Abstract
Background
In mammals, nucleostemin (NS), a nucleolar GTPase, is involved in stem cell proliferation, embryogenesis and ribosome biogenesis. Arabidopsis NUCLEOSTEMIN-LIKE 1 (NSN1) has previously been shown to be essential for plant growth and development. However, the role of NSN1 in cell proliferation is largely unknown.
Results
Using nsn1, a loss-of-function mutant of Arabidopsis NSN1, we investigated the function of NSN1 in plant cell proliferation and cell cycle regulation. Morphologically, nsn1 exhibited developmental defects in both leaves and roots, producing severely reduced vegetative organs with a much smaller number of cells than those in the wild type. Dynamic analysis of leaf and root growth revealed a lower cell proliferation rate and slower cell division in nsn1. Consistently, the transcriptional levels of key cell cycle genes, including those regulating the transition of G1-S and G2-M, were reduced drastically in nsn1. The introduction of CYCLIN B1::GUS into nsn1 resulted in confined expression of GUS in both the leaf primordia and root meristem, indicating that cell proliferation was hampered by the mutation of NSN1. Upon subjection to treatment with bleomycin and methyl methanesulfonate (MMS), nsn1 plants exhibited hypersensitivity to the genotoxic agents. In the nucleus, NSN1 interacted with nucleosome assembly protein1 (AtNAP1;1), a highly conserved histone chaperone functioning in cell proliferation. Notably, the N-terminal conserved domains of Arabidopsis NSN1 were critical for the physical interaction.
Conclusions
As a conserved homolog of mammalian nucleostemin, Arabidopsis NSN1 plays pivotal roles in embryogenesis and ribosome biogenesis. In this study, NSN1 was found to function as a positive regulator in cell cycle progression. The interaction between NSN1 and histone chaperone AtNAP1;1, and the high resemblance in sensitivity to genotoxics between nsn1 and atnap1;1 imply the indispensability of the two nuclear proteins for cell cycle regulation. This work provides an insight into the delicate control of cell proliferation through the cooperation of a GTP-binding protein with a nucleosome assembly/disassembly protein in Arabidopsis.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1289-2) contains supplementary material, which is available to authorized users.
Keywords: Cell cycle, Cell proliferation, Nucleostemin-like1, Nucleosome assembly protein1
Background
In multicellular organisms, organogenesis requires tight control of cell proliferation. Nucleostemin (NS), a nucleolar GTP-binding protein, was first identified in stem cells and some cancer cell lines in mouse [1]. It has been implicated in stem cell proliferation, cell cycle maintenance, ribosome biogenesis and embryogenesis [1–4]. For example, depletion or overexpression of NS in mouse caused arrest of G1-S and G2-M transition by inhibiting the activity of Murine Double Minute 2 (MDM2), an E3 ubiquitin ligase targeting p53 for proteasome-mediated degradation, from activating p53 [1, 3, 5, 6]. NS was found in a large protein complex containing pre-rRNA processing related nucleolar proteins, such as Pescadillo (Pes1), DDX21 (also known as RHII/Guα) and EBNA1 binding protein 2 (EBP2) [7], suggesting a key role in ribosome biogenesis.
As a member of the conserved GTPases present in prokaryotes and eukaryotes, NS belongs to the YlqF/YawG GTPase family [8, 9]. The human NS family consists of nucleostemin, Guanine Nucleotide binding protein-like 3 (GNL3L) and Ngp-1, and all the three members have been shown experimentally to bind GTP in vivo, which is consistent with the presence of the GTP binding domain in these proteins [3, 10, 11]. In addition, NS also contains an acidic amino acid (AAA) domain at the C-terminus, and a basic amino acid (BAA) domain and a coiled-coil (CC) domain at the N-terminus [1]. The latter two domains contribute predominantly to its biological functions as a nucleolar protein in cell cycle progression of both animals and plants [1, 7, 12–14].
Unlike in mammals, only two subfamilies of NS genes (nucleostemin and Ngp-1-like) have been identified in Arabidopsis with the absence of GNL3L-like family genes [15]. Nucleostemin-like1 (NSN1) of Arabidopsis has been found to posess the common domains of mamalian NS and GFP-fused NSN1 has been mainly localized to the nucleolus in tobacco BY-2 cells [15]. The nsn1 mutant exhibits defects in embryogenesis and in the development of both leaf and flower organs. Consistently, NSN1 is highly expressed in the developing embryos, floral and shoot apical meristems, and organ primordia [15, 16]. The observation of reduced expression of meristem genes including WUS, CLV3, STM and ANT in nsn1 suggests that NSN1 plays critical roles in embryogenesis [15]. Recently, a role of NSN1 in plant growth and senescence by modulating ribosome biogenesis has been reported based on the observations that silencing NSN1 leads to growth retardation and premature senescence in Arabidopsis and tobacco [14]. Several ribosomal proteins including Pescadillo and EBP2 have been found to interact with NSN1 and depletion of NSN1 has been shown to repress global translation probably by delaying the biogenesis of the 60S ribosome subunit [14]. Taken together, Arabidopsis NSN1 plays important roles in plant growth and development although the exact mechanism is not clear.
NUCLEOSOME ASSEMBLY PROTEINS1 (NAP1), a histone chaperone in nucleosome assembly/disassembly, is highly conserved from yeast to human [17–20]. NAP1 deficiency is known to cause perturbed expression of around 10% of nuclear genes in yeast [17, 20], and embryonic lethality in mammals and fruit fly [20, 21]. In plants, NAP1 has been identified in several species including rice, soybean, tobacco and Arabidopsis [18, 19, 22, 23]. In Arabidopsis, NAP1 is a multi-gene family containing 4 members: AtNAP1;1, AtNAP1;2, AtNAP1;3 and AtNAP1;4. Phenotypically the individual mutant resembles wild type plants under normal growth conditions, suggesting the functional redundance of the paralogs [18, 24]. Two alleles (m123-1 and m123-2) of the triple mutant Atnap1;1 Atnap1;2 Atnap1;3 exhibit hypersensitivity to DNA damage caused by UV-C irradiation [18], revealing a role of AtNAP1 genes in nucleotide excision repair of DNA. Research on AtNAP1;1 has revealed that like its orthologs in rice and tobacco, some AtNAP1;1-GFP fusion proteins are targeted to the nucleus at an early stage of leaf development whereas the abundance of the fusion proteins are retained in the cytoplasm [22]. Its function in promoting cell division or expansion and the farnesylation status of the protein has been found to be coupled with its subcellular localization [24]. In a bimolecular fluorescence complementation (BiFC) assay, as well as a pull-down experiment, AtNAP1;1 has been demonstrated to interact with ribosomal protein S6 (RPS6), supporting its positive regulatory role in plant rDNA transcription [25]. Analysis of a truncated protein of AtNAP1;3, which lacks 34 amino acids at the C-terminus, reveals altered Arabidopsis responses to abscisic acid (ABA) treatment and salt stress by functioning as a dominant negative factor in ABA responses [23].
