Abstract
Previous anti-EGFR trials in unselected gastroesophageal adenocarcinoma (GEA) patients were resoundingly negative. We identified EGFR amplification in 5% (19/363) of patients at the University of Chicago, including 6% (8/140) who were prospectively screened with intention-to-treat using anti-EGFR therapy. Seven pts received ≥1 dose of treatment: three first line FOLFOX plus ABT-806, one second line FOLFIRI plus cetuximab, and three third/fourth line cetuximab alone. Treatment achieved objective response in 58% (4/7) and disease control in 100% (7/7) with a median progression-free survival of 10 months. Pre and post-treatment tumor NGS, serial plasma ctDNA NGS, and tumor IHC/FISH for EGFR revealed pre-existing and/or acquired genomic events including EGFR negative clones, PTEN deletion, KRAS amplification/mutation, NRAS, MYC and HER2 amplification, and GNAS mutations serving as mechanisms of resistance. Two evaluable patients demonstrated interval increase of CD3+ infiltrate, including one who demonstrated increased NKp46+, and PD-L1 IHC expression from baseline, suggesting an immune therapeutic mechanism of action. EGFR amplification predicted benefit from anti-EGFR therapy, albeit until various resistance mechanisms emerged.
Keywords: EGFR amplification, gastroesophageal adenocarcinoma, cetuximab, ABT-806, heterogeneity
Introduction
Gastric (GC) and esophagogastric junction (EGJ) adenocarcinoma, together gastroesophageal adenocarcinoma (GEA), has the third highest incidence and second highest for cancer-related mortality, and it remains a significant global health problem (1). When routine screening is not conducted, most patients present with de novo metastatic disease or locally advanced disease with high risk of recurrence. Approximately 55-60% of patients recur within 5 years after curative intent resection despite perioperative therapy (2). Median overall survival of stage IV GEA is 11-12 months with optimal palliative chemotherapy, and increases to 16 months for patients with HER2 amplified tumors with the addition of trastuzumab to first line chemotherapy (3). To date, ramucirumab, an anti-VEGFR2 monoclonal antibody, is the only other approved second line biologic therapy for GEA as monotherapy or in combination with paclitaxel, with modest clinical benefit (4,5).
Numerous other targeted therapies have been evaluated in metastatic GEA in various lines and settings, but all of these have been uniformly negative. Recent examples include EGFR, MET, mTOR, and hedgehog pathway inhibitors – generally in genomically unselected patients (6–13).
Epithelial growth factor receptor (EGFR) is a well-recognized mediator of oncogenic phenotype. EGFR inhibitors including monoclonal antibodies (cetuximab, panitumumab, necitumumab) and tyrosine kinase inhibitors (erlotinib, gefitinib, afatinib, lapatinib, osimertinib) have been approved for various cancers including lung, head and neck, and colon. Early phase clinical trials had suggested potential benefit in unselected patients with GEA (14–26). These results supported further evaluation in larger phase III trials - EXPAND (cetuximab plus chemotherapy, first line), REAL-3 (panitumumab plus chemotherapy, first line), and COG (gefitinib monotherapy, second-fourth line)(6–8). Disappointingly, each of these phase III trials was negative, and panitumumab actually resulted in worse survival compared to the control; evaluation of EGFR inhibition was abruptly abandoned for GEA.
If HER2-targeted therapeutic development was devised in the same biologically unselected manner, it likely would have suffered the same fate as anti-EGFR therapy– to the detriment of that subset of patients with HER2 amplified tumors who we now understand to derive significant benefit from this targeted approach. In the registration TOGA trial, nearly 4000 patients were screened in order to identify the necessary number of HER2 amplified patients (n= 584) to adequately power the study. Despite this patient selection hurdle representing only ~15% of GEA, screening for HER2 amplification by FISH enriched for those most likely to benefit based on strong pre-clinical rationale (3). Analogously, EGFR amplification reportedly occurs in ~4% of GEA in TCGA (CbioPortal) (27,28), yet prospective EGFR amplification screening in stage IV patients and targeting therapy in this biologically relevant population is currently lacking.
TCGA as well as other sequencing efforts identified a high degree of chromosomal instability in GEA (27–37). This instability generates additional oncogenic drivers, including gene amplifications of well-known receptor tyrosine kinases including HER2, MET, FGFR2 and EGFR. Pre-clinical and clinical evidence suggest benefit of EGFR inhibitors for EGFR genomically-driven tumors. In lung cancer, patients with EGFR-mutated tumors derive a greater response and survival than EGFR wild type patients (38,39). More relevantly, analyses were reported from two large phase III trials for squamous cell lung cancer evaluating the subset of patients with EGFR amplification or increased gene copy number. In this ‘EGFR positive’ subgroup of patients, the addition of cetuximab to chemotherapy in the SWOG S0819 trial increased median overall survival from 6.4 to 11.8 months (p=0.007) (40), and in the SQUIRE trial (41), this same molecular subgroup, trended towards a significant benefit with necitumumab (PFS/OS: HR 0.71/0.70)(41,42). Similarly, phase II evaluation of second-line or greater icotinib in advanced squamous esophageal cancer patients with either high expression by IHC or amplification by FISH demonstrated a 16.7% objective response rate and 46.3% disease control rate (43). Most relevant, however, are studies focusing on EGFR gene copy and benefit from anti-EGFR therapy in GEA samples and patients. Pre-clinical xenograft models of GEA demonstrated that all responders possessed ≥4 EGFR copies and suggested an even smaller population of those with ‘high’ gene copy number having the highest probability of benefit (44). A post-hoc subset analysis of a phase II trial of FOLFOX with cetuximab in GEA confirmed an association between EGFR amplification and overall survival (p=0.011) (45). In the TRANS-COG correlative study of the prospective phase III COG trial of gefitinib monotherapy, 16.3% of patients possessed ‘EGFR positive’ tumors, and the smaller subset of these patients with true gene amplification (6.1%, 18/294) derived a statistically significant survival benefit with the addition of gefitinib in a post-hoc analysis (HR 0.19, p=0.007) (46). The EXPAND trial demonstrated survival benefit in the small subset with extremely high EGFR H-Score expression (6,47), which was possibly attributable to an underlying subset of EGFR amplified tumors, but this has yet to be confirmed.
EGFR monoclonal antibodies reportedly act by preventing activating ligands from binding the extracellular domain and/or receptor internalization/degradation of the receptor, but induction of antibody-dependent cell-mediated cytotoxicity (ADCC) via the Fc portion of the antibody may serve as an additional mechanism of action (48,49). This has been reported with multiple IgG1 monoclonal antibodies including rituximab, trastuzumab, and cetuximab and is believed to be mediated by NK cells with resultant dendritic cell and CD8+ T-cell priming (48–51).
Due to these pre-clinical and clinical subset analyses suggesting potential benefit of EGFR inhibitors for EGFR-amplified GEA patients, we sought to first describe the incidence of EGFR amplification in a large cohort of GEA patients across stages, and to evaluate for a direct correlation of gene copy number with protein expression levels. We then prospectively screened 140 stage IV GEA clinic patients (in any line of therapy) over 27 months at our center for EGFR amplification and, when present and otherwise eligible, treated with EGFR monoclonal antibody therapy under IRB approved protocols, when appropriate. We report their clinical responses and disease control to EGFR blockade as well as characterization of pre- and post-treatment tumor biopsies and serial circulating tumor DNA (ctDNA) next generation sequencing (NGS) in attempt to explain clinical outcomes, mechanisms of resistance, and to evaluate the contribution of NK-cell dependent ADCC to anti-tumoral effect.
Results
EGFR Gene Amplification and Protein Overexpression in GEA Cell Lines
EGFR/CEP7 FISH ratio and EGFR-SRM (selected reaction monitoring) mass spectrometry expression were assessed, as previously described (52), in 24 GEA cancer lines, lymphoblast and breast cancer negative controls, as well as two positive control head and neck cancer cells lines (HN5, SQ20B) both known to harbor EGFR amplification. (Figure 1, Supplemental Table S1). EGFR was only amplified (FISH ratio EGFR/CEP7 ≥2) in the two head and neck cell lines (Figure 1A). EGFR-SRM ranged from <100 to 41383 amol/μg (median 575 amol/μg) (Figure 1C). EGFR/CEP7 ratio ≥2 and SRM values ≥ 4000 amol/μg were strongly correlated (Fisher exact test p=0.002) in the cell lines. No EGFR expression ≥4000 amol/μg was observed in cell lines in the absence of EGFR amplification by FISH (Supplemental Table S1).
Figure 1.
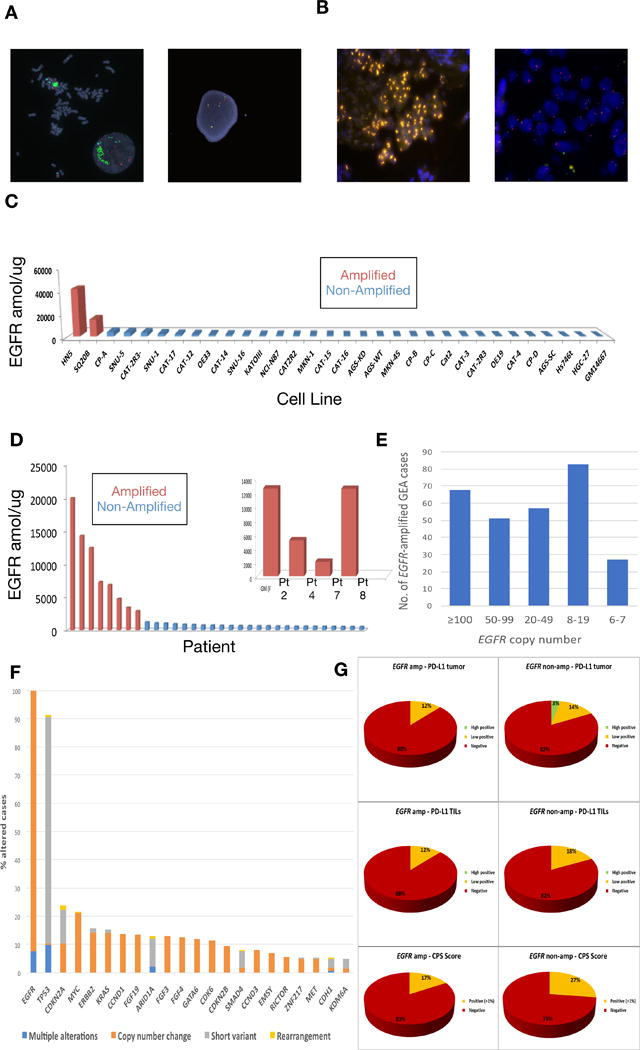
EGFR amplification and expression assessment in cancer cell lines and tissues. (A) FISH demonstrating EGFR amplification of HN5 cells (left) and lack of EGFR amplification in SNU-5 cells (right) (Polysomy with 4 copies of both EGFR (green) and CEP7 (orange) control probes). (B) FISH demonstrating EGFR amplification (orange) within the gastric primary tumor of patient 2 (left) and absence of EGFR amplification within a liver metastasis of patient 5 (right). (C) EGFR selected reaction monitoring (SRM) by amplification status in selected cell lines (red amplified by FISH ratio ≥2, blue not amplified). (D) EGFR selected reaction monitoring (SRM) by amplification status across retrospective cohort and overlay including EGFR amplified patients from the prospective cohort when adequate tissue was available for analysis (red amplified by FISH ratio ≥2 and/or NGS copy ≥8, blue not amplified). (E) Of 4645 patients undergoing Foundation One testing at Foundation Medicine, 259 (5.6%) demonstrated EGFR amplification, displayed by copy number ranges >100, 50-90, 20-49, and 8-19. Although clinical reports do note ‘equivocal’ amplification with 6-7 copies, these were considered negative for this study. (F) Concurrent genomic alterations occurring in ≥5% of EGFR amplified tumors (≥8 EGFR gene copies) within the Foundation Medicine cohort (N=259). (G) PD-L1 IHC testing (Ventana SP142, see methods) of 632 GEA samples performed at Foundation Medicine in the entire cohort, by anatomical location (proximal esophagogastric junction (EGJ) versus distal gastric) and EGFR amplification status, reported as percent tumor positivity score (TPS), percent tumor infiltrating lymphocytes (TILs) positivity, and combination of these for a combined positivity score (CPS).
