mg, current | 6 | (19%) | ||
|
Histology-RA Synovial Biopsy (n=30) | ||||
| % Synovial Tissue | 36.0 | ± | 5.6 | |
| CD45 score (0-4) | 1.7 | ± | 0.2 | |
| CD68 score (0-4) | 1 | ± | 0.1 | |
|
RNA-seq Stats | ||||
| Whole Tissue Samples (RA=9, OA=9)
| ||||
| RIN# | RA | 6.7 | ± | 0.5 |
| OA | 8.0 | ± | 0.2 | |
| # aligned reads (Million) | RA | 9.2 | ± | 0.7 |
| OA | 12.9 | ± | 1.1 | |
| % of total reads aligned | RA | 81 9 | ± | 1.2 |
| OA | 85.3 | ± | 1.2 | |
| % Complexity | RA | 54.8 | ± | 1.7 |
| OA | 56.2 | ± | 1.9 | |
|
Sorted Macrophage Samples (RA=15, OA=9) | ||||
| Cells sorted | RA | 1642 | ± | 1178 |
| OA | 77414 | ± | 26413 | |
| # aligned reads (Million) | RA | 21.0 | ± | 4.2 |
| OA | 21.1 | ± | 6.8 | |
| % of total reads aligned | RA | 53 9 | ± | 6.0 |
| OA | 65.2 | ± | 1.5 | |
| % Complexity | RA | 37.2 | ± | 2.7 |
| OA | 38.1 | ± | 2.2 | |
Figure 4.
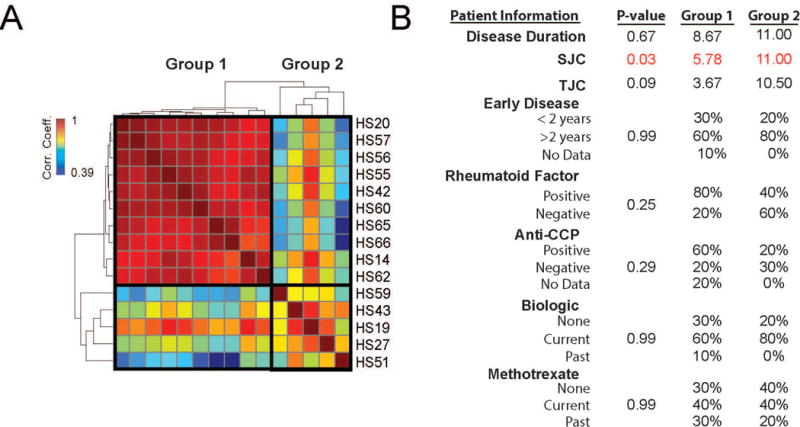
Analysis of global gene expression profiles in sorted macrophages from RA. A. Pairwise Pearson correlation of gene expression between individual patient samples. Samples are organized by hierarchical clustering based on their Pearson coefficients across samples forming 2 groups. B. Table of association analysis between patients in Group 1 and Group 2 as defined in A and clinical parameters. Values reflect either the group average (continuous variables) or percent of patients (categorical variables) in each group with given criteria. P-values for disease duration, SJC and TJC were determined by Student’s t-test and for early disease duration, rheumatoid factor, anti-CCP antibody and treatment status were determined by Fisher’s exact test. Significant values are shown in red. TJC=tender joint count; SJC=swollen joint count; CCP=cyclic citrullinated peptide
Results
Patient Demographics
The REASON consortium was created to adopt the ultrasound-guided synovial biopsies for research purposes in the United States. Over the two years, we recruited 41 patients with RA from REASON sites for ultrasound-guided synovial biopsies. Patient demographics, clinical data and other pertinent information are presented in Table 1.
Ultrasound-guided Synovial Biopsies are Safe and Well Tolerated
Real-time ultrasound images were utilized to guide placement of the needle device for biopsy within the synovium of the dorsal wrist. (Figure 1A-B). Thirty-one of the forty-one patients responded with a complete pre- and post-procedure VAS assessment of pain, stiffness and swelling of the biopsied wrist. There were no differences in the pre- and post- procedure scores in these patients (Figure 1C). Patients were then asked to rate their likelihood of agreeing to repeat the procedure. Over ninety percent (90.3%) of the patients reported that they would be very likely (VL) or somewhat likely (SL) to repeat the biopsy, while only 6.5% stated that they would be somewhat (SU) or very unlikely (VU) to have a repeat biopsy (Figure 1C).
Figure 1.
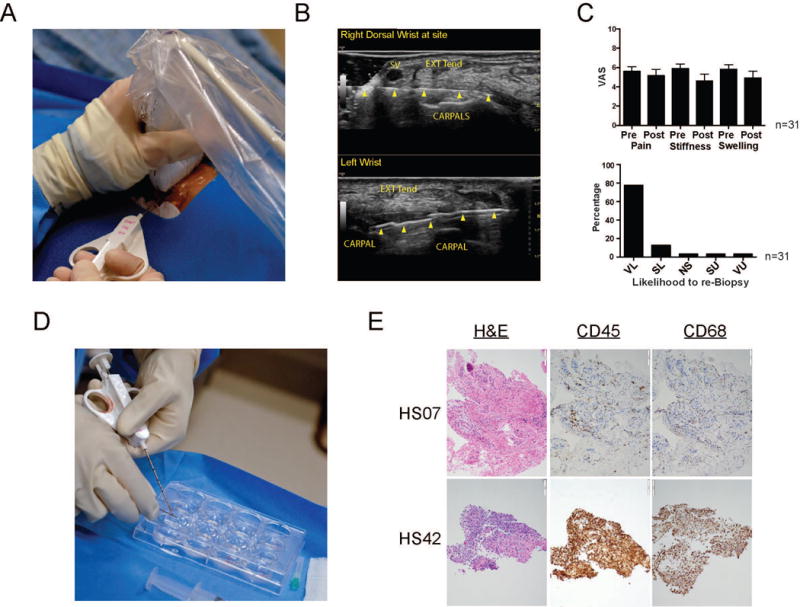
Acquisition of synovial tissue from patients with RA. A. Ultrasound guided synovial biopsy from inflamed wrist with an 18-gauge by 1.5-inch needle. B. Dorsal transverse (axial) view of a right wrist (top) with a 25-gauge by 1.5-inch needle (arrowheads) and left wrist with an 18-gauge by 9 cm needle biopsy device with a 10-mm throw opened (2nd and 3rd arrows from left). SV = superficial vein, EXT TEN = extensor tendon complex, CARPALS = proximal row of carpal bones. C. Patients were asked prior to and following the procedure to complete a visual analogue score (VAS) assessing their pain, stiffness and swelling on a scale of 1-10. Post procedure patients were also asked their likelihood to agree to a subsequent procedure: (VL) very likely, (SL) somewhat likely, (NS) not sure, (SU) somewhat unlikely, and (VU) very unlikely. Error bars display SEM. D. Synovial tissue is removed from biopsy device and placed into PBS on ice until processed. E. Histomorphological features of synovial biopsy obtained from two representative RA patients. Representative photomicrographs of sections stained with Hematoxylin and Eosin (H&E), anti-CD45 (hematopoietic cells), and anti-CD68 antibodies (macrophages).