In addition to NAP1, Arabidopsis has two NAP1-related proteins, AtNRP1 and AtNRP2. Both are primarily localized in the nucleus, and the double mutant Atnrp1-1Atnrp2-1 exhibits short roots and increased sensitivity to the genotoxic treatment [2]. A recent study using reporter constructs of homologous recombination (HR) has demonstrated reduced levels of HR in the double mutant Atnrp1-1Atnrp2-1 [26] and the AtNAP1 triple mutant m123-1, suggesting that NRP and AtNAP1 act in parallel pathways to synergistically promote somatic HR.
In this study, we focused on the function of NSN1 in cell cycle regulation. Molecular, histochemical and genetic analysis demonstrated that the growth retardation of dwarf mutant nsn1 is attributed to its arrested cell cycle. The interplay between NSN1 and AtNAP1;1, their involvement in cell cycling progression and the hypersensitivity of nsn1 and atnap1;1 to the genotoxic agents imply that in Arabidopsis NSN1 regulates cell cycling and DNA damage repair by cooperating with histone chaperone protein AtNAP1;1.
Results
Growth defects of nsn1 mutant
In previous studies, Nucleostemin-like1 (NSN1) in Arabidopsis has been demonstrated to be essential for the growth of both vegetative and reproductive organs by participation in regulation of embryogenesis and meristem development [14–16]. Here, we investigated the role of NSN1 in cell proliferation and division in both leaf and root organs. Consistent with the findings that cotyledons of nsn1, a loss-of-function mutant of NSN1, are initiated late at the embryo stage [15, 16], delayed emergence of the first leaf in nsn1 dwarf plant was observed (Fig. 1a) with the first leaf emerging at 9th day after germination (DAG), compared with the 5th DAG in the wild type plants (Table 1). For the duration of leaf expansion, at the 9th day after initiation (DAI), the fifth leaf of wild type reached maximum leaf area, while the counterpart leaf of nsn1 fully expended and achieved the maximum area at the 21st DAI (Table 1). Therefore, nsn1 displayed severe growth retardation during the life cycle. The fifth leaf pair was used in this study unless otherwise indicated. Analysis of the leaf area revealed that the size of cotyledons and the fifth leaf of nsn1 were about 50 and 40% of their counterparts in wild type, respectively (Fig. 1b, d). As a result, nsn1 with smaller size of leaves exhibited dwarfism (Fig. 1c). Similar developmental retardation was observed in roots of nsn1 plants, whose primary root length was about 40% of that of wild type (Fig. 1e, f). Therefore, both aerial and underground organs of nsn1 plants displayed growth defects, suggesting the indispensability of NSN1 for plant growth.
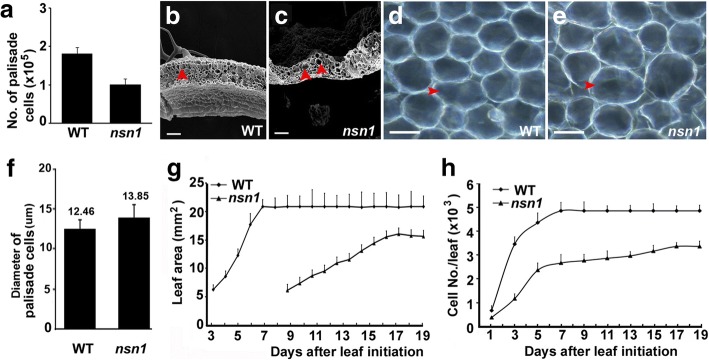
Fig. 1.
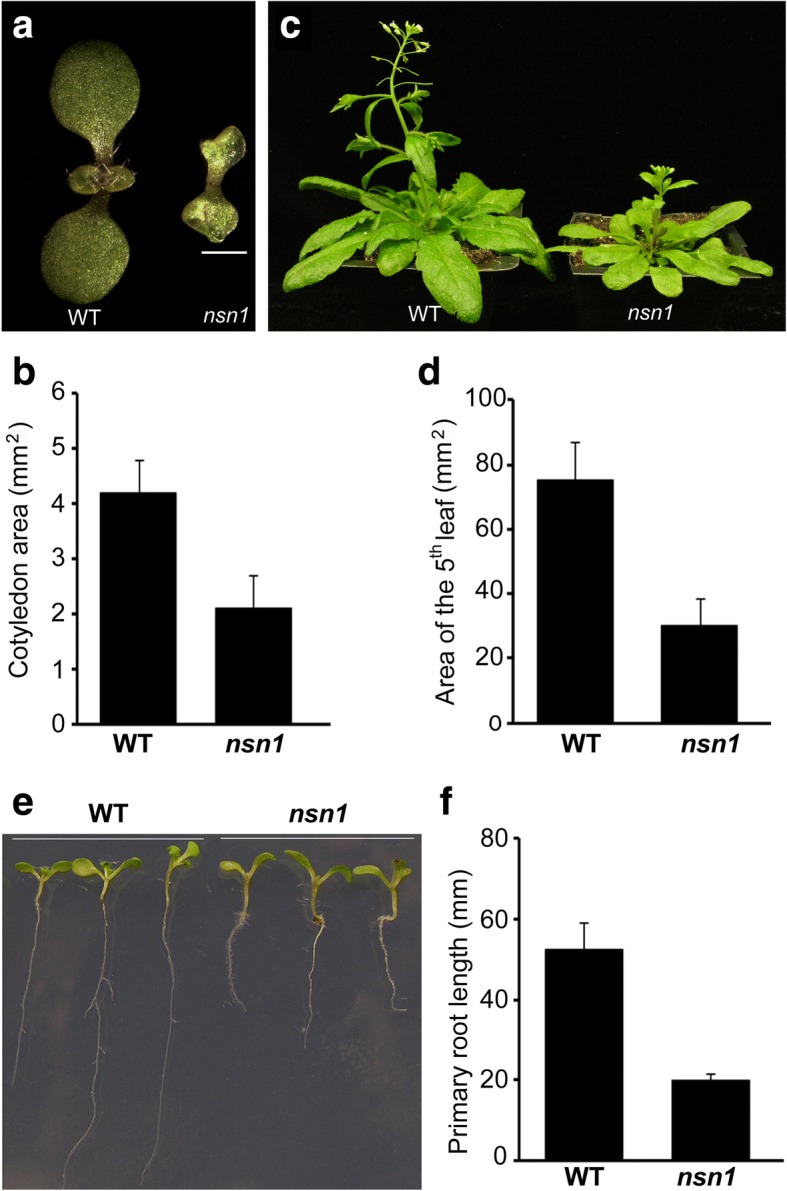
Retarded growth of leaf and root organs in nsn1. a Eight-day-old seedlings of wild type and nsn1, showing reduced size of cotyledons and late initiation of the first leaf in nsn1. Scale bar: 1 mm. b Statistical analysis of cotyledon area of 8-day-old seedlings. c Twenty-one-day-old wild type and nsn1 plants. d Leaf area analysis of the fifth leaf from 21-day-old plants. e Ten-day-old seedlings grown on MS medium showing retarded root growth in nsn1 as compared with the wild type. f Statistical analysis of the primary root length of 10-day-old seedlings. For the statistical analysis, three biological replicates were conducted with 15 plants measured for each experiment
Table 1.