EGFR Amplification and Overexpression in GEA FFPE Tissues
Five hundred and two samples from 363 patients in the University of Chicago GEA tumor bank underwent NGS by Foundation One (Cambridge, MA) and/or selected reaction monitoring mass spectrometry (SRM) by Nantomics (Rockville, MD) and were included in the overall cohort (Table 1A, Supplemental Table S2A). Among these cases, 18, 183, and 292 patients underwent EGFR FISH (Figure 1B), EGFR-SRM (Figure 1D), and NGS, respectively. One hundred twelve patients had both SRM and NGS, and 11 patients underwent testing by all three modalities. There was a statistically significant linear correlation between EGFR copy number and EGFR expression by SRM (Pearson correlation = 0.87, p<2.2×10−16), with a trend to significance when evaluating binary ‘presence’ or ‘absence’ of amplification versus expression (p=0.08). EGFR amplification was identified in 19/363 (5%) of overall patients across all disease stages in both the retrospective and prospective cohorts (Table 1A), 10/144 (7%) of retrospectively evaluated stage IV patients only (Table 1B), and 8/140 (6%) of prospectively screened stage IV patients only (Table 1C). Only one EGFR amplified case was identified in the absence of EGFR expression ≥1200amol/ug in FFPE samples (Supplemental Tables S2A–C). All cases with EGFR expression ≥1200amol/ug were EGFR amplified.
Table 1A.
Demographics at diagnosis by EGFR amplification status across the entire U of C cohort.
| Characteristic | EGFR Amplified | EGFR Non-Amplified | P-Value |
|---|---|---|---|
| Number of patients (%) | 19 (5) | 344 (95) | <2.2×10-16 |
| Median age yrs. (range) | 64 (37–77) | 63 (16–91) | 0.97 |
| Male Sex – no. (%) | 16(84) | 238 (69) | 0.20 |
| Disease Site no. (%) | |||
| Esophagus/GEJ | 12 (63) | 183 (53) | 0.54 |
| Gastric | 7 (37) | 161 (47) | |
| Tumor Grade no. (%) | |||
| Well Differentiated | 0 (0) | 13 (4) | 0.557 |
| Moderately Differentiated | 3 (16) | 89 (26) | |
| Poorly Differentiated | 15(79) | 229 (67) | |
| Unknown | 1 (5) | 13 (4) | |
| Stage at Diagnosis no. (%) | |||
| I | 0 (0) | 35 (10) | 0.11 |
| II | 0 (0) | 43 (13) | |
| III | 4 (21) | 93 (27) | |
| IV | 15(79) | 172 (50) | |
| Unknown | 0 (0) | 1 (0) | |
| HER2 – no. (%) | |||
| Positive | 5 (26) | 81 (24) | 0.91 |
| Negative | 12 (63) | 212 (62) | |
| Equivocal | 0 (0) | 17 (5) | |
| Unknown | 2 (11) | 34 (10) | |
| Race | |||
| Caucasian | 15 (79) | 250 (73) | 0.68 |
| African American | 2 (11) | 58(17) | |
| Asian | 1 (5) | 27 (8) | |
| Hispanic | 1 (5) | 9 (3) |
Table 1B.
Demographics by EGFR amplification status across all stage IV or recurrent esophagogastric cases in retrospective U of C cohort excluding those prospectively screened between 9/2014 and 12/2016.
| Characteristic | EGFR Amplified | EGFR Non-Amplified | P-Value |
|---|---|---|---|
| Number of patients (%) | 10 (7) | 134 (93) | P<2.2×10-16 |
| Median age yrs. (range) | 64.5 (37–77) | 62 (22–83) | 0.83 |
| Male Sex – no. (%) | 9 (90) | 93 (69) | 0.28 |
| Disease Site no. (%) | |||
| Esophagus/GEJ | 6 (60) | 63 (47) | 0.52 |
| Gastric | 4 (40) | 71 (53) | |
| Tumor Grade no. (%) | |||
| Well Differentiated | 0 (0) | 4 (3) | 0.76 |
| Moderately Differentiated | 1 (10) | 32 (24) | |
| Poorly Differentiated | 8 (80) | 97 (72) | |
| Unknown | 1 (10) | 1 (1) | |
| Stage at Diagnosis no. (%) | |||
| I | 0 (0) | 9 (7) | 0.94 |
| II | 0 (0) | 12 (9) | |
| III | 2 (20) | 30 (22) | |
| IV | 8 (80) | 83(62) | |
| Unknown | 0 (0) | 0 (0) | |
| HER2 – no. (%) | |||
| Positive | 3 (30) | 30 (22) | 0.74 |
| Negative | 6 (60) | 83 (62) | |
| Equivocal | 0 (0) | 11 (8) | |
| Unknown | 1 (10) | 10 (11) | |
| Race | |||
| Caucasian | 7 (70) | 104 (78) | 0.57 |
| African American | 2 (20) | 21 (16) | |
| Asian | 1 (10) | 9 (7) | |
| Hispanic | 0 (0) | 0 (0) |
Table 1C.
Demographics by EGFR amplification status across all stage IV or recurrent esophagogastric cases prospectively screened at U of C for anti-EGFR therapy between 9/2014 and 12/2016.
| Characteristic | EGFR Amplified | EGFR Non-Amplified | P-Value |
|---|---|---|---|
| Number of patients (%) | 8 (6) | 132 (94) | P<2.2×10-16 |
| Median age yrs. (range) | 61.5 (48–74) | 61.5 (19–91) | 0.68 |
| Male Sex – no. (%) | 6(75) | 95 (72) | 1 |
| Disease Site no. (%) | |||
| Esophagus/GEJ | 5 (63) | 73 (55) | 1 |
| Gastric | 3 (38) | 59 (45) | |
| Tumor Grade no. (%) | |||
| Well Differentiated | 0 (0) | 4 (3) | 1 |
| Moderately Differentiated | 2 (25) | 27 (20) | |
| Poorly Differentiated | 6 (75) | 93 (70) | |
| Unknown | 0 (0) | 8 (6) | |
| Stage at Diagnosis no. (%) | |||
| I | 0 (0) | 3 (2) | 0.79 |
| II | 0 (0) | 11 (8) | |
| III | 1 (13) | 29 (22) | |
| IV | 7 (88) | 88 (67) | |
| Unknown | 0 (0) | 1 (1) | |
| HER2 – no. (%) | |||
| Positive | 2 (25) | 29 (22) | 1 |
| Negative | 6 (75) | 90 (68) | |
| Equivocal | 0 (0) | 2 (2) | |
| Unknown | 0 (0) | 11 (8) | |
| Race | |||
| Caucasian | 7 (88) | 91 (69) | 0.43 |
| African American | 0 (0) | 18 (14) | |
| Asian | 0 (0) | 14 (11) | |
| Hispanic | 1 (13) | 9 (7) |
Incidence of EGFR Amplification and Concurrent Genomic Aberrations in Metastatic GEA
To further define the 6-7% EGFR amplification incidence observed in our stage IV cohorts as compared to the incidence noted from the TCGA (4%) comprising earlier stage tumors, we queried the Foundation Medicine database for the incidence of EGFR amplification amongst all unique GEA patients (N=4645) sequenced with the Foundation One test between 2012-2017. These samples were considered, for the most part, to be from advanced metastatic GEA patients, however detailed staging information was unavailable. EGFR amplification was identified in 5.6% of GEA patients, with a higher rate of 7.1% in proximal EGJ tumors as compared to 3.7% in distal gastric tumors (Table 2A). The median EGFR gene copy number was 40 copies with a range of 8-375 copies (Table 2A, Figure 1E). Forty-six percent of EGFR amplified GEA samples (2.6% of all GEA samples) had ≥50 EGFR gene copies. Concurrent genomic aberrations occurring in >5% of EGFR amplified samples in this dataset were mostly short variant events in tumor suppressors including TP53, CDKN2A, ARID1A, SMAD4, and CDH1, and amplifications of various oncogenes including MYC, ERBB2, KRAS, CCND1 and others (Figure 1F).
Table 2A. EGFR.
amplification incidence and characteristics in Foundation Medicine GEA database August 2012- November 2017
| All GEA cases | EGJ | Gastric | |
|---|---|---|---|
| No. patients | 4,645 | 2,534 | 2,111 |
| EGFR amp (%) | 259 (5.6) | 181 (7.1) | 78 (3.7) |
| Median EGFR CN (range) | 40 (8- 375) | 39 (8- 375) | 44 (8-281) |
| EGFR ≥ 50 copies | 119 (46%; 2.6% overall) | 82 | 37 |
| Median Age (range) for EGFR amp | 63 years (27- 90) | 63 years (32- 84) | 62.5 years (27- 90) |
| Gender for EGFR amp | 47 F: 212 M | 19 F: 162 M | 28 F: 50 M |
EGFR Amplification is Associated with Lower PD-L1 Expression by Immunohistochemistry
Given the growing interest and importance of programmed-death-1 (PD-1) and programmed-death-1-ligand (PD-L1) checkpoint inhibition in GEA, we also assessed the incidence of PD-L1 positivity by tumor positivity score (TPS), tumor infiltrating lymphocytes (TILs) and combined positivity score (CPS) by EGFR amplification status (Table 2B, Figure 1G). Of the 632 GEA patients in the Foundation Medicine database for whom PD-L1 IHC was performed (N=632), 26% of samples were CPS score positive (≥1%) (see Materials and Methods). EGFR amplified tumors had lower incidence of CPS positivity (17%) compared to non-amplified tumors (27%), that was not statistically significant.
Table 2B.
PD-L1 expression in GEA Foundation Medicine cohort, by EGFR amplification status
| All GEA | EGJ | Gastric | GEA EGFR WT | GEA EGFR AMP | |
|---|---|---|---|---|---|
|
| |||||
| No. cases with PD-L1 IHC | 632 | 377 | 255 | 591 | 41 |
|
| |||||
| PD-L1 tumor (%) | |||||
| High positive | 20 (3) | 12 (3) | 8 (3) | 20 (3) | 0 |
| Low positive | 89 (14) | 56 (15) | 33 (13) | 84 (14) | 5 (12) |
| Negative | 523 (83) | 309 (82) | 214 (84) | 487 (82) | 36 (88) |
|
| |||||
| PD-L1 TILs (%) | |||||
| High positive | 1 (0.2) | 0 | 1 (0.4) | 1 (0.2) | 0 |
| Low positive | 110 (17) | 72 (19) | 38 (15) | 105 (18) | 5 (12) |
| Negative | 521 (82) | 305 (81) | 216 (85) | 485 (82) | 36 (88) |
|
| |||||
| PD-LI CPS (%) | |||||
| Positive (≥1%) | 166 (26) | 106 (28) | 60 (24) | 159 (27) | 7 (17) |
| Negative | 466 (74) | 271 (72) | 195 (76) | 432 (73) | 34 (83) |
Clinicopathologic Characteristics of EGFR amplification
In the overall University of Chicago cohort (N=363), which comprised of 223 retrospectively accrued and 140 prospectively accrued patients, there was no statistically significant difference in gender, race, age, stage, tumor grade, primary tumor location, or HER2 positivity between patients with EGFR amplification versus those without amplification (Table 1A). However, amongst the EGFR amplified patients, 63% had esophageal or junctional tumors while 37% had distal gastric tumors, as compared to 53% and 47% in the non-amplified patients, respectively. When evaluating only patients having stage IV and recurrent disease, no statistical differences between gender, race, age, tumor grade, tumor location, or HER2 status were identified in the retrospective (Table 1B) nor the prospective cohorts (Table 1C).
