Abstract
Purpose
Survivin is an inhibitor of apoptosis protein (IAP) that is highly expressed in many cancers and represents an attractive molecule for targeted cancer therapy. Although primarily regarded as an intracellular protein with diverse actions, survivin has also been identified in association with circulating tumor exosomes.
Experimental Design
We have reported that active specific vaccination with a long peptide survivin immunogen leads to the development of survivin-specific CD8-mediated tumor cell lysis and prolongation of survival in tumor-bearing mice. In addition to cellular antitumor responses, circulating anti-survivin antibodies are detected in the serum of mice and human glioblastoma patients following vaccination with the survivin immunogen.
Results
Here we demonstrate that survivin is present on the outer cell membrane of a wide variety of cancer cell types, including both murine and human glioma cells. In addition, antibodies to survivin that are derived from the immunogen display antitumor activity against murine GL261 gliomas in both flank and intracranial tumor models and against B16 melanoma as well.
Conclusion
In addition to immunogen-induced, CD8-mediated tumor cell lysis, antibodies to the survivin immunogen have antitumor activity in vivo. Cell-surface survivin could provide a specific target for antibody-mediated tumor immunotherapeutic approaches.
Keywords: antibody, glioblastoma, survivin
INTRODUCTION
Survivin (BIRC5, API4) is a protein that belongs to a family of apoptosis inhibitors and acts in concert with the mitotic spindle apparatus to regulate cell division and chromosomal segregation [1]. It is expressed during the G2/M phase of the cell cycle in association with the spindle microtubule organizing center [2]. Survivin functions in critical roles at other cellular loci to regulate the cell cycle and to inhibit apoptotic cell death. It is frequently expressed by cancer cells of many different types, but its expression is uncommon in normal adult fully differentiated tissues [3].
Expression of survivin in gliomas and other cancers is associated with a poor prognosis [4–7] and refractoriness to chemotherapy [8]. Epitopes of survivin are presented by MHC I complexes on the surface of tumor cells making them immunologically targetable by specific cytotoxic CD8+ T cells. Patients with cancer have anti-survivin antibodies [9] and survivin-specific T cells [10] in their peripheral blood. Therefore, survivin is immunogenic and its immunogenicity may be enhanced with the aid of survivin mimics [11, 12]. Several studies have looked at active, specific vaccination against survivin to treat various cancers [13–15], including malignant gliomas [16].
Although it was initially defined as an intracellular molecule, more recently, survivin has been identified in association with exosomes produced by cervical and prostatic carcinoma cells [17,18]. While the intracellular functions of survivin have been characterized extensively, less well appreciated is its presence and function on the outer membrane of tumor cells where it is potentially targetable using antibody-mediated immunotherapeutic strategies. Here we identify survivin expression on the surface of a wide range of cancer cell types using complementary techniques and demonstrate that monoclonal antibodies derived from immunization with an engineered survivin peptide have direct antitumor properties in different mouse tumor models.
METHODS
Cell lines and culture conditions
GL261, U87, HT-29, HeLa, A2780, A1207, B16f1, Jurkat, Raji, MCF7, PC-3 and HEK293T cells were grown in complete Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum, 5,000 units of penicillin/streptomycin, 50 μM 2-mercaptoethanol, 25mM HEPES, and 1x non-essential amino acids at 37°C in 5% CO2 with media changes two to three times per week. For animal studies, GL261 and B16f1 were obtained from the Division of Cancer Treatment and Diagnosis (DCTD) repository at the National Cancer Institute, National Institutes of Health. Additional cell lines were obtained through American Type Culture Collection (ATCC). No further authentication studies were done.
Antibodies
Anti-c-Myc Tag (9E10) was purchased from Thermo Fisher Scientific. Anti-Caveolin-1 and anti-CD71 (D7G9X) were purchased from Cell Signaling Technology. Anti-Flotillin-1 (18/Flotillin-1) was purchased from BD Transduction Laboratories. Anti-GST was purchased from ABM labs. Anti-survivin (60.11) purchased from Novus. Anti-Gα q/11/14 (G7), anti-EGFR (R-1) and anti-GAPDH (FL-335) were purchased from Santa Cruz.
Peptide synthesis and antibody production
Survivin peptide synthesis was performed using Fmoc chemistry and solid support resin and followed by conjugation to keyhole limpet hemocyanin (KLH) for immunization (Genscript USA, Piscataway, NJ). Additional survivin peptides, including N-terminal biotinylated survivin were synthesized by CS Bio (Menlo Park, CA). Hybridomas were generated under contract with Genscript USA. Ten C57BL/6 mice were pre-bled at −4 days and followed with immunization s.c. with SVN53-67/M57-KLH on day 0. Mice were boosted on days 14, 28 and 56 days and test bled at 21 and 35 days. Cells were fused to create hybridomas on day 60. Immune responses were tested by ELISA and Western Blot with target and irrelevant tagged protein 7 days after each boost immunization. Based on the test bleed result, two animals were selected for electrofusion. A fusion efficiency of 1 hybridoma per 2,500 splenocytes was expected. An average of 1 × 108 B cells from each spleen of immunized mouse provides an anticipated recovery of 2 × 104 hybridoma clones. All fused cells from each cell fusion were plated into 15 × 96-well plates. Supernatants were screened by indirect ELISA with antigen peptide. Twenty positive clones were seeded into 24-well plates and 5 positive primary clones were subcloned by limiting dilution. The clones were carried for a maximum of 3 generations. Subcloned cells were again screened by indirect ELISA with both wild type (SVN53-67) and engineered (SVN53-67/M57) peptides. Two stable subclonal cell lines of each primary clone were chosen for cryopreservation based on confirmed antigen-recognition and normal doubling time. Isotype identification for all the subcloned cell lines was performed. Subclone cultures were scaled up for production of purified IgG.
DNA constructs
To generate hSurvivin-Myc/FLAG, human survivin fused to Myc and FLAG tags at its C-terminus was first excised from a commercial cDNA clone (OriGene). The survivin-Myc/FLAG insert was then used as the template in a PCR reaction with primers hSVN-EcoRI-F (5′-GCGATCGAATTCGTCGACTGG-3′) and hSVN-XbaI-R (5′-ACTCCTCTAGAAAACCTTATCGTCGTC-3′). Digested insert was then ligated into pcDNA3.1 (Thermo Fisher Scientific) and the construct was validated by sequencing. To generate the GST-hSurvivin plasmid used for protein purification, the survivin ORF (but not Myc or FLAG tag sequence) was excised from a commercial cDNA clone (OriGene) using EcoRI and NotI, and ligated into pGEX-5X-2 (GE Healthcare Life Sciences).
Purification of GST-hSurvivin
Overnight cultures of BL21-CodonPlus (DE3)-RIL (Agilent) containing GST-hSurvivin or the empty pGEX-5X-2 vector (500 μl) were used to inoculate into 10 mL of Terrific Broth containing ampicillin and 80 μM ZnCl2 which was then grown to mid-log phase at 37° with shaking. Expression was induced with 0.2 mM IPTG for 5 h at 30°, after which bacteria were pelleted, resuspended in buffer containing 50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 80 μM ZnCl2, 1 mM DTT plus protease inhibitors, and lysed by freeze/thaw and sonication. Supernatants were incubated with Glutathione Agarose (Thermo Fisher Scientific) for 4 h at 4°, then beads were collected and washed. GST and GST-Survivin were eluted in buffer containing 50 mM Tris-Cl, pH 8.0, 20 mM reduced L-Glutathione and 0.1 % Triton-X by incubating 10 minutes at RT, then elutions were concentrated using 10 KD MWCO concentrator columns (Thermo Fisher Scientific). Protein concentrations were measured by bicinchoninic acid (BCA) assay and adjusted by adding buffer without glutathione or Triton-X.
