Abstract
Mdm2 and Mdmx, both major repressors of p53 in human cancers, are predominantly localized to the nucleus and cytoplasm, respectively. The mechanism by which subcellular localization of Mdmx is regulated remains unclear. In this study, we identify the E3 ligase Peli1 as a major binding partner and regulator of Mdmx in human cells. Peli1 bound Mdmx in vitro and in vivo and promoted high levels of ubiquitination of Mdmx. Peli1-mediated ubiquitination was degradation-independent, promoting cytoplasmic localization of Mdmx which in turn resulted in p53 activation. Consistent with this, knockdown or knockout Peli1 in human cancer cells induced nuclear localization of Mdmx and suppressed p53 activity. Myc-induced tumorigenesis was accelerated in Peli1-null mice and associated with downregulation of p53 function. Clinical samples of human cutaneous melanoma had decreased Peli1 expression which was associated with poor overall survival. Together, these results demonstrate that Peli1 acts as a critical factor for the Mdmx-p53 axis by modulating the subcellular localization and activity of Mdmx, thus revealing a novel mechanism of Mdmx deregulation in human cancers.
Keywords: p53, Mdm2, Mdmx, Peli1
Introduction
The tumor suppressor p53 acts as the major sensor for a regulatory circuit that monitors signaling pathways from diverse sources, including DNA damage, oncogene activation, ribosomal stress, and other abnormal cellular stresses (1, 2). p53 mainly functions as a transcriptional factor regulating cell cycle, apoptosis, senescence, metabolism and DNA damage repair to maintain genome stability and prevent tumorigenesis (1, 3). The important roles of p53 in tumor suppression have been well established by the fact that more than 50% of human tumors have p53 mutation or loss of heterozygosity (1, 4). In addition, mice or humans harboring germline p53 mutations have the predisposition for early tumorigenesis and death (5, 6).
The p53 activities are fine-tuned in vivo by its two major repressors, Mdm2 and Mdmx (7–10). The critical roles of Mdm2 and Mdmx in regulating p53 are best demonstrated by studies carried out in mice where inactivation of p53 was shown to completely rescue the embryonic lethality caused by the loss of either Mdm2 or Mdmx (11, 12). Both proteins bind to the p53 transcriptional activation domain and suppress p53-dependent transcription in normal and cancer cells (10, 12). In addition, Mdm2 functions as a Ring domain E3 ubiquitin ligase to promote p53 degradation by poly-ubiquitination and nuclear export by mono-ubiquitination (13, 14). Although Mdmx does not exhibit detectable E3 ligase activity, the heterodimerization of Mdm2 and Mdmx through the Ring domains is essential for Mdm2 stabilization and promotes its ubiquitin ligase activity toward p53 degradation (15–17). Nevertheless, further studies from Mdmx Ring domain mutant mice indicate that the Mdm2/Mdmx interaction is dispensable for modulating Mdmx-mediated effects on p53 at later stages of development and adult tissues (18, 19). Moreover, it has been reported that Mdmx is amplified or overexpressed in several types of human tumors that retain wild-type p53 without Mdm2 amplification (7–8, 20). Thus, Mdmx regulates p53 functions in both Mdm2-dependent and Mdm2-independent manners.
Notably, in contrast to the nuclear localization of Mdm2, Mdmx is predominantly localized in the cytoplasm. Nevertheless, the mechanism by which the subcellular localization of Mdmx is regulated remains unclear. Here, we identify a novel Mdmx regulator named Peli1 in tumor cells by biochemical purification. We found that Peli1 induces Mdmx ubiquitination without promoting its degradation, which leads to the cytoplasmic localization of Mdmx and subsequent activation of p53 function. Moreover, we have provided evidence indicating that the Peli1-Mdmx interaction is critical for tumorigenesis by regulating p53 functions both in mouse model and human tumors.
Materials and Methods
Cell culture and stable lines
All the cell lines were purchased from American Type Culture Collection (ATCC) in February 2010 and have been proven to be negative for mycoplasma contamination. The cell lines were freshly thawed from the purchased seed cells and cultured for no more than 2 months. The cells were maintained in a 37°C incubator with 5% CO2. All media used were supplemented with 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin (all from Gibco). A375, H1299, U2OS and 293T cells were maintained in DMEM medium. To obtain an FLAG and HA double tagged Mdmx (FH-Mdmx) A375 melanoma stable cell line, the cells were transfected with pCIN4-FLAG-HA-Mdmx expression constructs and selected for 2 weeks with 1 mg/ml G418 (Gibco). To generate inducible stable lines, Peli1 cDNA was cloned into a modified tet-on pTRIPZ inducible expression vector (Thermo Open Biosystems). Cells were selected and maintained with puromycin (1 μg/ml) in DMEM medium containing 10% tetracycline-free FBS. To induce the expression of Peli1, 0.1 μg/ml of doxycycline was added to the culture medium. To generate Peli1 U2OS CRISPR cas9 knock out cells, two target guide RNA sequences were designed at the website (http://crispr.mit.edu/) as follows: guide RNA 1: Forward: 5′-GATCAGGAGAAAACATGAGCT-3′, Reverse, 5′-AGCTCATGTTTTCTCCTGATC-3′; guide RNA 2: Forward: 5′-TCTAAAGCACCAGTAAAATA-3′, Reverse, 5′-TATTTTACTGGTGCTTTAGA-3′. The sequences were cloned into pGL3-U6-sgRNA-PGK-puromycin vector according to the manufacturer’s instruction. The expression constructs for pST1374-Cas9 and two guide RNAs were co-transfected into U2OS cells. The cells were selected with puromycin (1 μg/ml) and blasticidin (5 μg/ml) in DMEM medium for 4–6 days. Clones with Peli1 knock-out were screened and acquired after continuing to culture 2–3 weeks without selective antibiotics.
Purification of Mdmx complexes from human cutaneous melanoma cells
The double epitope-tagging strategy was used to isolate Mdmx-containing protein complexes from human cells as previously described with some modifications (21). A375 parental and FH-Mdmx A375 stable cells were chosen to expand for complex purification. The cells were lysed in cold BC100 buffer (20 mM Tris-HCl, pH 7.9, 100 mM NaCl, 10% glycerol, 0.2 mM EDTA, 0.2% Triton X-100, and freshly supplemented protease inhibitor). The cell extracts were incubated with the anti-FLAG monoclonal antibody conjugated to M2 agarose beads (FLAG/M2, Sigma) at 4°C overnight. After five washes with the lysis buffer, the bound proteins were eluted with FLAG-peptide (Sigma) in BC100 buffer for 2 h at 4°C. The acquired elutes were further affinity purified by anti-HA antibody-conjugated agarose (Sigma). The final elutes from the HA-beads with TFA elution buffer (0.1% TFA and 50% Acetonitrile) were lyophilized and then resolved by SDS-PAGE on a 4–20% gradient gel (Invitrogen) for silver staining analysis or directly subjected to mass-spectrometry peptide sequencing.
Plasmids and transfection
The full-length Mdmx and Peli1 were amplified by PCR and cloned into pCIN4-FLAG-HA or pCMV-Myc expression vectors. For the different Mdmx and Peli1 deletion mutant constructs, DNA sequences corresponding to different regions were amplified by PCR from the above constructs and cloned into pCIN4-FLAG-HA or pCMV-Myc expression vectors. Myc-Mdmx and Mdmx Δ200–274 constructs were obtained from Jiandong Chen’s laboratory. Myc-Peli1 C395/398A mutant was generated by Quick Change Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s protocol. The expression vectors for HA-Mdmx-ubiquitin and HA-Mdmx-Sumo fusion proteins were constructed by amplifying and ligating Mdmx and ubiquitin or sumo cDNA into the expression vectors. The other plasmids were generated in our lab and described as before (14, 22). All sequences have been confirmed by DNA sequencing before transfection using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol.
Western blotting and antibodies
For Western blot analysis, cells were lysed in cold BC100 or FLAG buffer (50 mM Tris-HCl, pH 7.9, 137 mM NaCl, 10 mM NaF, 1 mM EDTA, 1% Triton X-100, 0.2% Sarkosyl, 10% glycerol, and freshly supplemented protease inhibitor cocktail) and then total protein concentrations were determined by Bradford method using the Bio-Rad protein assay kit. The cell extracts were boiled in SDS loading buffer, and then equally loaded and separated in polyacrylamide gels. Proteins were then transferred to Hybond ECL membrane (GE healthcare) and incubated overnight with primary antibodies. Rabbit polyclonal Mdmx antibody (A300-287A) was purchased from Bethyl Laboratories. Mouse monoclonal antibody for Peli1 (F-7), p53 (DO-1), Mdm2 (SMP14) and Myc (9E10) and rabbit polyclonal antibodies for p21 (C19) and Puma (H-136) were purchased from Santa Cruz Biotechnology. Rat anti-HA monoclonal antibody (3F10) was purchased from Roche. Mouse anti-GFP (JL-8) and β-actin (AC-15) monoclonal antibodies were purchased from Clontech and Sigma, respectively. HRP-conjugated secondary antibodies were purchased from GE Healthcare. Western blot signals were detected on auto-radiographic films after incubating with ECL (GE healthcare) or Super Signal West Dura reagents (Thermo scientific).