Anti-EGFR antibody therapy for EGFR amplified patients
Eight of the 140 screened patients (6%) during the prospective screening period (9/2014-12/2016) demonstrated baseline tumor tissue EGFR amplification (defined as ≥8 copies by NGS) ranging from 54 to 167 EGFR gene copies by NGS (Figure 2A). Evaluation of each patients’ samples for plasma EGFR ctDNA (Figure 2B), EGFR/CEP7 FISH (Figure 2C), along with EGFR IHC and PD-L1 IHC (Figure 2D) was performed. Seven of these eight patients ultimately underwent at least one dose of EGFR-directed therapy (Supplemental Table S3) – three patients received first line FOLFOX plus ABT-806 (investigational EGFR monoclonal antibody inhibitor as part of the PANGEA trial) (53), one received second line with FOLFIRI plus cetuximab, two received third line cetuximab monotherapy, and one received fourth line cetuximab monotherapy (for patient details, see Supplemental Data File S1). The eighth patient, who had concurrent MET and HER2 amplification, was not eligible for EGFR-directed therapy due to poor clinical condition after failure of first line FOLFOX therapy and enrollment in hospice.
Figure 2.
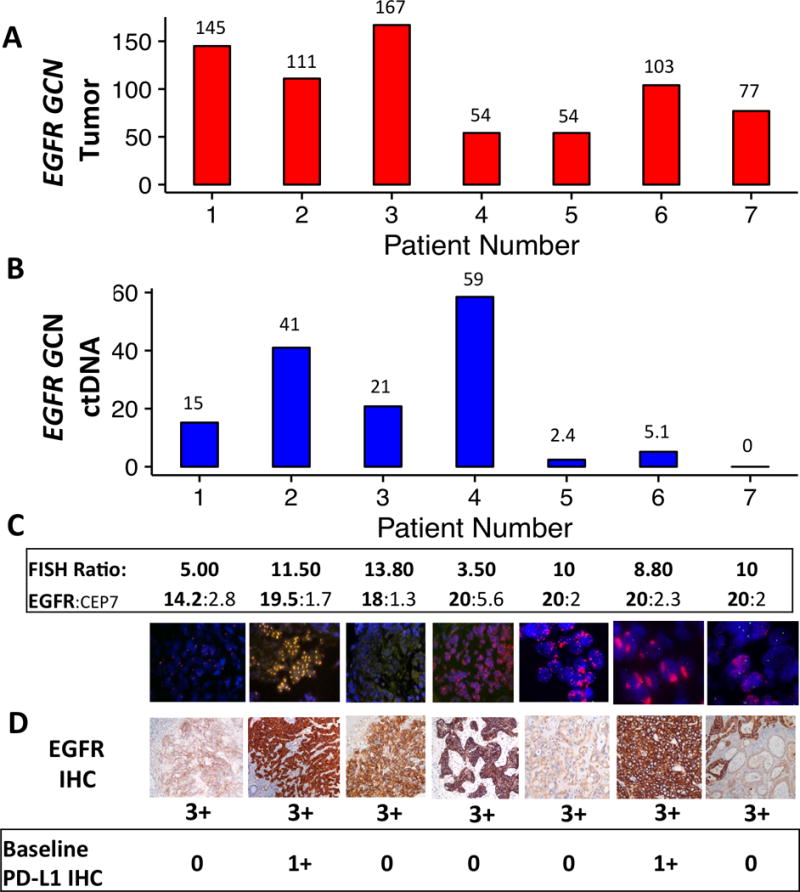
Assessment and correlation of baseline tumor tissue EGFR amplification and expression as well as EGFR plasma ctDNA amplification in 7 patients treated with anti-EGFR therapy. EGFR status assessment by (A) tumor NGS, (B) ctDNA NGS, (C) EGFR/CEP7 FISH, and (D) EGFR IHC, as well as PD-L1 IHC (Abcam antibody, see methods). All samples demonstrated concordant tumor/ctDNA (n=5 > 90th percentile, n=1 > 50th percentile in patient 5, where the metastatic biopsy was not EGFR amplified) except patient 7, whose metastatic biopsy was also not EGFR amplified (see Table 2).
Objective best response, the primary endpoint, was observed in 57% (4/7) of patients, including complete responses in 43% (3/7), partial response in 14% (1/7), and disease control in the remaining 43% (3/7) (Figure 3A,B). Complete responses included one patient (pt 3) receiving third line cetuximab monotherapy who had a durable response of 14 months with resolution of his symptomatic (cough) pulmonary metastases (Figure 3C). Median progression-free survival was 10 months (range 0.5+ to 14) (Figure 3B). Among the 7 patients treated, all four radiographic responses were seen in patients with baseline plasma-detected EGFR amplification over the 50th percentile (2.4 copies in plasma), and the degree of plasma copy number amplification correlated with objective RECIST response, with the mean ctDNA copy number being 2.5 in non-responders and 33.9 in responders, and mean difference 31.4 copies between responders and non-responders (p=0.049, 95% CI 0.25-62.5) (Figure 3A). Notably however, patients 5 and 7, both having EGFR amplification observed only in their primary tumors and not metastases, had clinically significant improvements in their dysphagia/dyspepsia only once ABT-806 was added to their chemotherapy (Supplemental Data File 1, Supplemental Figure S1).
Figure 3.
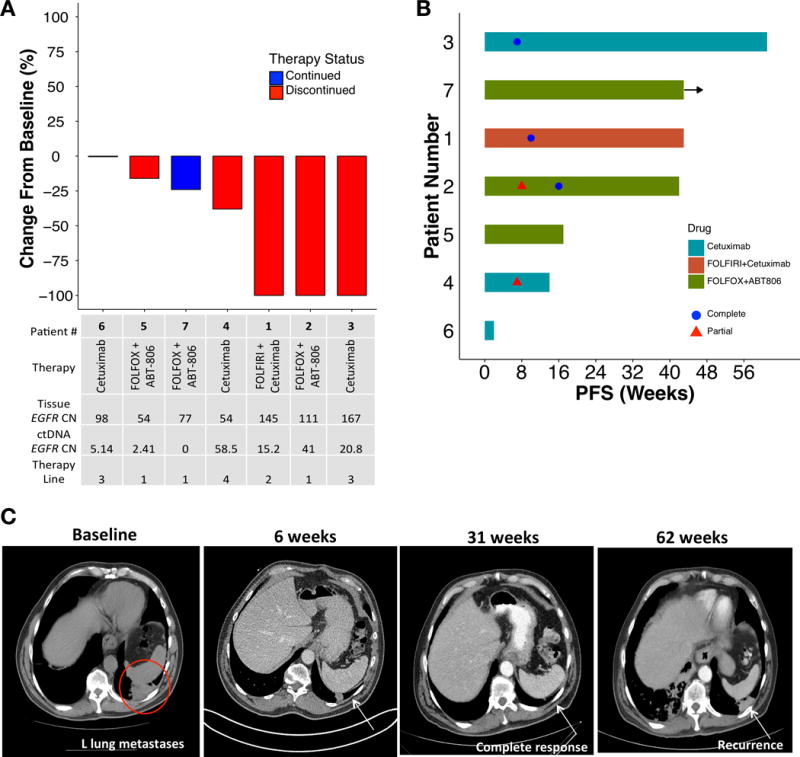
Clinical outcomes of EGFR amplified patients treated with anti-EGFR therapy. (A) Waterfall plot of 7 evaluable pts. (B) Swimmer plot of 7 treated patients. PFS, progression free survival. (C) Computed tomography demonstrating radiographic resolution of lung metastasis in patient 3.
All 4 patients receiving cetuximab developed a stereotypical acneiform rash (which interestingly continued during treatment benefit, yet resolved by the time of disease progression), whereas the 3 patients receiving ABT-806 did not; this was consistent with low rash frequency in phase I evaluation of ABT-806 (54). There were no new safety signals with the addition of EGFR monoclonal antibody therapy as monotherapy or in combination with chemotherapy.
Mechanisms of resistance to EGFR blockade
Underlying baseline and acquired mechanisms of resistance to therapy were evaluated using baseline and serial tumor tissue NGS in parallel with baseline and serial plasma ctDNA NGS in all treated patients, with confirmation by IHC/FISH, when applicable (Table 3, Figure 4A). Likely mechanisms of resistance existing prior to treatment initiation were identified in 7/8 patients, and included intra- and/or inter-tumoral EGFR amplification heterogeneity in 5/7 patients (n= 5, pts 1, 2, 5, 7 and 8), as observed by areas with and without EGFR amplification within the primary tumor itself, and/or across different tumor sites anatomically. Additional baseline mechanisms of resistance included co-amplification of HER2 (n=3, pts 2,4,8), NRAS (n=1, pt 4), KRAS (n=1, pt 6), MYC (n=4, pts 1,2,4,6) or CCNE1 (n=2, pts 4,6), as well as mutation in KRAS (n=1, pt 5) or mutation of another stimulatory G-protein alpha subunit, GNAS (n=1, pt 6) (Table 3, Figure 4A).
Table 3.
Observed mechanisms of resistance by somatic and ctDNA NGS in pre-, on-, and post-therapy samples.
| Patient Number | Baseline | Acquired |
|---|---|---|
| 1 | EGFR amp heterogeneity (only in metastasis, not in primary tumor), MYC amp | MYC amp, PTEN deletion tissue, PIK3CA ctDNA |
| 2 | EGFR/HER2 co-amp primary tumor: 50% HER2 amp, other 50% EGFR amp; EGFR/MYC amp in ctDNA | Residual HER2 amp & now absent EGFR amp in residual primary tumor; KRAS amp clone in ctDNA |
| 3 | None identified | Absent EGFR amp in residual primary tumor and recurrent lung metastases. New BRAF/MET/MYC amp in ctDNA |
| 4 | K-NRAS/HER2/MYC/CCNE1/CCND1 co-amp in primary tumor and ctDNA | Increase in NRAS/HER2/MYC amp and de novo GNAS mutated clone in ctDNA |
| 5 | MET/EGFR co-amp in tumor and ctDNA, but KRAS mutated liver biopsy but no EGFR amp | Increasing KRAS mutated ctDNA, absent EGFR amp in tissue and ctDNA |
| 6 | KRAS/MYC/CCNE1 amp and GNAS mutated clone in ctDNA | KRAS/MYC/CCNE1 amp and GNAS mutated clone in ctDNA |
| 7 | EGFR amp heterogeneity (only in 10% of primary tumor, not metastasis or ctDNA) | None yet identified |
| 8 | EGFR/MET/HER2 co-amp in primary tumor (different clones by FISH) | Never treated |
Figure 4.