Measurement of antibody affinity
Antibody binding to survivin peptides was measured using the AlphaScreen reagent Mouse IgG Detection Kit (Perkin-Elmer, Waltham, MA). Reactions were carried out in volumes of 50 μl in wells of an opaque, half-area 96-well plate, in buffer consisting of PBS pH 7.4, 0.1% BSA, and 0.01% Tween-20. Each well contained a final concentration of 20 μg/mL Acceptor Beads, 20 μg/mL Donor Beads, 0.5 nM antibody (2C2 or 30H3) and 0.02 – 20 nM biotinylated survivin peptide. Briefly, antibody was incubated with anti-mouse IgG Acceptor Beads plus dilutions of biotinylated survivin peptide for 30 min at RT, after which Streptavidin Donor Beads were added. The reactions were incubated another 30 minutes in the dark at RT and read on an EnVision Excite Multilabel Reader (PerkinElmer). Data was fit using Graphpad Prism 7, and Kd values were extrapolated from saturation curves.
Immunofluorescence microscopy
Cultured cells were seeded on sterile glass coverslips in complete media overnight. Cells were washed with PBS, fixed for 15 min at RT in 4% paraformaldehyde and (if noted) permeabilized for 10 min using 0.05 % Triton-X. Coverslips were incubated with the indicated survivin antibodies for 2 hr at RT, washed with PBS, incubated with secondary antibodies for 1 hr at RT, then washed in PBS overnight at 4° before mounting on glass slides and analyzing by fluorescence microscopy.
Immunohistochemistry
Formalin-fixed paraffin sections from a multi-tumor tissue microarray and one multi-organ normal tissue array (FDA999n; Biomax) (0.6mm per core) were cut at 4 μm, placed on charged slides, and dried at 60°C for one hr. Slides were de-paraffinized at RT through a series of xylene and graded ethanol baths. Slides were pretreated in citrate buffer (BioCare Medical CB910) for 60 min in a steamer and then cooled for 20 min. Slides were quenched in 0.3% H2O2 for 10 min, followed by protein block (X909, Dako) for 5 min. Slides were incubated with 2C2 antibody (1:100; 2 μg/ml) in a humidity chamber for 16 hr. Slides were loaded onto the Dako Autostainer PLUS and mouse Envision HRP was applied for 30 min (Dako K4007). Diaminobenzidine (Dako) was applied for 10 min. Slides were counterstained with hematoxylin for 2 min, dehydrated, cleared and coverslipped.
Flow cytometry staining of tumor cell lines
Aliquots of anti-survivin 2C2 (0.2 μg each) were pre-incubated with 100 μg of immunizing survivin peptide or scrambled peptide for 30 min at RT. Cells were trypsinized, washed and suspended in staining buffer. Pre-incubated antibody was added, and cells were incubated at RT for 1 hr. Cells were washed with PBS and stained with anti-mouse-FITC antibody for 30 min at RT, washed again and data acquired with a BD Fortessa flow cytometer running FACSDiva software with final analysis using FCS Express v4.0.
Protease treatment of cells
Cultured cells were collected either by gently dislodging with PBS or by limited trypsinization, then washed with PBS and resuspended in DMEM to a concentration of 106/mL. Cells were treated with either trypsin (1 mg/mL) or proteinase K (0.5 mg/mL) for 20 minutes at 37°, then proteases were inactivated by the addition of either 1 mg/mL Soybean trypsin inhibitor (for trypsin) or protease inhibitor diluted to 1X and 1 mM PMSF. Cells were washed twice with PBS, then resuspended in Cell Staining Buffer (Biolegend) or Cell Staining Buffer plus Fixation Buffer (Biolegend) plus 0.1 % triton-X. Cells were then stained with 0.2 μg of 2C2 for 1 hr at RT, washed once with PBS, incubated with FITC goat anti-mouse antibody and analyzed by flow cytometry.
Isolation of lipid raft fractions
Lipid rafts were isolated following the detergent-free method of Macdonald and Pike, 2005 [19]. Briefly, cells were scraped from 3–4 15 cm culture plates into PBS, collected, and resuspended in 1 mL of Base Buffer containing 20 mM Tris-Cl, pH 7.8, 250 mM sucrose, 1 mM CaCl2, 1 mM MgCl2, plus protease inhibitors. In the case of HEK293T, cells in 15 cm dishes were transfected with the hSVN-Myc/FLAG construct using Lipofectamine 2000 (Thermo Fisher Scientific) 24 h prior to collection. After 15 minutes on ice, cells in Base Buffer were homogenized through a 22-gauge needle 20 times and centrifuged at 1,000 x g for 10 min. The post-nuclear supernatant was collected on ice, and the nuclear pellet was subjected to homogenization and extraction again in 1 mL Base Buffer. Both post-nuclear supernatants were pooled and stored at −20°. Upon thawing, the post-nuclear supernatant was mixed with 2 mL of 50 % Optiprep (Stemcell Technologies) and dispensed into a 12 mL tube. 2 mL each of 20%, 15% and 10% Optipep solutions were then layered above the post-nuclear supernatant, followed by 2 mL base buffer. Gradients were centrifuged at 52,000 x g for either 2 hr (U87) or 3 hr (A1207 and HEK293T) in a Beckman Coulter L9-55M Ultracentrifuge equipped with SW28 rotor. After centrifugation, fractions were collected as 0.67 mL, and 25 ul of each was loaded onto 12 % SDS-PAGE for analysis by western blot.
Lipid raft visualization
Lipid rafts in GL261 cells were labeled using the Vybrant Lipid Raft Labeling Kit (Molecular Probes) as recommended by the manufacturer. Briefly, cells were detached from culture dishes and resuspended in cold complete media containing 1 μg/mL Alexa Fluor 594 dye-labeled CT-B, incubated for 10 min at 4°, washed in cold PBS, and then resuspended in anti-CT-B antibody solution (anti-CT-B diluted 1:200 in compete media) for 15 min at 4°. Cells were washed twice with PBS, then resuspended in Cell Staining Buffer (Biolegend) and stained with 0.2 μg of 2C2 for 45 min at RT, washed with PBS, incubated with FITC goat anti-mouse antibody and analyzed by imaging flow cytometry on an ImageStreamX Mark II Imaging Flow Cytometer (AMNIS/Millipore, Billerica, MA).
Western blotting
Cells were lysed in RIPA buffer and protein concentration was measured using Pierce BCA Assay Reagent. Aliquots of 20 μg total protein were mixed with Laemmli buffer, boiled, and separated on 12 % SDS-PAGE gels, transferred to PVDF membranes. Then blocked with 5 % non-fat milk in TBST for 1 hr. Antibodies were applied in blocker overnight at dilutions of 1:1000 (60.11) or 1:100 (2C2 and 30H3), after which membranes were washed, incubated with HRP-conjugated secondary antibody, washed again, incubated with ECL reagent and exposed to X-ray film. For western blotting of purified protein, 0.2 μg of GST or GST-Survivin were loaded in triplicate onto a 12 % SDS-PAGE gel, transferred, and blots were processed as above using anti-GST at 1:1000 and 2C2/30H3 at 1:500. ECL chemiluminescence was captured using the Biorad ChemiDoc Touch Imaging System.
Cell growth and apoptosis
GL261 and B16 cells were plated on sterile 96-well plates (2×103 cells per well). Cells were treated with 2C2 and 30H3 antibodies in DMEM at 10μg/mL in quadruplicate. Daily, cells were washed with PBS and fixed with 100% methanol at 4°C for 10 min. Fixed cells were stained with crystal violet for 10 min, washed and allowed to dry. Crystal violet was solubilized in 10% acetic acid and absorbance measured on a Synergy (TM) HTX multi-mode microplate reader at 595nm. Data were analyzed with GraphPad PRISM.
