Abstract
Understanding brain circuit organization and function requires systematic dissection of its cellular components. With vast cell number and diversity, mammalian nervous systems present a daunting challenge for achieving specific and comprehensive cell type access – prerequisite to circuit analysis. Genetic approaches in the mouse have relied on germline engineering to access marker-defined cell populations. Combinatorial strategies that engage marker intersection, anatomy and projection pattern (e.g. antero- and retro-grade viral vectors), and developmental lineage substantially increase the specificity of cell type targeting. While increasing number of mouse cell types are becoming experimentally accessible, comprehensive coverage requires larger coordinated efforts with strategic infrastructural and fiscal planning. CRISPR-based genome editing may enable cell type access in other species, but issues of time, cost and ethics remain, especially for primates. Novel approaches that bypass the germline, such as somatic cell engineering and cell surface-based gene delivery, may reduce the barrier of genetic access to mammalian cell types.
Introduction
Understanding the organization, function and assembly of neural circuits requires systematic dissection of their basic elements, groups of nerve cells that share similar anatomical and physiological properties, i.e. cell types. To reliably identify these cell types - individual nodes of the brain circuit connectome, and to measure and manipulate their activity in the context of behavior, we need a comprehensive toolkit that provides experimental access to a large set of these circuit elements. Indeed, a dream of many neuroscientists is to be able to readily identify, monitor and manipulate every cell type in the circuits that they investigate.
The broad challenges of cell type access are several-fold [1]. The first and foremost is specificity at an appropriate granularity. Precise targeting of a cell type in neural circuit analysis is, in a sense, analogous to successful cloning of a gene in genetics – it will provide the clarity in answers that cannot be obtained otherwise. Studying an unknown mixture of cells with non-specific tools is not only uninformative but often generates confusion. The second is comprehensiveness. It is highly desirable to be able access most if not all cell types in a given neural circuit to discover its precise organization and operation. The third is systematic coverage. With a vast number of cell types across brain regions, it is necessary to build broad collections of tools to cover many of these brain systems. The fourth is robustness of toolgenes (i.e. markers, sensors, transducers etc.) that allow easy and effective observation and manipulation.
Not surprisingly, the ease of accessing cell types is often correlated with the complexity of the nervous system and brain region in genetic model organisms (Table 1). In the round worm C.elegans with 302 neurons of exactly 118 types, thousands of cell type transgenic lines are available and each neuron type is covered on average by ~32 transgenic lines [2]. In Drosophila melanogaster with a total of ~105 neurons in the adult brain, over 2×104 driver lines have been generated to cover cell populations in most brain regions, with increasingly number of intersectional lines that target highly specific cell types[3]. And there are ample examples where every cell type in a brain circuit is recognized and targeted [4]. Together these cell type tools bestow unparalleled experimental versatility and have transformed the study of worm and fly neurobiology.
Table 1.
Comparison of cell type tools across several model organisms
| Species | Neurons | Neuron types | Cell type drivers | Drivers/cell type or population* |
|---|---|---|---|---|
| C. elegans | 302 | 118 | >103 | ~30 (4-150) |
| Drosophila | ~105 | Likely thousands | ~4×104 | Many cell types have multiple drivers |
| Mouse | ~108 | Unknown | ~5×102 | 1 and none for most |
| Rat | ~108 | Unknown | ~2×101 | None for most |
| Macaque | ~1010 | Unknown | None yet | None |
| Marmoset | ~109 | Unknown | None yet | None |
The numbers include those that are specific to a cell type and those that are expressed in a cell type/population as well as in other cell types/populations.
With over 108 neurons of vexing diversity and an unknown number of types in the mouse and most mammalian brains (Table 1), the challenge of cell type access is not only technical but also conceptual -the very definition of neuron type in many brain regions is often contentious [1,5,6]. Recent advances in single cell analysis, especially single cell genomics, present unprecedented opportunities for understanding, discovering, and accessing cell types in the mammalian brain. Furthermore, programmable nucleases-based site-specific genome editing techniques, such as clustered regularly inter-spaced short palindromic repeats-Cas9 (CRISPR-Cas9), transcription activator-like effector nuclease (TALEN), and zinc-finger nucleases (ZFNs), facilitates germline and somatic engineering in broader mammalian species for cell access, especially in primate brains with orders of magnitudes more neurons (~109 in marmoset and ~1010 in rhesus macaque). Here we highlight recent progress in cell type tools in the mouse, consider the requirement and prospect of more broad and comprehensive cell access in this genetic model organism, discuss the opportunities and challenges in other mammalian species, and call for innovation of novel approaches in parallel to germline engineering.
Cell transcriptomes provide unprecedented opportunities for discovering and targeting cell types
Molecular markers are the starting point for genetic access to cell populations defined by gene expression. Until recently, cell type markers in the mammalian brain were very sparse and were mostly discovered in a serendipitous and piecemeal manner. This situation has fundamentally changed in the past two years, with innovations in massively parallel single cell and single nuclei RNA sequencing (scRNAseq, snRNAseq). Thousands to millions of single cells are sequenced in multiple brain regions and species [7*–13*]. Unsupervised statistical clustering have identified increasing number of “transcriptional types” with distinct expression profiles, many may serve as single or combinatorial markers.
Furthermore, new generation of mRNA in situ techniques such as multiplexed error-robust fluorescence in situ hybridization (MERFISH)[14*] and in situ transcriptome profiling (seq-FISH)[15*,16**] enable cellular resolution spatial detection of dozens to hundreds mRNAs. These methods promise to reveal the precise spatial localization of transcriptional cell clusters, an important step towards identifying cell types. Together, the rapid accumulation of markers for transcriptional cell types and their spatial distribution pattern provide unprecedented opportunities for genetic access. Indeed, an increasingly restrictive bottleneck is the generation of recombinase driver lines and their characterization. It should be made clear though that progress in single cell transcriptomics by itself does not and cannot address the fundamental issue of how to define a cell type, a problem that can only be addressed by multi-faceted analyses of orthogonal cell features, which require reliable experimental access.