Immunoprecipitation assays
Co-immunoprecipitation assays were performed as described previously with some modifications (23). The cells were lysed in BC100 lysis buffer. The cell extracts were incubated with the antibody or control IgG at 4°C overnight. Protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) were added and incubated for 2 hours at 4°C. After five washes with the lysis buffer, the bound proteins were eluted by boiling with SDS sample buffer. For immunoprecipitation of ectopically expressed FLAG-tagged proteins, the cell extracts were incubated with the FLAG/M2 beads at 4°C overnight. After five washes with the lysis buffer, the bound proteins were eluted with FLAG-peptide in BC100 for 2 h at 4°C. The eluted material was resolved by SDS-PAGE and immunoblotted with antibodies as indicated.
GST pull-down assays
GST and GST-tagged fusion proteins were purified using GST resin (Novagen) according to the manufacturer’s instruction. To purify the FH-Mdmx and FH-Peli1 proteins, the 293T cells transfected with the respective expression constructs were lysed in the stringent BC300 buffer (20 mM Tris-HCl, pH 7.9, 300 mM NaCl, 10% glycerol, 0.2 mM EDTA, 0.2% Triton X-100, and freshly supplemented protease inhibitor) and then subjected with FLAG/M2 immunoprecipitation. The bound proteins were eluted with BC100 buffer with FLAG peptide. GST or GST-tagged fusion proteins were incubated with the purified FH-Mdmx and FH-Peli1 overnight at 4 °C in BC100 buffer. GST resins (Novagen) were then added, and the solution was incubated at 4 °C for 3 h. After washing five times, the bound proteins were eluted for 2 h at 4 °C in elution buffer (20 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0) and subjected to Western blot analysis.
Cell-based ubiquitination assay
Cell-based ubiquitination assays were performed essentially as described (23) with some modifications. H1299 cells were transfected with expression constructs as indicated. 10% of the cells were lysed with FLAG lysis buffer and the cell extracts were saved as input. The rest of the cells were lysed with phosphate/guanidine buffer (6 M guanidine-HCl, 0.1 M Na2HPO4, 6.8 mM NaH2PO4, 10 mM Tris-HCl, pH 8.0, 0.2% Triton X-100, freshly supplemented with 10 mM β-mercaptoethanol and 20 mM imidazole) with mild sonication and subjected to Ni-NTA (Qiagen) pulldown for 3 hours. Ni-NTA-captured fractions were then washed with phosphate/guanidine buffer two times and urea wash buffer (8 M urea, 0.1 M Na2HPO4, 6.8 mM NaH2PO4, 10 mM Tris-HCl, pH 8.0, 0.2% Triton X-100, freshly supplemented with 10 mM β-mercaptoethanol and 20 mM imidazole) two times, and further washed there times with PBS buffer. Precipitates were eluted by 30 min of incubation with elution buffer (0.3 M imidazole, 0.125 M DTT) and resolved by SDS-PAGE. For Mdmx mono-ubiquitination analysis, H1299 cells were lysed with FLAG lysis buffer. The cell extracts were subjected to FLAG/M2 immunoprecipitation and western blot analysis with anti-Mdmx polyclonal antibody.
siRNA-mediated ablation of Peli1, p53 and Mdmx
Ablation of Peli1 was performed by transfection of U2OS cells with siRNA duplex oligonucleotides synthesized by Dharmacon as follows: Peli1 siRNAi-1 sense sequence: 5′-GGAAAUCAAUGCAGCACGAUU-3′; antisense sequence: 5′-UCGUGCUGCAUUGAUUUCCUU-3′; Peli1 siRNA-2 sense sequence: 5′-AAUCAUAUGUGAACGGAAU; antisense sequence: 5′-AUUCCGUUCACAUAUGAUU-3′. Ablation of p53 and Mdmx was performed by transfection of U2OS cells with siRNA duplex oligo sets (On-Target-Plus Smart pool L00332900 for p53 and L-00653600 for Mdmx, Dharmacon). Control siRNA (On-Target-Plus siControl non-targeting pool D00181010, Dharmacon) was also used for transfection.
RNA extraction and Real-time PCR
Frozen lymphoma tissue samples were homogenized using a pestle (Axygen) in 1.5ml Eppendorf tube. Total RNA was isolated from tissues or cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Two micrograms of total RNA was reverse transcribed by Super Script™ IV VILO™ Master Mix (Invitrogen) following the manufacturer’s protocol. Quantitative PCR (Real-time PCR) was performed in triplicate using SYBR green mix (Applied Biosystems) and a 7500 Fast Real-Time PCR System (Applied Biosystems) under the following conditions: 15 min at 95 °C followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. To determine p53 mutational status, RNA was extracted and PCR was performed to amplify mouse p53 cDNA from mouse lymphoma samples. Full length p53 PCR product was gel-purified and sequenced using standard procedures. Primers for quantitative PCR are as follows: human Mdm2 forward 5′-CGATGAATCTACAGGGACGCCATCG-3′, human Mdm2 reverse 5′-TCCTGATCCAACCAATCACCTG-3′; human p21forward 5′-CCATGTGGACCTGTCACTGTCTT-3′, human p21 reverse 5′-CGGCCTCTTGGAGAAGATCAGCCG-3′; human GAPDH forward, 5′-ATCAATGGAAATCCCATCACCA-3′, human GAPDH reverse 5′-GACTCCACGACGTACTCAGCG-3′; mouse Mdm2 forward 5′-GGACTCGGAAGATTACAGCCTGA-3′, mouse Mdm2 reverse 5′-TGTCTGATAGACTGTGACCCG-3′; mouse p21 forward 5′-AGATCCACAGCGATATCCAGAC-3′, mouse p21 reverse 5′-ACCGAAGAGACAACGGCACACT-3′; mouse Puma forward 5′-ACGACCTCAACGCGCAGTACG-3′, mouse Puma reverse 5′-GAGGAGTCCCATG AAGAGATTG-3′; mouse GAPDH forward, 5′-AACTTTGGCATTGTGGAAGG-3′, mouse GAPDH reverse 5′-ACACATTGGGGGTAGGAACA-3′.
Immunofluorescent staining
For immunofluorescent staining, the cells were plated on 35 mm dishes with a 20 mm bottom well (In Vitro Scientific, Sunnyvale, California) 24 hours before cells were transfected with the expression constructs. The cells on the bottom well were washed three times with PBS and then fixed for 20 min with 4% paraformaldehyde on ice, rehydrated for 5 min in serum-free DMEM, and permeabilized for 10 min with 0.2% Triton X-100 (Fisher). The cells were blocked in 1% BSA (Sigma)/PBS (Gibco) 30 min and incubated in 1% BSA/PBS with primary rat anti-HA or mouse anti-Myc monoclonal antibodies for 45 mins at room temperature. After extensive washing, the cells were incubated in 1% BSA/PBS with Alexa Fluor 488 (green) conjugated anti-mouse and Alexa Fluor 568 (red) conjugated anti-rat fluorescent secondary antibody (Invitrogen) for 30 min at room temperature. Finally, cells were counterstained with DAPI (Sigma) to stain the nuclei.
Nuclear cytoplasmic fractionation
The U2OS cells grown in 60 or 100 mm dishes were scraped into cold PBS after the cells were washed two times. The cell pellets were re-suspended in hypotonic lysis buffer (10 mM Tris, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, and freshly supplemented protease inhibitor), placed on ice 15 minutes and followed by treatment with NP-40 at a final concentration of 0.5% with vortex for 10 seconds. The supernatant was acquired as cytoplasmic extracts after centrifuging 1 minute at full speed. The pellets were further treated with extraction buffer (20 mM Tris, pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, and freshly supplemented protease inhibitor) 2 hours on ice with vortex every 5–10 minutes. The supernatant from extraction buffer was acquired as nuclear extracts.