Comparison of leaf development between wild type and nsn1
| Emergence of 1st. leaf (DAG) | Full expansion of 5th. leaf (DAI) | |
|---|---|---|
| Wild type | 5.1 ± 1.2 | 9.3 ± 1.8 |
| nsn1 | 9.5 ± 1.9 | 21.2 ± 2.4 |
Note: DAG days after germination; DAI days after initiation. Value represents mean ± SD. Three biological replicates were performed with at least 15 plants measured for each experiment
Deficient cell proliferation in nsn1 leaf
Plant organ size is genetically determined by both the number and the size of its constituent cells. To investigate the cause of dwarfism in nsn1, the number and size of palisade cells of the fifth leaf pair were measured and analyzed. In agreement with the dwarf statue of nsn1, the amount of the palisade cells was reduced to 56% of that of the wild type (Fig. 2a). In terms of cell size, although cells in the palisade cell layer from the cross-section of nsn1 leaves were slightly larger than those of the wild type (Fig. 2b-e), statistically the diameter difference of the palisade cells between the two genotypes was not significant (P > 0.05) (Fig. 2f), suggesting that the cell size is not the major source for the dwarfism in nsn1. Hence, the reduction of the overall leaf size of nsn1 mutant is attributed to the decrease of cell numbers rather than cell size alteration.
Fig. 2.
Inefficient cell proliferation in nsn1 leaves. The fifth leaf pair was used for measurement and analysis. a Statistical analysis of the number of palisade cells from fully expanded leaves. Scanning electron microscope (SEM) images of fully expanded leaves of wild type (b) and nsn1 (c). Arrow heads indicate the palisade cells with similar size from the two genotypes. The palisade cells of fully expanded leaves of wild type (d) and nsn1 (e). f Statistical analysis of the diameter of palisade cells from fully expanded leaves. g Dynamics of leaf expansion of wild type and nsn1. h Dynamic analysis of cell number/leaf of wild type and nsn1. The difference between the two datasets was analyzed by Student’s t test (P < 0.01). Scale bars: 50 μm for (b) and (c); 10 μm for (d) and (e)
To search for the cause of cell reduction in nsn1, dynamics of leaf area and cell proliferation rate were examined and compared with the wild type plants. Kinetic analysis of leave growth showed that the leaf area of wild type increased dramatically after initiation and reached the maximum at 7th DAI, while the progressive leaf growth of nsn1 was relatively slow and the maximum value was achieved at 17th DAI (Fig. 2g). The long duration of cell proliferation in nsn1 indicated a slower cell proliferation rate in the dwarf mutant. Synchronically, dynamics of cell number/leaf demonstrated a significant difference of the maximum cell number between nsn1 and wild type, with about 3.4 × 103 at 17th DAI for nsn1 and 4.9 × 103 cells at 7th DAI for wild type (Fig. 2h), respectively. Taken together, the hampered leaf growth in nsn1 was resulted from the malfunction of NSN1 in cell proliferation. Additionally, in wild type the palisade cell layer and spongy cell layer are easily differentiated and the longitudinal axial of elongated palisade cell is vertical to the leaf epidermis (Fig. 2b). In nsn1, however, abnormal palisade cells with irregular patterning and highly variable size and shape were observed (Fig. 2c), suggesting that besides regulating cell proliferation and division, NSN1 may function in leaf development as well.
Inefficient cell proliferation in nsn1 roots
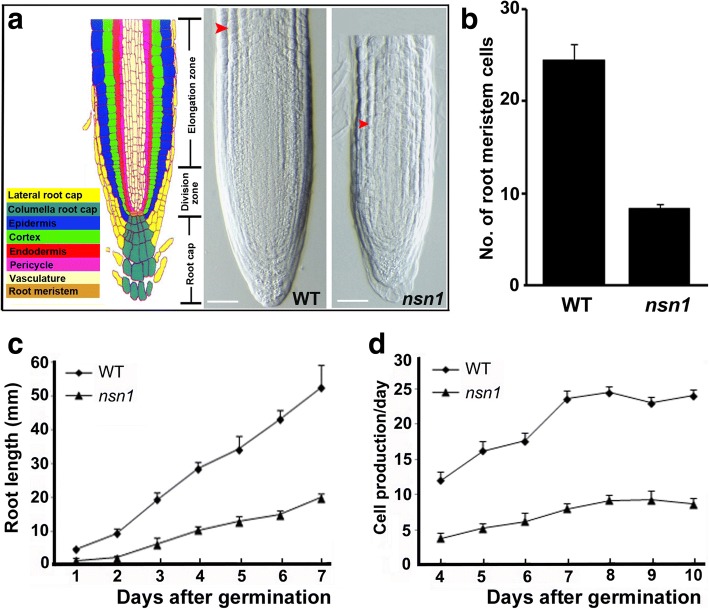
In agreement with the retardation of root growth in nsn1 (Fig. 1e, f), 10-day-old mutant exhibited shorter primary roots and a smaller number of meristematic cells were present in the root meristem region of nsn1 than in wild type (Fig. 3a, b). In terms of root growth rate, root length was monitored during the first week after germination. The length of primary root of nsn1 was about 25–35% of wild type within the first 7 days (Fig. 3c), and on day 10, nsn1 root reached 40% of wild type (Fig. 1e, f). Consistent with the measurement of meristem cells, the number of cells produced by nsn1 roots per day was about one third of that of the wild type roots (Fig. 3d). Therefore, our analysis indicates that slower root elongation and less daily root cell production of nsn1 account for the observed growth defects in the mutant.