Toward an overarching and mechanistic definition of cardinal neuron types
Nerve cells are, in a quite real sense, individual micro-organisms living in a highly connected brain cell society. They manifest multi-modal and multi-dimensional phenotypes that are extraordinarily difficult to describe and measure. These include morphology, connectivity pattern, physiological properties, gene expression profiles, developmental history, and ultimately circuit function. A fundamental conceptual and technical challenge is establishing an overarching and mechanistic framework of cell type identity that integrates multi-modal cell phenotypes. Combining genetic targeting, high-resolution single cell transcriptomics and computational analysis, a recent study discovered that the transcriptional architecture of synaptic communication delineates cortical GABAergic neuron identities [17**] (Figure 1). This architecture comprises 6 categories of ~40 gene families including cell adhesion molecules, transmitter-modulator receptors, ion channels, signaling proteins, neuropeptides and vesicular release components, and transcription factors. Combinatorial expression of select members across families shapes a multi-layered molecular scaffold along cell membrane that may customize synaptic connectivity patterns and input-output signaling properties. Transcriptional signatures of synaptic communication may integrate anatomical, physiological, functional and developmental genetic features that together define neuronal identity. This discovery provides an overarching and mechanistic framework for cell type definition, discovery, and cataloging. Future studies will evaluate whether this synaptic communication scheme apply to the definition of other neuron types, especially projection neurons whose input-output connectivity constitutes basic circuit elements of information processing and relay in global networks and brain systems[18]. Several methods begin to link orthogonal cell features with transcription profiles, such as physiological properties (e.g. patch-seq) [19,20], connectivity (e.g. MAPseq) [21] and activity (e.g. Act-seq)[22].
Figure 1.
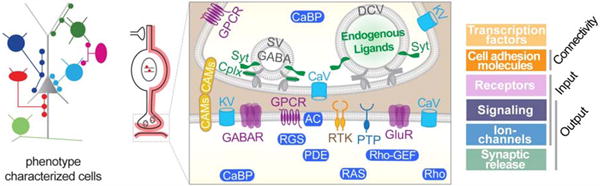
Six cardinal types of GABAergic neurons (left) are delineated by their transcription profiles mainly consisting of 6 functional gene categories encoding a molecular scaffold that mediate synaptic input-output communication (right). Modified from Paul et al 2017.
General approaches to cell type access
In broad terms, there are two approaches to access cell types (Figure 2). The first is through gene expression. These most often involve germline engineering but also include viral vectors with short promoters, and CRISPR mediated gene editing in neural progenitors or somatic cells. The second approach is to access neuron types according to their projection targets or cell surface properties. These include retrograde viral infection of axon terminals, viral particles or nanoparticles that recognize specific cell membrane proteins or lipids. Whereas much emphasis and progress have been made with the first approach, the second approach has been out of the spot light but may be crucial for future progress.
Figure 2.
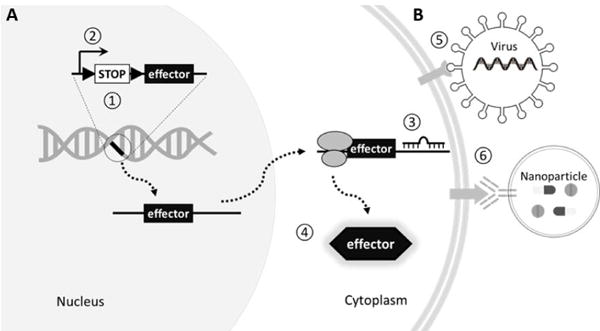
Two major approaches to access cell types. A. Utilizing gene expression regulatory mechanisms. Toolgene (“effector”) can be inserted into host genome via genetic engineering, or delivered into cells on a viral vector or plasmids. Its expression can be regulated on the DNA level by a binary system such as Cre-loxP(①), on the transcriptional level promoter and enhancers(②), on the post-transcription level by miRNAs (③), or on the protein level through modulating protein stability or localization(④). B. Recognizing cell surface molecules. Viral infection is mediated by interaction between capsid protein and membrane receptors (⑤). Nanobody recognizing cell surface antigen, or engineered ligands recognizing membrane receptors, can be used to decorate virus or non-viral vehicles such as nanoparticles which modulates neuron activity or delivers drugs (⑥).
Genetic cell type access in the mouse
As the most advanced mammalian genetic model organism, germline engineering has been the primary approach to capture cell types in the mouse. Although BAC transgenic[23] and enhancer trap[24,25] methods have generated useful tools, gene knockin (i.e. homologous recombination in embryonic stem cells) has proven to be the most specific and reliable approach to target marker defined cell populations. To date, on the order of ~200 Cre, Flp, Dre, and tTA driver lines have been characterized that target neuronal populations and cell types in multiple brain regions [26–29**](Allen Brain Institute transgenic atlas, http://connectivity.brain-map.org/transgenic). However, these are far from a systematic coverage of cell populations across brain regions and systems. Clearly, orders of magnitude more driver lines are needed[30].
Furthermore, single gene-driven recombinase lines often mark relatively broad and mixed cell populations. More specific targeting is feasible by combining multiple driver-reporter alleles with viral vectors to engage a spectrum of cell-defining features that include lineage, birth time, marker genes, and anatomy (Figure 3). A recent study demonstrates that combinatorial genetic and viral approaches can target highly restricted GABAergic subpopulations and cell types characterized by distinct laminar location, morphology, axonal projection, and electrophysiological properties [28**]. Intersectional embryonic transcription factor drivers allow finer fate mapping of progenitor pools that give rise to distinct GABAergic populations, including laminar cohorts. Conversion of progenitor fate restriction signals to constitutive recombinase expression enables viral targeting of cell types based on their lineage and birth time. Properly designed intersection, subtraction, conversion, and multi-color reporters enhance the precision and versatility of drivers and viral vectors. These strategies and tools should apply to other brain regions and facilitate cell type access throughout the mouse brain.
Figure 3.