Mice
Mice were maintained in a specific pathogen-free facility, and all animal experiments were in accordance with protocols approved by the IACUC (Institutional Animal Care and Use Committee) of Columbia University. Peli1 null mice were a kind gift from Dr. Shao-Cong Sun at the University of Texas MD Anderson Cancer Center. Peli1 mice were bred and genotyped as described (24). The Eμ-Myc mice were purchased from The Jackson Laboratory. The offspring of Eμ-Myc mice were genotyped using a mouse primer set as follows: Eμ-Myc forward, 5′-AGACGTCAGGTGGCACTTTT-3′, Eμ-Myc reverse, 5′-AGCAAAAACAGGAAGGCAAA-3′. Peli1 null mice were crossed with the C57BL/6 background mice to generated Peli1 heterozygous mice. Mice heterozygous for Peli1 deletion (Peli1+/-) were bred to generate age-matched mice homozygous for Peli1 deletion (Peli1-/-) and wild-type mice. These mice were crossed with the Eμ-Myc mice to generate mice with Peli1 wild-type (Peli1+/+) and Peli1 null (Peli1-/-) mice in the Eμ-Myc background. Animals were observed two times each week for signs of morbidity and tumor development. Tumor development was monitored by palpation of the abdomen and cervical, axillary, and inguinal regions. Mice were euthanized humanely while reaching tumor-specific endpoints. Lymphoma tissues were fixed in formalin for histopathology or snap frozen for protein and RNA extraction.
Patients
To determine the Peli1 expression in patients with cutaneous melanoma, we analyzed and compared the data from GEO profiles (https://www.ncbi.nlm.nih.gov/geoprofiles/13276984). The data were from expression profiling by array from human samples including 7 human normal skins, 11 benign nevi and 45 cutaneous malignant melanomas (25). To understand the association of Peli1 with overall survival in patients with malignant cutaneous melanoma, we made use of another set of publicly available clinical information provided by cBioportal for Cancer Genomics databases (http://www.cbioportal.org/), which included basic demographical and clinical information, p53 status and overall survival (OS). We extracted a total of 250 patients from the tumor cohort with higher and lower quartiles for Peli1 expression for the survival analysis.
Statistical analysis
Comparisons of mean values between the groups were analyzed using GraphPad Prism software (GraphPad Software Inc., San Diego, CA). Statistical significance of the differences was analyzed using unpaired Student’s t-test for comparisons of two groups or by One-way analysis of variance for comparisons of multiple groups. Survival curves were plotted using the Kaplan-Meier method and compared by the log-rank test using GraphPad Prism. All p values were two sided and the level of statistical significance was set at <0.05.
Results
Identification of Mdmx-associated complexes in tumor cells
Previous studies indicated that p53 is not frequently mutated in human melanomas, partially caused by Mdmx amplification (20, 26). To further elucidate the mechanism of Mdmx regulation in tumorigenesis, we sought to identify specific regulators of Mdmx in human melanoma cells. To this end, we generated an N terminal FLAG and HA double tagged Mdmx (FH-Mdmx) protein in human cutaneous malignant melanoma A375 cells. To isolate Mdmx-associated complexes, the cell extracts from the stable cells were sequentially subjected to affinity purification by FLAG antibody conjugated M2 beads (FLAG/M2) and an HA-affinity column. The bound proteins were then fractionated by SDS-PAGE and visualized by silver staining (Fig. 1A). The whole elutes were also subjected to mass spectrometry analysis to identify novel components of the Mdmx-associated complexes. As expected, FH-Mdmx, CK1α, 14-3-3, p53, Mdm2 and p73 were identified as major components of the complexes (Fig. 1A, Supplementary Fig. 1A and Supplementary Table 1), indicating that ectopic Mdmx can form stable complexes with these endogenous binding partners. Interestingly, an E3 ligase named Pelino1 (Peli1) was identified as a novel major cellular protein in the Mdmx-associated complexes from A375 cells (Fig. 1A, Supplementary Fig. 1A and Supplementary Table 1). Peli1 is a 46KD cellular adaptor protein interacting with Drosophila Pelle, a protein highly homologous with the IRAKs (Interlukin-1 receptor-associated kinase) (27). Peli1 regulates innate and adaptive immune signaling pathways by ubiquitination (28, 29). Although it has been shown that Peli1 is involved in NF-κB activation and inflammatory diseases, the precise roles of Peli1 in tumorigenesis await to be clarified.
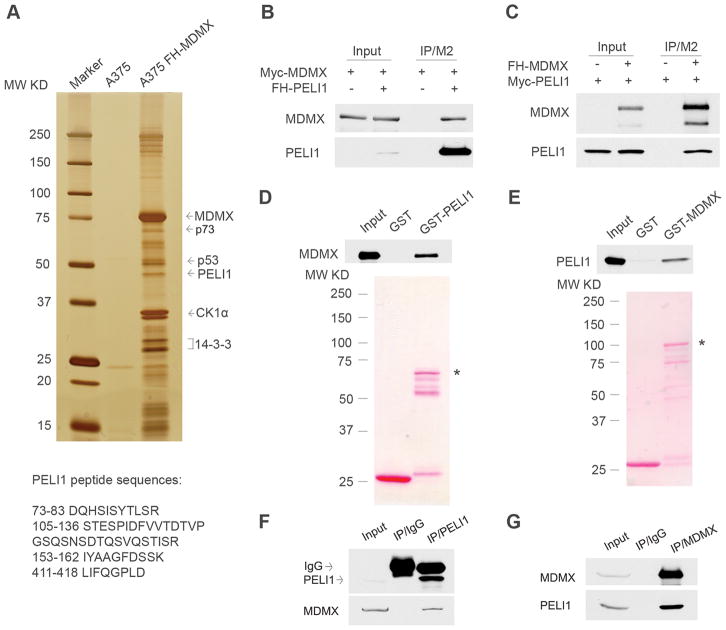
Figure 1. Identification of Peli1 as a bona fide Mdmx interacting protein in cancer cells.
(A) Silver staining of affinity-purified Mdmx complexes from the cell extracts of the FH-Mdmx A375 stable cells and the parental A375 cells. Mdmx-interacting partners were identified by mass spectrometry, and the Peli1 peptide sequences are presented.
(B) Co-immunoprecipitation of FH-Peli1 with Myc-Mdmx from H1299 cells. Whole cell extracts or immunoprecipitates with FLAG/M2 beads from H1299 cells transfected with Myc-Mdmx alone or with Myc-Mdmx and FH-Peli1 together were subjected to western blot with anti-Myc (top) and anti-HA antibody (lower).
(C) Co-immunoprecipitation of FH-Mdmx with Myc-Peli1 from H1299 cells. Whole cell extracts or immunoprecipitates with FLAG/M2 beads from H1299 cells transfected with Myc-Peli1 alone or with Myc-Peli1 and FH-Mdmx together were subjected to western blot with anti-HA (top) and anti-Myc antibody (lower).
(D) Direct interactions of GST-Peli1 with Mdmx. The GST-Peli1 fusion protein and GST alone were used in a GST pull-down assay with the FH-Mdmx protein purified from H1299 cells. Elutes from GST beads were subjected to western blot with anti-HA antibody (top) after Ponceau S staining (lower). Asterisk indicates GST-Peli1 fusion protein.
(E) Direct interactions of GST-Mdmx with Peli1. The GST-Mdmx fusion protein and GST alone were used in a GST pull-down assay with the FH-Peli1 protein purified from H1299 cells. Elutes from GST beads were subjected to western blot with anti-HA antibody (top) after Ponceau S staining (lower). Asterisk indicates GST-Mdmx fusion protein.
(F) Co-immunoprecipitation of endogenous Mdmx with Peli1 from A375 melanoma cells. Western blot analysis of whole cell extract (lane 1) and immunoprecipitates with a control IgG (lane 2) or a Peli1-specific antibody (lane 3) by anti-Peli1 monoclonal antibody (top) or anti-Mdmx polyclonal antibody (lower).
(G) Co-immunoprecipitation of endogenous Peli1 with Mdmx from A375 melanoma cells. Western blot analysis of whole cell extract (lane 1) or immunoprecipitates with control IgG (lane 2) or anti-Mdmx polyclonal antibody (lane 3) by anti-Mdmx polyclonal antibody (top) or anti-Peli1 monoclonal antibody (lower).