GL261 and B16 flank tumor models
C57BL6 and nude (nu/nu) mice were implanted with murine GL261 glioma cells or B16f1 melanoma cells. For flank tumors, a suspension of 2 × 107 Gl261 glioma cells or 1 × 106 B16f1 melanoma cells in 100μl of PBS was injected subcutaneously in male C57Bl/6 (immunocompetent) mice (Charles River, Wilmington, MA) and in nude (immunocompromised) NCr-nu/nu mice (Charles River, Wilmington, MA). After 3 days, mice were randomly assigned to groups and were given intraperitoneal (i.p.) injections of 10 μg of anti-survivin antibody 2C2 or 10 μg of IgG control antibody. Antibodies were administered every 5 days for a total of 4 doses. Tumor growth was measured daily using calipers and volumes were calculated according to the formula V = {a × b2}/2, where V is volume and a and b are perpendicular diameters of the tumor. Subcutaneous tumor volume significance was measured using Wilcoxon matched pairs signed rank test. All mouse flank tumor model experiments were approved by the Roswell Park Institutional Animal Care and Use Committee (IACUC).
Intracerebral GL261 tumor model
Male C57BL/6 mice (Charles River, Wilmington, MA) were anesthetized with isofluorane and fixed in a stereotactic head frame (David Kopf Instruments, Tujunga, CA). A midline scalp incision was made and the bregma was identified. Stereotactic coordinates were measured (2.0 mm lateral, and 1.2 mm anterior to the bregma). A bur hole was drilled at this point and 1 × 105 GL261 cells suspended in 2.5μl of DMEM were injected through a Hamilton syringe with a fixed, 25-gauge needle at a depth of 3.0 mm relative to the dura mater. Injections were performed at 1 μl/min. After 3 days, mice were randomized into groups and injected with 10μg anti-survivin antibody or 10μg IgG for control. Antibodies were administered subcutaneously every 5 days for a total of 5 doses. SurVaxM (100 μg) was administered in Montanide ISA 51 with 100ng mGM-CSF. Mice were followed for signs of neurological deficits as an indicator of tumor growth and were sacrificed according to established criteria. Significance was assessed by Kaplan-Meir methodology and Mantel-Cox test for all groups. All mouse intracranial implantation experiments were approved by the Roswell Park Institutional Animal Care and Use Committee (IACUC).
Antibody dependent cell-mediated cytotoxicity (ADCC)
ADCC was assessed using a mFcγRIV ADCC Reporter Bioassay Kit (Promega, Madison, WI). For experiments using pre-bound target cells, U87 cells were trypsinized, washed with complete media, re-suspended at a concentration of 106 cells/mL and recovered at 37° for 2 hr. Cells were collected and re-suspended in assay buffer and incubated with nonspecific mouse IgG or mAb 2C2 for 30 min at 37°. Cells were washed to remove unbound antibody, diluted and used as directed for the assay. Effector cells were diluted in assay buffer according to the manufacturer’s instructions. In 96 well opaque culture plates, 25 μl effectors were either added to either 50 μl of pre-bound U87 cells, 50 μl of assay buffer containing increasing concentrations of 2C2, or to 25 μl of peptides at increasing concentrations, after which 25 μl 2C2 was added to a final concentration of 4 μg/mL. Plates were covered and incubated at 37° in a humidified, 5% CO2 incubator for 5–6 h, after which luminescence was measured using Bio-Glo Reagent (Promega, Madison, WI).
RESULTS
Mouse monoclonal antibodies detect wild type and engineered survivin peptides
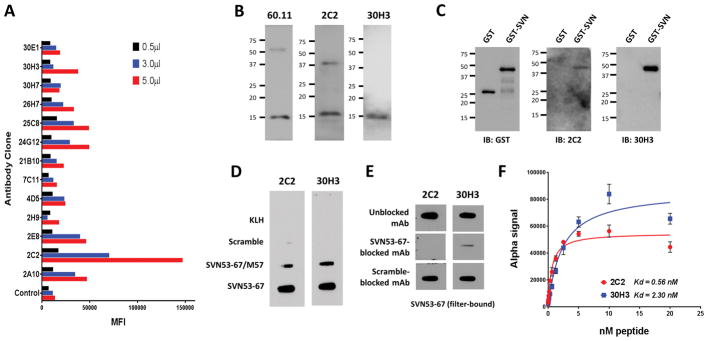
A panel of 19 murine hybridoma cell lines were developed against the engineered survivin peptide (SVN53-67/M57). Supernatants were screened, and high-affinity IgG monoclonal antibodies were produced. Purified antibodies were characterized for target recognition and specificity. Cross-reactivity of individual hybridoma culture supernatants to wild type survivin peptide SVN53-67 from 13 representative cell lines are shown in Figure 1A. We chose to focus on two of these clones (2C2 and 30H3) with different affinities and isotype. Antibodies were compared to a commercially available survivin antibody (60.11; Novus) in Western blots of total protein derived from human embryonic kidney 293T cells (Figure 1B). All three antibodies detected a 16.5 kDa survivin species with limited minor bands. To measure antibody affinity, we utilized the fluorescence proximity-based AlphaLISA™ assay in which binding Kd was measured from the saturation of antibodies by biotinylated wild type survivin peptide SVN53-67, (Figure 1F). Anti-survivin antibody 2C2 (IgG2a) displayed approximately 4-fold greater binding affinity (Kd = 0.56 nM) than mAb 30H3 (IgG1) (Kd = 2.3 nM) for wild type survivin (Figure 1F).
Figure 1.
(A) Antibody panel raised following vaccination of mice with survivin peptide (SVN53-67/M57) present in SurVaxM. Relative binding of different hybridoma clones to immobilized survivin peptide (ELISA) is displayed with indicated amount of supernatant volume (MFI = mean fluorescence index). (B) Western blot of whole cell lysates from human embryonic kidney 293T cells with indicated antibodies demonstrating the dominant 16.5 kD survivin species. (C) Western blot of purified recombinant GST or GST-survivin proteins detected by the indicated antibodies (GST, glutathione-S-transferase). (D) Slot blots detecting filter-immobilized KLH carrier protein and scrambled, mutant and wild type survivin peptides with indicated survivin antibodies. (E) Competition blocking of monoclonal antibodies pre-incubated with peptides (left) and then used to detect filter-immobilized, wild type SVN53-67 peptide. (F) AlphaLISA assay performed to determine relative affinity of purified 2C2 and 30H3 antibodies forSVN53-67 peptide.
Antibody specificity and detection of survivin in human cancers
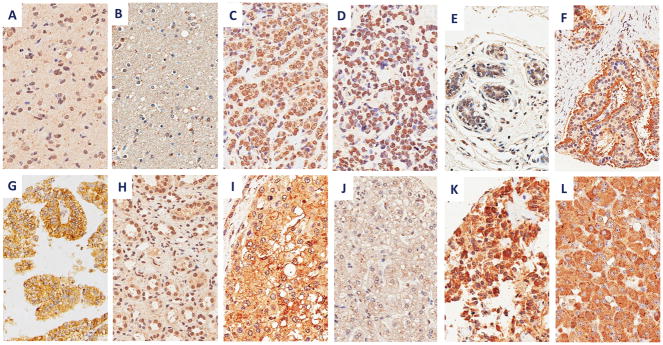
Both antibodies 2C2 and 30H3 detected filter-bound survivin peptides SVN53-67 and SVN 53-67/M57 with little reactivity to scrambled peptide or KLH carrier protein (Figure 1D). The addition of wild-type survivin peptide to the antibodies during immunoblotting effectively blocked their detection of survivin peptide, demonstrating high antibody specificity (Figure 1E). Antibody 2C2 detected strong survivin expression in a wide range of human cancers, including: glioblastoma, invasive ductal breast carcinoma, lobular breast carcinoma, carcinoid tumor, clear cell renal carcinoma, hepatocellular carcinoma, medullary thyroid carcinoma and pheochromocytoma in a tissue microarray (Figure 4). As determined by the pathology co-investigator (J.Q.), patterns of survivin expression included nuclear, cytoplasmic, apical and membranous. Normal human brain, breast and liver revealed no specific survivin expression, although modest nuclear staining was seen in renal tubular epithelium (Figure 4H).