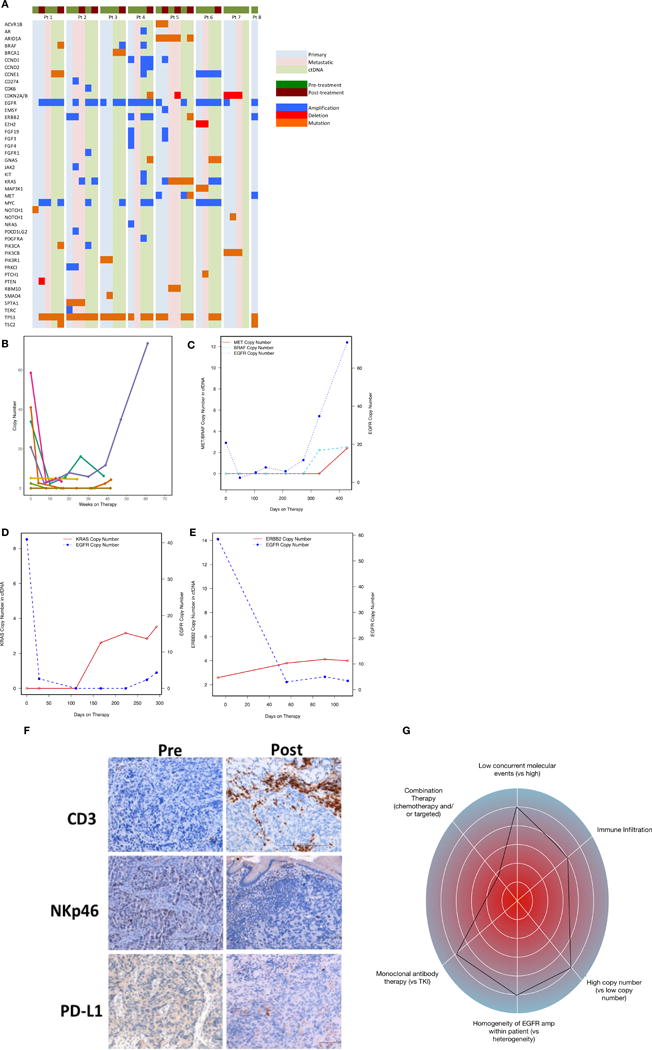
Molecular and immunological correlatives of EGFR amplified samples treated with anti-EGFR therapy. (A) Comparison of intra-patient and inter-patient significant genomic alterations in primary tumor DNA, metastatic tumor DNA, and circulating tumor DNA pre/post anti-EGFR therapy. (B) Serial ctDNA EGFR copy number for all treated patients. (C) Serial ctDNA for pt #3 demonstrating decline of EGFR copy number when cetuximab was begun followed by rise in EGFR copy number when MET and BRAF amplification arose at the time of radiographic progression. (D) Serial ctDNA for pt #2 revealing steep decline in EGFR copy number after FOLFOX and later ABT-806 initiation with cycle 3 followed by rise in KRAS copy number at the time of occult peritoneal disease growth leading to hydronephrosis. (E) Serial ctDNA for pt #4 demonstrating sharp decline in EGFR copy number after cetuximab administration with concomitant rise in ERBB2 copy number at the time of clinical progression and progressive liver failure. (F) IHC assessment for CD3+, NKp46+, and PD-L1+ cells pre-/post anti-EGFR therapy demonstrating increased inflammation within primary tumor from pt #1. (G) “Genogram” figure of patient 3. A framework detailing predictive factors favoring response to genomic targeted therapy are towards the periphery including i) intrapatient homogeneity of EGFR amplification therapy, ii) higher EGFR copy number, iii) combination of chemotherapy + EGFR antagonist, iv) fewer concurrent molecular events, v) monoclonal EGFR antibody use, and vi) increased CD3 infiltration of tumor and stroma. This patient 3 demonstrated homogeneous and high EGFR amplification at baseline, with moderate immune infiltrate, and without concurrent resistance mechanisms. He therefore would be predicted to derive significant benefit, despite being in the third line setting and with monotherapy; he had a complete response lasting 61 weeks.
There were two observed groups of patients upon disease progression - those with retained and those without retained tissue EGFR amplification/overexpression (Figure 4B–E). Serial ctDNA demonstrated a steep decline in EGFR copy number with EGFR-directed therapy in all evaluable patients including monotherapy anti-EGFR antibody, but eventual recovery and increase was seen in some patients (pts 1 and 3) upon disease progression and development of resistance mechanisms, which correlated with rise in serum CA19-9 (Figure 4B, Supplemental Figure S2 A,C). In a patient with retained EGFR amplification in tissue, acquired PTEN deletion contributed to resistance, along with de novo PIK3CA mutation identified in ctDNA (pt 1). In contrast, loss of EGFR amplification/expression (likely a selection of pre-existing but not previously identified EGFR non-amplified clones) was seen in 3 cases (pts 2,3,5), as well as pt 1 in a separate large region of the primary tumor, while pt 7 never harbored systemic EGFR amplification in his metastatic biopsy nor ctDNA. Patient 3 demonstrated persistent EGFR amplification by ctDNA, but his post-treatment biopsies (new lung, residual primary tumor) were not EGFR amplified and he developed BRAF, MET, and MYC co-amplification in ctDNA (Figure 4C). He also developed new brain metastases after disease progression that were not biopsied.
Immune IHC evaluation pre- and post-anti-EGFR therapy
Pre-treatment tissue IHC revealed weak tumoral and stromal CD3 staining in all patients with available tissue (Figure 4F, Supplemental Table S4). Both tumoral and/or stromal CD3 baseline staining persisted in all five available pre/post therapy biopsy pairs (pts 1-5). Four of the five post-treatment biopsies were performed on treatment while clinically stable, and one at the time of clinical progression.
At baseline, six of seven patients also had NK cells present in the stroma, but only one had baseline tumoral NKp46 cell staining (patient 2). CD3 stromal staining in patient 1 increased from 2+ to 4+ with concomitant increased NKp46 and PD-L1 staining in a biopsy taken during therapeutic response (obtained prior to receiving anti-PD-L1 and anti-CTLA4 combination therapy previously, because a post-IO and pre-anti-EGFR biopsy could not be obtained). (Supplementary Table S4). Increased intra-tumoral CD3 staining was also observed in patient 2. Conversely, patient 2 demonstrated decreased NKp46 stromal cell staining and no tumoral staining in a biopsy obtained after disease progression on anti-EGFR ABT-806 plus FOLFOX therapy.
Of the 4 biopsies performed on treatment during disease stability, only 2 had persistent EGFR expression and EGFR amplification. Both of these persistent EGFR amplified tumors (pts 1,4) also expressed PD-L1, whereas patients now lacking EGFR amplification (pt 3) or post-progression and no longer EGFR amplified (pt 5), did not demonstrate PD-L1 expression. Patient 2 also exhibited PD-L1 expression, but only in the moderately differentiated HER2+ region of his primary tumor both pre- and post-treatment, yet again not in the poorly differentiated residual HER2-/EGFR- non-amplified component post-treatment, where EGFR amplified clones were no longer detected. A biopsy of progressing peritoneal carcinomatosis, which was EGFR non-amplified, harbored stromal CD3+ staining, but absent PD-L1 and NKp46 expression (Supplementary Table S4).
These observations suggest that while deriving clinical benefit from therapy, anti-EGFR ADCC may have elicited a reflexive upregulation of PD-L1 expression in tumor cells, a so-called interferon alpha/gamma T cell-induced immune response (55), yet over time as tumor response occurred (ie EGFR amplified clones eradicated in patients 2, 3, 5 at post-treatment biopsies) the immune response appeared to have dissipated, and therefore PD-L1 expression subsequently downregulated.
Discussion
Herein we quantified the incidence of EGFR amplification and consequent significant EGFR overexpression in 24 GEA cell lines and 502 samples from 363 GEA patients within the University of Chicago Gastrointestinal Tumor Bank, as well as from a large commercial NGS database of 4645 GEA patients. We observed no statistically significant differences in clinicopathologic characteristics in patients with EGFR amplification versus those without amplification, other than a higher proportion in proximal EGJ tumors compared to distal gastric tumors, consistent with the known higher incidence of chromosomal instable (CIN) tumors proximally in the TCGA cohort. We then prospectively screened patients for EGFR amplification and treated them with EGFR-targeting agents when possible. As expected, in this relatively large cohort of 140 stage IV patients screened, only 6% of patients were found to be EGFR amplified, slightly higher than the TCGA 4% incidence. Notwithstanding, a demonstrable and robust treatment response and disease control to EGFR antagonists was observed in this select population. Notably, with monotherapy in heavily pretreated patients, two significant responses (one of which was a complete response) were observed, and a third patient had disease control. Moreover, using tumor NGS in parallel with ctDNA NGS allowed identification and understanding of multiple likely baseline and acquired resistance mechanisms, often concurrently within the same patient across and within tumor sites.
None of the 24 available GEA cell lines demonstrated EGFR amplification or extremely high expression by mass spectrometry, as compared to the EGFR amplified head and neck cell line controls. An effort to establish more EGFR amplified cell lines and xenografts is needed in order to enhance understanding of this molecular subset of the disease. Notably, none of the patients identified for treatment in this report had peritoneal ascites or pleural effusions – both recognized metastatic sites that are easily accessible and conducive for establishing tumor cell lines (56). Finally, we demonstrated that tissue EGFR amplification correlated well with protein expression by immunohistochemistry and mass spectrometry in cell lines and tissues analyzed from the same time point and anatomical location, with the caveat of stromal cellular and tumoral molecular heterogeneity affecting this relationship, as previously described with MET and HER2 (57–59).
Amongst University of Chicago tissue samples analyzed, EGFR amplification incidence ranged from 5-7% across all stages and cohorts, which is consistent with previous reports (28,46,60). Regarding incidence of EGFR amplification specifically in metastatic patients, this was similar in the retrospective and prospective stage IV patients (6%) suggesting a reflective prospective cohort. In the overall population, EGFR amplification trended to be more commonly observed in stage IV patients compared to non-amplified patients (79% vs 50% p=0.11, Table 1A). As such, EGFR amplification incidence was slightly higher in our study than the 4% seen in the TCGA GEA cohort, which was based entirely upon early stage resected specimens. This disparity may also reflect the difference in incidence between esophageal/esophagogastric junction and gastric primary tumor location, as 14% of TCGA esophageal adenocarcinoma cases were EGFR amplified (28). Accordingly, our study was comprised with a majority of proximal tumors (54%) and EGFR amplification incidence was consistent with the TRANS-COG analysis comprised exclusively of esophageal cancers (46). Our findings are also consistent with previous work demonstrating that CIN GEAs, which are more likely to harbor amplifications, tend to have proximal locations (27,28). The higher incidence in EGJ versus distal gastric adenocarcinoma and the generally higher incidence compared to TCGA was corroborated in the larger Foundation Medicine database of 4645 GEA samples undergoing Foundation One testing (Table 2A). All other pertinent positive/negative clinicopathological findings (age, grade, HER2 status, gender, site, and race) were similar regardless of EGFR status.
Despite amplification of EGFR being found in only ~5-7% of GEAs, with the high global incidence of distal gastric cancer alone, this may represent nearly fifty thousand patients diagnosed each year with EGFR amplified GEA. ALK-positive non-small cell lung cancer represents a similar paradigm with a 3-5% ALK-translocation frequency that has led to the approval of crizotinib, ceritinib, alectinib, and brigatinib (61–64). In this report, from 140 patients prospectively screened at one treatment center, we identified and treated seven patients with GEA with extreme amplification of EGFR (54-167 gene copies) in tissue biopsies. By chance, we did not encounter any patients with EGFR amplification with tissue gene copies between 8-53, but these patients are not uncommon as demonstrated in the Foundation Medicine GEA cohort (Figure 1E). As such, clinical benefit, or differential clinical benefit, in this “lower level tissue copy number subset” cannot be determined from our study. However we did observe that higher plasma ctDNA copy number did correlate with response within our treated cohort.
We demonstrated clinical benefit with anti-EGFR targeted therapy in clinical scenarios that historically have poor response rates to conventional therapies. In particular, three patients were treated with third/fourth line monotherapy after exhausting all standard therapies. The observed best objective response rate by RECIST was 4 of 7 (57%) patients across multiple lines of therapy, including a complete and durable response lasting 14 months with monotherapy in the third line setting (pt 3). We noted a median progression-free survival of 10 months overall (nearly double the mPFS of standard first line chemotherapy). In comparison, the first line ToGA trial combined trastuzumab with chemotherapy in treatment naïve HER2 positive patients and achieved a 6.7 month mPFS, and the second line RAINBOW study of ramicirumab plus paclitaxel achieved a 4.4 month mPFS (3). In our treated cohort, tumor reduction with clinical benefit was observed in all cases, and the best disease control rate was 100%. Furthermore, as highlighted in the patient therapy summaries (Supplemental Data File 1), subjective quality of life improvements were seen – even in those with the shortest duration of benefit and those with ‘primary-tumor-only’ EGFR amplification.