Fig. 3.
Inefficient cell proliferation in nsn1 root. a Longitudinal sections of the primary root from 10-day-old wild type and nsn1. Illustration of the tissue organization of the Arabidopsis primary root apex (left in color) was adapted from [42]. Arrowhead indicated the cortex cells in elongation zone. Scale bar: 100 μm. b Statistical analysis of cell numbers in the root meristem region of 10-day-old plants. c Comparison of the dynamics of the primary root length in wild type and nsn1 within the first week after germination. d Analysis of the daily cell production rate in the primary root of wild type and nsn1. The number of cortex cells at the root tip including elongation and cell division zones was counted under microscope daily from the 4th day to the 10th day after germination. Cortex cells on one side as indicated by arrowheads in (a) were calculated
Impaired cell cycling progression in nsn1
Given that the dwarfism of nsn1 was resulted from defective cell proliferation and division, we investigated whether the nsn1 mutation affected the transcriptional levels of five cell cycle marker genes, including three cyclin family genes CYCA2;3, CYCB1;1, and CYCD3;1, and two S-phase specific genes HISTONE H4 and Ribonucleotide reductase (RNR) [27]. For the three cyclin genes, CYCD3;1 has been reported as a key gene in G1-S transition by controlling cell division rate [28], while the remaining two genes, CYCB1;1 and CYCA2;3, have been found to affect G2/M transition in cell cycle [29, 30]. Quantitative analysis revealed that in nsn1, the expression level of the cyclin genes was about 30–40% of that in wild type, and the transcriptional level of HISTONE H4 and RNR, a key enzyme in the DNA synthesis pathway [27], decreased to approximately 60 and 50% of those in wild type, respectively (Fig. 4a). The notable down-regulation of these cell cycle regulating genes in nsn1 implied the disruption of both G1-S and G2-M transition.
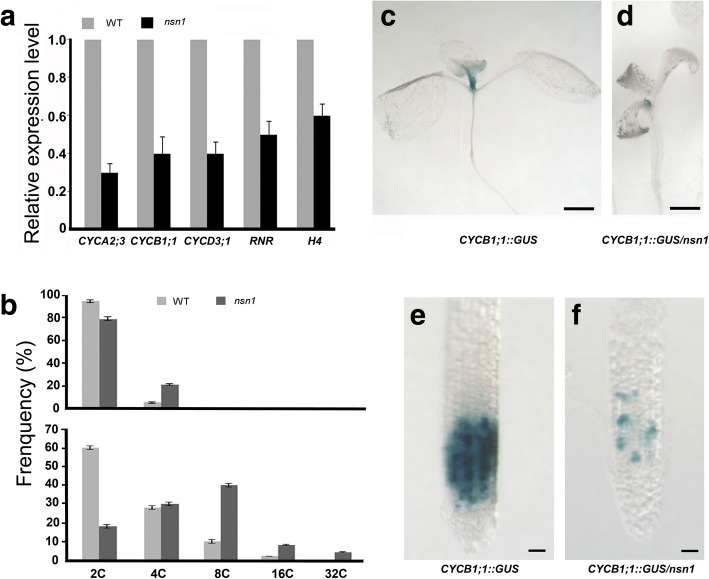
Fig. 4.
Analysis of cell cycle defects in nsn1. a Relative transcriptional levels of cell cycle genes in nsn1 as measured by q-RT-PCR. Ten-day-old seedlings shown in Fig. 1e were used for total RNA extraction after the root length measurement. Three biological replicates were conducted with 15 plants for each experiment. b Flow cytometry assay of nuclei isolated from leaves of 4-days after initiation (upper panel) and of fully-expanded fifth leaves (lower panel). c-f Histochemical analysis of GUS signal in ten-day-old seedlings of indicated genotypes. GUS signal in the shoot apical meristem (c) and root meristem (e) of CYCB1;1::GUS expressing wild type plants stained with X-glux solution. GUS signal in the shoot apical meristem (d) and root meristem (f) of CYCB1;1::GUS expressing nsn1 plants stained with X-glux solution. Scale bars: 0.5 mm for (c) and (d); 100 μm for (e) and (f)
To further dissect the involvement of NSN1 in cell cycle regulation, flow cytometry analysis was performed. Leaves from two developmental stages (4-day-old and mature) were subjected to nuclei isolation. Our results showed a lower percentage of 2C nuclei in nsn1 than wild type at both the young and mature phases: 79 vs. 95% and 18 vs. 60%, respectively (Fig. 4b). Consistently, compared with the wild type, in nsn1, 16% more of 4C nuclei were detected in young leaves, and 42% more of polyploidy including 4C, 8C, 16C and 32C nuclei were observed in mature leaves (Fig. 4b). The detection of higher percentage of polyploidy indicated a condensed DNA content in nsn1 due to the malfunction of nsn1 mutation in cell division.
To visualize the effect of NSN1 on cell cycle progression, transgenic plants expressing CYCLIN B1;1::GUS, which expresses the reporter β-glucuronidase (GUS) fused to the mitotic destruction sequence (D-box) under the promoter of the cyclin CYCB1;1 [31], were crossed with nsn1 mutant to obtain CYCLIN B1;1::GUS-containing nsn1 lines. The GUS activity marked cells in G2 and early M phase [31] because the fusion gene was expressed upon entry into G2 (via the CYCB1:1 promoter) and its protein product was degraded upon exit from metaphase (via D-box) [32]. Histochemical analysis showed that in the wild type background, GUS-positive cells were predominantly detected in the regions of leaf primordia (Fig. 4c). When CYCLIN B1;1::GUS was introduced into nsn1, however, the expression of GUS was confined to a much smaller area containing meristematic cells of the shoot apical region (Fig. 4d). In roots, a clearly lower number of GUS-positive cells were observed in nsn1 plants than those in the control plant (Fig. 4e, f). The restricted CYCLIN B1;1::GUS signal in both the root and shoot apical meristems of nsn1 plants provided genetic evidence of arrested cell cycling in nsn1. Therefore, our molecular and genetic data revealed that the cell cycle progression in both leaves and roots of nsn1 was impaired by the nsn1 mutation.