Figure 4.
Survivin immunohistochemistry with 2C2 mAb in human cancers and normal tissues with indicated staining patterns, including: (A) glioblastoma (nuclear), (B) normal brain (negative), (C) invasive ductal breast carcinoma (nuclear), (D) lobular breast carcinoma (nuclear), (E) normal breast tissue (negative), (F) typical carcinoid tumor (apical), (G) clear cell renal cell carcinoma (membranous and cytoplasmic), (H) normal kidney (nuclear), (I) hepatocellular carcinoma (membranous and cytoplasmic), (J) normal liver (negative), (K) medullary thyroid carcinoma (cytoplasmic), and (L) pheochromocytoma (cytoplasmic). Images are shown at 400x.
Survivin expression on the surface of tumor cells
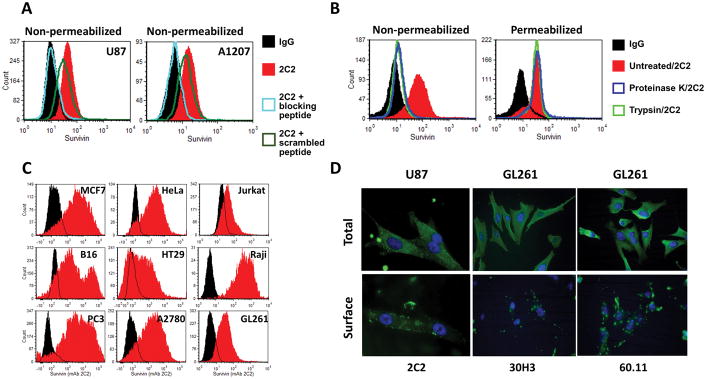
Analysis of non-permeabilized U87 and A1207 human glioma cells by flow cytometry confirmed that cell-surface binding was present and was competed out by wild type survivin peptide, but not scrambled peptide (Figure 2A). Survivin was not detected on the surface of non-permeabilized cells that had been treated with either trypsin or proteinase K (Figure 2B). Treatment with proteases did not impair the ability of antibody to detect intracellular survivin following subsequent permeabilization. Flow cytometry analysis using anti-survivin 2C2 identified survivin expression on the surface of a diverse set of human and murine cancer cell lines, including: those derived from gliomas (U87, A1207 and GL261), ovarian (A2708), breast (MCF7), prostate (PC3), colon (HT29), skin (B16 melanoma) and cervical (HeLa) cancers, leukemia (Raji) and lymphoma (Jurkat) (Figure 2C). Survivin was also detected in permeabilized tumor cell lines with 2C2 and 30H3 by immunofluorescence microscopy (Figure 2D), and similar results were obtained with a commercially available survivin antibody (60.11) to an undefined survivin sequence. Non-permeabilized cells showed significant surface binding of survivin antibodies with focal concentrations of fluorescence on the surface of the cell membranes of both murine and human cancer cell lines.
Figure 2.
(A) Flow cytometric detection of cell-surface survivin in unfixed (i.e. non-permeabilized) human U87 and A1207 glioma cells. Purified anti-survivin 2C2 mAb was used to stain cells with or without pre-incubation with SVN53-67 peptide, or a scrambled peptide to assess specificity through blocking. (B) Loss of cell surface survivin binding by 2C2 in non-permeabilized A1207 cells treated with proteinase K or trypsin (left panel) and detection of intracellular survivin in protease-stripped and permeabilized cells (right panel). (C) Flow cytometric measurement of cell-surface survivin (red) staining compared to IgG control (black) in non-permeabilized cancer cells lines, including: MCF7 (human breast carcinoma), HeLa (human cervical carcinoma), Jurkat (human T cell leukemia), B16f1 (mouse melanoma), HT-29 (human colon carcinoma), Raji (human Burkitt’s lymphoma), PC3 (human prostate carcinoma), A2780 (human ovarian carcinoma) cells and GL261 (murine glioma cells). (D) Immunofluorescence microscopic detection of total (permeabilized) and cell-surface (non-permeabilized) survivin in human (U87) and murine (GL261) glioma cells with 2C2, 30H3 and commercially available 60.11 anti-survivin mAbs.
Cell membrane and lipid raft localization of survivin
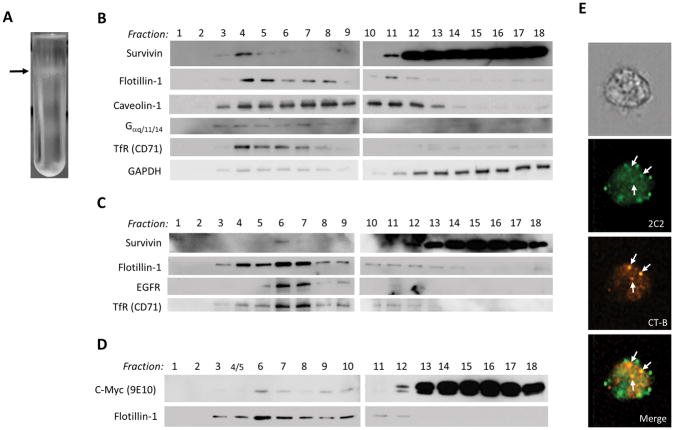
To test whether survivin is contained within lipid rafts of the cell membrane we performed cell fractionation. Western blotting of fractionated lysates from U87 (Figure 3B) and A1207 (Figure 3C) human glioblastoma cells revealed the presence of survivin in the low-density lipid raft fraction (Figure 3A) along with known lipid raft markers flotillin-1, EGFR and caveolin-1. Cells transfected with an expression vector encoding a c-Myc/FLAG-tagged survivin construct also revealed detectable recombinant myc-survivin within the lipid raft fractions (Figure 3D). Similarly, imaging flow cytometry showed co-localization of survivin and bound cholera toxin B (CT-B) (a lipid raft marker) in some, but not all, lipid rafts (Figure 3E).
Figure 3.
(A) Photograph of gradient separation in U87 cells with arrow indicating low density lipid raft band corresponding to survivin-containing fraction #4. Western blot detection of lipid raft-associated proteins and survivin in subcellular fractions derived from U87 (B) and A1207 (C) glioblastoma cells. (D) Transiently expressed recombinant c-Myc-tagged human survivin in association with lipid raft protein fractions in HEK293T cells detected with an anti-c-Myc-tag antibody (9E10). (E) Imagestream imaging flow cytometric visualization of co-localized bound cholera toxin B (CT-B) and survivin in lipid rafts.
Effects of survivin antibodies on tumor growth in vivo
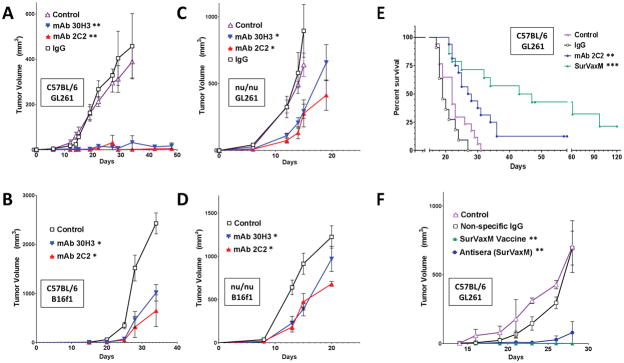
Both 2C2 and 30H3 anti-survivin antibodies inhibited growth of GL261 gliomas and B16 melanomas in flank tumor models compared to irrelevant IgG control antibody (Figure 5A–D). There was no significant difference between 2C2 and 30H3 with respect to tumor inhibition, despite their difference in affinity for the target antigen. Although the inhibition of tumor growth by 2C2 and 30H3 was more effective in immunocompetent mice (Figure 5, A and B), both survivin antibodies inhibited GL261 and B16 tumor growth in immunodeficient (nude) mice as well (Figure 5, C and D). Vaccination of immunocompetent (C57BL/6) mice with SVN53-67/M57-KLH (SurVaxM) produced effective suppression of tumor growth (Figure 5F). In comparison, treatment of tumor-bearing mice with pooled serum from vaccinated mice was also effective at suppressing tumor growth in naïve GL261 tumor-bearing mice, although less so than vaccine (Figure 5F; p < 0.05). Survivin antibody (2C2) also significantly prolonged the survival of C57BL/6 mice with orthotopic (intracerebral) GL261 gliomas compared to either nonspecific PBS or IgG control (Figure 5E; p = 0.004). There was no significant difference between nonspecific IgG and PBS controls (p = 0.089). In these experiments, treatments with vaccine and antibodies began 4 days after tumor cell implantation.