Peli1 directly interacts with Mdmx both in vitro and in vivo
To confirm interaction between Peli1 and Mdmx, the expression vectors for FLAG and HA tagged Peli1 (FH-Peli1) and Myc tagged Mdmx (Myc-Mdmx) were transfected into H1299 cells. Western blot analysis of FLAG/M2 immunoprecipitates from cell extracts revealed that Mdmx was specifically detected in Peli1-associated immunoprecipitates (Fig. 1B). Further, the expression constructs for FH-Mdmx and Myc tagged Peli1 (Myc-Peli1) were transfected into H1299 cells. Reciprocal immunoprecipitation indicated that Peli1 was also in Mdmx complexes (Fig. 1C). To test the direct interaction between Peli1 and Mdmx in vitro, we performed GST pull-down assays. Purified GST and GST-Peli1 were incubated with purified FH-Mdmx protein. Western blot analysis of elutes from GST resin showed Mdmx strongly binds to immobilized GST-Peli1 but not GST alone (Fig. 1D). Further, reciprocal GST pull-down assay revealed that Peli1 specifically binds to GST-Mdmx (Fig. 1E). To investigate the interaction between endogenous Peli1 and Mdmx, cell extracts from A375 cells were immunoprecipitated with a control IgG or a Peli1-specific antibody. As expected, western blot analysis revealed endogenous Peli1 was immunoprecipitated by the Peli1 antibody. Importantly, Mdmx was clearly detected in the immunoprecipitations obtained from the anti-Peli1 antibody but not the control IgG (Fig. 1F). Conversely, endogenous Peli1 was readily immunoprecipitated with the Mdmx specific antibody but not with a control antibody (Fig. 1G). Similar results were acquired from endogenous immunoprecipitations in U2OS cells (Supplementary Fig. 1B and 1C). Taken together, these results indicate that Peli1 is a bona fide Mdmx interacting protein in tumor cells.
Mapping of interaction regions between Mdmx and Peli1
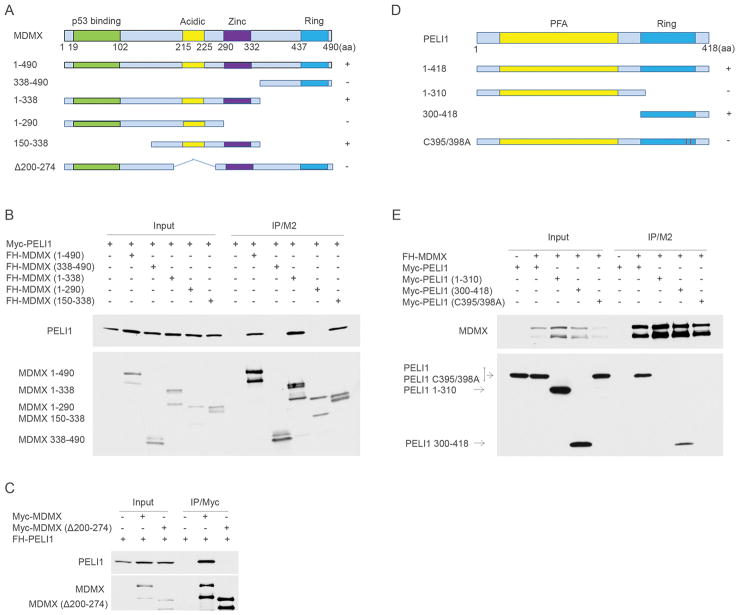
Next, we determined the functional domains required for the interaction between Mdmx and Peli1. The expression constructs for FH-Mdmx and deletion mutants were co-transfected with Myc-Peli1 into H1299 cells. Western blot analysis of FLAG/M2 immunoprecipitates revealed that middle region in Mdmx (150–338 amino acids) is sufficient for its interaction with Peli1 (Fig. 2A and 2B). Notably, Mdmx Δ200–274 which has a small internal deletion covering the acidic domain but retains all the other functional domains completely abrogates the interaction between Mdmx and Peli1, suggesting that both acidic and zinc domains in Mdmx protein are required for the Peli1 binding (Fig. 2A and 2C). Peli1 consists of an N-terminal PFA core region and an atypical Ring-like domain in the C-terminus, which contains an intrinsic E3 ligase activity (Fig. 2D). To map Peli1 domains for Mdmx binding, Myc-Peli1 and deletion mutants were co-transfected with FH-Mdmx in H1299 cells. Western blot analysis of FLAG/M2 immunoprecipitates revealed that Ring domain (300–418) in Peli1 is required for the interaction with Mdmx (Fig. 2D and 2E). Interestingly, Peli1 C395/398A mutant with the Ring domain disrupted completely abrogated the binding to Mdmx (Fig. 2D and 2E). Finally, we found that Peli1 cannot form a homodimer although it strongly binds to Mdmx by the Ring domain in cells (Supplementary Fig. 2).
Figure 2. Mapping of interaction regions between Mdmx and Peli1.
(A) A schematic representation of Mdmx and the corresponding mutants (+: binding positive to Peli1; -: binding negative to Peli1).
(B) FH-Mdmx and the corresponding deletion mutants were individually transfected with Myc-Peli1 in H1299 cells. Whole cell lysates and immunoprecipitates with FLAG/M2 beads were subjected to western blot with anti-Myc (top) and anti-HA (lower) monoclonal antibodies.
(C) Myc-Mdmx and the Myc-Mdmx Δ200–274 mutant were individually transfected with FH-Peli1 in H1299 cells. Whole cell lysates and immunoprecipitates with anti-Myc antibody were subjected to western blot with anti-HA (top) and anti-Myc (lower) monoclonal antibodies.
(D) A schematic representation of Peli1 and the corresponding mutants (+: binding positive to Mdmx; -: binding negative to Mdmx).
(E) Myc-Peli1 and the corresponding mutants were individually transfected with FH-Mdmx in H1299 cells. Whole cell lysates and immunoprecipitates with FLAG/M2 beads were subjected to western blot with anti-HA (top) and anti-Myc (lower) monoclonal antibodies.
Peli1 interacts specifically with Mdmx and promotes Mdmx ubiquitination
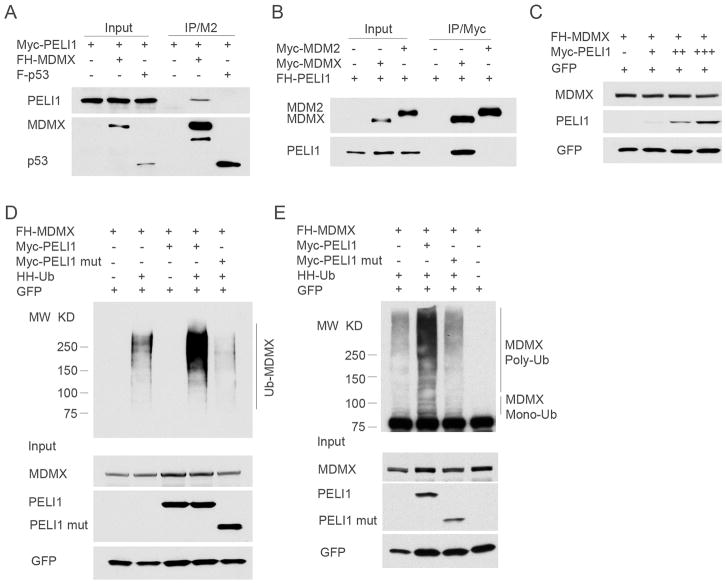
Accumulating evidence indicates that Mdmx interacts with both p53 and Mdm2 and that the activities of Mdmx are intimately linked with either p53 or Mdm2 in human cancer cells (7, 8). To understand the mechanism of the interaction between Peli1 and Mdmx in modulating the p53 pathway, we first tested whether this interaction is specific for Mdmx among the three major components of this pathway. To this end, the expression constructs for FH-Mdmx or FLAG tagged p53 are co-transfected with Myc-Peli1 into H1299 cells. Western blot analysis of FLAG/M2 immunoprecipitates from cell extracts revealed that Peli1 was specifically detected in Mdmx but not p53-associated immunoprecipitates although the approximately equal amounts of Mdmx and p53 are immunoprecipitated (Fig. 3A). Further, Myc tagged Mdm2 and Mdmx expression constructs were co-transfected with FH-Peli1 into H1299 cells. Western blot analysis revealed the same amount of Mdm2 and Mdmx were immunoprecipitated by the anti-Myc antibody. Importantly, Peli1 was clearly detected in the immunoprecipitations obtained from the Mdmx but not Mdm2 complexes (Fig. 3B). Since Peli1 has an intrinsic E3 ligase activity to catalyze the formation of both K48 and K63 ubiquitin chains (30, 31), we next examined whether Peli1 promotes Mdmx ubiquitination and degradation. Although Peli1 cannot cause any significant degradation of Mdmx (Fig. 3C), Peli1 promotes both mono- and poly-ubiquitination of Mdmx by using an in vivo ubiquitination assay (Fig. 3D, 3E and Supplementary Fig. 3B). However, Peli1 mutant (1–310 amino acids) with Ring domain deleted has no effects on Mdmx ubiquitination (Fig. 3D and E). Lysine 48 (K48) linked poly-ubiquitination chain targets protein for destruction by 26S proteasome whereas other types of poly-ubiquitination, for example, K63 linked poly-ubiquitination is not associated with protein degradation. As shown in Supplementary Figure 3A, although Peli1 induced K63-linked poly-ubiquitination by using Ub-K63 only (lysine to arginine mutations on all other lysine residues except K63), Peli1 was also able to promote Mdmx poly-ubiquitination with either K48R-ub or K63R-ub (lysine to arginine mutation only at either K48 or K63, respectively). Notably, as shown in Supplementary Figure 3B, Mdm2 mostly induced Mdmx ubiquitination with high molecular weight (long ubiquitin chains) whereas Peli1 promoted its ubiquitination mostly with medium ranges of molecular weight, suggesting that Peli1-mediated ubiquitination mode is different from the one induced by Mdm2, which promotes Mdmx degradation. These findings indicate that Peli1 induced Mdmx poly-ubiquitination with branched ubiquitin chains containing mixed linkage types. The non-canonical ubiquitination mode may be the reason that Peli1 cannot promote Mdmx degradation.