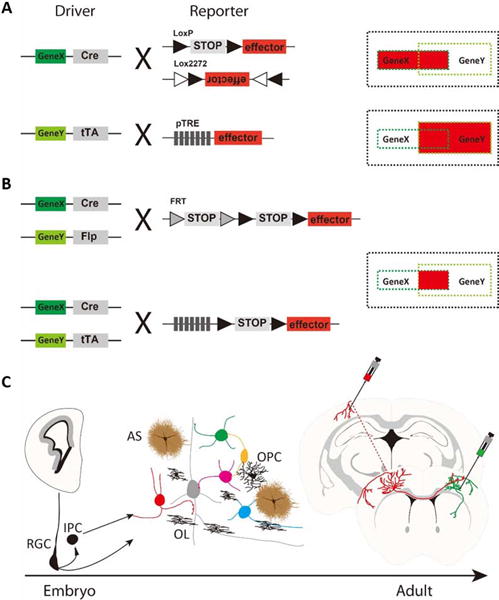
Genetic access to cell types in the mouse brain. A. Binary system for cell type targeting using single driver. B. Intersectional targeting paradigms. C. Besides marker gene expression, other cellular features including developmental lineage, anatomical location and connectivity can also be utilized to target specific cell types within broad classes of neural stem cells, neurons and glia. For example, retrograde virus can be combined with Cre driver mice to label cells expressing a specific marker and also projecting to a specific target brain region.
Binary gene expression systems have proven to be a powerful and versatile strategy to cell access in both mouse (Figure 3) and Drosophila. While the driver component of this system provide cell specificity, the reporter/responder component supplies the toolgenes (markers, sensors, transducers) for observing and manipulating cell phenotypes and function. The robustness of reporter gene expression is thus crucial for the success of cell type and circuit analysis. The Allen Institute of Brain Science has pioneered efforts with sustained progress in the improvement of reporter lines at the Rosa26 and TIGRE loci[29**,31]. In particular, the most recent TIGRE2.0 allele features order of magnitude higher expression level than other reporter lines and will significantly facilitate neural circuit analysis[29**].
A major goal of cell type access is to understand their function in circuit operations and behavior. In this context, marker defined cell types can be functionally heterogeneous; thus it is highly desirable to target neuron types or ensembles based on their activity pattern and history during behavior. Significant progress has been made through innovative combination of activity regulated gene expression and viral methods to access activated neurons. These were covered in a recent review[32]. Newly developed calcium- and light-gated transcriptional control systems enable the detection and manipulation of activity tagged neuronal subtypes with high spatiotemporal precision [33*,34*].
In summary, the technical issues of specificity and robustness of germline based cell access in the mouse have largely been solved. However, the issues of systematic and comprehensive cell coverage remain and will require much larger coordinated efforts with strategic infrastructural and fiscal planning. The Drosophila community has addressed this challenge by a large scale organized project at HHMI/Janelia Farm campus[3]. In the mouse, thousands if not tens of thousands driver lines are likely needed to exert a decisive impact on mammalian circuit neuroscience. Recognizing the cell type issue as the number one priority in the US BRAIN Initiative, NIH has recently launched the BRAIN Initiative Cell Census Network (BICCN) with the goal of establishing a Mouse Brain Cell Atlas[35] (NIH websites). Cell type discovery and access are crucial components of this effort. Currently no conventional scientific institutions and infrastructure (even the Allen Institute) is particularly suited for the task. A strategic balance of broad and sparse coverage vs more focused dense coverage will be necessary to fuel the next phase of progress.
Genetic cell access in the rat
Several rat Cre driver lines and reporter lines have been generated by traditional[36] or BAC transgenic[37–41], and classical[42], ZFN[43] or CRISPR-Cas9[44,45] assisted knock-in methods. They allowed labelling and optogenetic manipulation of restricted neuronal populations. For example, optogenetic stimulation of dopamine neurons in the ventral tegmental area induced positive reinforcement[37]. Currently, the number of transgenic rat drivers and reporter is very limited and they have yet to be widely used in studying neurobiological questions.
Genetic engineering in primates
While mice and rats are essential models in many areas of neuroscience, there are aspects of higher brain function that cannot be adequately modeled in rodents[46]. Similarly, many brain disorders affect higher cognitive functions that have no clear parallels in rodents[47]. There is thus an urgent need for extending circuit neuroscience to primates which are phylogenetically closer to humans. Recent advances in genome-editing technologies have made it feasible to generate primate genetic models[48]. For example, in cynomolgus monkeys, lentiviral-based transgenesis was used to overexpress the Methyl-CpG binding protein 2 gene (MeCP2) to model autism [49], and TALEN-based genome editing was used to mutate MeCP2 gene to model the Rett syndrome [50]. Recently, two groups have generated base-precision knockin cynomogus monkeys expressing reporter genes by CRISPR/Cas9-assisted homologous recombination[51,52]. However, given the complexity of the primate brain and the difficulty and cost involved in primate germline manipulation, it is difficult to conceive that we will be able to map and manipulate many cell types in a primate using traditional germline approaches. Currently, cell access in the primate brain mainly relied on viral vectors carrying short promoter elements. Broad cell populations such as glutamatergic excitatory neurons[53–56], GABAergic inhibitory neurons [57], dopaminergic neurons[58,59], cerebellar Purkinje cells[60] and astrocytes[61] have been targeted with adeno-associated virus (AAV) or lentivirus (Table 2), but there is a large gap between the immense cellular diversity within primate nervous system and the very limited number of currently available cell type specific viral tools.
Table 2.
Viral strategies for targeting specific cell types in non-human primate brain
| Cell type | Promoter | Viral Vectors | Species | Reference |
|---|---|---|---|---|
| Ubiquitous | elongation factor 1α (Ef1α) chicken β-actin (CAG) | AAV, Lentivirus | Macaque | [81,82] |
| CMV | AAV | Macaque Marmoset | [54,83] | |
| Neuron | human Synapsin-1 hThy-1 | AAV, Lentivirus | Macaque | [56,82] |
| Excitatory neuron | mouse calcium-calmodulin kinase-2α(CamK2α) | AAV, Lentivirus | Macaque Marmoset | [51–54] |
| GABAergic neuron | Mouse distalless homeobox 5 and 6 enhancer | AAV | Marmoset | [55] |
| Dopaminergic neuron | 300-bp fragment of the 5′ tyrosine hydroxylase(TH) promoter | AAV | Macaque | [57] |
| Dopaminergic/noradrenergic neuron | a 3.1 kb proximal promoter fragment of the rhesus monkey tyrosine hydroxylase promoter | Lentivirus | Macaque | [56] |
| Cerebellar Purkinje cell | 1 kb L7/Pcp2 promoter | AAV | Macaque | [58] |
| Astrocyte 0.3-kb | Callithrix jacchus GFAP (cjGFAP) promoter | AAV | Marmoset | [59] |
Non-germline approaches to cell access
Diverse neurotropic viruses have evolved mechanisms to transduce nerve cells and can be engineered to achieve regulated and high level gene expression. Although gene regulatory promoter and enhancer elements have been incorporated into viral vectors to drive toolgene expression, the cell specificity of this approaches is quite limited, in part because most enhancer elements are not well defined and viral vectors have limited capacity to include large genomic fragments. MicroRNA binding sites can also be incorporated to regulated gene expression post-transcriptionally for tissue or cell type specific targeting [62–64].