Histological Assessment of Synovial Tissue
The quality of the synovial biopsies obtained from patients with RA was first quantified by histological assessment including tissue structure, presence of synovial lining and leukocytes (Table 1, Figure 1D-E). Importantly, all but five samples contained synovial tissue (>10%) with eleven biopsies containing ≥50% synovial tissue. The abundance of CD45- and CD68-positive cells in each sample was scored on modified scale of 0-4 (35) revealing a substantial enrichment of hematopoietic cells and more specifically macrophages in most biopsy samples (Table 1).
RNA-seq Analysis of Whole Tissue Synovial Biopsies
The fidelity of the cDNA library created from whole synovial tissue from RA synovial biopsy or from osteoarthritis (OA) patients following whole joint replacement was assessed by a variety of criteria (Table 1). The quality of the RNA-seq data for whole tissue samples was determined by plotting the number of genes at each Fragments Per Kilobase per Million (FPKM) value for all samples. There was no significant difference between the number of expressed genes in RA versus OA samples (Figure 2A). The read density over the length of genes revealed that all samples had comparable coverage across the genome (Figure 2B). Nine RA synovial biopsies and nine OA tissue samples produced high quality RNA-seq libraries and were used for further analyses, concentrating on a set of 9366 expressed genes. The global gene expression profiles from synovial tissue samples were heterogeneous across the RA samples and did not clearly cluster apart from OA (Figure 2C). The variability among patients highlights the complexity in the presentation of RA in patients using whole tissue and points to a shift in gene expression in individual cell types that could be associated with disease activity status and therapy at the time of biopsy.
Figure 2.
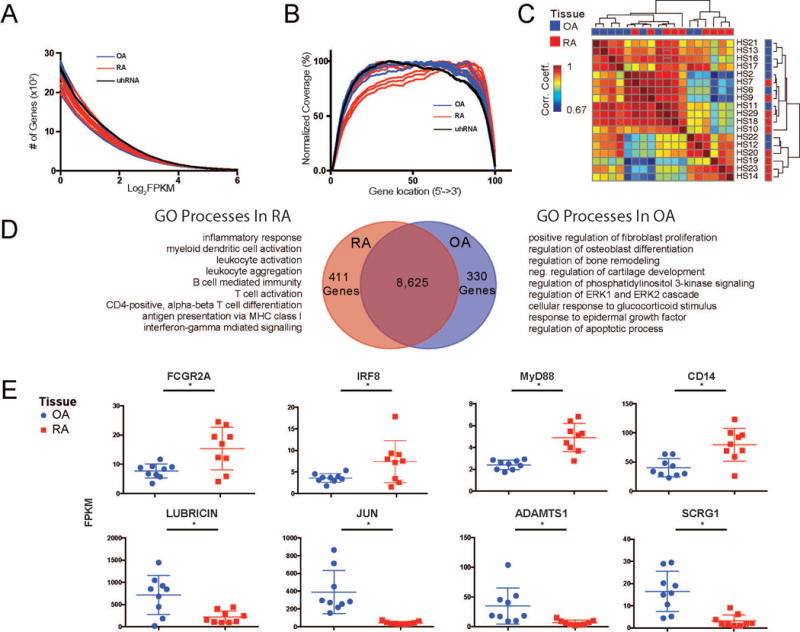
Analysis of whole tissue RNA-seq libraries. A. The number of genes greater than a given expression level (x-axis - Log2FPKM [Fragments Per Kilobase of transcript per Million reads]) are displayed for each sample. [uhRNA = universal human RNA control] B. Gene coverage plot displays the average read density across genes from 5′ to 3′. C. Pairwise Pearson correlations of gene expression between individual patient samples. Samples are organized by hierarchical clustering based on their Pearson coefficients across samples. The tissue type is indicated by red (RA) and blue (OA) squares on the top and right of the matrix. The patient number for each sample is indicated to the right of the matrix. D. Venn diagram of the genes expressed in RA and OA samples. The differential analysis (adj. p-value <0.01) revealed 411 and 330 genes that were preferentially expressed in RA (red) and OA (blue), respectively. Select processes from GO enrichment analysis on genes preferentially expressed in each tissue type are listed. E. Whisker plots indicating normalized gene expression of individual genes between RA (red) and OA (blue) samples. * indicates differential expression (adj. p-value<0.01).
Differential expression analysis identified genes that were specific to either RA or OA synovial tissue samples. We found 411 RA-specific genes and 330 OA-specific genes (Figure 2D). Gene Ontology (GO) analysis revealed distinct pathways enriched with genes from RA or OA. RA-specific genes were associated with a wide range of immune processes – including “leukocyte activation”, “T cell activation”, and “B cell mediated immunity”, while OA-specific genes were associated with more homeostatic processes such as “osteoblast differentiation”, “bone remodeling” and “epidermal growth factor receptor signaling pathway.” Specifically, we found several macrophage-related genes (FcγR2A, IRF8, MyD88 and CD14) that were significantly upregulated in RA relative to OA synovial tissue (Figure 2E). Genes that were preferentially expressed in OA, such as lubricin (57), JUN (58), ADAMTS1 (59), and SCRG1(60) have been previously linked to differences in synovial tissue function in OA. The broad range of pathways involved in RA at the whole tissue level highlights the need for a cell-type-specific approach to better understand the role of particular cell populations.