Figure 3. Peli1 interacts specifically with Mdmx and promotes Mdmx ubiquitination.
(A) Peli1 cannot bind to p53. Whole cell extracts or immunoprecipitates with FLAG/M2 beads from H1299 cells transfected with Myc-Peli1 alone or together with FH-Mdmx or FLAG-p53 were subjected to western blot with anti-Myc (top) and anti-FLAG antibody (lower).
(B) Peli1 cannot bind to Mdm2. Whole cell extracts or immunoprecipitates with anti-Myc antibody from H1299 cells transfected with FH-Peli1 alone or together with Myc-Mdmx or Myc-Mdm2 were subjected to western blot with anti-Myc (top) and anti-FLAG antibody (lower).
(C) Peli1 cannot promote Mdmx degradation. Western blot analysis of cell extracts from the H1299 cells transfected with FH-Mdmx alone or together with increased Myc-Peli1 with anti-HA (top) and anti-Myc antibody (middle). GFP was used as loading control.
(D) Peli1 promotes Mdmx poly-ubiquitination. FH-Mdmx was transfected alone (lane 1), or with HA-His-Ubiquitin (HH-Ub, lane 2), or with Myc-Peli1 (lane 3), or with Myc-Peli1 and HH-Ub (lane 4), or with Myc-Peli1 mutant (1–310aa) and HH-Ub (lane 5) in H1299 cells. The cell lysates were immunoprecipitated with Ni-NTA agarose followed by western blot with anti-Mdmx polyclonal antibodies (upper panel). The crude cell extracts were also detected with anti-HA (lower panel, top) and anti-Myc (lower panel, middle) monoclonal antibodies. GFP was used as loading control.
(E) Peli1 promotes Mdmx mono-ubiquitination. FH-Mdmx was transfected alone (lane 4), or with HH-Ub (lane 1), or with Myc-Peli1 and HH-Ub (lane 2), or with Myc-Peli1 mutant (1–310aa) and HH-Ub (lane 3) in H1299 cells. The crude cell extracts and immunoprecipitates from FLAG/M2 beads were subjected to western blot with anti-Mdmx polyclonal antibody (upper panel), anti-HA (lower panel, top) and anti-Myc (lower panel, middle) monoclonal antibodies. GFP was used as loading control.
Peli1 promotes p53 transcriptional activity in an Mdmx-dependent manner
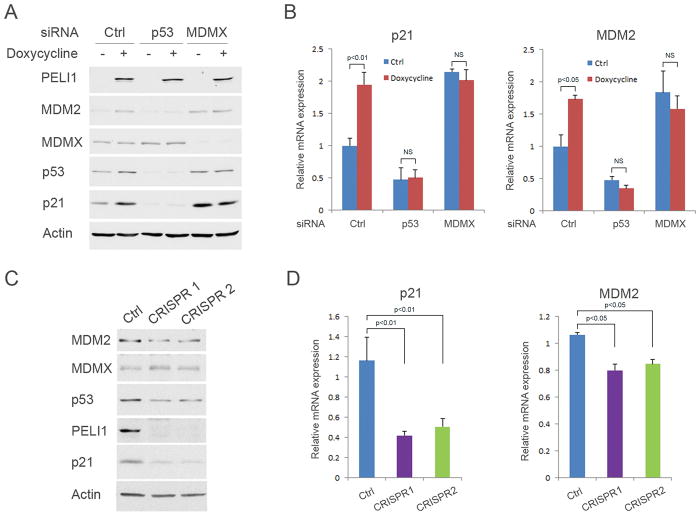
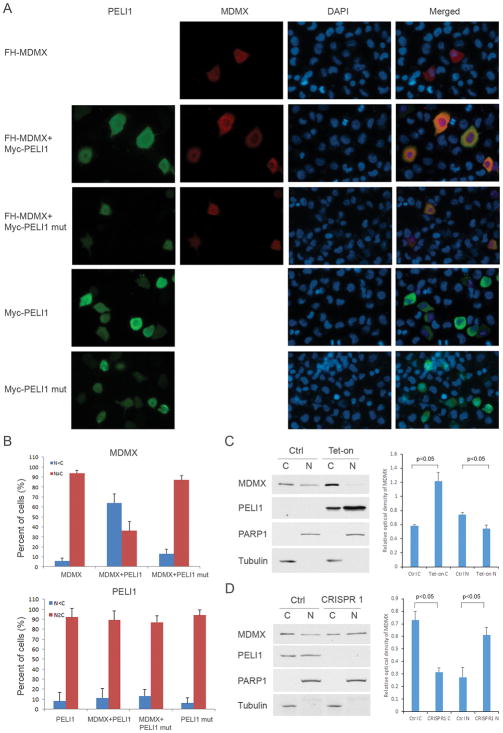
It has been well established that Mdmx is crucial to repress p53-mediated transcriptional activity both in embryonic development and tumor formation (7, 19). To evaluate the functional consequence of the interaction between Peli1 and Mdmx, we next assessed the roles of Peli1 in regulating p53 transcriptional activity mediated by Mdmx. To this end, we generated a tetracycline controlled (tet-on) Peli1-inducible U2OS cell line. Western blot (Fig. 4A and Supplementary Fig. 4A) and real-time PCR (Fig. 4B) analysis showed that p21 and Mdm2 expression is significantly increased after Peli1 is induced by doxycycline in U2OS cells. However, p21 and Mdm2 expression remains constant after p53 and Mdmx were knocked down by the respective specific siRNAs from these cells (Fig. 4A and B), suggesting that Peli1 induced p21 and Mdm2 expression in a p53 and Mdmx dependent manner. Next, we investigated cell growth using the Peli1-inducible U2OS cell line. The results showed that cell growth is markedly slower when Peli1 is overexpressed in U2OS cells (Supplementary Fig. 4B). We also generated Peli1 CRISPR-Cas9 knock-out U2OS cells to further assess Peli1 effects on p53 transcriptional activity. Western blot analysis revealed that p21 expression is markedly inhibited in two Peli1 knock-out cell lines (Fig. 4C). Although Mdm2 protein expression is modestly decreased (Fig. 4C), real-time qPCR analysis indicated that both p21 and Mdm2 messenger RNA (mRNA) levels are significantly decreased in these cells (Fig. 4D). The similar results were obtained when the endogenous Peli1 expression was knocked down by two specific siRNA sequences in U2OS cells (Supplementary Fig. 4C). We analyzed cell proliferation from the control parental and Peli1 knock-out U2OS cells. The results demonstrated that Peli1 null cells grow faster than the control cells (Supplementary Fig. 4D). Notably, it is well known that Mdmx is degraded upon DNA damage stress. Our data indicate that Peli1-mediated effect on p21 and Mdm2 expression is Mdmx-dependent (Figure 4A and 4B). Thus, it is very likely that Peli1-mediated effect will be abrogated when Mdmx is degraded upon DNA damage. To this end, we examined p21 and Mdm2 expression in both wild type and Peli1 knock-out cells under DNA damage stress conditions. Consistently, we observed basal p21 and Mdm2 levels are decreased in Peli1 knockout cells. However, we did not detect significant differences between the control and Peli1 knock-out cells upon DNA damage (Supplementary Fig. 4E). These results suggested that Peli1-mediated p53 regulation is abrogated under DNA damage conditions.
Figure 4. Peli1 promotes p53 transcriptional activity in an Mdmx-dependent manner.
(A–B) U2OS Peli1 inducible stable line cells were transfected with control siRNA or siRNA against p53 or Mdmx. After 24 hours, the cells were without treatment or treated with 0.1 μg/ml of doxycycline 48 hours. (A) Western blot analysis of cell extracts with antibody against Peli1, Mdm2, Mdmx, p53, p21 and β-actin. (B) p21 and Mdm2 mRNA levels were determined by real-time PCR in these cells. (C) Western blot analysis of cell extracts from two U2OS Peli1 CRISPR-Cas9 knock-out cell lines (CRISPR 1 and 2) with antibody against Mdm2, Mdmx, p53, Peli1, p21 and β-actin. Parental U2OS cells were used as the control. (D) p21 and Mdm2 mRNA levels were determined by real-time PCR in U2OS Peli1 knock-out cell lines. Parental U2OS cells were used as the control.