Figure 5.
Subcutaneous (A–D and F) and intracranial (E) tumor models in C57BL/6 (A, B, E and F) and nude (C and D) mice with GL261 glioma (A, C, E and F), and B16f1 melanoma (B and D) cells. Mice were given the indicated treatments once every 5 days beginning 3 days following tumor cell implantation (A, C n = 8–10 per group; B, D n = 5 per group). Subcutaneous tumors (A–D and F) were measured and tumor volumes were calculated as described. (E) Mice with GL261 intracranial tumors were treated with PBS (control), nonspecific IgG, mAb 2C2 at a dose of 10 or 20 μg, or SurVaxM vaccine (n = 11–17 per group) and survival was determined by the Kaplan-Meier method. Median survival time and range; control=22 days (range 17–31 days), nonspecific IgG = 19 days (range 17–27 days), mAb 2C2 = 28 days (range 21–58+ days), SurVaxM = 45 days (range 21–120+ days). (F) Mice with GL261 flank tumors were treated with conjugated survivin vaccine (SVN53-67/M57-KLH; SurVaxM), pooled antiserum derived from non-tumor-bearing mice that had been vaccinated with SurVaxM, or nonspecific IgG control antibody (n = 4 per group). Statistical significance in A–D and F was assessed using the Wilcoxon matched-pairs test; *p<0.05; **p<0.0014. Statistical significance in E was assessed using the Logrank Mantel-Cox test **p=0.0041 and ***p<0.0001.
Antibody dependent cytotoxicity and survivin expression on effector cells
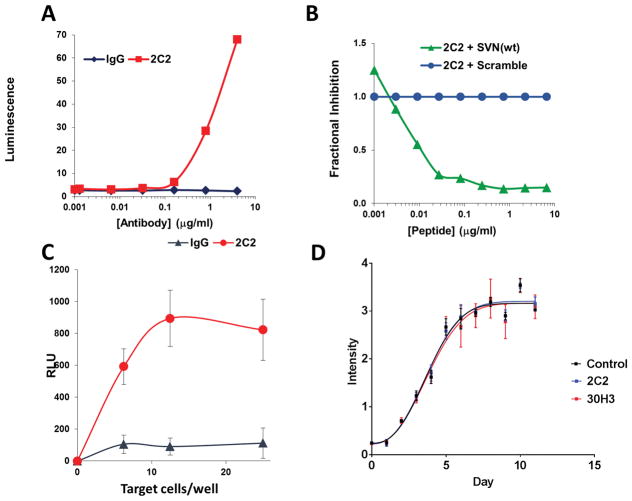
To assess one potential mechanism by which anti-survivin antibodies could mediate antitumor effects, we measured for antibody dependent cell-mediated cytotoxicity (ADCC) in vitro. Preliminary experiments using splenocytes did not demonstrate measurable ADCC. To improve sensitivity, ADCC was assessed using a reporter assay that measures bioluminescent signal from engineered Jurkat effector cells expressing a FcγRIV-luciferase construct. We also found that Jurkat cells express survivin on their cell surface (Figure 2C). Consequently, 2C2 antibody triggers FcγRIV-mediated luciferase expression, through binding Jurkat effector cells even in the absence of target cells (Figure 6A). In addition, competition using free wild type survivin peptide disrupts the 2C2-mediated, auto-activation of effector cells; whereas, scrambled peptide does not. (Figure 6B). Due to antibody recognition of survivin by effector (Jurkat) cells, subsequent assays were performed using U87 target cells that had been pre-incubated with 2C2 antibody, followed by washing to remove unbound antibody (Figure 6C). Target U87 cells pre-incubated with normal mouse IgG did not trigger effector cell luminescence; whereas, target cells pre-incubated with 2C2 stimulated mFcγRIV engagement and luciferase expression. In contrast to the results with 2C2, mAb 30H3 did not increase luciferase activity due to the fact that mouse IgG1 does not bind mFcγRIV [20, 21]. In order to rule out any direct non-immunological effect upon cell viability, we assayed cell growth in the presence of antibody. Incubation of GL261 cells with 2C2 and 30H3 antibodies had no detectable effect upon in vitro cell viability (Figure 6D).
Figure 6.
Monoclonal antibody 2C2 produces FcγRIV activation in luciferase reporter assays (A) Jurkat lymphoma cells as both effector cells and reporter targets. Jurkat cells have cell-surface survivin expression (Figure 2C) leading to antibody (2C2) dose-dependent activation of luciferase reporter via mFcγRIV. Dilutions of murine anti-survivin mAb (2C2) or anti-IgG were added to effector cells and luminescence was measured. (B) Blocking of Fc-mediated activation of luciferase in Jurkat effector cells by SVN53-67 peptide binding to 2C2 compared to scrambled peptide. (C) U87 human glioma cells engage mAb 2C2 with activation of FcγRIV in Jurkat effector cells (RLU = relative luminescence units). (D) Growth curve of Gl261 cells, in vitro, exposed to 10μg of indicated antibody over 11 days.
DISCUSSION
Survivin is expressed by many different types of cancer cells. Cancer patients have been shown to have anti-survivin antibodies in their circulation, even without active, specific vaccination [9]. The presence of anti-survivin antibodies and survivin-specific T cells [9,10] in cancer patients indicates that survivin is released systemically by tumor cells and that it is immunogenic to some degree. Survivin peptide sequences, including those derived from the wild type survivin molecule, and proteins encoded by various survivin splice variants, have been used to develop different cancer vaccine strategies [16, 22, 23–25]. In these studies, the assumption that survivin is largely an intracellular protein target has led to a focus on T cell-mediated antitumor mechanisms in these vaccine-based approaches. Since survivin has been regarded as an intracellular molecule, its presentation as an immunological target is postulated to be of processed survivin epitopes displayed by MHC-I molecules for TCR recognition [10, 11, 15].
In the present study, we found that the survivin protein is present on the outer surface of a wide range of human and murine tumor cell lines. In addition, we developed mouse monoclonal antibodies that inhibited tumor growth in several murine tumor models, perhaps via targeting of survivin associated with the outer surface of the tumor cell membrane. Our work shows, for the first time, that antibodies generated through survivin vaccination can also have antitumor effects. Thus, the finding of surface expression of survivin on tumor cells changes the paradigm for survivin-targeted immunotherapy to include antibody-mediated approaches as well.
Although the origin of circulating anti-survivin antibodies in cancer patients is unclear, survivin protein has been identified in patient serum [26–29] and in association with tumor-derived exosomes [18]. We have recently reported that the survivin protein is detectable by imaging flow cytometry on the outer surface of glioma-derived exosomes present in the circulation of patients with malignant gliomas, but not in normal non-cancer controls. Moreover, vaccination of glioma patients using the SVN53-67/M57-KLH peptide vaccine may affect levels of survivin-bearing exosomes in the bloodstream [30] The current study suggests that antibodies could arise from an immunologic response to tumor membrane survivin expression as well. It is also possible that exosomal survivin is derived from lipid raft survivin during the process of exosome generation and release [30].