Synovial Macrophage Digestion and RNA-seq Analysis
Multi-parameter flow cytometry was used to isolate macrophages from RA and OA synovial tissue (Figure 3A). Macrophages were gated via the inclusion of singlets, live, CD45+, and HLA-DR+ cells. Macrophages were further isolated by excluding DCs and gating on the remaining CD11b+ cells to identify CD206+ macrophage populations. An optimized digestion protocol was developed to isolate viable macrophages from synovial tissue. The effectiveness of tissue digestion was assessed by maximizing the number of viable, CD45+, and CD11b+ cells within a given single cell suspension (Figure 3B). An average of 1642 (SEM±1178) macrophages were isolated from digested biopsies and prepared for RNA-seq (Table 1). An identical digestion protocol was used to isolate cell populations from OA synovial tissue as described above, resulting in an average of 77,414 macrophages per sample (Table 1).
Figure 3.
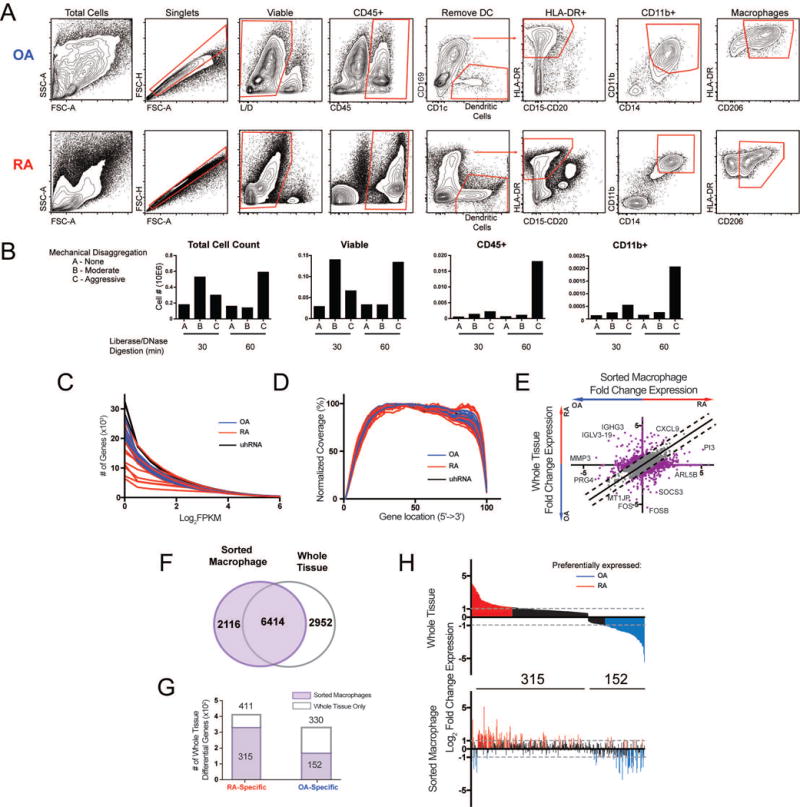
Isolation of synovial macrophages and macrophage-specific RNA-seq. A. Gating strategy used to identify synovial macrophages in both RA and OA tissue. B. Optimization of the synovial tissue processing procedure. The success of the tissue processing was evaluated by the number of viable, CD45+, and CD11b+ cells identified by flow cytometry. C. Number of genes with expression greater than a given FPKM value for each sample. D. Gene coverage plot displays the average read density across genes from 5′ to 3′. E. Log2 fold change (Log2FC) of gene expression between RA and OA from whole tissue (y-axis) and macrophages (x-axis). Lines indicate differences >1. Select genes are displayed. F. Venn diagram comparing genes from sorted macrophages (purple) and whole tissue (grey outline). G. Bar graph of 741 differential genes from whole tissue (Figure 2D) that are also detected in sorted macrophages (purple). H. The Log2FC in expression of 467 differential genes from G detected in macrophages in whole tissue (top) and sorted macrophages (bottom). In both plots, genes are ordered along the x-axis by decreasing fold-change in whole tissue. Genes preferentially expressed in RA tissue (Log2FC>1) and OA tissue (Log2FC<−1) are colored in red and blue, respectively.
Isolated macrophages from 15 RA biopsies and 9 OA tissue samples were collected and passed sequencing quality control for further analysis. The percent alignment of reads (average 54%) was lower than in whole tissue (average 82%) likely due to the increased noise of low input. There was a minimal difference in the number of detected genes between RA and OA macrophages, (Figure 3C). RA macrophages demonstrated comparable complexity (Table 1) and gene coverage (Figure 3D) to OA samples even though the number of RA macrophages was 45× less than sorted OA macrophages.
To determine whether macrophage-specific RNA-seq data provided additional information that could not be gleaned from the whole tissue data, we investigated the ‘macrophage-specific’ gene expression profiles. A comparison of fold-change in gene expression between RA and OA revealed differentially expressed genes that were detected only by ‘whole tissue’ or only by macrophage-specific RNA-seq (Figure 3E). For example, genes associated with inflammatory arthritis, such as and PI3 (61) and MMP-3 (62) were preferentially expressed in RA and OA macrophages, respectively, while no change in expression was observed in whole tissue RNA-seq. Further comparison revealed that 6414 genes were expressed in both datasets, while 2952 genes were exclusively found in whole tissue and 2116 genes were macrophage-specific (Figure 3F). Genes exclusively expressed by whole tissue were likely associated with other cell types, while macrophage-specific genes were likely below the limit of detection from whole tissue RNA-seq. Of the 411 RA-specific and 330 OA-specific genes from whole tissue (Figure 2D), 315 and 152 were expressed in the macrophage-specific dataset (Figure 3G). However, many of these 467 genes were not differentially expressed in the same direction in the macrophage-specific data set (Figure 3H).