As viral infection rely on the interaction of viral capsid protein with receptors on cell membrane, selective transduction can be achieved by engineered tropism. For example, EvnA is widely used to psuedotype virus such as rabies virus to infect TVA expressing neurons for cell type specific retrograde tracing[65]. Screening of naturally existing or directed engineered viral capsid variants with higher cell type specific is a currently underexplored but worth-noting future direction.
As many projection neuron types can be defined, to the first approximating, by their axon projection targets, application of retrograde viral infection at axon terminals is an effective approach of targeting cell types. Canine adenovirus type 2[66], rabies virus[67], herpes simplex virus[68], and the recently developed retrograde AAV (rAAV2-retro)[69**] have all proven to be effective. In particular, the ease in the construction of rAAV2-retro with diverse payloads enables infection of multiple projection targets, which enhance cell type specificity and allow more flexible observation and manipulation of these cell types.
Retrovirus like Maloney murine leukemia virus can only transduce proliferating cells including neural stem cells and glia but not post-mitotic neurons. Using this type of virus as carrier, fluorescent or enzymatic markers can be integrated into neural stem cell genome for lineage tracing[70]. Clonal analysis from single progenitors can further be achieved by titrating the virus to infect very few stem cells, or tagging different stem cells with highly diverse DNA barcodes[71,72].
Combing AAV and CRISPR-Cas9, efficient genome editing via homology-directed repair (HDR) can be achieved in vivo in postmitotic neurons and neural progenitors to tag endogenous proteins. Targeting CamKIIα gene locus, selective labeling of neurons but not glia were observed in the all injected brain area including cerebral cortex, hippocampus, amygdala and striatum. This virus-mediated single-cell labeling of endogenous proteins via HDR (vSLENDR) is widely applicable to any brain area, cell type, and age, and could be adapted to other mammalian species like non-human primates. [73**]
Cell type targeting can also utilize ligand-receptor interaction on the cell surface. For example, gold nanoparticles conjugated to high-avidity ligands to membrane proteins of specific neuronal cell type such as dorsal root ganglion neurons enabled optical triggering of action potentials in these neurons[74*]. Antibody-antigen interaction is another strategy for targeted delivery of effector molecules or toolgenes. In tumor therapy, camelid nanobody-based approaches have been developed to block cell signaling or to exert therapeutic activity through its conjugated toxin and nano-sized drug carriers[75*]. Nanobodies can also be used to decorate viral vectors for cell specific transduction[75*]. In principal, these approaches can also be applied in the nervous system to deliver toxin, drug and transgenes into selective cell types.
Although nanobody based targeted deliver into neurons has yet to be achieved, nanobody against GFP has been used for cell type specific analysis in the nervous system. For example, the retoTRAP method captures translating mRNAs from neurons defined by their axon projection target through retrogradely infection of a viral vector expressing a GFP-tagged ribosomal protein [76**–78*]. Further, GFP-dependent transcription factor (T-DDOGs)[79] and Cre recombinase (CRE-DOG)[80] methods were used to regulate transcription and recombination activity, respectively. In the future, nanobodies recognizing endogenous cellular marker proteins could be developed as an alternative to viral approaches for cell type targeting in mammalian species.
Summary and perspectives
Cell types are fundamental building blocks that underlie system level operations across organs, much beyond the brain, and contribute to the well-being of the individual organism. A broad and fundamental need in current biomedical research is to be able to systematically identify the diverse cell types in body systems and to manipulate the function of each type in order to understand tissue and system level function and dysfunction. With the massive scale and throughput of single cell genomics that define transcriptional cell clusters, experimental access to cell types is rapidly becoming one of the most rate-limiting bottlenecks in biomedical research.
In the mouse with sophisticated and well established genome engineering system, major technical hurdles in achieving specificity and robustness of cell type access have been overcome. It will be very useful to achieve multiplexed and routine targeting of two or more cell types in the same animal. Novel recombinases for orthogonal gene regulation with Cre, Flp, Dre, tTA will be desirable[81], and improved intersection/subtraction viral vectors[82*] will be especially needed to achieve flexibility and multiplex cell type access. Perhaps the most pressing need is to expand the coverage of cell type access across the mouse brain. Even with BICCN scale projects, only a modest number of new driver lines (~100) have been proposed (https://www.nih.gov/news-events/news-releases/nih-brain-initiative-launches-cell-census). Large-scale germline engineering and breeding present significant challenges in organization, coordination, infrastructure, and fiscal planning.
CRISPR based genome editing presents opportunities for germline manipulation and cell type targeting in other mammalian species. However, issues of time, cost, scale, and ethics remain, especially for primates. Indeed it is hard to conceive even a small scale project for a germline based approach to cell type access in primates with broader impact. Therefore in addition to specificity and comprehensiveness, what is most needed is the ability to access cell types in a way that is fast (hours and days), inexpensive, and general (applicable across mammalian species). Novel non-germline approach will be the game changer. Innovative methods such as somatic cell engineering and cell surface-based gene delivery may reduce the barrier of genetic access to mammalian cell types.
Highlights.