Treatment of mice with purified anti-survivin antibodies or with pooled antisera from survivin-vaccinated mice inhibited tumor growth. Vaccination was more effective in preventing tumor progression than injection of antibody alone, and tumor growth inhibition was greater in syngeneic, immunocompetent, C57BL/6 mice than in immunodeficient (nu/nu) mice, suggesting that cellular immunological elements may be necessary for antibody mediated tumor inhibition. While some ADCC activity is observed in a highly sensitive luciferase reporter systems, it is unclear if ADCC is a major component of the anti-tumor effect of survivin antibodies in vivo. Nevertheless, both antibodies were similarly effective suggesting that the biological actions of anti-survivin antibodies are more complex in vivo than in cell culture and that ADCC probably does not account solely for the antitumor effects of anti-survivin antibodies in vivo.
For this study, we specifically selected antibodies generated in response to vaccination that were cross-reactive to the wild type survivin sequence so that high affinity for the target antigen on tumor cells would be assured. In addition to activity in immunocompetent mice, these antibodies have efficacy against GL261 and B16 tumors in immuno-compromised mice. In fully immunocompetent mice, vaccine was more effective than monoclonal antibodies in inhibiting tumor growth, consistent with the fact that cytotoxic T cells and helper support are important components of vaccine action against tumors [11]. Since nude mice retain B cells, NK cells, macrophages and dendritic cells, which can all potentially mediate ADCC and other immunologic processes, additional studies will be required to define the mechanism underlying antibody-mediated antitumor effects. For example, we cannot exclude the possibility that antitumor effects of survivin antibodies in vivo are either fully, or in-part, the result of opsonization, cell-mediated tumor killing or interference with survivin-containing exosomes in the tumor microenvironment [29].
As previously shown with SurVaxM vaccine [11] and replicated here, antibody 2C2 prolongs the survival of immunocompetent mice with orthotopic (cerebral) GL261 gliomas indicating that the blood brain barrier (BBB) does not fully prevent survivin-specific, antibodies and activated effector cells from gaining access to cerebral tumors. Moreover, it has been shown that the GL261 glioma model produces some disruption of the BBB as measured by high molecular weight tracers such as Evans Blue [32]. Thus, cellular and antibody-mediated, survivin immunotherapies may have utility in treating cerebral tumors.
Although our work highlights the potential clinical importance of membrane-bound survivin, the cellular mechanics of surface expression and the biological role of survivin on the cell surface both remain to be defined. In one study, the protein encoded by solute carrier gene family 5a, member 8 (SLC5A8) was shown to translocate survivin to the plasma membrane in cancer cells by way of direct protein-protein interaction [33]. Beyond that, survivin lacks a transmembrane domain and it is not known to interact with specific cell membrane components; however, it does possess amphipathic alpha helical structure [34]. The focal pattern of expression observed by immunofluorescence microscopy suggests the likelihood that cell-surface survivin may be specifically associated with membranous lipid rafts. Packaging of exosomal survivin has previously been reported to be dependent on lipid rafts and to be associated with Hsp70, Hsp90 and PRDX1 proteins [35]. If, as several studies suggest, exosomal survivin has tumor-promoting properties in the microenvironment, antibodies to survivin could also interfere with the action of these nanobodies, thus inhibiting tumor growth.
There are at least six survivin isoforms that appear to be self-regulating and may have distinct functions [36, 37]. Survivin splice variants are generated through combinations of exons 1–4 with two alternate exons 2B and 3B [36–38]. Survivin-2B is formed by the addition of an alternate exon 2; whereas, survivin-ΔEx3 contains a deletion of exon 3 that results in a truncated protein due to a frame shifted stop codon [36]. A high ratio of survivin-ΔEx3 to wild type survivin has been identified in aggressive cancers. In contrast, when survivin-2B is predominant over wild type survivin, a reduced anti-apoptotic potential is observed [39–46]. High expression of survivin-ΔEx3 is also associated with high proliferative activity and tumor recurrence, while survivin-2B is associated with a lower rate of tumor recurrence [47–49].
Survivin variants can differentially traffic within the cell and can have variable subcellular localization [50, 51]. For example, survivin-2B has been reported to preferentially localize in the cytoplasm; whereas, survivin-ΔEx3 preferentially accumulates in the nucleus [50, 52]. Wild type survivin and survivin-ΔEx3 have also been observed to interact within the mitochondria [40]. Such observations demonstrate the multi-faceted nature of survivin biology and raise the possibility that the unique survivin epitopes present in these isoforms may be differentially targetable via specific antibodies. It is possible that a surface-survivin isoform or conformational variant of survivin on the cell membrane is being detected by 2C2 and 30H3. In our study, the survivin epitope from which 2C2 and 30H3 antibodies are derived is conserved in all survivin isoforms described to date. Therefore, we are unable to make any conclusions about which specific isoforms are present on the outer surface of tumor cells. Such information could help guide the development of highly specific antibody-mediated survivin targeting tumor therapies.
CONCLUSION
While cancer vaccines provide one method for anti-survivin cancer immunotherapy, there are several potential advantages to antibody mediated approaches. Unlike with peptide vaccines, the effectiveness of monoclonal antibodies is not HLA-dependent. Antibody therapy could potentially also have more immediate action against cancer cells than vaccines which may take weeks to generate active immune responses and which may not work well in patients who are immunocompromised by their tumors or other treatments. Finally, with the presence of survivin protein on the surface of cancer cells, surface-survivin (API4s) antibody-armed, CAR T-cells could have broad applicability for treating survivin-expressing cancers.
TRANSLATIONAL RELEVANCE.
Survivin is one of the most specific and ubiquitous cancer molecules identified to date. Attention has been directed to targeting this protein via small molecule inhibitors and specific immunologic approaches. To date, vaccine therapy has been the primary immunotherapeutic strategy studied, based on extensive research which has focused on survivin as an intracellular molecule. Thus, proteasomal processing of the molecule leads to presentation of survivin epitopes on the surface of tumor cells where they are subject to immune recognition via T-cell receptor-mediated processes. Our data show for the first time that the survivin protein is present on the surface of a wide variety of tumor cell types and that anti-survivin monoclonal antibodies have significant therapeutic antitumor effects in vivo. Thus, antibody-mediated, immunotherapeutic strategies that target survivin could offer promising treatment approaches in the future.
Acknowledgments
Grant Support: This project was funded by research grants from: American Cancer Society RSG-11-153-01-LIB (to M.J. Ciesielski), the Roswell Park Alliance Foundation, Mr. Philip H. Hubbell and the Linda Scime Endowment. This work was supported by Roswell Park Comprehensive Cancer Center and National Cancer Institute (NCI) grant P30CA016056.
Footnotes
Authors’ Contributions
Conception and design: M.J. Ciesielski, R.A. Fenstermaker, S. Figel
Development of methodology: M.J. Ciesielski, R.A. Fenstermaker, S. Figel, P.M. Galbo, J. Qiu
Acquisition of data: T. Barone, M.J. Ciesielski, S.S. Dharma, S. Figel, P.M. Galbo, E. Winograd, J. Qiu
Analysis and interpretation of data: M.J. Ciesielski, R.A. Fenstermaker, S. Figel, J. Qiu
Writing, review, and/or revision of the manuscript: M.J. Ciesielski, R.A. Fenstermaker, S. Figel
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M.J. Ciesielski, R.A. Fenstermaker, S. Figel
Study supervision: M.J. Ciesielski, R.A. Fenstermaker
R.A. Fenstermaker and M.J. Ciesielski are co-inventors listed on patents regarding the survivin vaccine and are co-founders of MimiVax, LLC, which has licensed such patents from Roswell Park Cancer Institute. The other authors report no conflicts of interest.