Next, we compared the global gene expression profiles from synovial macrophages across patients. We identified two distinct groups of RA patients based on the correlation between patients (Figure 4A). Group 1 consisted of two-thirds of the patients (10/15) with highly similar gene expression profiles while Group 2 contained the remaining 5 patients with divergent gene expression. Patients in Group 2 exhibited significantly higher SJC than Group 1 (p>0.03). These findings demonstrate the possibility for cell-type-specific transcriptional profiles of RA patients to inform on disease severity.
Modules of co-regulated genes may represent pathways that perform specific functions in disease. To identify gene modules within the macrophage transcriptional profiles, we defined a subset of 553 genes as differentially expressed across 15 RA patients after accounting for noise and outliers (see Materials and Methods). These genes were clustered into 6 modules (Modules 1-6) based on the similarity in their patterns of expression across patients (Figure 5A). We calculated the enrichment or depletion of gene module within the expression profile of each patients to determine which patients were driving these modules (Figure 5B). Despite the limited statistical power of 15 patients, we were able to identify associations between expression of these gene modules in patients and clinical parameters (Figure 5C-D, Supplemental Table 2). For example, expression of Module 2 genes, including CCR1 and TREM 2 (Figure 5E), were significantly increased (1.31 fold or 0.39 Log2Fold-Change (FC); p-value=0.05 Figure 5D) in patients who had stopped taking methotrexate. In addition, expression of genes in Module 3 were negatively correlated with disease severity as measured by SJC (p-value=0.007) and had 1.7-fold (Log2FC=0.78; p-value=0.04) higher expression in patients recently diagnosed of RA (Figure 5D). Module 3 genes, such as NFKB-1A and TIMP1, are involved in cellular response to IL-1 as determined by GO enrichment (Supplemental Figure 1). Expression of Module 4 genes were 1.7-fold (Log2FC=0.78; p-value=0.01) more highly expressed in patients who were not taking a biologic medication at the time of biopsy. Module 4 genes were enriched with immune response genes such as TNF and MAFB (Figure 5E). Expression of Module 5 genes, such as MIF and HMGB2 were positively associated with disease severity as measured by TJC (p-value=0.03) (Figure 5D). Genes in Module 6, such as CD83 and CXCR4, were 2.0-fold (Log2FC=1.0; p-value=0.03) higher in patients who were negative for rheumatoid factor. Taken together, our data demonstrate for the first time that transcriptional profiling of isolated synovial macrophages using ultrasound-guided synovial biopsies may be used to characterize patients in a biologically relevant manner.
Discussion
Recent advances in ultrasound technology have opened up a new opportunity for rheumatologists to perform minimally invasive ultrasound-guided synovial tissue biopsies (25). While arthroplasty has the ability to collect large pieces of synovial tissue, its utility as a vehicle to obtain tissue for research purposes from RA patients in the United States and in longitudinal manner is challenging (23, 29, 41, 45, 63–66). Moreover, the tissue obtained from arthroplasty is usually late stage and may not reflect the ongoing active disease, which may be obtained using ultrasound guided synovial biopsy. The fact that the ultrasound guided synovial biopsies may occur in the clinic without a surgical suite and require minimal to no recovery time for the patient outweighs the amount of tissue retrieved (23, 29, 41, 45, 63–66). In fact, in several countries in Europe, this technique is used to obtain synovial tissue for research purposes in a large number of patients without significant complications (34–36, 38–40, 67–72). The procedure itself is well accepted by both patients and referring rheumatologists at similar rates to that observed by our European colleagues (34, 36). Previous studies have already focused on access to the joint, intrajoint synovial variation, and reproducibility of measurements using the ultrasound guided synovial biopsy technique (34–36). Furthermore, these biopsies do not appear to alter subsequent clinical or ultrasound disease activity assessments, which is important for patients who might subsequently enroll in clinical trials (73). These European groups have also performed numerous studies to validate the needle biopsy and portal and forceps procedures and tissue sampling (34–36, 38–40, 67, 74, 75). Our data demonstrate that the ultrasound-guided synovial tissue biopsies obtained from patients with RA are sufficient for RNA-seq, distinguish differences between patients with RA and OA, and importantly sets the framework for the stratification of patients with RA according to the most prominent disease pathway. We also report an optimized digestion protocol for synovial tissue obtained by ultrasound-guided biopsies and demonstrate the ability to sort viable hematopoietic cells by FACS. Further, we show that only small numbers of cells (as few as 10 cells) are sufficient for generation of libraries for quality RNA-seq analysis. With our initial cohort of 41 patients we have been able to link the cell-type-specific transcriptional signatures with patients’ treatment regimen and clinical information.
Currently, the standard of care for rheumatologists is to prescribe biologic therapy to RA patients through a costly and time-consuming trial-and-error process. Therefore, the utility of a biomarker to identify how a patient will respond to a particular therapy cannot be overstated. While peripheral blood is attractive for identifying a potential biomarker due to its ease of attainability, this approach has not been fruitful. Early studies by Dr. Paul Tak and colleagues demonstrated the potential of obtaining synovial tissue as a means to determine a biomarker for responsiveness to therapy (25). In his seminal studies, he showed that a reduction in the number of synovial sublining macrophages correlates with a decrease in disease activity (i.e. DAS28) (25). The abundance of synovial sublining macrophages is currently a leading candidate for a viable biomarker of therapeutic response in RA (25). We posit that transcriptional signatures in macrophages regardless of location (sublining vs synovial lining) will predict responsiveness to specific non-biologic and/or biologic therapies. Our data suggest the existence of associations between the transcriptional signature of macrophages and treatment course (or compliance) of the patient. However, the current study is limited in its ability to predict therapeutic response because of the constraint of a single time point for each patient at different stages of disease. Future studies beyond the scope of this manuscript will entail collection of synovial biopsies from a larger cohort longitudinally, prior to and following therapy. Therefore, this study serves as proof of the principal that transcriptional analysis of synovial macrophages using ultrasound-guided synovial biopsies may function to uncover novel pathways underlying disease pathogenesis or response to therapy. Currently, studies are also underway in the Accelerating Medicines Partnership (AMP) which will take advantage of synovial biopsies for identification of molecular pathways (65).