Peli1 promotes Mdmx nuclear export
Although Peli1 increases p53 transcriptional activity, our findings indicate that Peli1 cannot promote Mdmx degradation. To understand the mechanism of the interaction between Peli1 and Mdmx in modulating the p53 pathway, we firstly considered whether Peli1 has effects on p53 and Mdmx binding. We performed co-immunoprecipitation experiments in both Peli1 inducible and knock-out cells. The results showed that Peli1 cannot affect Mdmx-p53 binding after it is overexpressed or knocked out in these cells (Supplementary Fig. 5A and B). Next, we investigated whether Peli1 promotes Mdmx nuclear export. Immunofluorescence staining showed that Mdmx distributes equally in both the nucleus and cytoplasm when a FH-Mdmx expression vector was transfected into H1299 cells. Interestingly, only wild-type Peli1 but not Peli1 C395/398A mutant promotes significant Mdmx cytoplasmic translocation when both constructs were co-transfected with FH-Mdmx into the cells (Fig. 5A and B). Peli1 and Peli1 mutants distribute equally in both nucleus and cytoplasm when the constructs were individually transfected into the cells (Fig. 5A and B). To further investigate the role of Peli1 in Mdmx nuclear export, we examined the localization of endogenous Mdmx by nuclear cytoplasmic fractionation. The results showed that Mdmx in U2OS cells is distributed in both the cytoplasm and nucleus, with slight preference for the cytoplasm (Fig. 5C). Mdmx cytoplasmic distribution became prominent whereas nuclear distribution was decreased after Peli1 was induced by doxycycline (Fig. 5C). Interestingly, Peli1 is distributed equally both in the nucleus and cytoplasm (Fig. 5C). We also used Peli1 CRISPR-Cas9 knock-out U2OS cells to further assess Peli1 effects on Mdmx subcellular localization. Consistently, Peli1 is distributed approximately equally in the nucleus and cytoplasm whereas Mdmx has slight preference for the cytoplasm in the parental U2OS cells (Fig. 5D). However, Mdmx cytoplasmic distribution is less and the nuclear distribution is more in Peli1 knock-out U2OS cells (Fig. 5D). Taken together, these results indicated that Peli1 promotes Mdmx nuclear export. Our previous study indicates that p53 mono-ubiquitination mediated by low levels of Mdm2 activity induces p53 nuclear export, providing an example of ubiquitination-mediated nuclear cytoplasmic shuttling (14). To further examine the effect of Mdmx ubiquitination in modulating its subcellular localization, we generated an HA-Mdmx-Ubiquitin expression construct mimicking ubiquitinated Mdmx (Supplementary Fig. 6A). Interestingly, HA-Mdmx-Ubiquitin fusion protein was mainly localized in the cytoplasm but wild-type FH-Mdmx and HA-Mdmx-Sumo distribute equally both in nucleus and cytoplasm (Supplementary Fig. 6B and 6C), suggesting ubiquitination of Mdmx modulates its subcellular localization although it remains possible that attaching ubiquitin or sumo to MDMX may potentially affect the RING domain conformation. Taken together, these data indicate that Peli1 induces the non-canonical ubiquitination of Mdmx, which leads to cytoplasmic localization but not proteasome degradation of Mdmx.
Figure 5. Peli1 promotes the cytoplasmic localization of Mdmx.
(A) FH-Mdmx was transfected alone, or with Myc-Peli1, or with Myc-Peli1 mutant (C395/398A) in H1299 cells. Myc-Peli1 or Myc-Peli1 mutant were also individually transfected into the cells. Twenty-four hours after transfection, the cells were fixed, permeabilized and then incubated with anti-Myc mouse antibody or anti-HA rat antibody followed by Alexa Fluor 488 (green) conjugated anti-mouse and Alexa Fluor 568 (red) conjugated anti-rat secondary antibody. DAPI was used for the nuclear staining. The representative images of Mdmx, Peli1, DAPI and merged staining are shown.
(B) Mdmx and Peli1 staining was scored as cytoplasmic staining stronger than nuclear (N<C) and nuclear staining stronger or equal to cytoplasmic (N≥C). At least 100 cells were counted for each group. Shown is the average from 3 independent experiments. Error bars represent the SD.
(C) Analysis of endogenous Mdmx distribution by subcellular fractionation. U2OS Peli1 inducible stable cells were without treatment or treated with 0.1 μg/ml of doxycycline 48 hours. Western blot analysis (left panel) of nuclear and cytoplasmic fractions was performed with antibody against Mdmx, Peli1, PARP1 and Tubulin. The relative optical density of MDMX to PARP1 (nuclear fraction marker) or Tubulin (cytoplasmic fraction marker) bands were analyzed by Quantity One software (Bio-Rad). Shown (right panel) is the average from 2 independent experiments. Error bars represent the SD.
(D) U2OS parental and Peli1 CRISPR-Cas9 knock-out cells were used to analyze endogenous Mdmx distribution by subcellular fractionation. Western blot analysis (left panel) of nuclear and cytoplasmic extracts from these cells was performed with antibody against Mdmx, Peli1, PARP1 and Tubulin. The relative optical density of MDMX (right panel) was analyzed and shown as in Figure 5C.
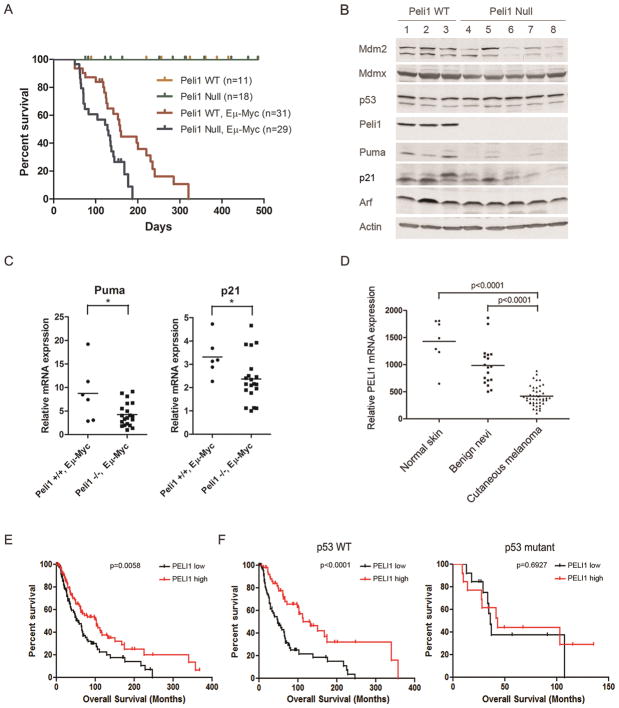
Myc-induced tumorigenesis is accelerated in Peli1 null mice
Loss of one Mdmx allele or Mdmx modifications affects the onset of c-Myc induced lymphomagenesis, suggesting Mdmx function is crucial for c-Myc induced tumorigenesis (32, 33). Since Peli1 is an Mdmx regulator, we next evaluated whether loss of Peli1 modulated oncogene-induced tumorigenesis using a mouse Eμ-Myc lymphoma model. Consistent with a previous study (24), we found that the Peli1 homozygous knockout (Peli1 null) mice were viable and did not display overt abnormalities in growth and survival (Fig. 6A). However, Peli1 null Eμ-Myc mice developed significant early-onset B-cell lymphoma compared with Peli1 wild-type Eμ-Myc mice (Fig. 6A). Median survival was 128 days for Peli1 null and 158 days for Peli wild-type mice in the Eμ-Myc background, respectively (Fig. 6A). Peli1 null Eμ-Myc mice with tumors presented with severe lymphadenopathy and hepatosplenomegaly (Supplementary Fig. 7A). Notably, the mesenteric, cervical, axillary, mediastinal and inguinal lymph nodes were markedly enlarged (Supplementary Fig. 7A). Histologically, high grade diffuse B-cell lymphoma developed in Peli1 null Eμ-Myc mice. The lymph node architecture was replaced with the solid sheets of the infiltrated middle size lymphocytes, the apoptotic lymphoma cells and the large multinucleated macrophages. The intermediate size lymphocytes displayed moderate to high anisocytosis, high mitotic index and abnormal mitosis (Supplementary Fig. 7B). It is well established that the p53 pathway is inactivated in Eμ-Myc lymphomas either by p53 mutation, loss of Arf, or overexpression of Mdm2. We found that the majority of Peli1 null Eμ-Myc lymphoma retained both wild-type p53 and Arf expression (Fig. 6B). Of note, we detected mutant p53 only from one of 20 Peli1 null mice with lymphomas. Thus, these data suggest that p53 mutation is not absolutely required for Peli1-null lymphoma tumorigenesis. To further clarify Peli1 effects on p53 pathway in lymphomagenesis, we compared p53 transcriptional activity between Peli1 wild-type and null Eμ-Myc lymphomas. The results revealed that Puma and p21 levels were significantly decreased although Mdm2 level was decreased modestly in Peli1 null Eμ-Myc lymphoma (Fig. 6B). Real-time PCR analysis demonstrated that mRNA levels for both Puma and p21 were markedly decreased in Peli1 null lymphoma tissues (Fig. 6C). Taken together, these findings indicate that Peli1-Mdmx interaction is implicated in mouse lymphomagenesis by inhibiting p53 transcriptional activity.