Baseline and serial ctDNA along with DNA from primary, metastatic, and serial tumor biopsies highlighted significant tumor heterogeneity and widespread potential of therapeutic resistance mechanisms in these patients. Resistance mechanisms included regions of tumors at baseline not harboring EGFR amplification, and regions without EGFR amplification at the time of clinical progression. Resistance mechanisms also included concomitant amplifications and mutations in genes putatively involved in circumventing EGFR signaling in the setting of anti-EGFR therapy (65–70), which we also observed in the larger Foundation Medicine GEA cohort (Figure 1F). Patient 2 was observed to harbor both EGFR and ERBB2 amplification with high expression of both, each within two independent regions (50:50 ratio) of his primary tumor, whereas the retroperitoneal lymph node and bone marrow biopsies, as well as ctDNA, at initial diagnosis harbored EGFR amplification and lacked ERBB2 amplification. For this patient, from a standard-of-care perspective, chemotherapy combined with trastuzumab would be indicated (3), but this would not likely have addressed his primarily HER2-negative metastatic burden. In this patient 2, clinical resistance and progression of peritoneal carcinomatosis after 10 months of anti-EGFR based therapy corresponded with the rise of a ctDNA KRAS amplified clone and KRAS amplification confirmed in the progressing peritoneal biopsy; the primary tumor at this time point was no longer EGFR amplified, nor KRAS amplified, but remained HER2 amplified in ~75% of the biopsy (Figure 4D, Supplemental Figure S2B). Pt 1, who had both baseline EGFR amplified and non-amplified regions of tumor, derived benefit from treatment with FOLFIRI and cetuximab, which together effectively controlled all disease including the EGFR amplified clone for a period of time. However, as demonstrated on repeat biopsy, this therapy eventually selected for an EGFR amplified clone with concurrent downstream PTEN exon 6 deletion, along with persistence of the previously identified EGFR non-amplified region (Supplemental Figure S2A), as well as a de novo PIK3CA mutation in the ctDNA. Cetuximab-resistance via loss of PTEN has been previously demonstrated in colorectal cancer (71). Of note, this deletion was not detected by ctDNA, but rather by endoscopic biopsy and tumor NGS, and this highlights a potential challenge of detecting larger segment deletions in ctDNA. Pt 3 initially had homogenously EGFR amplified disease, but after 14 months developed a combination of EGFR non-amplified regions along with presumably EGFR amplified and concurrent BRAF, MET and MYC amplified clones that circumvented EGFR inhibition (Figure 4C, Supplemental Figure S2C). His persistent EGFR amplification by ctDNA despite absence of amplification identified within post-progression biopsies (of both the residual primary tumor and new lung metastasis) suggests additional non-biopsied sites harboring EGFR amplification, potentially within new brain metastases which were not biopsied. Similarly, selective pressure with cetuximab led to expansion of pre-existing HER2, NRAS, and MYC amplified subclones and emergence of de novo GNAS mutation in pt 4, which all likely conferred therapeutic resistance in various sites within the patient (Figure 4E, Supplemental Figure S2D). Pt 5 had baseline MET co-amplification as well as KRAS mutation in different anatomical locations. In this patient, significant clinical benefit of anti-EGFR therapy was reported despite lack of RECIST response, with improved local esophagogastric symptoms of pain/dyspepsia, which was potentially explained by the EGFR amplification identified only in the primary tumor. Notably this patient’s post-treatment residual primary tumor biopsy no longer identified an EGFR amplified region. Again, as in pt 1 above, this heterogeneity highlights the benefit of concurrent combination chemotherapy in suppressing other pre-existing or acquired resistant clones that would otherwise progress at an accelerated rate if treated with anti-EGFR monotherapy. Pt 6 was unable to obtain drug after cycle 1 due to insurance denial, but presumably would have a more limited benefit in the face of pre-existing KRAS/MYC/CCNE1 amplifications and GNAS mutation in the ctDNA at baseline. Her best response was short-lived stable disease after one dose of cetuximab, yet somewhat impressively after disease progression on first line FOLFOX and second line paclitaxel/ramucirumab. Finally, pt 7 further highlights the intratumoral and intertumoral heterogeneity of EGFR amplification as only a fraction of his primary tumor exhibited EGFR amplification, but not in the liver metastasis nor ctDNA. The patient did however derive significant benefit, similar to pt 5 above, as demonstrated with improved dysphagia only after anti-EGFR therapy was added after 4 cycles of ineffective standard FOLFOX chemotherapy (see Supplementary File S1). These seven cases highlight the utility of a composite of tumor and ctDNA sequencing in tailoring therapy for patients with GEA and using anti-EGFR, cytotoxic therapy, and other targeted and immuno-oncologic agents combined for optimal tumor control.
A limitation of this study in terms of defining benefit from anti-EGFR therapy is the combination of anti-EGFR therapy with chemotherapy in 4 of the 7 patients. The individual contribution of cetuximab/ABT-806 in combination with chemotherapy is therefore difficult to discern in this small cohort. However, the median progression-free survival with FOLFIRI in second line is only 4-5 months (72). Also, clinical benefit in patient 7 was not experienced with four cycles of FOLFOX, and only after addition of anti-EGFR ABT806 antibody at cycle 5 (when biologic grouping was determined on PANGEA study – see Supplemental Figure S1) was dysphagia dramatically improved, which avoided further intervention including stent and or palliative radiation. Data regarding the prognostic significance and natural progression of EGFR amplification remain unknown, but EGFR amplification and EGFR over-expression has been associated with shortened survival in some reports (73,74). Strikingly however, all three patients who received late-line cetuximab monotherapy began their therapy approximately two years after initial stage IV diagnosis – double the median overall survival in this cancer (Supplementary Table S3). This may suggest that EGFR amplification portends a relatively favorable prognosis, but further larger studies will need to sort this out. It should be noted that patient 3 refused surgery while locally advanced, and therefore the duration of ‘first line’ therapy in this case is distorted (he was stage IV to the lung by the time of initiating ‘third line’ monotherapy cetuximab). Despite this, these late-line patients 3, 4, and 6 all demonstrated clinical benefit from cetuximab monotherapy, suggesting that this is indeed an actionable alteration. Notably, each of these three patients had identified EGFR amplification in their original stage IV diagnostic samples as well as their profiling just prior to anti-EGFR therapy in later lines, suggesting stability (and dependence) over time of this aberration with standard therapies. In contrast and interestingly, most patients treated in early lines in combination with chemotherapy in our cohort had evidence of loss of the aberration in all or at least some of their tissue/plasma samples and/or acquisition of likely concurrent resistance mechanisms (e.g. PTEN deletion) after experiencing disease progression on anti-EGFR therapy. This again confirms the specific targeting of anti-EGFR therapy towards EGFR amplified clones, with consequent EGFR amplified clonal eradication and/or pressure to select for concurrent circumventing alterations.
Previous phase II and III trials (including COG with gefitinib and EXPAND using cetuximab) demonstrated an overall survival benefit in the small subset of patients with EGFR amplified (TRANS-COG) or over-expressed (EXPAND) GEA. This was despite an unimpressive response rate (15,46), particularly in the TRANS-COG analysis. Of 13 patients in TRANS-COG with EGFR amplification who received gefitinib, none had objective response. Our results from 7 EGFR amplified patients treated with anti-EGFR monoclonal antibodies suggest a similar benefit with four patients having durable progression-free survival of over 6 months, but also a high response rate, even in those treated with monotherapy (2/3, 66%), with minimal adverse drug reactions. The difference in response rates between TKI and antibodies is intriguing, and could be explained by ADCC and/or receptor internalization/downregulation, which is not seen with TKIs. Antibody therapy was intentionally chosen for treatment in our study due to potential for ADCC via NK cells seen with cetuximab and other IgG1 monoclonal antibodies, such as ABT-806, as well as less toxicity in combination with chemotherapy relative to TKIs. On the other hand, panitumumab, an IgG2 antibody, may act via myeloid cell lineage ADCC (75,76), and differences/similarities between these two IgG classes with respect to ADCC is not well delineated, certainly so for EGFR amplified GEA.
Regardless, in head and neck cancer, cetuximab stimulates NK cell recruitment and interferon γ (IFNγ) secretion, which mediates dendritic cell maturation and cross presentation to cytotoxic T lymphocytes against EGFR (77). IFNγ and its associated genes are currently under evaluation as a predictive biomarker of response to PD-1 and PD-L1 antagonists due to their association with a T cell-inflamed tumor environment (78–82). Interestingly, from a large Foundation Medicine cohort of GEA samples undergoing PD-L1 IHC testing, we observed a slightly lower rate of positivity by TPS, TILs, and CPS scoring in EGFR amplified tumors as compared to non-amplified tumors (Table 2B, Figure 1G). A limitation of this analysis is the use of the Ventana SP142 PD-L1 antibody as opposed to the 22C3 pharmDx companion antibody, which was recently approved for pembrolizumab in PD-L1 expressing patients. SP142 has lower sensitivity and therefore possibly underestimates PD-L1 expression (83–85). Regardless, relatively lower frequency of PD-L1 expression by EGFR amplified tumors compared to non-amplified tumors as we observed in the large Foundation Medicine cohort, if confirmed, may correspond to lower responses to anti-PD-1/anti-PD-L1 checkpoint inhibitor monotherapy in EGFR-amplified patients. This requires further investigation.
To evaluate the effect of anti-EGFR antibodies on the tumor immune environment, including evidence of ADCC, we evaluated pre- and post-treatment tumor biopsies for EGFR, NKp46, CD8 and PD-L1 expression when possible. In this study, results from “during therapy” biopsies imply that treatment with EGFR-directed monoclonal antibodies led to increased tumoral infiltration by CD3+ T cells and NKp46+ NK cells as well as increased PD-L1 expression, which suggested that consequent ADCC may create, or ‘trigger’, a reflexive immunosuppressed tumor environment. On-treatment PD-L1 expression appeared more common in cases with persistent EGFR amplification, which also supports this proposed mechanism. Furthermore, pt 1 had previously received, though not responded to, CTLA-4 and PD-L1 combination inhibition, and so we cannot detail if his increased post-therapy CD3/PD-L1 staining represents a delayed effect from prior immunotherapy alone, anti-EGFR ADCC alone, sequential immunotherapy and targeted monoclonal antibodies, or spatial heterogeneity. Therefore, although suggestive, these results are limited due to the low sample size and temporospatial biopsy variability. These hypothesis-generating findings merit further prospective investigation to tease out the individual contributions of canonical EGFR ligand binding inhibition and receptor internalization/degradation versus ADCC/immune phenomena. Should further studies confirm an upregulation of PD-L1 in anti-EGFR antibody treated EGFR amplified tumors, combination with PD-1 checkpoint blockade would be an appealing combination strategy.
In summary, we report EGFR amplification with overexpression in 5% (19/363) of a large GEA patient cohort. Prospectively, 6% (8/140) of stage IV advanced patients demonstrated EGFR amplification, of which seven patients were successfully treated with at least one dose of anti-EGFR monoclonal antibody therapy. A 57% objective response rate and 100% disease control rate was observed. Within our cohort, elevated plasma-based ctDNA NGS copy number estimation correlated with objective response by RECIST criteria. This is consistent with a similar-sized prospective study of HER2 amplification in plasma associated with an 80% response rate to targeted therapy in GEA (86,87). It is likely that response to anti-EGFR therapy will be optimized for depth and duration with the following contributing factors (quite analogous to anti-HER2 for HER2 amplification and likely also to anti-MET for MET amplification and anti-FGFR2 for FGFR2 amplification): i) homogeneity of EGFR gene amplification within and across all sites of a patient’s tumor burden versus heterogeneous ‘EGFR-negative’ sites, ii) higher EGFR gene copy number versus lower, iii) concurrent chemotherapy for synergy on EGFR amplified clones as well as simultaneous suppression of EGFR-negative clones versus anti-EGFR monotherapy, iv) lack of baseline genomic resistance mechanisms versus ‘molecular chaos’, v) ADCC mechanism of monoclonal antibodies as compared to TKI lacking this mechanism of action, with the patient’s general immune status playing an important role, and vi) addition of concurrent PD-1/PD-L1 checkpoint blockade to increase immune response. This has been conceptualized in a ‘genogram’, or ‘EGFR ampligram’, akin to the recently suggested ‘immunogram’ (55) to serve as a framework to predict clinical benefit from anti-EGFR therapy in EGFR amplified tumors (Figure 4G, Supplementary Figure S3). The degree to which each of these variables contributes to predicted response and response duration will require further investigation. Further assessment of anti-EGFR treatment for EGFR amplification is warranted. Given the relatively low frequency of EGFR amplification, not to mention the issues with intra-patient heterogeneity, evaluation in a traditional phase III study has been elusive and remains difficult, as demonstrated in all phase III EGFR-directed GEA trials to date having only small subsets to evaluate this event, without definitive practice changing results. Novel trial designs, such as PANGEA, a type II expansion platform design trial in GEA, tests a treatment strategy of cytotoxic therapy plus matched targeted therapies across a number of biologic subgroups, including EGFR amplification. This design may optimally identify and treat these low incidence aberrations as well as addressing the various mechanisms of resistance at baseline and progression over time (53,59,88,89).