Hypersensitivity of nsn1 to genotoxic agents
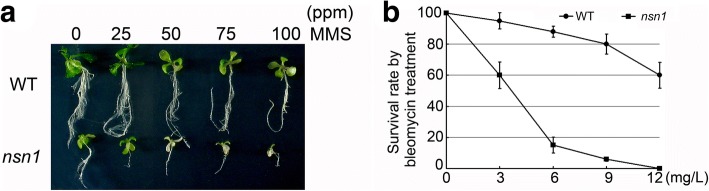
Given that NSN1 is essential for proper cell cycle progression, plant sensitivity to DNA damaging agents methyl methanesulfonate (MMS) and bleomycin was tested. MMS is known to result in alkylated DNA, which is poorly replicated by DNA polymerases [33], while treatment with bleomycin, a radiomimetic drug, causes multiple types of molecular damages including double strand breaks (DSBs) [34]. Four-day-old Arabidopsis plantlets were subjected to treatment with MMS and bleomycin. When nsn1 seedlings were exposed to 25 ppm MMS, rosette leaves turned yellow and root growth was severely inhibited (Fig. 5a). MMS of 50 ppm inhibited the growth of leaf and root organs, resulting in leaf etiolation and plant death. The damages caused by MMS was dosage-dependent (Fig. 5a). In contrast, the growth of wild type was not obviously affected by MMS at 25 ppm, and for treatment of 100 ppm MMS, no plant death was observed although rosette leaves were yellowish and root growth was badly inhibited (Fig. 5a). These results indicated that nsn1 is hypersensitive to MMS treatment in comparison with the wild type. The survival rate of nsn1 plants was dramatically decreased by bleomycin treatment, especially at dosages higher than 6 mg/L (Fig. 5b). At the dosage of 12 mg/L, all nsn1 plantlets died after being treated for 1 week, while about 60% of wild type plants survived the treatment (Fig. 5b). Hence, nsn1 displayed more pronounced sensitivity to the two genotoxic agents than the wild type plants, implying the involvement of NSN1 in cell cycle progression in plants.
Fig. 5.
Comparison of sensitivity to genotoxic agent treatment. a Four-day-old seedlings were treated with MMS at the indicated concentrations for 1 week. b Four-day-old seedlings were treated with bleomycin at the different concentrations for 3 weeks. Three biological replicates were performed with at least 15 plants tested per experiment
Interaction of NSN1 with AtNAP1;1 in vitro and in vivo
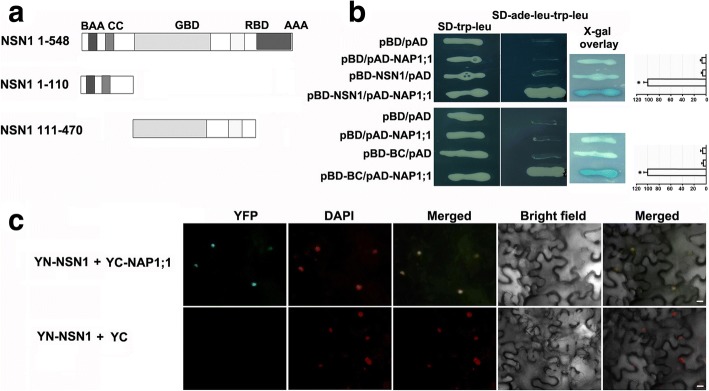
Like mammalian NS, Arabidopsis NSN1 interacts with several nucleolar proteins involved in ribosomal biogenesis [14]. To search for potential plant-specific protein(s) physically interacting with NSN1, a yeast two-hybrid screen was carried out. We screened a cDNA library of Arabidopsis flower from ABRC (stock CD4-30) constructed in pAD-GAL4-2.1. NSN1 cDNA was cloned into pBD-GAL4 (Strategy, USA) and served as bait. Empty vectors of pBD-GAL4 and pAD-GAL4-2.1 were used as negative controls for verification of candidate clones.
Around a million yeast colonies were screened and candidate colonies were verified to eliminate false positives. One colony showed consistent growth on the selective media, and the colony was identified as NAP1;1 (At4g26110.2), a member of the NUCLEOSOME ASSEMBLY PROTEINS1 (NAP1) [24]. Further analysis demonstrated that the yeast colony expressing the full length coding sequence (CDS) of AtNAP1;1 grew healthily on the selective media and turned blue on X-gal indicator plates (Fig. 6b), suggesting an interaction between the encoded protein and NSN1. For the control vectors, as expected, yeast hardly grew on the selective media with extremely faint blue in X-gal overlay assay (Fig. 6b upper panel). Quantification of the interaction strength revealed a significantly strong signal in the cells expressing both NSN1 and AtNAP1;1 (Fig. 6b upper panel), indicating that the presence of both NSN1 and AtNAP1;1 is required for the observed interaction. Interestingly, similar to NSN1, AtNAP1;1 has also been demonstrated to regulate cell proliferation and leaf development [24].
Fig. 6.
Interaction between NSN1 and NAP1;1 in yeast and tobacco. a Illustration of NSN1 fragments used for yeast two-hybrid assays. NSN1 protein consists of 548 amino acids. Two fragments tested in the yeast two-hybrid assay are the N-terminal fragment (amino acids 1–110) including the basic amino acid (BAA) domain and coiled-coil (CC) domain, and the fragment of amino acids 111–470 representing the GTP binding domain (GBD) and RNA binding domain (RBD). b Analysis of in vitro interaction between NAP1;1 and NSN1 with or without a domain deletion using the yeast two-hybrid system. The full length NSN1 was cloned into pBD vector as bait to screen a library of flower from ABRC (upper panel). A positive interaction colony was confirmed to contain AtNAP1;1 (At4g26110.2) by DNA sequencing. pAD-NAP1;1 represents the full length CDS of AtNAP1;1 cloned into pAD vector. The upper panel displays the interaction between AtNAP1;1 and NSN1; The lower panel shows the interactions between AtNAP1;1 and the N-terminal fragment of NSN1 including the basic amino acid (BAA) domain and coiled-coil (CC) domain, which was abbreviated as BC. Empty vectors pBD and pAD were used as negative controls. X-gal overlay assays and the quantification of ß-galactosidase activity were shown (right panels). Significant difference (p < 0.001) was evaluated by Student’s t test. c Analysis of in vivo interaction between NSN1 and NAP1;1 using biomolecule fluorescence complementation (BiFC) in tobacco leaves. An enhanced GFP (eGFP) was used [35] to test the interaction by fusing the coding sequence of NSN1 and AtNAP1;1 with the N-terminal (YFPN-NSN1) and the C-terminal fragment of GFP (YFPC-NAP1;1), respectively. The two constructs were co-expressed in tobacco leaves by Agrobacterium-infiltration. Infiltrated leaf epidermal cells were checked with confocal laser scanning. The co-expression of YFPN-NSN1 and no-fusion pSPYCE (M) 155 (YC) was used as a negative control. Nuclei were stained with DAPI. Scale bars: 10 μm
NSN1 contains the common conserved domains of nucleostemin, including the basic amino acid domain (BAA), coiled-coil domain (CC), GTP-binding domain (GBD), RNA binding domain (RBD) and acidic amino acid domain (AAA) (Fig. 6a). To investigate which domains of NSN1 were indispensable for the in vitro interaction with AtNAP1;1, two adjacent combined domains, including BC standing for the N-terminal BAA and CC domains, and GR for the GBD and RBD domains, were tested (Fig. 6a). Domain deletion analysis demonstrated that with the BC domains, yeast growth and strong blue signal were observed, while the controls comprising either of the tested peptides with an empty vector were negative (Fig. 6b), suggesting that the presence of the two N-terminal domains, BAA and CC, of NSN1 is essential for the interaction (Fig. 6b). No physical interaction was detected between AtNAP1;1 and the two middle domains of NSN1, e.g. GBD and RBD (data not shown). These results indicated that the N-terminal two conserved domains (BAA and CC), but not the middle two domains (RBD and AAA), were essential for the physical interaction between NSN1 and AtNAP1;1.