-
➢
Genetic access to cell types aims to achieve specificity and comprehensive coverage
-
➢
Cell transcriptomes provide ample molecular markers for accessing cell types
-
➢
Combinatorial germline and viral approaches in mice enable specific cell targeting
-
➢
Novel non-germline methods are critical for accessing cell types in other species
Acknowledgments
M.H. is supported by funds from the National Natural Science Foundation of China (31771196, 31471037, 91432106, 81428010), Shanghai Science and Technology Commission Innovation Fund (17JC1401500) and Shanghai Rising-Star Program. Z.J.H. is supported by funds from the National Institute of Health (U19MH114823-01, 5R01MH109665-02, 5R01MH101268-05) and Robertson Neuroscience Fund of the Cold Spring Harbor Laboratory.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* Of special interest
**Of outstanding interest
- 1.Huang ZJ, Zeng H. Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci. 2013;36:183–215. doi: 10.1146/annurev-neuro-062012-170307. [DOI] [PubMed] [Google Scholar]
- 2.Hobert O, Glenwinkel L, White J. Revisiting Neuronal Cell Type Classification in Caenorhabditis elegans. Curr Biol. 2016;26:R1197–R1203. doi: 10.1016/j.cub.2016.10.027. [DOI] [PubMed] [Google Scholar]
- 3.Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, et al. The neuronal architecture of the mushroom body provides a logic for associative learning. Elife. 2014;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeFelipe J, Lopez-Cruz PL, Benavides-Piccione R, Bielza C, Larranaga P, Anderson S, Burkhalter A, Cauli B, Fairen A, Feldmeyer D, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013;14:202–216. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng H, Sanes JR. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat Rev Neurosci. 2017;18:530–546. doi: 10.1038/nrn.2017.85. [DOI] [PubMed] [Google Scholar]
- 7*.Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. scRNAseq in the mouse somatosensory cortex and hippocampal CA1 region identified 47 transcriptomic cell types comprising all known major cell types and a layer I interneuron expressing Pax6 and a distinct postmitotic oligodendrocyte subclass marked by Itpr2. [DOI] [PubMed] [Google Scholar]
- 8.Usoskin D, Furlan A, Islam S, Abdo H, Lonnerberg P, Lou D, Hjerling-Leffler J, Haeggstrom J, Kharchenko O, Kharchenko PV, et al. Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18:145–153. doi: 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 9*.Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. scRNAseq in mouse visual cortex identified 49 transcriptomic cell types and confirmed some of the transciptomic cell types can be associated with electrophysiological and axon projection properties. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Shekhar K, Lapan SW, Whitney IE, Tran NM, Macosko EZ, Kowalczyk M, Adiconis X, Levin JZ, Nemesh J, Goldman M, et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 2016;166:1308–1323 e1330. doi: 10.1016/j.cell.2016.07.054. scRNAseq in mouse retina identified 15 transcriptomic cell types including all previously known bipolar cell types and two novel types, one of which has a non-canonical morphology and position. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11*.La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell. 2016;167:566–580 e519. doi: 10.1016/j.cell.2016.09.027. scRNAseq during ventral midbrain development in human and mouse identified 25 and 26 transcriptomic cell types respectively. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Bruggen D, Agirre E, Castelo-Branco G. Single-cell transcriptomic analysis of oligodendrocyte lineage cells. Curr Opin Neurobiol. 2017;47:168–175. doi: 10.1016/j.conb.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 13*.Kee N, Volakakis N, Kirkeby A, Dahl L, Storvall H, Nolbrant S, Lahti L, Bjorklund AK, Gillberg L, Joodmardi E, et al. Single-Cell Analysis Reveals a Close Relationship between Differentiating Dopamine and Subthalamic Nucleus Neuronal Lineages. Cell Stem Cell. 2017;20:29–40. doi: 10.1016/j.stem.2016.10.003. scRNAseq in the the developing mouse ventral mesencephalic and diencephalic region that either express or do not express the transcription factor Lmx1a uncovered an unexpected strong relationship between mesDA and STN neural lineage development. [DOI] [PubMed] [Google Scholar]
- 14*.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. Describes multiplexed error-robust FISH (MERFISH), a single-molecule imaging method that allows thousands of RNA species to be imaged in single cells by using combinatorial FISH labeling with encoding schemes capable of detecting and/or correcting errors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. Describes sequential fluorescence in situ hybridization(seq-FISH) which uses a sequential barcoding scheme to identify and directly image multiple mRNAs in a single cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16**.Shah S, Lubeck E, Zhou W, Cai L. In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron. 2016;92:342–357. doi: 10.1016/j.neuron.2016.10.001. Describes an improved version of seq-FISH used to resolve the structural organization of the hippocampus with single-cell resolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Paul A, Crow M, Raudales R, He M, Gillis J, Huang ZJ. Transcriptional Architecture of Synaptic Communication Delineates GABAergic Neuron Identity. Cell. 2017;171:522–539 e520. doi: 10.1016/j.cell.2017.08.032. Discovered that the transcriptional architecture of synaptic communication delineates cortical GABAergic neuron identities. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lerner Talia N, Ye L, Deisseroth K. Communication in Neural Circuits: Tools, Opportunities, and Challenges. Cell. 2016;164:1136–1150. doi: 10.1016/j.cell.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuzik J, Zeisel A, Mate Z, Calvigioni D, Yanagawa Y, Szabo G, Linnarsson S, Harkany T. Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat Biotechnol. 2016;34:175–183. doi: 10.1038/nbt.