In summary, this study is the first in the United States to harness the potential of ultrasound-guided synovial biopsies as a method for obtaining synovial tissue from patients with RA. Based on the recent success of REASON using minimally invasive ultrasound-guided synovial biopsies, coupled with our ability to interrogate synovial tissue at multiple levels using cutting-edge technologies, we believe that future studies have the potential to provide critical information to rheumatologists in establishing precision medicine as a reality for our patients.
Supplementary Material
Acknowledgments
In loving memory of Colleen Pope Vitu. We thank the patients who underwent synovial biopsy for this study and acknowledge technical assistance at each site (UAB: Stephanie Ledbetter, MS; Laticia Woodruff, RN; Keith Wanzeck, BS). We also want to thank Drs. Terrance Peabody, Ali Shilatifard, Hussain Bhikhapurwala and Danette Ko for their assistance in the manuscript. The authors declare no competing financial interests. NIH grant AR064313 to CMC; Pfizer ASPIRE to RMP and; NIH grant AR064546, HL134375, AG049665, and UH2AR067687 and the United States-Israel Binational Science Foundation (2013247), the Rheumatology Research Foundation (Agmt 05/06/14) to HRP. HRP was also supported by the Mabel Greene Myers Professor of Medicine and generous donations to the Rheumatology Precision Medicine Fund. This project was supported in part by an ANRF grant awarded to DRW. Flow Cytometry Cell Sorting was performed in the Lurie Cancer Center Flow Cytometry Core Facility that is supported by NCI P30-CA060553 and on a BD FACSAria SORP system, purchased through the support of NIH 1S10OD011996-01. Histology services were provided by the Northwestern University Mouse Histology and Phenotyping Laboratory which is supported by NCI P30-CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. RNA sequencing was performed in the Division of Rheumatology and Pulmonary and Critical Care sequencing facility.
Footnotes
DR. S. LOUIS BRIDGES, JR. (Orcid ID : 0000-0003-3785-1389)
Disclosures
The authors declare no competing financial interests.
References
- 1.O’Dell JR, Curtis JR, Mikuls TR, Cofield SS, Bridges SL, Jr, Ranganath VK, et al. Validation of the methotrexate-first strategy in patients with early, poor-prognosis rheumatoid arthritis: results from a two-year randomized, double-blind trial. Arthritis Rheum. 2013;65(8):1985–94. doi: 10.1002/art.38012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saevarsdottir S, Wallin H, Seddighzadeh M, Ernestam S, Geborek P, Petersson IF, et al. Predictors of response to methotrexate in early DMARD naive rheumatoid arthritis: results from the initial open-label phase of the SWEFOT trial. Ann Rheum Dis. 2011;70(3):469–75. doi: 10.1136/ard.2010.139212. [DOI] [PubMed] [Google Scholar]
- 3.Bathon JM, Martin RW, Fleischmann RM, Tesser JR, Schiff MH, Keystone EC, et al. A comparison of etanercept and methotrexate in patients with early rheumatoid arthritis. N Engl J Med. 2000;343(22):1586–93. doi: 10.1056/NEJM200011303432201. [DOI] [PubMed] [Google Scholar]
- 4.Breedveld FC, Weisman MH, Kavanaugh AF, Cohen SB, Pavelka K, van Vollenhoven R, et al. The PREMIER study: A multicenter, randomized, double-blind clinical trial of combination therapy with adalimumab plus methotrexate versus methotrexate alone or adalimumab alone in patients with early, aggressive rheumatoid arthritis who had not had previous methotrexate treatment. Arthritis Rheum. 2006;54(1):26–37. doi: 10.1002/art.21519. [DOI] [PubMed] [Google Scholar]
- 5.Emery P, Breedveld FC, Hall S, Durez P, Chang DJ, Robertson D, et al. Comparison of methotrexate monotherapy with a combination of methotrexate and etanercept in active, early, moderate to severe rheumatoid arthritis (COMET): a randomised, double-blind, parallel treatment trial. Lancet. 2008;372(9636):375–82. doi: 10.1016/S0140-6736(08)61000-4. [DOI] [PubMed] [Google Scholar]
- 6.Moreland LW, O’Dell JR, Paulus HE, Curtis JR, Bathon JM, St Clair EW, et al. A randomized comparative effectiveness study of oral triple therapy versus etanercept plus methotrexate in early aggressive rheumatoid arthritis: the treatment of Early Aggressive Rheumatoid Arthritis Trial. Arthritis Rheum. 2012;64(9):2824–35. doi: 10.1002/art.34498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenberg JD, Reed G, Decktor D, Harrold L, Furst D, Gibofsky A, et al. A comparative effectiveness study of adalimumab, etanercept and infliximab in biologically naive and switched rheumatoid arthritis patients: results from the US CORRONA registry. Ann Rheum Dis. 2012;71(7):1134–42. doi: 10.1136/annrheumdis-2011-150573. [DOI] [PubMed] [Google Scholar]
- 8.Hetland ML, Christensen IJ, Tarp U, Dreyer L, Hansen A, Hansen IT, et al. Direct comparison of treatment responses, remission rates, and drug adherence in patients with rheumatoid arthritis treated with adalimumab, etanercept, or infliximab: results from eight years of surveillance of clinical practice in the nationwide Danish DANBIO registry. Arthritis Rheum. 2010;62(1):22–32. doi: 10.1002/art.27227. [DOI] [PubMed] [Google Scholar]
- 9.Jobanputra P, Maggs F, Deeming A, Carruthers D, Rankin E, Jordan AC, et al. A randomised efficacy and discontinuation study of etanercept versus adalimumab (RED SEA) for rheumatoid arthritis: a pragmatic, unblinded, non-inferiority study of first TNF inhibitor use: outcomes over 2 years. BMJ Open. 2012;2(6) doi: 10.1136/bmjopen-2012-001395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markenson JA, Gibofsky A, Palmer WR, Keystone EC, Schiff MH, Feng J, et al. Persistence with anti-tumor necrosis factor therapies in patients with rheumatoid arthritis: observations from the RADIUS registry. J Rheumatol. 2011;38(7):1273–81. doi: 10.3899/jrheum.101142. [DOI] [PubMed] [Google Scholar]