References
- 1.Kallio MJ, Nieminen M, Eriksson JE. Human inhibitor of apoptosis protein (IAP) survivin participates in regulation of chromosome segregation and mitotic exit. FASEB J. 2001;15(14):2721–3. doi: 10.1096/fj.01-0280fje. [DOI] [PubMed] [Google Scholar]
- 2.Rosa J, Canovas P, Islam A, Altieri DC, Doxsey SJ. Survivin modulates microtubule dynamics and nucleation throughout the cell cycle. Mol Biol Cell. 2006;17(3):1483–93. doi: 10.1091/mbc.E05-08-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adida C, Crotty PL, McGrath J, Berrebi D, Diebold J, Altieri DC. Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am J Pathol. 1998;152(1):43–9. [PMC free article] [PubMed] [Google Scholar]
- 4.Chakravarti A, Noll E, Black PM, Finkelstein DF, Finkelstein DM, Dyson NJ, et al. Quantitatively determined survivin expression levels are of prognostic value in human gliomas. J Clin Oncol. 2002;20(4):1063–8. doi: 10.1200/JCO.2002.20.4.1063. [DOI] [PubMed] [Google Scholar]
- 5.Kajiwara Y, Yamasaki F, Hama S, Yahara K, Yoshioka H, Sugiyama K, et al. Expression of survivin in astrocytic tumors: correlation with malignant grade and prognosis. Cancer. 2003;97(4):1077–83. doi: 10.1002/cncr.11122. [DOI] [PubMed] [Google Scholar]
- 6.Uematsu M, Ohsawa I, Aokage T, Nishimaki K, Matsumoto K, Takahashi H, et al. Prognostic significance of the immunohistochemical index of survivin in glioma: a comparative study with the MIB-1 index. J Neurooncol. 2005;72(3):231–8. doi: 10.1007/s11060-004-2353-3. [DOI] [PubMed] [Google Scholar]
- 7.Lv S, Dai C, Liu Y, Shi R, Tang Z, Han M, et al. The impact of survivin on prognosis and clinicopathology of glioma patients: a systematic meta-analysis. Mol Neurobiol. 2015;51(3):1462–7. doi: 10.1007/s12035-014-8823-5. [DOI] [PubMed] [Google Scholar]
- 8.Virrey JJ, Guan S, Li W, Schonthal AH, Chen TC, Hofman FM. Increased survivin expression confers chemoresistance to tumor-associated endothelial cells. Am J Pathol. 2008;173(2):575–85. doi: 10.2353/ajpath.2008.071079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soling A, Plugge EM, Schmitz M, Weigle B, Jacob R, Illert J, et al. Autoantibodies to the inhibitor of apoptosis protein survivin in patients with brain tumors. Int J Oncol. 2007;30(1):123–8. [PubMed] [Google Scholar]
- 10.Andersen MH, Pedersen LO, Capeller B, Brocker EB, Becker JC, thor Straten P. Spontaneous cytotoxic T-cell responses against survivin-derived MHC class I-restricted T-cell epitopes in situ as well as ex vivo in cancer patients. Cancer Res. 2001;61(16):5964–8. [PubMed] [Google Scholar]
- 11.Ciesielski MJ, Ahluwalia MS, Munich SA, Orton M, Barone T, Chanan-Khan A, et al. Antitumor cytotoxic T-cell response induced by a survivin peptide mimic. Cancer Immunol Immunother. 2010;59(8):1211–21. doi: 10.1007/s00262-010-0845-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.NoeDominguez-Romero A, Zamora-Alvarado R, Servin-Blanco R, Perez-Hernandez EG, Castrillon-Rivera LE, Munguia ME, et al. Variable epitope library carrying heavily mutated survivin-derived CTL epitope variants as a new class of efficient vaccine immunogen tested in a mouse model of breast cancer. Hum Vaccin Immunother. 2014;10(11):3201–13. doi: 10.4161/hv.29679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross S, Lennerz V, Gallerani E, Sessa C, Mach N, Boehm S, et al. First-in-human trial focusing on the immunologic effects of the survivin-derived multiepitope vaccine EMD640744. Journal of Clinical Oncology. 2011;29(15_suppl):2515. doi: 10.1200/jco.2011.29.15_suppl.2515. [DOI] [Google Scholar]
- 14.Berinstein NL, Karkada M, Oza AM, Odunsi K, Villella JA, Nemunaitis JJ, et al. Survivin-targeted immunotherapy drives robust polyfunctional T cell generation and differentiation in advanced ovarian cancer patients. Oncoimmunology. 2015;4(8):e1026529. doi: 10.1080/2162402X.2015.1026529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Becker JC, Andersen MH, Hofmeister-Muller V, Wobser M, Frey L, Sandig C, et al. Survivin-specific T-cell reactivity correlates with tumor response and patient survival: a phase-II peptide vaccination trial in metastatic melanoma. Cancer Immunol Immunother. 2012;61(11):2091–103. doi: 10.1007/s00262-012-1266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenstermaker RA, Ciesielski MJ, Qiu J, Yang N, Frank CL, Lee KP, et al. Clinical study of a survivin long peptide vaccine (SurVaxM) in patients with recurrent malignant glioma. Cancer Immunol Immunother. 2016;65(11):1339–52. doi: 10.1007/s00262-016-1890-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan S, Jutzy JM, Aspe JR, McGregor DW, Neidigh JW, Wall NR. Survivin is released from cancer cells via exosomes. Apoptosis. 2011;16(1):1–12. doi: 10.1007/s10495-010-0534-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan S, Jutzy JM, Valenzuela MM, Turay D, Aspe JR, Ashok A, et al. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One. 2012;7(10):e46737. doi: 10.1371/journal.pone.0046737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macdonald JL, Pike LJ. A simplified method for the preparation of detergent-free lipid rafts. Journal of Lipid Research. 2005;46(5):1061–7. doi: 10.1194/jlr.D400041-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Michaelsen TE, Kolberg J, Aase A, Herstad TK, Hoiby EA. The four mouse IgG isotypes differ extensively in bactericidal and opsonophagocytic activity when reacting with the P1. 16 epitope on the outer membrane PorA protein of Neisseria meningitidis. Scand J Immunol. 2004;59(1):34–9. doi: 10.1111/j.0300-9475.2004.01362.x. [DOI] [PubMed] [Google Scholar]
- 21.Bruhns P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood. 2012;119(24):5640–9. doi: 10.1182/blood-2012-01-380121. [DOI] [PubMed] [Google Scholar]
- 22.Tsuruma T, Iwayama Y, Ohmura T, Katsuramaki T, Hata F, Furuhata T, et al. Clinical and immunological evaluation of anti-apoptosis protein, survivin-derived peptide vaccine in phase I clinical study for patients with advanced or recurrent breast cancer. J Transl Med. 2008;6:24. doi: 10.1186/1479-5876-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyazaki A, Kobayashi J, Torigoe T, Hirohashi Y, Yamamoto T, Yamaguchi A, et al. Phase I clinical trial of survivin-derived peptide vaccine therapy for patients with advanced or recurrent oral cancer. Cancer Sci. 2011;102(2):324–9. doi: 10.1111/j.1349-7006.2010.01789.x. [DOI] [PubMed] [Google Scholar]
- 24.Nitschke NJ, Bjoern J, Met O, Svane IM, Andersen MH. Therapeutic Vaccination against A Modified Minimal Survivin Epitope Induces Functional CD4 T Cells That Recognize Survivin-Expressing Cells. Scand J Immunol. 2016;84(3):191–3. doi: 10.1111/sji.12456. [DOI] [PubMed] [Google Scholar]
- 25.Gross S, Lennerz V, Gallerani E, Mach N, Bohm S, Hess D, et al. Short Peptide Vaccine Induces CD4+ T Helper Cells in Patients with Different Solid Cancers. Cancer Immunol Res. 2016;4(1):18–25. doi: 10.1158/2326-6066.CIR-15-0105. [DOI] [PubMed] [Google Scholar]