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, et al. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat Biotechnol. 2016;34:199–203. doi: 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kebschull JM, Garcia da Silva P, Reid AP, Peikon ID, Albeanu DF, Zador AM. High-Throughput Mapping of Single-Neuron Projections by Sequencing of Barcoded RNA. Neuron. 2016;91:975–987. doi: 10.1016/j.neuron.2016.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu YE, Pan L, Zuo Y, Li X, Hong W. Detecting Activated Cell Populations Using Single-Cell RNA-Seq. Neuron. 2017;96:313–329 e316. doi: 10.1016/j.neuron.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 23.Gerfen CR, Paletzki R, Heintz N. GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silberberg SN, Taher L, Lindtner S, Sandberg M, Nord AS, Vogt D, McKinsey GL, Hoch R, Pattabiraman K, Zhang D, et al. Subpallial Enhancer Transgenic Lines: a Data and Tool Resource to Study Transcriptional Regulation of GABAergic Cell Fate. Neuron. 2016;92:59–74. doi: 10.1016/j.neuron.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shima Y, Sugino K, Hempel CM, Shima M, Taneja P, Bullis JB, Mehta S, Lois C, Nelson SB. A Mammalian enhancer trap resource for discovering and manipulating neuronal cell types. Elife. 2016;5:e13503. doi: 10.7554/eLife.13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.Harris JA, Hirokawa KE, Sorensen SA, Gu H, Mills M, Ng LL, Bohn P, Mortrud M, Ouellette B, Kidney J, et al. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits. 2014;8:76. doi: 10.3389/fncir.2014.00076. Characterization of Cre driver mice commonly used in neuroscience research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28**.He M, Tucciarone J, Lee S, Nigro MJ, Kim Y, Levine JM, Kelly SM, Krugikov I, Wu P, Chen Y, et al. Strategies and Tools for Combinatorial Targeting of GABAergic Neurons in Mouse Cerebral Cortex. Neuron. 2016;91:1228–1243. doi: 10.1016/j.neuron.2016.08.021. Combinatorial genetic targeting of highly specific GABAergic neurons in the mouse neocortex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29**.Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, Takeno MM, Huang L, Gu H, Larsen R, et al. A suite of transgenic driver and reporter mouse lines with enhanced brain cell type targeting and functionality. bioRxiv. 2017 doi: 10.1016/j.cell.2018.06.035. Describes a large set of driver and reporter transgenic mouse lines, including 23 new driver lines targeting a variety of cortical and subcortical cell populations and 26 new reporter lines expressing an array of molecular tools. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang ZJ. Toward a genetic dissection of cortical circuits in the mouse. Neuron. 2014;83:1284–1302. doi: 10.1016/j.neuron.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeNardo L, Luo L. Genetic strategies to access activated neurons. Curr Opin Neurobiol. 2017;45:121–129. doi: 10.1016/j.conb.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Lee D, Hyun JH, Jung K, Hannan P, Kwon HB. A calcium- and light-gated switch to induce gene expression in activated neurons. Nat Biotechnol. 2017;35:858–863. doi: 10.1038/nbt.3902. One of the two recently developed methods which integrate calcium and light sensing for selective labelling of and gene expression regulation in an active population of neurons with high temporal precision. [DOI] [PubMed] [Google Scholar]
- 34*.Wang W, Wildes CP, Pattarabanjird T, Sanchez MI, Glober GF, Matthews GA, Tye KM, Ting AY. A light- and calcium-gated transcription factor for imaging and manipulating activated neurons. Nat Biotechnol. 2017;35:864–871. doi: 10.1038/nbt.3909. The other of the two recently developed methods which integrate calcium and light sensing for selective labelling of and gene expression regulation in an active population of neurons with high temporal precision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ecker JR, Geschwind DH, Kriegstein AR, Ngai J, Osten P, Polioudakis D, Regev A, Sestan N, Wickersham IR, Zeng H. The BRAIN Initiative Cell Census Consortium: Lessons Learned toward Generating a Comprehensive Brain Cell Atlas. Neuron. 2017;96:542–557. doi: 10.1016/j.neuron.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schonig K, Weber T, Frommig A, Wendler L, Pesold B, Djandji D, Bujard H, Bartsch D. Conditional gene expression systems in the transgenic rat brain. BMC Biol. 2012;10:77. doi: 10.1186/1741-7007-10-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Witten IB, Steinberg EE, Lee SY, Davidson TJ, Zalocusky KA, Brodsky M, Yizhar O, Cho SL, Gong S, Ramakrishnan C, et al. Recombinase-driver rat lines: tools, techniques, and optogenetic application to dopamine-mediated reinforcement. Neuron. 2011;72:721–733. doi: 10.1016/j.neuron.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Kaneko R, Yanagawa Y, Saito Y. The vestibulo- and preposito-cerebellar cholinergic neurons of a ChAT-tdTomato transgenic rat exhibit heterogeneous firing properties and the expression of various neurotransmitter receptors. Eur J Neurosci. 2014;39:1294–1313. doi: 10.1111/ejn.12509. [DOI] [PubMed] [Google Scholar]
- 39.Ikeda K, Takahashi M, Sato S, Igarashi H, Ishizuka T, Yawo H, Arata S, Southard-Smith EM, Kawakami K, Onimaru H. A Phox2b BAC Transgenic Rat Line Useful for Understanding Respiratory Rhythm Generator Neural Circuitry. PLoS One. 2015;10:e0132475. doi: 10.1371/journal.pone.0132475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Igarashi H, Koizumi K, Kaneko R, Ikeda K, Egawa R, Yanagawa Y, Muramatsu S, Onimaru H, Ishizuka T, Yawo H. A Novel Reporter Rat Strain That Conditionally Expresses the Bright Red Fluorescent Protein tdTomato. PLoS One. 2016;11:e0155687. doi: 10.1371/journal.pone.0155687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh Y-M, Karube F, Takahashi S, Kobayashi K, Takada M, Uchigashima M, Watanabe M, Nishizawa K, Kobayashi K, Fujiyama F. Using a novel PV-Cre rat model to characterize pallidonigral cells and their terminations. Brain Structure and Function. 2017;222:2359–2378. doi: 10.1007/s00429-016-1346-2. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi T, Kato-Itoh M, Yamaguchi T, Tamura C, Sanbo M, Hirabayashi M, Nakauchi H. Identification of rat Rosa26 locus enables generation of knock-in rat lines ubiquitously expressing tdTomato. Stem Cells Dev. 2012;21:2981–2986. doi: 10.1089/scd.2012.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z, Brown A, Fisher D, Wu Y, Warren J, Cui X. Tissue Specific Expression of Cre in Rat Tyrosine Hydroxylase and Dopamine Active Transporter-Positive Neurons. PLoS One. 2016;11:e0149379. doi: 10.1371/journal.pone.0149379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma Y, Yu L, Pan S, Gao S, Chen W, Zhang X, Dong W, Li J, Zhou R, Huang L, et al. CRISPR/Cas9-mediated targeting of the Rosa26 locus produces Cre reporter rat strains for monitoring Cre/loxP-mediated lineage tracing. FEBS J. 2017 doi: 10.1111/febs.14188. [DOI] [PubMed] [Google Scholar]