- 11.Rendas-Baum R, Wallenstein GV, Koncz T, Kosinski M, Yang M, Bradley J, et al. Evaluating the efficacy of sequential biologic therapies for rheumatoid arthritis patients with an inadequate response to tumor necrosis factor-alpha inhibitors. Arthritis Res Ther. 2011;13(1):R25. doi: 10.1186/ar3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmick CG, Felson DT, Lawrence RC, Gabriel S, Hirsch R, Kwoh CK, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheum. 2008;58(1):15–25. doi: 10.1002/art.23177. [DOI] [PubMed] [Google Scholar]
- 13.Myasoedova E, Crowson CS, Kremers HM, Therneau TM, Gabriel SE. Is the incidence of rheumatoid arthritis rising?: results from Olmsted County, Minnesota, 1955-2007. Arthritis Rheum. 2010;62(6):1576–82. doi: 10.1002/art.27425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chamberlain ND, Kim SJ, Vila OM, Volin MV, Volkov S, Pope RM, et al. Ligation of TLR7 by rheumatoid arthritis synovial fluid single strand RNA induces transcription of TNFalpha in monocytes. Ann Rheum Dis. 2013;72(3):418–26. doi: 10.1136/annrheumdis-2011-201203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Q, Pope RM. Toll-like receptor signaling: a potential link among rheumatoid arthritis, systemic lupus, and atherosclerosis. J Leukoc Biol. 2010;88(2):253–62. doi: 10.1189/jlb.0310126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duffau P, Menn-Josephy H, Cuda CM, Dominguez S, Aprahamian TR, Watkins AA, et al. Promotion of Inflammatory Arthritis by Interferon Regulatory Factor 5 in a Mouse Model. Arthritis Rheumatol. 2015;67(12):3146–57. doi: 10.1002/art.39321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang QQ, Koessler RE, Birkett R, Perlman H, Xing L, Pope RM. TLR2 deletion promotes arthritis through reduction of IL-10. J Leukoc Biol. 2013;93(5):751–9. doi: 10.1189/jlb.0912473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer G, Chobaz V, Talabot-Ayer D, Taylor S, So A, Gabay C, et al. Assessment of the efficacy of different statins in murine collagen-induced arthritis. Arthritis and Rheumatism. 2004;50(12):4051–9. doi: 10.1002/art.20673. [DOI] [PubMed] [Google Scholar]
- 19.Nuki G, Bresnihan B, Bear MB, McCabe D. Long-term safety and maintenance of clinical improvement following treatment with anakinra (recombinant human interleukin-1 receptor antagonist) in patients with rheumatoid arthritis: Extension phase of a randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2002;46(11):2838–46. doi: 10.1002/art.10578. [DOI] [PubMed] [Google Scholar]
- 20.Genovese MC, Bathon JM, Martin RW, Fleischmann RM, Tesser JR, Schiff MH, et al. Etanercept versus methotrexate in patients with early rheumatoid arthritis: two-year radiographic and clinical outcomes. Arthritis Rheum. 2002;46(6):1443–50. doi: 10.1002/art.10308. [DOI] [PubMed] [Google Scholar]
- 21.De Rycke L, Baeten D, Foell D, Kruithof E, Veys EM, Roth J, et al. Differential expression and response to anti-TNFalpha treatment of infiltrating versus resident tissue macrophage subsets in autoimmune arthritis. J Pathol. 2005;206(1):17–27. doi: 10.1002/path.1758. [DOI] [PubMed] [Google Scholar]
- 22.Barland P, Novikoff AB, Hamerman D. Electron microscopy of the human synovial membrane. J Cell Biol. 1962;14:207–20. doi: 10.1083/jcb.14.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filkova M, Cope A, Mant T, Galloway J. Is there a role of synovial biopsy in drug development? BMC Musculoskelet Disord. 2016;17(1):172. doi: 10.1186/s12891-016-1028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sieberts SK, Zhu F, Garcia-Garcia J, Stahl E, Pratap A, Pandey G, et al. Crowdsourced assessment of common genetic contribution to predicting anti-TNF treatment response in rheumatoid arthritis. Nat Commun. 2016;7:12460. doi: 10.1038/ncomms12460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Udalova IA, Mantovani A, Feldmann M. Macrophage heterogeneity in the context of rheumatoid arthritis. Nat Rev Rheumatol. 2016;12(8):472–85. doi: 10.1038/nrrheum.2016.91. [DOI] [PubMed] [Google Scholar]
- 26.de Hair MJ, van de Sande MG, Ramwadhdoebe TH, Hansson M, Landewe R, van der Leij C, et al. Features of the synovium of individuals at risk of developing rheumatoid arthritis: implications for understanding preclinical rheumatoid arthritis. Arthritis Rheumatol. 2014;66(3):513–22. doi: 10.1002/art.38273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerlag D, Tak PP. Synovial biopsy. Best Pract Res Clin Rheumatol. 2005;19(3):387–400. doi: 10.1016/j.berh.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Gerlag DM, Tak PP. How useful are synovial biopsies for the diagnosis of rheumatic diseases? Nat Clin Pract Rheumatol. 2007;3(5):248–9. doi: 10.1038/ncprheum0485. [DOI] [PubMed] [Google Scholar]