To test whether NSN1 and AtNAP1;1 interact in vivo, the BiFC assay, a widely used tool for visualization of protein-protein interaction in living cells, was performed. The coding sequences of NSN1 and AtNAP1;1 were fused in frame with the N-terminal and C-terminal fragments of enhanced GFP (eGFP), respectively [35]. The fusion proteins (YFPN-NSN1 and YFPC-NAP1;1) were co-expressed in tobacco leaves by Agrobacterium-infiltration. Leaf epidermal cells of infiltrated tobacco were checked with confocal laser scanning microscopy after 48 h. GFP fluorescence was observed in the nuclei of tobacco leaves transfected with YFPN-NSN1 and YFPC-NAP1;1 as indicated by DAPI-stained nuclei (Fig. 6c). This result is consistent with the previous studies that NSN1 was predominantly localized in nucleoli [14, 16]. In contrast, no YFP signal was detected when YFPN-NSN1 was co-expressed with no-fusion pSPYCE (M) 155 (YC), as the control, in tobacco cells (Fig. 6c). These results demonstrated that NSN1 interacted in vivo with AtNAP1;1, one of AtNAP1 proteins functioning as a histone chaperone in nucleosome assembly/disassembly.
Discussion
Plant Nucleostemin homologs have been identified in recent years from Arabidopsis and tobacco [14, 15]. It has been reported that Arabidopsis NSN1 plays critical roles in floral meristem development, floral organ identity, embryogenesis, leaf development and senescence [14–16]. The aim of this work was to functionally characterize NSN1 in cell cycle regulation in Arabidopsis. Based on the molecular and genetic analysis of the cell cycling components and the dynamics of cell proliferation in nsn1 mutant, we concluded that NSN1 is indispensable for correct cell proliferation control. The identification of AtNAP1;1, a histone chaperone, as an interacting partner of NSN1 implies that the two proteins might function in a nucleolar complex to regulate cell cycle progression.
Arabidopsis NSN1 is involved in plant growth and development by maintaining proper cell cycle progression. It has been documented that in animals when NS is deleted or overexpressed, cell cycling is arrested and the transition of G1-S and G2-M is stopped [3, 5, 6]. Given that the Arabidopsis loss-of-function mutant nsn1 exhibits dwarfism and NSN1 is localized primarily in the nucleolus where the key features of cell growth occur, NSN1 may affect plant cell cycle progression. This view is supported by the molecular evidence of a uniform down-regulation of core cell cycling genes, such as CYCA2;3, CYCB1;1, CYCD3;1, HISTONE H4 and RNR, in nsn1 (Fig. 4a). In addition to controlling cell-cycle transition, the core cell cycling components have been documented to function in coordinating cell division with differentiation and development in plants [36, 37]. The observations of higher percentages of polyploidy, severely reduced numbers of CYCLIN B1;1::GUS-expressing cells and slower cell proliferation in nsn1 suggest the deficiency in cell division. Consequently, growth retardation was exhibited in both leaves and roots of nsn1, and has also been observed in NbNSN1 silencing tobacco plants as well [14]. These results provided genetic and molecular evidence that like its homolog in tobacco, Arabidopsis NSN1 regulates cell cycling. Therefore, besides its function in embryogenesis and the development of leaf and flower organs [15, 16], the nuclear protein NSN1 is indispensable for maintaining proper cell proliferation in Arabidopsis.
It appears that nucleosome assembly protein AtNAP1;1 and NSN1 may function together as subunits of a functional protein entity. This view is supported by the facts that the two proteins share several common features, including involvement in cell cycle progression, nucleus-localization and the resemblance of their mutants in response to genotoxics. As conserved histone chaperones, plant NAP1s and NRPs have been documented in both monocot and dicot species [18, 19, 22, 23, 26, 38]. In Arabidopsis, for example, overexpression of AtNAP1;1 increases the expression of CYCB1;1 and shortenes G2 phase, thereby promoting cell division [24]. The double mutant nrp1-1 nrp2-1 exhibits arrested cell cycle progression at G2/M [38]. In tobacco, NAP1 genes have been observed with high expression levels at the G1/S transition [22]. Together with our findings that mutation of NSN1 caused defect of G1-S and G2-M transition, we concluded that both AtNAP1;1 and NSN1 play roles in the regulation of cell cycle progression.
Supportively, intracellular localization analysis of NSN1 and AtNAP1;1 has shown that both proteins reside in the nucleus [14, 15, 18]. The result that the N-terminal domain of NSN1 primarily determined its localization [14, 15] is in agreement with the findings of mammalian NS, whose nucleolar localization is specified by the N-terminal basic domain [1]. Based on our findings that the N-terminal domain of NSN1 was essential for its interplay with AtNAP1;1, we postulate that the two proteins interact in the nuclei. This postulation was supported by the BiFC experiment showing in vivo interaction between NSN1 and AtNAP1;1 in the nuclei of tobacco leaves. Interestingly, like NS in mammals [11, 13], AtNAP1;1 is localized in both the nucleus and the cytoplasm, and the protein appears to shuttle between the two organelles [18]. Studies of gain- or loss-of-function mutants have revealed that when localized to the nucleus, AtNAP1;1 promotes cell division during the early stage of leaf development [24]. The co-localization of NSN1 and AtNAP1;1 in the nucleus (Fig. 6) provides new evidence that both proteins may function as subunits of a complex in cell cycling regulation. Further investigation of the shuttling mechanism of the two proteins will provide valuable information for the functional characterization.