- 45.Ma Y, Zhang L, Huang X. Building Cre Knockin Rat Lines Using CRISPR/Cas9. Methods Mol Biol. 2017;1642:37–52. doi: 10.1007/978-1-4939-7169-5_3. [DOI] [PubMed] [Google Scholar]
- 46.Izpisua Belmonte JC, Callaway EM, Caddick SJ, Churchland P, Feng G, Homanics GE, Lee KF, Leopold DA, Miller CT, Mitchell JF, et al. Brains, genes, and primates. Neuron. 2015;86:617–631. doi: 10.1016/j.neuron.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jennings CG, Landman R, Zhou Y, Sharma J, Hyman J, Movshon JA, Qiu Z, Roberts AC, Roe AW, Wang X, et al. Opportunities and challenges in modeling human brain disorders in transgenic primates. Nat Neurosci. 2016;19:1123–1130. doi: 10.1038/nn.4362. [DOI] [PubMed] [Google Scholar]
- 48.Okano H, Kishi N. Investigation of brain science and neurological/psychiatric disorders using genetically modified non-human primates. Curr Opin Neurobiol. 2017;50:1–6. doi: 10.1016/j.conb.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 49.Liu Z, Li X, Zhang JT, Cai YJ, Cheng TL, Cheng C, Wang Y, Zhang CC, Nie YH, Chen ZF, et al. Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature. 2016;530:98–102. doi: 10.1038/nature16533. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y, Yu J, Niu Y, Qin D, Liu H, Li G, Hu Y, Wang J, Lu Y, Kang Y, et al. Modeling Rett Syndrome Using TALEN-Edited MECP2 Mutant Cynomolgus Monkeys. Cell. 2017;169:945–955 e910. doi: 10.1016/j.cell.2017.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui Y, Niu Y, Zhou J, Chen Y, Cheng Y, Li S, Ai Z, Chu C, Wang H, Zheng B, et al. Generation of a precise Oct4-hrGFP knockin cynomolgus monkey model via CRISPR/Cas9-assisted homologous recombination. Cell Res. 2018 doi: 10.1038/cr.2018.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao X, Liu Z, Wang X, Wang Y, Nie YH, Lai L, Sun R, Shi L, Sun Q, Yang H. Generation of knock-in cynomolgus monkey via CRISPR/Cas9 editing. Cell Res. 2018 doi: 10.1038/cr.2018.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Han X, Qian X, Bernstein JG, Zhou H-H, Franzesi GT, Stern P, Bronson RT, Graybiel AM, Desimone R, Boyden ES. Millisecond-Timescale Optical Control of Neural Dynamics in the Nonhuman Primate Brain. Neuron. 2009;62:191–198. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dai J, Brooks DI, Sheinberg DL. Optogenetic and electrical microstimulation systematically bias visuospatial choice in primates. Curr Biol. 2014;24:63–69. doi: 10.1016/j.cub.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 55.Lu Y, Truccolo W, Wagner FB, Vargas-Irwin CE, Ozden I, Zimmermann JB, May T, Agha NS, Wang J, Nurmikko AV. Optogenetically induced spatiotemporal gamma oscillations and neuronal spiking activity in primate motor cortex. J Neurophysiol. 2015;113:3574–3587. doi: 10.1152/jn.00792.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watakabe A, Ohtsuka M, Kinoshita M, Takaji M, Isa K, Mizukami H, Ozawa K, Isa T, Yamamori T. Comparative analyses of adeno-associated viral vector serotypes 1, 2, 5, 8 and 9 in marmoset, mouse and macaque cerebral cortex. Neuroscience Research. 2015;93:144–157. doi: 10.1016/j.neures.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi G-A, Guo L, Xu Q, Liu R, Lu C, Chu J, et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nature Neuroscience. 2016;19:1743–1749. doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lerchner W, Corgiat B, Der Minassian V, Saunders RC, Richmond BJ. Injection parameters and virus dependent choice of promoters to improve neuron targeting in the nonhuman primate brain. Gene Ther. 2014;21:233–241. doi: 10.1038/gt.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stauffer WR, Lak A, Yang A, Borel M, Paulsen O, Boyden ES, Schultz W. Dopamine Neuron-Specific Optogenetic Stimulation in Rhesus Macaques. Cell. 2016;166:1564–1571. doi: 10.1016/j.cell.2016.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El-Shamayleh Y, Kojima Y, Soetedjo R, Horwitz GD. Selective Optogenetic Control of Purkinje Cells in Monkey Cerebellum. Neuron. 2017;95:51–62. doi: 10.1016/j.neuron.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shinohara Y, Konno A, Takahashi N, Matsuzaki Y, Kishi S, Hirai H. Viral Vector-Based Dissection of Marmoset GFAP Promoter in Mouse and Marmoset Brains. PLOS ONE. 2016;11:e0162023. doi: 10.1371/journal.pone.0162023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie J, Xie Q, Zhang H, Ameres SL, Hung JH, Su Q, He R, Mu X, Seher Ahmed S, Park S, et al. MicroRNA-regulated, systemically delivered rAAV9: a step closer to CNS-restricted transgene expression. Mol Ther. 2011;19:526–535. doi: 10.1038/mt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karali M, Manfredi A, Puppo A, Marrocco E, Gargiulo A, Allocca M, Corte MD, Rossi S, Giunti M, Bacci ML, et al. MicroRNA-restricted transgene expression in the retina. PLoS One. 2011;6:e22166. doi: 10.1371/journal.pone.0022166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahmed SS, Li H, Cao C, Sikoglu EM, Denninger AR, Su Q, Eaton S, Liso Navarro AA, Xie J, Szucs S, et al. A single intravenous rAAV injection as late as P20 achieves efficacious and sustained CNS Gene therapy in Canavan mice. Mol Ther. 2013;21:2136–2147. doi: 10.1038/mt.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ciabatti E, González-Rueda A, Mariotti L, Morgese F, Tripodi M. Life-Long Genetic and Functional Access to Neural Circuits Using Self-Inactivating Rabies Virus. Cell. 2017;170:382–392. doi: 10.1016/j.cell.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hnasko TS, Perez FA, Scouras AD, Stoll EA, Gale SD, Luquet S, Phillips PEM, Kremer EJ, Palmiter RD. Cre recombinase-mediated restoration of nigrostriatal dopamine in dopamine-deficient mice reverses hypophagia and bradykinesia. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8858–8863. doi: 10.1073/pnas.0603081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ugolini G. Rabies virus as a transneuronal tracer of neuronal connections. Adv Virus Res. 2011;79:165–202. doi: 10.1016/B978-0-12-387040-7.00010-X. [DOI] [PubMed] [Google Scholar]