- 29.Gerlag DM, Tak PP. How to perform and analyse synovial biopsies. Best Pract Res Clin Rheumatol. 2013;27(2):195–207. doi: 10.1016/j.berh.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Haringman JJ, Vinkenoog M, Gerlag DM, Smeets TJ, Zwinderman AH, Tak PP. Reliability of computerized image analysis for the evaluation of serial synovial biopsies in randomized controlled trials in rheumatoid arthritis. Arthritis Res Ther. 2005;7(4):R862–7. doi: 10.1186/ar1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jahangier ZN, Jacobs JW, Kraan MC, Wenting MJ, Smeets TJ, Bijlsma JW, et al. Pretreatment macrophage infiltration of the synovium predicts the clinical effect of both radiation synovectomy and intra-articular glucocorticoids. Ann Rheum Dis. 2006;65(10):1286–92. doi: 10.1136/ard.2005.042333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraan MC, Reece RJ, Smeets TJ, Veale DJ, Emery P, Tak PP. Comparison of synovial tissues from the knee joints and the small joints of rheumatoid arthritis patients: Implications for pathogenesis and evaluation of treatment. Arthritis Rheum. 2002;46(8):2034–8. doi: 10.1002/art.10556. [DOI] [PubMed] [Google Scholar]
- 33.Koski JM, Hammer HB. Ultrasound-guided procedures: techniques and usefulness in controlling inflammation and disease progression. Rheumatology (Oxford) 2012;51(Suppl 7):vii, 31–5. doi: 10.1093/rheumatology/kes331. [DOI] [PubMed] [Google Scholar]
- 34.Humby F, Kelly S, Bugatti S, Manzo A, Filer A, Mahto A, et al. Evaluation of Minimally Invasive, Ultrasound-guided Synovial Biopsy Techniques by the OMERACT Filter–Determining Validation Requirements. J Rheumatol. 2016;43(1):208–13. doi: 10.3899/jrheum.141199. [DOI] [PubMed] [Google Scholar]
- 35.Humby F, Kelly S, Hands R, Rocher V, DiCicco M, Ng N, et al. Use of ultrasound-guided small joint biopsy to evaluate the histopathologic response to rheumatoid arthritis therapy: recommendations for application to clinical trials. Arthritis Rheumatol. 2015;67(10):2601–10. doi: 10.1002/art.39235. [DOI] [PubMed] [Google Scholar]
- 36.Kelly S, Humby F, Filer A, Ng N, Di Cicco M, Hands RE, et al. Ultrasound-guided synovial biopsy: a safe, well-tolerated and reliable technique for obtaining high-quality synovial tissue from both large and small joints in early arthritis patients. Ann Rheum Dis. 2015;74(3):611–7. doi: 10.1136/annrheumdis-2013-204603. [DOI] [PubMed] [Google Scholar]
- 37.Koski JM, Helle M. Ultrasound guided synovial biopsy using portal and forceps. Ann Rheum Dis. 2005;64(6):926–9. doi: 10.1136/ard.2004.027409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lazarou I, D’Agostino MA, Naredo E, Humby F, Filer A, Kelly SG. Ultrasound-guided synovial biopsy: a systematic review according to the OMERACT filter and recommendations for minimal reporting standards in clinical studies. Rheumatology (Oxford) 2015;54(10):1867–75. doi: 10.1093/rheumatology/kev128. [DOI] [PubMed] [Google Scholar]
- 39.Ramirez J, Celis R, Usategui A, Ruiz-Esquide V, Fare R, Cuervo A, et al. Immunopathologic characterization of ultrasound-defined synovitis in rheumatoid arthritis patients in clinical remission. Arthritis Res Ther. 2015;18:74. doi: 10.1186/s13075-016-0970-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scire CA, Epis O, Codullo V, Humby F, Morbini P, Manzo A, et al. Immunohistological assessment of the synovial tissue in small joints in rheumatoid arthritis: validation of a minimally invasive ultrasound-guided synovial biopsy procedure. Arthritis Res Ther. 2007;9(5):R101. doi: 10.1186/ar2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sitt JC, Griffith JF, Wong P. Ultrasound-guided synovial biopsy. Br J Radiol. 2016;89(1057):20150363. doi: 10.1259/bjr.20150363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Vugt RM, van Dalen A, Bijlsma JW. Ultrasound guided synovial biopsy of the wrist. Scand J Rheumatol. 1997;26(3):212–4. doi: 10.3109/03009749709065683. [DOI] [PubMed] [Google Scholar]
- 43.Filer A, de Pablo P, Allen G, Nightingale P, Jordan A, Jobanputra P, et al. Utility of ultrasound joint counts in the prediction of rheumatoid arthritis in patients with very early synovitis. Ann Rheum Dis. 2011;70(3):500–7. doi: 10.1136/ard.2010.131573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filkova M, Aradi B, Senolt L, Ospelt C, Vettori S, Mann H, et al. Association of circulating miR-223 and miR-16 with disease activity in patients with early rheumatoid arthritis. Ann Rheum Dis. 2013 doi: 10.1136/annrheumdis-2012-202815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pitzalis C, Kelly S, Humby F. New learnings on the pathophysiology of RA from synovial biopsies. Curr Opin Rheumatol. 2013;25(3):334–44. doi: 10.1097/BOR.0b013e32835fd8eb. [DOI] [PubMed] [Google Scholar]
- 46.Yeo L, Toellner KM, Salmon M, Filer A, Buckley CD, Raza K, et al. Cytokine mRNA profiling identifies B cells as a major source of RANKL in rheumatoid arthritis. Ann Rheum Dis. 2011;70(11):2022–8. doi: 10.1136/ard.2011.153312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO, 3rd, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010;62(9):2569–81. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- 48.Singh JA, Saag KG, Bridges SL, Jr, Akl EA, Bannuru RR, Sullivan MC, et al. 2015 American College of Rheumatology Guideline for the Treatment of Rheumatoid Arthritis. Arthritis Rheumatol. 2016;68(1):1–26. doi: 10.1002/art.39480. [DOI] [PubMed] [Google Scholar]
- 49.Singh JA, Saag KG, Bridges SL, Jr, Akl EA, Bannuru RR, Sullivan MC, et al. 2015 American College of Rheumatology Guideline for the Treatment of Rheumatoid Arthritis. Arthritis Care Res (Hoboken) 2016;68(1):1–25. doi: 10.1002/acr.22783. [DOI] [PubMed] [Google Scholar]
- 50.Wakefield RJ, Balint PV, Szkudlarek M, Filippucci E, Backhaus M, D’Agostino MA, et al. Musculoskeletal ultrasound including definitions for ultrasonographic pathology. J Rheumatol. 2005;32(12):2485–7. [PubMed] [Google Scholar]