Additionally, the phenotypic features of Atnap1;1 are highly reminiscent of those of nsn1 upon subjected to UV or other DNA damage agents. AtNAP1;1 belongs to a multi-gene family. Mutations of AtNAP1s individually or simultaneously barely cause visible phenotypes under standard laboratory conditions [23, 26], indicating the dispensability of these genes for plant growth and development under normal conditions. When exposed to bleomysin or UV treatment, nsn1 and Atnap1 mutants including single, double, triple and quadruple ones, together with Atnrp1 and Atnrp2, exhibit hypersensitivity to the genotoxic agents [18, 26, 38]. The similar response of NSN1 and AtNAP1s to genotoxic treatment suggests their involvement in the DNA-damage repair process although the functional mechanism is unclear.
It has been reported that in mammals NSN1 forms a large protein complex with DDX21, EBNA1 binding protein 2 (EBP2), Pescadillo (PES), and a subset of ribosomal proteins [7]. In Arabidopsis, AtEBP2 and AtPES, the orthologs of EBP2 and PES, respectively, have also been found to interact with NSN1, especially its N-terminal domains (1–174 amino acids) [14], suggesting that NSN1 and its interacting partners regulate plant growth by modulating ribosome biogenesis. In search for potential NSN1-interacting partners involved in cell cycling regulation, AtNAP1;1 (At4g26110.2) was identified by the yeast two-hybrid system (Fig. 6). In Arabidopsis, there are four AtNAP1 family members (AtNAP1;1, AtNAP1;2, AtNAP1;3 and AtNAP1;4) and two NRP proteins (AtNRP1 and AtNRP2) [18, 38], clustering in two clades in the phylogenetic tree (Additional file 1: Figure S1a). In AtNAP1 subgroup, AtNAP1;1 shares a sequence identity of 80.3, 72.4 and 46.4% with AtNAP1;3, AtNAP1;2 and AtNAP1;4, respectively (Additional file 2: Table S1). The interaction of AtNAP1;1 with NSN1 might somehow attribute to the divergent C-terminus of AtNAP1;1 (Additional file 1: Figure S1b). Another possible reason why AtNAP1;1 was singled out from the screening might be that the coverage of the flower library used as pray in this experiment was not sufficient to contain low expression genes such as AtNAP1;3, whose expression level in flower is about half of AtNAP1;1 (Additional file 1: Figure S1c) according to the data from Arabidopsis eFP Browser (http://bar.utoronto.ca/efp_arabidopsis) [39]. Further investigation by deleting the domain(s) of AtNAP1;1 will provide information on the domain(s) of AtNAP1;1 that is required for interaction with NSN1. Future study will focus on the nsn1 Atnap1;1 double mutant to unveil the molecular functions of NSN1 and AtNAP1;1 in regulating plant growth and development as interacting partners.
Conclusions
The biological functions of Arabidopsis NSN1 in maintaining proper cell cycle progression were characterized by molecular and genetic approaches. Our results provide direct evidence that the dwarfism of nsn1 is resulted from the improper cell proliferation in the meristems of both shoot and roots of the mutant plant. As a positive cell cycle regulator, Arabidopsis NSN1 is co-localized and interacts with AtNAP1;1, a nucleosome assembly protein, in the nucleus. The two proteins are involved in regulation of cell cycle progression and their mutants, nsn1 and Atnap1;1, are hypersensitive to treatments of genotoxic agents. We propose that NSN1 and AtNAP1;1 act together as subunits of a functional protein complex in regulation of cell cycling progression. Hence, like its homologs in mammals, NSN1 in Arabidopsis functions conservatively in regulating plant cell cycling. This study sheds new light on the crosstalk between NSN1 and AtNAP1;1 in cell cycle regulation in plants.
Methods
Plant materials and growth conditions
The nsn1-1 T-DNA insertion line (SALK_029201) was obtained from the Arabidopsis Biological Resource Center (ABRC) [16], and the CYCLIN B1::GUS expressing plant was a gift from Prof. Peter Doerner (University of Edinburg, UK). Plants were grown in soil under normal conditions with 16 h light/8 h darkness at 21 °C.
Plant treatment
Sterilized Arabidopsis (Col-0 and nsn1) seeds were treated at 4 °C for 2 days before moving to normal conditions for germination on half strength MS plates. Plates were placed vertically during seed germination and 4 days after germination, seedlings were transferred to half strength MS plates (control) or half strengh MS plates supplemented with methyl methanesulfonate (MMS) at concentrations of 25, 50, 75 and 100 ppm, or with bleomycin at 3, 6, 9, and 12 mg/L. Images were captured and survival rates were recorded after genotoxic treatment with MMS and bleomycin for one and 3 weeks, respectively.
Constructs and Agrobacterium-mediated transient expression in N. benthamiana
For the biomolecule fluoresence complementation (BiFC) experiment, the coding region of NSN1 cDNA was amplified by RT-PCR using primers pBD-NSN F: 5′-GTC GAC AGA TGG TGA AAC GGA GTA AAA AGA G-3′ and pBD-NSN R: 5′-GTC GAC TTT TTC TTC GGC AAA AGT CCA G-3′, and cloned into vector pCR2.1 (Invitrogen). To express YFPN-NSN1, the NSN1 cDNA fragment was ligated into vector pSPYNE (R) 173 [35] after digestion by Sal I. For construction of NAP1;1-cYFP, the NAP1;1 (At4g26110.2) coding region was amplified by RT-PCR using primers pNAP1;1 F: 5′-GAA TTC ATG AGC AAC GAC AAG GAT AGC -3′ and pNAP1;1 R: 5′- GTC GAC ACA AAT AAA CTT TAG TTC TGA AAG G-3′. The PCR product was cloned into vector pCR2.1, and then sub-cloned into pSPYCE (M) 155.
For infiltration of tobacco leaf, the two constructs were transformed into Agrobacterium tumefaciens GV3101 and infiltration was carried out as described [35].
Scanning electronic microscopy (SEM) and microscopy
SEM was carried out as described [15]. Briefly, plant tissues were fixed overnight in 50 mM phosphate buffer containing 2% glutaraldehyde and 2% paraformaldehyde. Fixed tissues were rinsed three times with 50 mM phosphate buffer and kept in osmium tetroxide (1%) at 4 °C overnight. After being rinsed three times with 50 mM phosphate buffer, the tissues were dehydrated in a 30, 50, 70, 95 and 100% alcohol gradient. Dehydrated tissues were coated with gold and observed under SEM.