- 68.Zemanick MC, Strick PL, Dix RD. Direction of transneuronal transport of herpes simplex virus 1 in the primate motor system is strain-dependent. Proc Natl Acad Sci U S A. 1991;88:8048–8051. doi: 10.1073/pnas.88.18.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69**.Tervo DG, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron. 2016;92:372–382. doi: 10.1016/j.neuron.2016.09.021. Describes in vivo directed evolution to screen out an robust retrograde recombinant AAV, rAAV2-retro, for retrograde targeting of projection neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma J, Shen Z, Yu YC, Shi SH. Neural lineage tracing in the mammalian brain. Curr Opin Neurobiol. 2017;50:7–16. doi: 10.1016/j.conb.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mayer C, Jaglin XH, Cobbs LV, Bandler RC, Streicher C, Cepko CL, Hippenmeyer S, Fishell G. Clonally Related Forebrain Interneurons Disperse Broadly across Both Functional Areas and Structural Boundaries. Neuron. 2015;87:989–998. doi: 10.1016/j.neuron.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harwell CC, Fuentealba LC, Gonzalez-Cerrillo A, Parker PR, Gertz CC, Mazzola E, Garcia MT, Alvarez-Buylla A, Cepko CL, Kriegstein AR. Wide Dispersion and Diversity of Clonally Related Inhibitory Interneurons. Neuron. 2015;87:999–1007. doi: 10.1016/j.neuron.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73**.Nishiyama J, Mikuni T, Yasuda R. Virus-Mediated Genome Editing via Homology-Directed Repair in Mitotic and Postmitotic Cells in Mammalian Brain. Neuron. 2017;96:755–768 e755. doi: 10.1016/j.neuron.2017.10.004. An efficient genome engineering method for non-germline cells which can be used for cell type targeting in the brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74*.Carvalho-de-Souza JL, Treger JS, Dang B, Kent SB, Pepperberg DR, Bezanilla F. Photosensitivity of neurons enabled by cell-targeted gold nanoparticles. Neuron. 2015;86:207–217. doi: 10.1016/j.neuron.2015.02.033. Targeting of high-avidity ligands conjugated gold nanoparticles to selective neuron populations for activity modulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75*.Hu Y, Liu C, Muyldermans S. Nanobody-Based Delivery Systems for Diagnosis and Targeted Tumor Therapy. Front Immunol. 2017;8:1442. doi: 10.3389/fimmu.2017.01442. Nanobody-based delivery system which could potentially by applied in the nervous system. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76**.Ekstrand MI, Nectow AR, Knight ZA, Latcha KN, Pomeranz LE, Friedman JM. Molecular profiling of neurons based on connectivity. Cell. 2014;157:1230–1242. doi: 10.1016/j.cell.2014.03.059. Describes a method of ribosome tagging by a camelid nanobody raised against GFP to selectively capture translating mRNAs from neurons retrogradely labeled with GFP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nectow AR, Ekstrand MI, Friedman JM. Molecular characterization of neuronal cell types based on patterns of projection with Retro-TRAP. Nature Protocols. 2015;10:1319–1327. doi: 10.1038/nprot.2015.087. [DOI] [PubMed] [Google Scholar]
- 78*.Nectow AR, Moya MV, Ekstrand MI, Mousa A, McGuire KL, Sferrazza CE, Field BC, Rabinowitz GS, Sawicka K, Liang Y, et al. Rapid Molecular Profiling of Defined Cell Types Using Viral TRAP. Cell Reports. 2017;19:655–667. doi: 10.1016/j.celrep.2017.03.048. Describes viral mediated Cre-dependent expression of an EGFP-tagged ribosomal protein for translational profiling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tang JCY, Szikra T, Kozorovitskiy Y, Teixiera M, Sabatini BL, Roska B, Cepko CL. A Nanobody-Based System Using Fluorescent Proteins as Scaffolds for Cell-Specific Gene Manipulation. Cell. 2013;154:928–939. doi: 10.1016/j.cell.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang JCY, Rudolph S, Cepko CL. Viral Delivery of GFP-Dependent Recombinases to the Mouse Brain. Methods Mol Biol. 2017;1642:109–126. doi: 10.1007/978-1-4939-7169-5_8. [DOI] [PubMed] [Google Scholar]
- 81.Weinberg BH, Pham NTH, Caraballo LD, Lozanoski T, Engel A, Bhatia S, Wong WW. Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nature Biotechnology. 2017;35:453–462. doi: 10.1038/nbt.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82*.Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. Describes a versatile single-AAV system with engineered introns for selective expression conditional upon multiple cell-type features using Boolean logical operations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Galvan A, Hu X, Smith Y, Wichmann T. In Vivo Optogenetic Control of Striatal and Thalamic Neurons in Non-Human Primates. PLOS ONE. 2012;7:e50808. doi: 10.1371/journal.pone.0050808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li M, Liu F, Jiang H, Lee TS, Tang S. Long-Term Two-Photon Imaging in Awake Macaque Monkey. Neuron. 2017;93:1049–1057 e1043. doi: 10.1016/j.neuron.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 85.Inoue K, Takada M, Matsumoto M. Neuronal and behavioural modulations by pathway-selective optogenetic stimulation of the primate oculomotor system. Nat Commun. 2015;6:8378. doi: 10.1038/ncomms9378. [DOI] [PMC free article] [PubMed] [Google Scholar]