Materials and Methods
GEA Clinical Samples and Cell Lines
Retrospective and prospective GEA patient samples, with linked clinical and pathological correlates, were obtained from the University of Chicago (Chicago, IL) under institutional review board approved tissue banking protocols. This work was conducted in full concordance with the principles of the Declaration of Helsinki. All patients provided written informed consent, where applicable. The human GEA lines and lymphoblast/breast cancer negative controls were obtained and cultured as previously described (57,90). The genetic identity of parental cell lines was authenticated by short tandem repeat profiling (Cell ID System; Promega) at 10 different loci not fewer than 2 months before profiling and experiments. Cell lines tested negative for Mycoplasma contamination with the VenorGeM Classic Kit (Minerva Biolabs). These included AGS, CAT-2, CAT-3, CAT-4, CAT11B, CAT12, CAT13, CAT14A, CAT15pl, CP-A, CP-B, CP-C, CP-D, GM14667, HGC-27, Hs746T, KATO III, MKN-1, MKN-45, NCI-N87, OE19, OE33, SNU-1, SNU-16, SNU-5, ZR-75-30 obtained between 2008-2012. The head and neck cancer lines (HN5, SQ20B) were graciously provided by Dr. Ezra Cohen (UCSD) in 2012. CAT lines were established between 2009-2016 from malignant ascites or pleural effusion aspirates from patients at the University of Chicago under pre-approved guidelines and IRB protocols.
EGFR Fluorescence in situ hybridization (FISH)
FISH results for cell lines and retrospective samples included mean EGFR and CEP7 copies/nucleus and EGFR/CEP7 ratio as previously described (90–92), and prospectively screened patients using Clarient Diagnostics Services Inc (Aliso Veijo, CA). FISH amplification was defined as EGFR/CEP7 ratio ≥2 in all settings.
Sample Preparation and EGFR-SRM Assay
Laser microdissection isolated tumor cells were obtained from FFPE tumor sections as previously described (52,57,58,92,93). Total protein content for lysates was measured using Micro-BCA assay (Thermo Fisher Scientific Inc, Rockford, IL). EGFR-SRM assay followed previously described methods and quantified expression in attomols/microgram (amol/ug) (93).
EGFR copy number by Tissue Next-Generation Sequencing (NGS)
All NGS results were obtained through routine clinical testing using Foundation One (Cambridge, MA) (94); EGFR amplification was defined as EGFR copy ≥8. Equivocal amplification as noted in the clinical report (copy number 6-7) was considered EGFR non-amplified in this study.
PD-L1 Immunohistochemistry (IHC) from Foundation Medicine Cohort
GEA samples having PD-L1 testing through Foundation Medicine were identified for analysis (N=632). PD-L1 testing was performed using the Ventana antibody (SP142) as previously described (95), and was scored three ways: % tumor positivity score (TPS), % tumor infiltrating lymphocytes (TILs), and combination of these two for a combined positivity score (CPS) given recent approval for third line therapy with pembrolizumab for PD-L1 positive GEA tumors as defined by CPS score ≥1% (83–85).
Quantitative Analysis and Validation of EGFR in Clinical GEA Tissues and Cell lines
EGFR-SRM for 225 retrospective GEA FFPE samples and 28 cell lines was performed by Nantomics (Rockville, MD) and expression was calculated from the ratio of area under the curve (AUC) for the endogenous and isotopically-labeled standard peptide multiplied by the known amount of isotopically-labeled standard peptide spiked into the sample before analysis, as previously described (93).
Identification and treatment of EGFR amplified GEA patients with anti-EGFR therapy
Patients at the University of Chicago with metastatic GEA (any line of therapy) were prospectively screened for EGFR amplification between September 2014 to December 2016 with NGS using the Foundation One test (Foundation Medicine, Cambridge, MA). When remaining tissue was available, EGFR amplification identified by NGS was confirmed by FISH (Clarient Diagnostics Services Inc, Aliso Veijo, CA), and EGFR overexpression was confirmed with immunohistochemistry (IHC; Invitrogen, Clone 31G7, Ventana Ultra View Detection Kit, Ventana XT), and EGFR-SRM through Nantomics (Rockville, MD) (93). All assays used were CLIA certified.
Treatment Assignment
Newly diagnosed metastatic, or recurrent after previous curative intent surgery, first line patients were treated on the PANGEA protocol, (NCT02213289) (53) with anti-EGFR antibody ABT-806, in combination with FOLFOX per protocol (Supplementary Figure S1). Otherwise, patients were treated with off-label cetuximab 500mg q2 weeks IV (96), in combination with FOLFIRI in the second line, or as monotherapy in the 3rd and 4th line settings.
Clinical Outcome Assessment
The primary objective of this study was clinical response as assessed by CT using RECIST 1.1 (97). Measurements were performed independently by clinical interpreting radiologists. Secondary endpoints included progression-free survival (PFS), and toxicity.
Circulating cell-free DNA NGS
Circulating free DNA (cfDNA) sequencing was obtained at baseline prior to anti-EGFR therapy and serially monitored by Guardant 360 (Redwood City, CA) in order to correlate cfDNA levels and genomic findings with initial response outcomes and for potential mechanisms of resistance over time (98–100). Absolute plasma copy number was determined utilizing the mode of the normalized number of cell-free DNA fragments covering each gene to estimate the fragment number corresponding to two copies to derive a baseline diploid value. All values of unique fragments for each gene were then normalized by this baseline value. The baseline derivation was informed by molecule counts data from a large set of normal samples from healthy donors’ plasma. Note that the plasma copy number was related to two variables - the copy number in the tissue, and the amount of shedding of tumor DNA into the blood where the tumor DNA - and thus the copy number, was expected to be diluted by abundant leukocyte-derived EGFR fragments, the latter having a copy number of 2.0. Centiles of EGFR copy number reported in the clinical G360 results were denoted by a ‘+’ for absolute plasma copy number greater than 2.14 (<50th percentile), ‘++’ for copy number greater than 2.4 but less than 4 (<90th percentile), or ‘+++’ for copy number greater than 4 (≥90th percentile). In this study we reported absolute plasma copy number, not these percentiles.
Antibody Dependent Cell-Mediated Cytotoxicity (ADCC)
The contribution of ADCC was assessed in the prospectively identified anti-EGFR treated EGFR amplified cohort by IHC using pre- and post-treatment immunohistochemistry for CD3 (Agilent A0452, Santa Clara, CA), NKp46 (R&D Systems, Clone 195314, Minneapolis, MN) and PD-L1 (Abcam ab205921, Cambridge, MA) in order to evaluate for treatment-related tumor-stroma modulation.
Statistical Analysis
Comparisons between EGFR amplified and non-amplified cases were performed using chi-square testing or a two-sided Fisher’s exact test. The relationship between EGFR amplification and expression by SRM or RECIST response was evaluated using the Student’s t-test and by linear regression. Progression-free survival was estimated by the Kaplan-Meier method.
Supplementary Material
Statement of Significance.
This paper highlights the role of EGFR inhibitors in EGFR amplified GEA – despite negative results in prior unselected phase III trials. Using serial ctDNA and tissue NGS, we identified mechanisms of primary and acquired resistance in all patients, as well as potential contribution of antibody-dependent cell-mediated cytotoxicity (ADCC) to their clinical benefit.
Acknowledgments
The authors would like to acknowledge Kyle Holen, Brian Panzl, James Ward, Vincent Blot, Earl Bain from Abbvie Pharmaceuticals. This work was supported by the NIH (K23 CA178203 and P30 CA014599), UCCCC (University of Chicago Comprehensive Cancer Center) Award in Precision Oncology, LLK (Live Like Katie) Foundation Award, Castle Foundation Award, and the Sal Ferrara II Fund for PANGEA (to D.V.T.C).
Financial support: This work was supported by the NIH K23 award (CA178203-01A1), UCCCC (University of Chicago Comprehensive Cancer Center) Award in Precision Oncology—CCSG (Cancer Center Support Grant) (P30CA014599), Castle Foundation Award, LLK (Live Like Katie) Foundation Award, and the Sal Ferrara II Fund for PANGEA.
Competing interests: DVTC has received research funding from Genentech/Roche, Amgen, Nantomics and honoraria from Genentech/Roche, Amgen, Eli Lilly, Five Prime, Merck, BMS, Nantomics, Guardant Health, Foundation Medicine.