Figure 6. Peli1 plays a crucial role in suppressing tumorigenesis.
(A) Kaplan-Meier survival curves of Peli1 wild-type (n=11), Peli1 null (n=18), Peli1 wild-type and Eμ-Myc (n=31) and Peli1 null and Eμ-Myc mice (n=29). Statistical significance was determined using log-rank test (p<0.01; Peli1 wild-type versus null mice in Eμ-Myc background).
(B) Western blot analysis of cell extracts from subcutaneous or mesenteric lymphoma tissues of 3 Peli1 wild-type and 5 Peli1 null mice in Eμ-Myc background with antibodies against Mdm2, Mdmx, p53, Peli1, Puma, p21, Arf and β-actin.
(C) Puma and p21 mRNA levels were determined from 6 Peli1 wild-type and 20 Peli1 null Eμ-Myc lymphoma tissues by real-time PCR. Lines represent means of each group. Each circle or square represents a different tumor tissue. Asterisk represents p<0.05 from student’s t test.
(D) Peli1 is downregulated in human cutaneous melanoma. Relative Peli1 mRNA levels in human normal skin (n=7), benign nevi (n=18) and cutaneous melanoma (n=45) were analyzed from the GEO database. Lines represent means of each group. Each circle, square, or triangle represents an individual patient. The p values from one-way ANOVA are given for each comparison.
(E) Peli1 correlates with overall survival in patients with cutaneous melanoma. Kaplan-Meier survival curves for overall survival of higher Peli1 (n=117) and lower Peli1 (n=116) expression in patients with cutaneous melanoma using cBioportal for Cancer Genomics databases. The tumor patients from higher and lower quartiles for Peli1 expression were extracted and analyzed by a log-rank test (p=0.0058).
(F) Kaplan-Meier survival curve of the same cohort from (E) stratified according to the p53 status and Peli1 expression level. Left panel: higher Peli1 expression with wild-type p53 (n=60) and lower Peli1 expression with wild-type p53 (n=63). Right panel: higher Peli1 expression with mutant p53 (n=16) and lower Peli1 expression with mutant p53 (n=6). Statistical significance was determined using log-rank test (p<0.0001 for left panel and p=0.6927 for right panel).
Peli1 is down-regulated and correlates with overall survival in patients with cutaneous melanoma
Cutaneous melanoma is the leading cause of skin cancer-related deaths. p53 remains wild-type in cutaneous melanoma and reactivation of p53 is as therapeutic intervention for melanoma. Accumulating evidence indicates that Mdmx is upregulated and plays a crucial role in inhibiting p53 response in cutaneous melanoma (20, 26). This prompted us to investigate the functional consequence of the interaction between Peli1 and Mdmx in human cutaneous melanoma. We found that Peli1 is significantly downregulated in human cutaneous melanoma compared with human normal skin and benign nevi when Peli1 expression data from the GEO database was analyzed (Fig. 6D). Next, we analyzed the correlation between Peli1 expression and overall survival in a cohort of primary cutaneous melanoma patients. The demographic and clinical characteristics were shown (Supplementary Table 2) in these patients. Interestingly, we found that Peli1 expression is correlated with p53 status in these patients (Supplementary Table 2). Importantly, the patients with elevated Peli1 levels showed significantly longer overall survival as compared to the patients bearing tumors that express lower levels of Peli1 transcript (Fig. 6E). Finally, we stratified these patients according to p53 status. The results showed that the patients with higher Peli1 expression have a better overall survival in comparison with those with lower Peli1 expression in the wild-type p53 population (Fig. 6F). We did not observe significant difference in overall survival between the patients with higher and lower Peli1 expression in p53 mutant population (Fig. 6F). Taken together, these findings indicated that Peli1 functional consequences are highly p53 dependent in human cutaneous melanoma.
Discussion
Numerous studies indicate that the role of Mdmx in repressing p53 activity is as critical as that of Mdm2 in tumorigenesis (7–9). Nevertheless, the mechanisms by which Mdmx suppresses the activities of p53-mediated transcription are not completely understood. Here, we identified Peli1 as a major component of Mdmx associated complexes in tumor cells. Peli1 specifically interacts with Mdmx both in vivo and in vitro and promotes Mdmx ubiquitination without degradation. Further, Peli1 promotes p53-mediated transcriptional activation through targeting Mdmx nuclear export. Importantly, deletion of Peli1 accelerates oncogene induced mouse lymphomagenesis. In accordance, Peli1 expression is downregulated in human cutaneous melanoma and lower expression correlated with a worse overall survival in these patients. Thus, by establishing Peli1 as a key regulator in modulating the p53-Mdmx axis, these results reveal a novel mechanism by which Mdmx controls p53 function in tumorigenesis.
The previous studies from our group and others (34, 35) indicate both p53 and Mdm2 mainly localize in nucleus in U2OS cells, suggesting that Mdm2 and Mdmx heterodimer functions in nucleus. Accumulating evidence also indicates that Mdm2 and Mdmx functional dimer has a stronger E3 ligase activity toward p53 degradation (15–17). We found that Peli1 targets Mdmx nuclear export, which will separate Mdmx from the functional heterodimer and therefore slightly promotes p53 stabilization. Peli1 cannot affect Mdmx binding to p53 although it forms a complex with Mdmx. These results suggested that Mdmx nuclear export induced by Peli1 expression is sufficient to activate p53 activity. In fact, the Mdmx, Mdm2 and p53 axis is more complicated than previously anticipated. The increased Mdmx activity from Peli1 deletion causes p53 repression and therefore loss of Mdm2 transcription, which counteracted the increased Mdmx activity. In this setting, cells establish a new balance with overall loss of p53 activity. This may provide an explanation for Peli1 function in oncogene-induced tumorigenesis. Notably, a recent report demonstrated Peli1 pro-tumorigenic role in a Peli1 transgenic mouse (36). However, Peli1 is randomly integrated into the genome and under the control of strong promoters in the transgenic mouse. Our study using Peli1 deficient mice provides more physiological outcome to evaluate the roles Peli1 plays in tumorigenesis.
Many previous studies indicate that Peli1 catalyzes ubiquitination of substrate proteins by K48 and/or K63-linked poly-ubiquitin chains (28, 37). We found that Peli1 promotes Mdmx both K48 and K63 linked poly-ubiquitination, suggested that Peli1 mediated ubiquitination is non-canonical and degradation-independent. Although the functional consequence of Mdmx poly-ubiquitination requires further investigation, Peli1 also promotes Mdmx mono-ubiquitination. Our previous study indicates that p53 mono-ubiquitination mediated by low level Mdm2 contributes to its nuclear export (14). Consistently, we found that Mdmx shuttles to the cytoplasm while Peli1 is overexpressed in cells. Further, an Mdmx and ubiquitin fusion protein mimicking Mdmx mono-ubiquitination status has a predominant trend to localize to cytoplasm. Moreover, Peli1 is equally distributed in nucleus and cytoplasm although Mdmx shuttle to the cytoplasm. These findings suggested that Mdmx nuclear export is mediated by ubiquitination. While the Ring domain is essential from mediating Peli1-induced ubiquitination of Mdmx, it is also indispensable for the Peli1-Mdmx interactions. Indeed, the Peli1 truncation and C395/398A mutants that disrupted the C-terminal Ring domain are not capable to binding to Mdmx. Therefore, we cannot exclude the possibilities that Mdmx cytoplasmic localization is mediated by binding between Peli1 and Mdmx.