- 26.Guney N, Soydine HO, Derin D, Tas F, Camlica H, Duranyildiz D, et al. Serum and urine survivin levels in breast cancer. Med Oncol. 2006;23(3):335–9. doi: 10.1385/mo:23:3:335. [DOI] [PubMed] [Google Scholar]
- 27.Shehata HH, Abou Ghalia AH, Elsayed EK, Ziko OO, Mohamed SS. Detection of survivin protein in aqueous humor and serum of retinoblastoma patients and its clinical significance. Clin Biochem. 2010;43(4–5):362–6. doi: 10.1016/j.clinbiochem.2009.10.056. [DOI] [PubMed] [Google Scholar]
- 28.Hong JY, Ryu KJ, Park C, Hong M, Ko YH, Kim WS, et al. Clinical impact of serum survivin positivity and tissue expression of EBV-encoded RNA in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Oncotarget. 2017;8(8):13782–91. doi: 10.18632/oncotarget.14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren YQ, Zhang HY, Su T, Wang XH, Zhang L. Clinical significance of serum survivin in patients with pancreatic ductal adenocarcinoma. Eur Rev Med Pharmacol Sci. 2014;18(20):3063–8. [PubMed] [Google Scholar]
- 30.Lancaster GI, Febbraio MA. Exosome-dependent trafficking of HSP70: a novel secretory pathway for cellular stress proteins. J Biol Chem. 2005;280(24):23349–55. doi: 10.1074/jbc.M502017200. Epub 2005 Apr 12. [DOI] [PubMed] [Google Scholar]
- 31.Galbo PM, Jr, Ciesielski MJ, Figel S, Maguire O, Qiu J, Wiltsie L, et al. Circulating CD9+/GFAP+/survivin+ exosomes in malignant glioma patients following survivin vaccination. Oncotarget. 2017;8:114722–35. doi: 10.18632/oncotarget.21773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leten C, Struys T, Dresselaers T, Himmelreich U. In vivo and ex vivo assessment of the blood brain barrier integrity in different glioblastoma animal models. J Neurooncol. 2014;119(2):297–306. doi: 10.1007/s11060-014-1514-2. [DOI] [PubMed] [Google Scholar]
- 33.Coothankandaswamy V, Elangovan S, Singh N, Prasad PD, Thangaraju M, Ganapathy V. The plasma membrane transporter SLC5A8 suppresses tumour progression through depletion of survivin without involving its transport function. Biochem J. 2013;450(1):169–78. doi: 10.1042/BJ20121248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verdecia MA, Huang H, Dutil E, Kaiser DA, Hunter T, Noel JP. Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement. Nat Struct Biol. 2000;7(7):602–8. doi: 10.1038/76838. [DOI] [PubMed] [Google Scholar]
- 35.Valenzuela MMA, Bennit HRF, Osterman CJD, Khan S, Casiano C, Wall NR. Survivin packaging into exosomes is lipid raft-dependent. AACR. 2014 [Google Scholar]
- 36.Wesierska-Gadek J, Schmid G. Transcriptional repression of anti-apoptotic proteins mediated by the tumor suppressor protein p53. Cancer Ther. 2007;5:203–12. [Google Scholar]
- 37.Activities at the Universal Protein Resource (UniProt) Nucleic Acids Research. 2014;42(D1):D191–D8. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng W, Ma X, Wei D, Wang T, Ma Y, Yang S. Molecular cloning and bioinformatics analysis of a novel spliced variant of survivin from human breast cancer cells. DNA Seq. 2005;16(5):321–8. doi: 10.1080/10425170500226490. [DOI] [PubMed] [Google Scholar]
- 39.Krieg A, Mahotka C, Krieg T, Grabsch H, Muller W, Takeno S, et al. Expression of different survivin variants in gastric carcinomas: first clues to a role of survivin-2B in tumour progression. Br J Cancer. 2002;86(5):737–43. doi: 10.1038/sj.bjc.6600153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caldas H, Jiang Y, Holloway MP, Fangusaro J, Mahotka C, Conway EM, et al. Survivin splice variants regulate the balance between proliferation and cell death. Oncogene. 2005;24(12):1994–2007. doi: 10.1038/sj.onc.1208350. [DOI] [PubMed] [Google Scholar]
- 41.Vivas-Mejia PE, Rodriguez-Aguayo C, Han HD, Shahzad MM, Valiyeva F, Shibayama M, et al. Silencing survivin splice variant 2B leads to antitumor activity in taxane--resistant ovarian cancer. Clin Cancer Res. 2011;17(11):3716–26. doi: 10.1158/1078-0432.CCR-11-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling X, Cheng Q, Black JD, Li F. Forced expression of survivin-2B abrogates mitotic cells and induces mitochondria-dependent apoptosis by blockade of tubulin polymerization and modulation of Bcl-2, Bax, and survivin. J Biol Chem. 2007;282(37):27204–14. doi: 10.1074/jbc.M705161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atlasi Y, Mowla SJ, Ziaee SA. Differential expression of survivin and its splice variants, survivin-DeltaEx3 and survivin-2B, in bladder cancer. Cancer Detect Prev. 2009;32(4):308–13. doi: 10.1016/j.cdp.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Cheng Z, Hu L, Fu W, Zhang Q, Liao X. Expression of survivin and its splice variants in gastric cancer. J Huazhong Univ Sci Technolog Med Sci. 2007;27(4):393–8. doi: 10.1007/s11596-007-0411-8. [DOI] [PubMed] [Google Scholar]
- 45.Mahotka C, Wenzel M, Springer E, Gabbert HE, Gerharz CD. Survivin-deltaEx3 and survivin-2B: two novel splice variants of the apoptosis inhibitor survivin with different antiapoptotic properties. Cancer Res. 1999;59(24):6097–102. [PubMed] [Google Scholar]
- 46.Li F. Role of survivin and its splice variants in tumorigenesis. Br J Cancer. 2005;92(2):212–6. doi: 10.1038/sj.bjc.6602340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takashima H, Nakajima T, Moriguchi M, Sekoguchi S, Nishikawa T, Watanabe T, et al. In vivo expression patterns of survivin and its splicing variants in chronic liver disease and hepatocellular carcinoma. Liver Int. 2005;25(1):77–84. doi: 10.1111/j.1478-3231.2004.0979.x. [DOI] [PubMed] [Google Scholar]
- 48.Moore AS, Alonzo TA, Gerbing RB, Lange BJ, Heerema NA, Franklin J, et al. BIRC5 (survivin) splice variant expression correlates with refractory disease and poor outcome in pediatric acute myeloid leukemia: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2014;61(4):647–52. doi: 10.1002/pbc.24822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cho GS, Ahn TS, Jeong D, Kim JJ, Kim CJ, Cho HD, et al. Expression of the survivin-2B splice variant related to the progression of colorectal carcinoma. J Korean Surg Soc. 2011;80(6):404–11. doi: 10.4174/jkss.2011.80.6.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahotka C, Liebmann J, Wenzel M, Suschek CV, Schmitt M, Gabbert HE, et al. Differential subcellular localization of functionally divergent survivin splice variants. Cell Death Differ. 2002;9(12):1334–42. doi: 10.1038/sj.cdd.4401091. [DOI] [PubMed] [Google Scholar]
- 51.Mahotka C, Krieg T, Krieg A, Wenzel M, Suschek CV, Heydthausen M, et al. Distinct in vivo expression patterns of survivin splice variants in renal cell carcinomas. Int J Cancer. 2002;100(1):30–6. doi: 10.1002/ijc.10450. [DOI] [PubMed] [Google Scholar]
- 52.Mull AN, Klar A, Navara CS. Differential localization and high expression of SURVIVIN splice variants in human embryonic stem cells but not in differentiated cells implicate a role for SURVIVIN in pluripotency. Stem Cell Res. 2014;12(2):539–49. doi: 10.1016/j.scr.2014.01.002. [DOI] [PubMed] [Google Scholar]