- 51.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28(16):2184–5. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 53.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–97. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruan MZ, Erez A, Guse K, Dawson B, Bertin T, Chen Y, et al. Proteoglycan 4 expression protects against the development of osteoarthritis. Sci Transl Med. 2013;5(176):176ra34. doi: 10.1126/scitranslmed.3005409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Yuan Z, Cui S. Identifying candidate genes involved in osteoarthritis through bioinformatics analysis. Clin Exp Rheumatol. 2016;34(2):282–90. [PubMed] [Google Scholar]
- 59.Nagase H, Kashiwagi M. Aggrecanases and cartilage matrix degradation. Arthritis Res Ther. 2003;5(2):94–103. doi: 10.1186/ar630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ochi K, Derfoul A, Tuan RS. A predominantly articular cartilage-associated gene, SCRG1, is induced by glucocorticoid and stimulates chondrogenesis in vitro. Osteoarthritis Cartilage. 2006;14(1):30–8. doi: 10.1016/j.joca.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 61.Li J, Hsu HC, Mountz JD. Managing macrophages in rheumatoid arthritis by reform or removal. Curr Rheumatol Rep. 2012;14(5):445–54. doi: 10.1007/s11926-012-0272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han Z, Boyle DL, Manning AM, Firestein GS. AP-1 and NF-kappaB regulation in rheumatoid arthritis and murine collagen-induced arthritis. Autoimmunity. 1998;28(4):197–208. doi: 10.3109/08916939808995367. [DOI] [PubMed] [Google Scholar]
- 63.Humby FC, Al Balushi F, Lliso G, Cauli A, Pitzalis C. Can Synovial Pathobiology Integrate with Current Clinical and Imaging Prediction Models to Achieve Personalized Health Care in Rheumatoid Arthritis? Front Med (Lausanne) 2017;4:41. doi: 10.3389/fmed.2017.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orr C, Vieira-Sousa E, Boyle DL, Buch MH, Buckley CD, Canete JD, et al. Synovial tissue research: a state-of-the-art review. Nat Rev Rheumatol. 2017;13(10):630. doi: 10.1038/nrrheum.2017.161. [DOI] [PubMed] [Google Scholar]
- 65.van de Sande MG, Gerlag DM, Lodde BM, van Baarsen LG, Alivernini S, Codullo V, et al. Evaluating antirheumatic treatments using synovial biopsy: a recommendation for standardisation to be used in clinical trials. Ann Rheum Dis. 2011;70(3):423–7. doi: 10.1136/ard.2010.139550. [DOI] [PubMed] [Google Scholar]
- 66.Wechalekar MD, Smith MD. Utility of arthroscopic guided synovial biopsy in understanding synovial tissue pathology in health and disease states. World J Orthop. 2014;5(5):566–73. doi: 10.5312/wjo.v5.i5.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Najm A, Orr C, Heymann MF, Bart G, Veale DJ, Le Goff B. Success Rate and Utility of Ultrasound-guided Synovial Biopsies in Clinical Practice. J Rheumatol. 2016;43(12):2113–9. doi: 10.3899/jrheum.151441. [DOI] [PubMed] [Google Scholar]
- 68.Sitt JC, Griffith JF, Lai FM, Hui M, Chiu KH, Lee RK, et al. Ultrasound-guided synovial Tru-cut biopsy: indications, technique, and outcome in 111 cases. Eur Radiol. 2016 doi: 10.1007/s00330-016-4545-6. [DOI] [PubMed] [Google Scholar]
- 69.De Groof A, Ducreux J, Humby F, Nzeusseu Toukap A, Badot V, Pitzalis C, et al. Higher expression of TNFalpha-induced genes in the synovium of patients with early rheumatoid arthritis correlates with disease activity, and predicts absence of response to first line therapy. Arthritis Res Ther. 2016;18:19. doi: 10.1186/s13075-016-0919-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kelly S, Bombardieri M, Humby F, Ng N, Marrelli A, Riahi S, et al. Angiogenic gene expression and vascular density are reflected in ultrasonographic features of synovitis in early Rheumatoid Arthritis: an observational study. Arthritis Res Ther. 2015;17:58. doi: 10.1186/s13075-015-0567-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frank-Bertoncelj M, Trenkmann M, Klein K, Karouzakis E, Rehrauer H, Bratus A, et al. Epigenetically-driven anatomical diversity of synovial fibroblasts guides joint-specific fibroblast functions. Nat Commun. 2017;8:14852. doi: 10.1038/ncomms14852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luukkonen J, Pascual LM, Patlaka C, Lang P, Turunen S, Halleen J, et al. Increased amount of phosphorylated proinflammatory osteopontin in rheumatoid arthritis synovia is associated to decreased tartrate-resistant acid phosphatase 5B/5A ratio. PLoS One. 2017;12(8):e0182904. doi: 10.1371/journal.pone.0182904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lazarou I, Kelly S, Humby F, Di Cicco M, Zou L, Rocher-Ros V, et al. Ultrasound-guided synovial biopsy of the wrist does not alter subsequent clinical or ultrasound disease activity assessments: a prospective study for incorporation of imaging in clinical trials. Clin Exp Rheumatol. 2016;34(5):802–7. [PubMed] [Google Scholar]
- 74.Choi IY, Karpus ON, Turner JD, Hardie D, Marshall JL, de Hair MJH, et al. Stromal cell markers are differentially expressed in the synovial tissue of patients with early arthritis. PLoS One. 2017;12(8):e0182751. doi: 10.1371/journal.pone.0182751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Filer A, Ward LSC, Kemble S, Davies CS, Munir H, Rogers R, et al. Identification of a transitional fibroblast function in very early rheumatoid arthritis. Ann Rheum Dis. 2017 doi: 10.1136/annrheumdis-2017-211286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