Eμ-Myc lymphoma mice are an ideal model to monitor Mdmx-p53 axis in vivo. The c-Myc oncogene is induced under the control of the Eμ immunoglobulin enhancer in B cell lineage (38). C-Myc overexpression can activate p53 by inducing Arf expression, which antagonizes Mdm2 mediated p53 repression and leads to p53 activation (39, 40). Deletion of either p53 or Arf expression in Eμ-Myc mice markedly accelerates the onset of lymphoma (41, 42). In contrast, loss of one copy of Mdm2 delays the tumor formation (27, 43), whereas overexpression of Mdm2 accelerates the onset of B cell lymphoma in Eμ-Myc mice (44). Recently, losing one copy of Mdmx also exhibited the increased lymphoma latency, suggesting Mdmx crucial roles in Myc-induced lymphomagenesis (32). Considering that Peli1 functions in regulating Mdmx-p53 axis, our findings that Peli1 deletion contributes to Myc-induced lymphomagenesis, provides an example for Mdmx regulation in tumor suppression. Although either deletion of Arf or p53 mutations is frequently observed in Eμ-Myc lymphoma (41), the majority of Peli1 null Eμ-Myc lymphoma exhibits a wild type p53 and intact Arf expression. Importantly, we observed that p53 downstream targets are markedly inhibited in these Eμ-Myc lymphomas. These findings suggest that Peli1 depletion blunts p53 responses mainly by inhibiting p53 transcriptional activity.
Although the p53 remains wild-type, it is inactivated in the majority of cutaneous melanomas as a result of up-regulated expression of Mdmx (20, 26). This prompted us to investigate the consequence of the interaction between Peli1 and Mdmx in human cutaneous melanoma. We found that Peli1 expression is closely related with p53 status, suggesting Peli1 is an alternative regulator for p53 functions in cutaneous melanoma. Interestingly, Peli1 levels decreased markedly in cutaneous melanoma compared with normal skin or nevi. It is conceivable that the increased Mdmx function caused by down-regulated Peli1 levels inhibits p53 activity, which constitutes an alternative mechanism for melanoma formation. Importantly, higher Peli1 expression predicts a better overall survival in patients with cutaneous melanoma, especially in these patients with wild-type p53. Although these findings highlight the important roles of Peli1-Mdmx interaction in the pathogenesis of cutaneous melanoma, the significance is required to be further assessed in more human tumor types retaining wild-type p53.
Supplementary Material
Significance.
Peli1-mediated regulation of Mdmx, a major inhibitor of p53, provides critical insight into activation of p53 function in human cancers.
Acknowledgments
We thank Dr. Jiandong Chen for providing important reagents for this study. Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number 5R01CA193890, 5RO1CA190477, 5RO1216884 and 5RO1CA085533 to W. Gu. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. O. Tavana was partially supported by NIH cancer biology training grant T32-CA09503.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary Data (Supplementary Figures and Tables) are included.
References
- 1.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 3.Helton ES, Chen X. p53 modulation of the DNA damage response. J Cell Biochem. 2007;100:883–96. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Zhang C, Hu W, Feng Z. Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett. 2015;356:197–203. doi: 10.1016/j.canlet.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gannon HS, Jones SN. Using Mouse Models to Explore MDM-p53 Signaling in Development, Cell Growth, and Tumorigenesis. Genes Cancer. 2012;3:209–18. doi: 10.1177/1947601912455324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varley JM. Germline TP53 mutations and Li-Fraumeni syndrome. Hum Mutat. 2003;21:313–20. doi: 10.1002/humu.10185. [DOI] [PubMed] [Google Scholar]
- 7.Eischen CM, Lozano G. The Mdm network and its regulation of p53 activities: a rheostat of cancer risk. Hum Mutat. 2014;35:728–37. doi: 10.1002/humu.22524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J. The Roles of MDM2 and MDMX Phosphorylation in Stress Signaling to p53. Genes Cancer. 2012;3:274–82. doi: 10.1177/1947601912454733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X, Cao B, Lu H. Negative auto-regulators trap p53 in their web. J Mol Cell Biol. 2017;9:62–68. doi: 10.1093/jmcb/mjx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–6. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 12.Marine JC, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. J Cell Sci. 2007;120:371–8. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 13.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–15. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono-versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–5. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 15.Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domain-mediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer Res. 2007;67:6026–30. doi: 10.1158/0008-5472.CAN-07-1313. [DOI] [PubMed] [Google Scholar]
- 16.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. EMBO J. 2007;26:90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–12. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pant V, Xiong S, Iwakuma T, Quintas-Cardama A, Lozano G. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci USA. 2011;108:11995–2000. doi: 10.1073/pnas.1102241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moyer SM, Larsson CA, Lozano G. Mdm proteins: critical regulators of embry ogenesis and homeostasis. J Mol Cell Biol. 2017 doi: 10.1093/jmcb/mjx004. pii: mjx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jochemsen AG. Reactivation of p53 as therapeutic intervention for malignant melanoma. Curr Opin Oncol. 2014;26:114–9. doi: 10.1097/CCO.0000000000000033. [DOI] [PubMed] [Google Scholar]
- 21.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–83. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Gu W. A critical role for noncoding 5S rRNA in regulating Mdmx stability. Mol Cell. 2011;43:1023–32. doi: 10.1016/j.molcel.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai C, Shi D, Gu W. Negative regulation of the acetyltransferase TIP60-p53 interplay by UHRF1 (ubiquitin-like with PHD and RING finger domains 1) J Biol Chem. 2013;288:19581–92. doi: 10.1074/jbc.M113.476606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang M, Jin W, Sun SC. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol. 2009;10:1089–95. doi: 10.1038/ni.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, et al. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005;11:7234–42. doi: 10.1158/1078-0432.CCR-05-0683. [DOI] [PubMed] [Google Scholar]
- 26.Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med. 2012;18:1239–47. doi: 10.1038/nm.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Z, Johnson HJ, Nie H, Qin J, Bird TA, Li X. Pellino 1 is required for interleukin-1 (IL-1)-mediated signaling through its interaction with the IL-1 receptor-associated kinase 4 (IRAK4)-IRAK-tumor necrosis factor receptor-associated factor 6 (TRAF6) complex. J Biol Chem. 2003;278:10952–56. doi: 10.1074/jbc.M212112200. [DOI] [PubMed] [Google Scholar]
- 28.Medvedev AE, Murphy M, Zhou H, Li X. E3 ubiquitin ligases Pellinos as regulators of pattern recognition receptor signaling and immune responses. Immunol Rev. 2015;266:109–22. doi: 10.1111/imr.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moynagh PN. The roles of Pellino E3 ubiquitin ligases in immunity. Nat Rev Immunol. 2014;14:122–31. doi: 10.1038/nri3599. [DOI] [PubMed] [Google Scholar]
- 30.Ordureau A, Smith H, Windheim M, Peggie M, Carrick E, Morrice N, et al. The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem J. 2008;409:43–52. doi: 10.1042/BJ20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler MP, Hanly JA, Moynagh PN. Kinase-active interleukin-1 receptor-associated kinases promote polyubiquitination and degradation of the Pellino family: direct evidence for PELLINO proteins being ubiquitin-protein isopeptide ligases. J Biol Chem. 2007;282:29729–37. doi: 10.1074/jbc.M704558200. [DOI] [PubMed] [Google Scholar]
- 32.Terzian T, Wang Y, Van Pelt CS, Box NF, Travis EL, Lozano G. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol. 2007;27:5479–85. doi: 10.1128/MCB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang YV, Leblanc M, Wade M, Jochemsen AG, Wahl GM. Increased radioresistance and accelerated B cell lymphomas in mice with Mdmx mutations that prevent modifications by DNA-damage-activated kinases. Cancer Cell. 2009;16:33–43. doi: 10.1016/j.ccr.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Chen L, Chen J. DNA damage induces MDMX nuclear translocation by p53-dependent and -independent mechanisms. Mol Cell Biol. 2002;22:7562–71. doi: 10.1128/MCB.22.21.7562-7571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruse JP, Gu W. MSL2 promotes Mdm2-independent cytoplasmic localization of p53. J Biol Chem. 2009;284:3250–63. doi: 10.1074/jbc.M805658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park HY, Go H, Song HR, Kim S, Ha GH, Jeon YK, et al. Pellino 1 promotes lymphomagenesis by deregulating BCL6 polyubiquitination. J Clin Invest. 2014;124:4976–88. doi: 10.1172/JCI75667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Humphries F, Moynagh PN. Molecular and physiological roles of Pellino E3 ubiquitin ligases in immunity. Immunol Rev. 2015;266:93–108. doi: 10.1111/imr.12306. [DOI] [PubMed] [Google Scholar]
- 38.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The E mu-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1998;167:353–71. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–33. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci USA. 1998;95:8292–97. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–69. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–77. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eischen CM, Alt JR, Wang P. Loss of one allele of ARF rescues Mdm2 haploinsufficiency effects on apoptosis and lymphoma development. Oncogene. 2004;23:8931–40. doi: 10.1038/sj.onc.1208052. [DOI] [PubMed] [Google Scholar]
- 44.Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene. 2008;27:1590–8. doi: 10.1038/sj.onc.1210788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.