References and Notes
- 1.Sehdev A, Catenacci DV. Gastroesophageal cancer: focus on epidemiology, classification, and staging. Discov Med. 2013;16:103–11. [PubMed] [Google Scholar]
- 2.Al-Batran S-E, Homann N, Schmalenberg H, Kopp H-G, Haag GM, Luley KB, et al. Perioperative chemotherapy with docetaxel, oxaliplatin, and fluorouracil/leucovorin (FLOT) versus epirubicin, cisplatin, and fluorouracil or capecitabine (ECF/ECX) for resectable gastric or gastroesophageal junction (GEJ) adenocarcinoma (FLOT4-AIO): A multicenter, randomized phase 3 trial. J Clin Oncol American Society of Clinical Oncology. 2017;35:4004. [Google Scholar]
- 3.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 4.Fuchs CS, Tomasek J, Yong CJ, Dumitru F, Passalacqua R, Goswami C, et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): an international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet. 2014;383:31–9. doi: 10.1016/S0140-6736(13)61719-5. [DOI] [PubMed] [Google Scholar]
- 5.Wilke H, Muro K, Van Cutsem E, Oh SC, Bodoky G, Shimada Y, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol. 2014;15:1224–35. doi: 10.1016/S1470-2045(14)70420-6. [DOI] [PubMed] [Google Scholar]
- 6.Lordick F, Kang YK, Chung HC, Salman P, Oh SC, Bodoky G, et al. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): a randomised, open-label phase 3 trial. Lancet Oncol. 2013;14:490–9. doi: 10.1016/S1470-2045(13)70102-5. [DOI] [PubMed] [Google Scholar]
- 7.Waddell T, Chau I, Cunningham D, Gonzalez D, Okines AF, Okines C, et al. Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): a randomised, open-label phase 3 trial. Lancet Oncol. 2013;14:481–9. doi: 10.1016/S1470-2045(13)70096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutton SJ, Ferry DR, Blazeby JM, Abbas H, Dahle-Smith A, Mansoor W, et al. Gefitinib for oesophageal cancer progressing after chemotherapy (COG): a phase 3, multicentre, double-blind, placebo-controlled randomised trial. Lancet Oncol. 2014;15:894–904. doi: 10.1016/S1470-2045(14)70024-5. [DOI] [PubMed] [Google Scholar]
- 9.Catenacci DVT, Tebbutt NC, Davidenko I, Murad AM, Al-Batran SE, Ilson DH, et al. Rilotumumab plus epirubicin, cisplatin, and capecitabine as first-line therapy in advanced MET-positive gastric or gastro-oesophageal junction cancer (RILOMET-1): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017 doi: 10.1016/S1470-2045(17)30566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah MA, Bang YJ, Lordick F, Alsina M, Chen M, Hack SP, et al. Effect of Fluorouracil, Leucovorin, and Oxaliplatin With or Without Onartuzumab in HER2-Negative, MET-Positive Gastroesophageal Adenocarcinoma: The METGastric Randomized Clinical Trial. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2016.5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohtsu A, Ajani JA, Bai YX, Bang YJ, Chung HC, Pan HM, et al. Everolimus for previously treated advanced gastric cancer: results of the randomized, double-blind, phase III GRANITE-1 study. J Clin Oncol. 2013;31:3935–43. doi: 10.1200/JCO.2012.48.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen DJ, Christos PJ, Kindler HL, Catenacci DVT, Bekaii-Saab TB, Tahiri S, et al. Vismodegib (V), a hedgehog (HH) pathway inhibitor, combined with FOLFOX for first-line therapy of patients (pts) with advanced gastric and gastroesophageal junction (GEJ) carcinoma: A New York Cancer Consortium led phase II randomized study. J Clin Oncol American Society of Clinical Oncology. 2013;31:4011. [Google Scholar]
- 13.Maron SB, Catenacci DV. Novel Targeted Therapies for Esophagogastric Cancer. Surg Oncol Clin N Am. 2017;26:293–312. doi: 10.1016/j.soc.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim C, Lee JL, Ryu MH, Chang HM, Kim TW, Lim HY, et al. A prospective phase II study of cetuximab in combination with XELOX (capecitabine and oxaliplatin) in patients with metastatic and/or recurrent advanced gastric cancer. Invest New Drugs. 2011;29:366–73. doi: 10.1007/s10637-009-9363-0. [DOI] [PubMed] [Google Scholar]
- 15.Lordick F, Luber B, Lorenzen S, Hegewisch-Becker S, Folprecht G, Woll E, et al. Cetuximab plus oxaliplatin/leucovorin/5-fluorouracil in first-line metastatic gastric cancer: a phase II study of the Arbeitsgemeinschaft Internistische Onkologie (AIO) Br J Cancer. 2010;102:500–5. doi: 10.1038/sj.bjc.6605521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinto C, Di Fabio F, Barone C, Siena S, Falcone A, Cascinu S, et al. Phase II study of cetuximab in combination with cisplatin and docetaxel in patients with untreated advanced gastric or gastro-oesophageal junction adenocarcinoma (DOCETUX study) Br J Cancer. 2009;101:1261–8. doi: 10.1038/sj.bjc.6605319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto C, Di Fabio F, Siena S, Cascinu S, Rojas Llimpe FL, Ceccarelli C, et al. Phase II study of cetuximab in combination with FOLFIRI in patients with untreated advanced gastric or gastroesophageal junction adenocarcinoma (FOLCETUX study) Ann Oncol. 2007;18:510–7. doi: 10.1093/annonc/mdl459. [DOI] [PubMed] [Google Scholar]
- 18.Woell E, Greil R, Eisterer W, Fridrik M, Grunberger B, Gattringer K, et al. Oxaliplatin, irinotecan and cetuximab in advanced gastric cancer: First results of a multicenter phase II trial (AGMT Gastric-2) J Clin Oncol American Society of Clinical Oncology. 2008;26:15587. [Google Scholar]
- 19.Yeh K, Hsu C, Hsu C, Lin C, Shen Y, Wu S, et al. Phase II study of cetuximab plus weekly cisplatin and 24-hour infusion of high-dose 5-fluorouracil and leucovorin for the first-line treatment of advanced gastric cancer. J Clin Oncol American Society of Clinical Oncology. 2009;27:4567. [Google Scholar]
- 20.Enzinger PC, Burtness BA, Niedzwiecki D, Ye X, Douglas K, Ilson DH, et al. CALGB 80403 (Alliance)/E1206: A Randomized Phase II Study of Three Chemotherapy Regimens Plus Cetuximab in Metastatic Esophageal and Gastroesophageal Junction Cancers. J Clin Oncol. 2016;34:2736–42. doi: 10.1200/JCO.2015.65.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dragovich T, McCoy S, Fenoglio-Preiser CM, Wang J, Benedetti JK, Baker AF, et al. Phase II trial of erlotinib in gastroesophageal junction and gastric adenocarcinomas: SWOG 0127. J Clin Oncol. 2006;24:4922–7. doi: 10.1200/JCO.2006.07.1316. [DOI] [PubMed] [Google Scholar]
- 22.Wainberg ZA, Lin LS, DiCarlo B, Dao KM, Patel R, Park DJ, et al. Phase II trial of modified FOLFOX6 and erlotinib in patients with metastatic or advanced adenocarcinoma of the oesophagus and gastro-oesophageal junction. Br J Cancer. 2011;105:760–5. doi: 10.1038/bjc.2011.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomblyn MB, Goldman BH, Thomas CR, Jr, Benedetti JK, Lenz HJ, Mehta V, et al. Cetuximab plus cisplatin, irinotecan, and thoracic radiotherapy as definitive treatment for locally advanced, unresectable esophageal cancer: a phase-II study of the SWOG (S0414) J Thorac Oncol. 2012;7:906–12. doi: 10.1097/JTO.0b013e31824c7bed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan JA, Blaszkowsky LS, Enzinger PC, Ryan DP, Abrams TA, Zhu AX, et al. A multicenter phase II trial of single-agent cetuximab in advanced esophageal and gastric adenocarcinoma. Ann Oncol. 2011;22:1367–73. doi: 10.1093/annonc/mdq604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adelstein DJ, Rodriguez CP, Rybicki LA, Ives DI, Rice TW. A phase II trial of gefitinib for recurrent or metastatic cancer of the esophagus or gastroesophageal junction. Invest New Drugs. 2012;30:1684–9. doi: 10.1007/s10637-011-9736-z. [DOI] [PubMed] [Google Scholar]
- 26.Gold PJ, Goldman B, Iqbal S, Leichman LP, Zhang W, Lenz HJ, et al. Cetuximab as second-line therapy in patients with metastatic esophageal adenocarcinoma: a phase II Southwest Oncology Group Study (S0415) J Thorac Oncol. 2010;5:1472–6. doi: 10.1097/JTO.0b013e3181e77a92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research N, Analysis Working Group: Asan U, Agency BCC Brigham Women’s H, Broad I et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–75. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murugaesu N, Wilson GA, Birkbak NJ, Watkins TB, McGranahan N, Kumar S, et al. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 2015;5:821–31. doi: 10.1158/2159-8290.CD-15-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–86. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nones K, Waddell N, Wayte N, Patch AM, Bailey P, Newell F, et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun. 2014;5:5224. doi: 10.1038/ncomms6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ross-Innes CS, Becq J, Warren A, Cheetham RK, Northen H, O’Donovan M, et al. Whole-genome sequencing provides new insights into the clonal architecture of Barrett’s esophagus and esophageal adenocarcinoma. Nat Genet. 2015;47:1038–46. doi: 10.1038/ng.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stachler MD, Taylor-Weiner A, Peng S, McKenna A, Agoston AT, Odze RD, et al. Paired exome analysis of Barrett’s esophagus and adenocarcinoma. Nat Genet. 2015;47:1047–55. doi: 10.1038/ng.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahle-Smith A, Stevenson D, Massie D, Murray GI, Dutton SJ, Roberts C, et al. Epidermal Growth Factor (EGFR) copy number aberrations in esophageal and gastro-esophageal junctional carcinoma. Mol Cytogenet. 2015;8:78. doi: 10.1186/s13039-015-0181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ali SM, Sanford EM, Klempner SJ, Rubinson DA, Wang K, Palma NA, et al. Prospective comprehensive genomic profiling of advanced gastric carcinoma cases reveals frequent clinically relevant genomic alterations and new routes for targeted therapies. Oncologist. 2015;20:499–507. doi: 10.1634/theoncologist.2014-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Wu Z, Wong G, Pectasides E, Nagaraja A, Stachler M, et al. CDK4/6 or MAPK blockade enhances efficacy of EGFR inhibition in oesophageal squamous cell carcinoma. Nat Commun. 2017;8:13897. doi: 10.1038/ncomms13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 39.Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 40.Herbst RS, Redman MW, Kim ES, Semrad TJ, Bazhenova L, Masters G, et al. Cetuximab plus carboplatin and paclitaxel with or without bevacizumab versus carboplatin and paclitaxel with or without bevacizumab in advanced NSCLC (SWOG S0819): a randomised, phase 3 study. Lancet Oncol. 2017 doi: 10.1016/S1470-2045(17)30694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thatcher N, Hirsch FR, Luft AV, Szczesna A, Ciuleanu TE, Dediu M, et al. Necitumumab plus gemcitabine and cisplatin versus gemcitabine and cisplatin alone as first-line therapy in patients with stage IV squamous non-small-cell lung cancer (SQUIRE): an open-label, randomised, controlled phase 3 trial. Lancet Oncol. 2015;16:763–74. doi: 10.1016/S1470-2045(15)00021-2. [DOI] [PubMed] [Google Scholar]
- 42.Hirsch F, Boyle T, Thatcher N, Paz-Ares L, Varella-Garcia M, Kowalewski A, et al. EGFR IHC and FISH Correlative Analyses (SQUIRE Trial): necitumumab plus gemcitabine-cisplatin vs gemcitabine-cisplatin in 1st-line squamous NSCLC. Journal of Thoracic Oncology. 2015 [Google Scholar]
- 43.Huang J, Fan Q, Lu P, Ying J, Ma C, Liu W, et al. Icotinib in Patients with Pretreated Advanced Esophageal Squamous Cell Carcinoma with EGFR Overexpression or EGFR Gene Amplification: A Single-Arm, Multicenter Phase 2 Study. J Thorac Oncol. 2016;11:910–7. doi: 10.1016/j.jtho.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, Yang J, Cai J, Song X, Deng J, Huang X, et al. A subset of gastric cancers with EGFR amplification and overexpression respond to cetuximab therapy. Sci Rep. 2013;3:2992. doi: 10.1038/srep02992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luber B, Deplazes J, Keller G, Walch A, Rauser S, Eichmann M, et al. Biomarker analysis of cetuximab plus oxaliplatin/leucovorin/5-fluorouracil in first-line metastatic gastric and oesophago-gastric junction cancer: results from a phase II trial of the Arbeitsgemeinschaft Internistische Onkologie (AIO) BMC Cancer. 2011;11:509. doi: 10.1186/1471-2407-11-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petty RD, Dahle-Smith A, Stevenson DAJ, Osborne A, Massie D, Clark C, et al. Gefitinib and EGFR Gene Copy Number Aberrations in Esophageal Cancer. J Clin Oncol. 2017;35:2279–87. doi: 10.1200/JCO.2016.70.3934. [DOI] [PubMed] [Google Scholar]
- 47.Lordick F, Kang Y-K, Salman P, Oh SC, Bodoky G, Kurteva GP, et al. Clinical outcome according to tumor HER2 status and EGFR expression in advanced gastric cancer patients from the EXPAND study. J Clin Oncol American Society of Clinical Oncology. 2013;31:4021. [Google Scholar]
- 48.Kurai J, Chikumi H, Hashimoto K, Yamaguchi K, Yamasaki A, Sako T, et al. Antibody-dependent cellular cytotoxicity mediated by cetuximab against lung cancer cell lines. Clin Cancer Res. 2007;13:1552–61. doi: 10.1158/1078-0432.CCR-06-1726. [DOI] [PubMed] [Google Scholar]
- 49.Kimura H, Sakai K, Arao T, Shimoyama T, Tamura T, Nishio K. Antibody-dependent cellular cytotoxicity of cetuximab against tumor cells with wild-type or mutant epidermal growth factor receptor. Cancer Sci. 2007;98:1275–80. doi: 10.1111/j.1349-7006.2007.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hatjiharissi E, Xu L, Santos DD, Hunter ZR, Ciccarelli BT, Verselis S, et al. Increased natural killer cell expression of CD16, augmented binding and ADCC activity to rituximab among individuals expressing the Fc{gamma}RIIIa-158 V/V and V/F polymorphism. Blood. 2007;110:2561–4. doi: 10.1182/blood-2007-01-070656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnould L, Gelly M, Penault-Llorca F, Benoit L, Bonnetain F, Migeon C, et al. Trastuzumab-based treatment of HER2-positive breast cancer: an antibody-dependent cellular cytotoxicity mechanism? Br J Cancer. 2006;94:259–67. doi: 10.1038/sj.bjc.6602930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hembrough T, Thyparambil S, Liao WL, Darfler MM, Abdo J, Bengali KM, et al. Application of selected reaction monitoring for multiplex quantification of clinically validated biomarkers in formalin-fixed, paraffin-embedded tumor tissue. J Mol Diagn. 2013;15:454–65. doi: 10.1016/j.jmoldx.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Catenacci DV. Next-generation clinical trials: Novel strategies to address the challenge of tumor molecular heterogeneity. Mol Oncol. 2015;9:967–96. doi: 10.1016/j.molonc.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cleary JM, Reardon DA, Azad N, Gandhi L, Shapiro GI, Chaves J, et al. A phase 1 study of ABT-806 in subjects with advanced solid tumors. Invest New Drugs. 2015;33:671–8. doi: 10.1007/s10637-015-0234-6. [DOI] [PubMed] [Google Scholar]
- 55.Blank CU, Haanen JB, Ribas A, Schumacher TN. CANCER IMMUNOLOGY. The “cancer immunogram”. Science. 2016;352:658–60. doi: 10.1126/science.aaf2834. [DOI] [PubMed] [Google Scholar]
- 56.Shepherd TG, Theriault BL, Campbell EJ, Nachtigal MW. Primary culture of ovarian surface epithelial cells and ascites-derived ovarian cancer cells from patients. Nat Protoc. 2006;1:2643–9. doi: 10.1038/nprot.2006.328. [DOI] [PubMed] [Google Scholar]
- 57.Catenacci DV, Liao WL, Thyparambil S, Henderson L, Xu P, Zhao L, et al. Absolute quantitation of Met using mass spectrometry for clinical application: assay precision, stability, and correlation with MET gene amplification in FFPE tumor tissue. PLoS One. 2014;9:e100586. doi: 10.1371/journal.pone.0100586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Catenacci DV, Liao WL, Zhao L, Whitcomb E, Henderson L, O’Day E, et al. Mass-spectrometry-based quantitation of Her2 in gastroesophageal tumor tissue: comparison to IHC and FISH. Gastric Cancer. 2015 doi: 10.1007/s10120-015-0566-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pectasides E, Stachler MD, Derks S, Liu Y, Maron S, Islam M, et al. Genomic Heterogeneity as a Barrier to Precision Medicine in Gastroesophageal Adenocarcinoma. Cancer Discov. 2018;8:37–48. doi: 10.1158/2159-8290.CD-17-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lennerz JK, Kwak EL, Ackerman A, Michael M, Fox SB, Bergethon K, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol. 2011;29:4803–10. doi: 10.1200/JCO.2011.35.4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963–71. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaw AT, Engelman JA. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:2537–9. doi: 10.1056/NEJMc1404894. [DOI] [PubMed] [Google Scholar]
- 63.Ou SH, Ahn JS, De Petris L, Govindan R, Yang JC, Hughes B, et al. Alectinib in Crizotinib-Refractory ALK-Rearranged Non-Small-Cell Lung Cancer: A Phase II Global Study. J Clin Oncol. 2016;34:661–8. doi: 10.1200/jco.2015.63.9443. [DOI] [PubMed] [Google Scholar]
- 64.Kim DW, Tiseo M, Ahn MJ, Reckamp KL, Hansen KH, Kim SW, et al. Brigatinib in Patients With Crizotinib-Refractory Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer: A Randomized, Multicenter Phase II Trial. J Clin Oncol. 2017;35:2490–8. doi: 10.1200/JCO.2016.71.5904. [DOI] [PubMed] [Google Scholar]
- 65.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–62. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 67.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–34. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 68.Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97:1139–45. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–7. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 70.Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013;3:658–73. doi: 10.1158/2159-8290.CD-12-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, Santini D, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol. 2009;27:2622–9. doi: 10.1200/JCO.2008.20.2796. [DOI] [PubMed] [Google Scholar]
- 72.Assersohn L, Brown G, Cunningham D, Ward C, Oates J, Waters JS, et al. Phase II study of irinotecan and 5-fluorouracil/leucovorin in patients with primary refractory or relapsed advanced oesophageal and gastric carcinoma. Ann Oncol. 2004;15:64–9. doi: 10.1093/annonc/mdh007. [DOI] [PubMed] [Google Scholar]
- 73.Nagatsuma AK, Aizawa M, Kuwata T, Doi T, Ohtsu A, Fujii H, et al. Expression profiles of HER2, EGFR, MET and FGFR2 in a large cohort of patients with gastric adenocarcinoma. Gastric Cancer. 2015;18:227–38. doi: 10.1007/s10120-014-0360-4. [DOI] [PubMed] [Google Scholar]
- 74.Terashima M, Kitada K, Ochiai A, Ichikawa W, Kurahashi I, Sakuramoto S, et al. Impact of expression of human epidermal growth factor receptors EGFR and ERBB2 on survival in stage II/III gastric cancer. Clin Cancer Res. 2012;18:5992–6000. doi: 10.1158/1078-0432.CCR-12-1318. [DOI] [PubMed] [Google Scholar]
- 75.Mellor JD, Brown MP, Irving HR, Zalcberg JR, Dobrovic A. A critical review of the role of Fc gamma receptor polymorphisms in the response to monoclonal antibodies in cancer. J Hematol Oncol. 2013;6:1. doi: 10.1186/1756-8722-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schneider-Merck T, Lammerts van Bueren JJ, Berger S, Rossen K, van Berkel PH, Derer S, et al. Human IgG2 antibodies against epidermal growth factor receptor effectively trigger antibody-dependent cellular cytotoxicity but, in contrast to IgG1, only by cells of myeloid lineage. J Immunol. 2010;184:512–20. doi: 10.4049/jimmunol.0900847. [DOI] [PubMed] [Google Scholar]
- 77.Srivastava RM, Lee SC, Andrade Filho PA, Lord CA, Jie HB, Davidson HC, et al. Cetuximab-activated natural killer and dendritic cells collaborate to trigger tumor antigen-specific T-cell immunity in head and neck cancer patients. Clin Cancer Res. 2013;19:1858–72. doi: 10.1158/1078-0432.CCR-12-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Muro K, Chung HC, Shankaran V, Geva R, Catenacci D, Gupta S, et al. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): a multicentre, open-label, phase 1b trial. Lancet Oncol. 2016 doi: 10.1016/S1470-2045(16)00175-3. [DOI] [PubMed] [Google Scholar]
- 79.Gajewski TF, Louahed J, Brichard VG. Gene signature in melanoma associated with clinical activity: a potential clue to unlock cancer immunotherapy. Cancer J. 2010;16:399–403. doi: 10.1097/PPO.0b013e3181eacbd8. [DOI] [PubMed] [Google Scholar]
- 80.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5:200ra116. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doi T, Piha-Paul SA, Jalal SI, Mai-Dang H, Saraf S, Koshiji M. Updated results for the advanced esophageal carcinoma cohort of the phase Ib KEYNOTE-028 study of pembrolizumab (MK-3475) J Clin Oncol. 2016;34 [Google Scholar]
- 82.Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17:956–65. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 83.Fuchs CS, D T, Jang RWJ, Muro K, Satoh T, Machado M, Sun W, Jalal SI, Shah MA, Metges JP, Garrido M. Pembrolizumab Monotherapy in Patients With Previously Treated Advanced Gastric and Gastroesophageal Junction Cancer: Phase 2 KEYNOTE-059 Study. JAMA Oncol. 2018 doi: 10.1001/jamaoncol.2018.0013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Joshi SS, Maron SB, Catenacci DV. Pembrolizumab for treatment of advanced gastric and gastroesophageal junction adenocarcinoma. Future Oncol. 2018;14:417–30. doi: 10.2217/fon-2017-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kang YK, Boku N, Satoh T, Ryu MH, Chao Y, Kato K, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:2461–71. doi: 10.1016/S0140-6736(17)31827-5. [DOI] [PubMed] [Google Scholar]
- 86.Kim ST, Banks KC, Lee S-H, Kim K, Park JO, Park SH, et al. Prospective Feasibility Study for Using Cell-Free Circulating Tumor DNA–Guided Therapy in Refractory Metastatic Solid Cancers: An Interim Analysis. JCO Precision Oncology. 2017;1:1–15. doi: 10.1200/PO.16.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liang DH, Ensor JE, Liu ZB, Patel A, Patel TA, Chang JC, et al. Cell-free DNA as a molecular tool for monitoring disease progression and response to therapy in breast cancer patients. Breast Cancer Res Treat. 2016;155:139–49. doi: 10.1007/s10549-015-3635-5. [DOI] [PubMed] [Google Scholar]
- 88.Catenacci DV. Expansion platform type II: testing a treatment strategy. Lancet Oncol. 2015;16:1276–8. doi: 10.1016/S1470-2045(15)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Joshi SSMS, Lomnicki S, Polite BN, Sharma M, Ibe J, Allen K, Racette C, Rampurwala MM, Amico AL, Shergill A, Kozloff M, Phillips BE, Suh GK, Narula S, Rayani S, Kipping-Johnson K, Wojak E, Kindler HL, Catenacci DVT. Personalized antibodies for gastroesophageal adenocarcinoma (PANGEA): A phase II precision medicine trial ( NCT02213289) J Clin Oncol American Society of Clinical Oncology. 2017;36 abstr TPS198. [Google Scholar]
- 90.Catenacci DV, Cervantes G, Yala S, Nelson EA, El-Hashani E, Kanteti R, et al. RON (MST1R) is a novel prognostic marker and therapeutic target for gastroesophageal adenocarcinoma. Cancer Biol Ther. 2011;12:9–46. doi: 10.4161/cbt.12.1.15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Catenacci DV, Henderson L, Xiao SY, Patel P, Yauch RL, Hegde P, et al. Durable complete response of metastatic gastric cancer with anti-Met therapy followed by resistance at recurrence. Cancer Discov. 2011;1:573–9. doi: 10.1158/2159-8290.CD-11-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sellappan S, Blackler A, Liao WL, O’Day E, Xu P, Thyparambil S, et al. Therapeutically Induced Changes in HER2, HER3, and EGFR Protein Expression for Treatment Guidance. J Natl Compr Canc Netw. 2016;14:503–7. doi: 10.6004/jnccn.2016.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hembrough T, Thyparambil S, Liao WL, Darfler MM, Abdo J, Bengali KM, et al. Selected Reaction Monitoring (SRM) Analysis of Epidermal Growth Factor Receptor (EGFR) in Formalin Fixed Tumor Tissue. Clin Proteomics. 2012;9:5. doi: 10.1186/1559-0275-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rimm DL, Han G, Taube JM, Yi ES, Bridge JA, Flieder DB, et al. A Prospective, Multi-institutional, Pathologist-Based Assessment of 4 Immunohistochemistry Assays for PD-L1 Expression in Non-Small Cell Lung Cancer. JAMA Oncol. 2017;3:1051–8. doi: 10.1001/jamaoncol.2017.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tabernero J, Pfeiffer P, Cervantes A. Administration of cetuximab every 2 weeks in the treatment of metastatic colorectal cancer: an effective, more convenient alternative to weekly administration? Oncologist. 2008;13:113–9. doi: 10.1634/theoncologist.2007-0201. [DOI] [PubMed] [Google Scholar]
- 97.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 98.Zill OBK, Fairclough S, Mortimer S, Vowles J, Mokhtari R, Gandara DR, Mack PC, Odegaard J, Nagy R, Baca A. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. bioRxiv. 2017:233205. doi: 10.1158/1078-0432.CCR-17-3837. [DOI] [PubMed] [Google Scholar]
- 99.Maron SBJS, Lomnicki S, Oliwa T, Landron S, Johnson J, Kiedrowski LA, Nagy RJ, Lanman RB, Janjigian YY, Kelly KJ, Kim ST, Lee J, Catenacci DVT. Circulating tumor DNA (ctDNA) landscape and prognostic implications in gastroesophageal adenocarcinoma (GEA) J Clin Oncol American Society of Clinical Oncology. 2018;36 Abstract 45. [Google Scholar]
- 100.Catenacci DVTNR, Braiteh FS, Kim S, Kwak EL, Maron SB, Banks KC, Lanman RB, Talasaz A, Lee J. Cell free circulating tumor DNA (ctDNA) landscape in patients with advanced gastroesophageal adenocarcinoma (GEC) J Clin Oncol American Society of Clinical Oncology. 2017;35 Abstract 47. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