Leaf initiation was defined as described [40]. For measurement of leaf area, the dissected fifth leaf was pictured daily and leaf area was measured using ImageJ. Cell number per leaf was calculated as leaf area divided by cell area. All dissected leaves were treated with chloral hydrate for 3 days. After clearing, photos of palisade cells were taken using Zeiss microscope with DIC lens. For individual leaf, area of 10 palisade cells was measured using ImageJ and an average area per palisade cell was calculated accordingly. For each time point, the fifth leaf from 5 plants was measured and three biological replicates were conducted.
For root length, 70 sterilized Arabidopsis seeds of wild type and nsn1 were germinated on 1/2 MS medium with 0.8% agar. Root tip was marked for daily image capture and root length was measured by ImageJ. For analysing the cell production per day, 4 days after germination, ten primary root tips grown during the last 24 h were cut and mounted daily on slide using buffer (chloral hydrate: glycerol: water = 8:3:1). DIC optics of a Zeiss confocal microscope (Zeiss Axioskop, Germany) was used for image capture. The number of cortex cells on one side of a root tip was counted.
Quantitative RT-PCR
Total RNA was isolated from 10-day-old seedlings of wild type and nsn1 using the RNeasy plant kit followed by treatment with RNase-free DNase I according to the manufacturer instructions (Qiagen, Germantown, MD). RNA of 2 μg was used as template for first strand cDNA synthesis using SuperScrit III (Invitrogen). cDNA was diluted 100 times and 5 μl of cDNA was used as PCR template. Real-time PCR was performed using a mix containing 10 μl of 2× SYBR Premix Ex Taq (Takara Bio Inc., Otsu, Shiga, Japan), 0.8 μl of forward and reverse primer mix (0.2 μM final concentration), 0.4 μl of 50 ROX Reference Dye II, and 3.8 μl of deionized water. PCR was run on an ABI 7500 fast real time PCR system (Applied Biosystems) using a 2-min initial denaturation at 95 °C, followed by 40 cycles of 95 °C (15 s) and 60 °C (40 s). Primers used for qRT-PCR were as follows: for CYCA2;3 (F: 5′-GGC TAA GAA GCG ACC TGA TG-3′ and R: 5′-TAC AAG CCA CAC CAA GCA AC-3′); for CYCB1;1 (F: 5′-AAG CTT CCA TTG CAG ACG A-3′ and R: 5′-AGC AGA TTC AGT TCC GGT C-3′); for CYCD3;1 (F: 5′-ACA ACT CTC GTG CAT TAA CAG GAA-3′ and R: 5′-GAA GAT TGG ATT TGG ATC TGT AAA C-3′); for H4 (F: 5′-TTA GGC AAA GGA GGA GCA AA-3′ and R: 5′-CTC CTC GCA TGC TCA GTG TA-3′); and for RNR (F: 5′- CAA GTG GCT CAG GAC TGT CA-3′ and R: 5′-TCC ATC AGG TCA ACA GCT TG-3′). Actin2 (At3g18780) was used as an internal control (F: 5′-TGG TGT CAT GGT TGG GAT G-3′ and R: 5′-CAC CAC TGA GCA CAA TGT TAC-3′). The relative expression level was calculated based on the value of ΔΔCt.
Flow cytometry
Flow cytometry was carried out as described [41] with minor modifications. Briefly, leaves (4-days after initiation or fully-expended leaves) were chopped with razor blade into fine strips in cold nuclei isolation buffer (Partec, Müster, Germany). After filtration, the extracts were kept on ice until measurement. The DNA content of nuclei was measured using FACS Caliber flow cytometry (BD Biosciences, USA). Cell nuclei were stained with 2 μg mL− 1 DAPI. Each sample was prepared three times and subjected to FACS Caliber cytometry independently. A total of around 10,000 nuclei were measured per analysis.
Yeast two-hybrid assay
Yeast two-hybrid experiments were carried out as described (Stratagene, USA). ABRC stock CD4-30, an Arabidopsis cDNA library of inflorescence meristem, floral meristem and floral buds up to stage 8, was used as pray. The cDNA library was cloned into the EcoR I – Xho I site of pAD-Gal4-2.1 (Stratagene), and transformed into yeast strain PJ69-4a (his, leu, ura; Gal1-HIS3, Gal2-ADE2, Gal7-LacZ). The full length NSN1 coding region and the fragment corresponding to the BC domain were amplified using primers pBD-NSN (F: 5′-GTC GAC AGA TGG TGA AAC GGA GTA AAA AGA G-3′; R: 5′-GTC GAC TTT TTC TTC GGC AAA AGT CCA G -3′) and pBD-BC (F: 5′-GTC GAC AGA TGG TGA AAC GGA GTA AAA AGA G-3′; R: 5′-GTC GAC CTC TTC ATG CTT ATT GGG ACC GGC-3′), respectively. The two fragments were cloned into pBD-GAL4 (Cam) and used as bait after transformation into yeast strain PJ69-4α (his, leu, ura; Gal1-HIS3, Gal2-ADE2, Gal7-LacZ). The full-length CDS of AtNAP1;1 (At4g26110.2) cloned into pAD-Gal4-2.1 was amplified with primers (pNAP1;1 F: 5′-GAA TTC ATG AGC AAC GAC AAG GAT AGC-3′; pAD-NAP1;1 R: 5′-CTC GAG AAT AAA CTT TAG TTC TGA AAG -3′). Cells of the two yeast strains were mated and selected on a plate supplemented with Glu-Trp-Leu-Ade. Yeast colonies were further selected for growth on an Ade Selection Plate (Glc-Ade-Trp-Leu + His). For X-gal overlay assay, agarose solution containing 1 mg/ml 5-bromo-4-chloro-3-indoly-galactopyranoside (X-Gal) in Z buffer was brought to 55 oC and poured to the cooled yeast colonies grown on the Ade selection plates.
Quantification of Y2H ß-Galactosidase assay
ß-Galactosidase activity was measured using Y2H ß-Galactosidase Kit (Molecular Biotechnology). Briefly, several yeast colonies picked from selection plate were inoculated into 1 ml of the appropriate selective medium and cultured to the late exponential phase. Yeast cells were collected and resuspended in lysis mixture containing Dye solution. Color development was monitored by spectrophotometer at 615 nm. Three independent biological replicates were performed and difference was evaluated by Student’s t test.
Additional files
Figure S1. Sequence analysis and expression profiles of NAP1 family members. a. Homology analysis of AtNAP1 and AtNRP proteins using DNAMAN version 7. Sequence accession number: AtNAP1;1 (AT4G26110.2); AtNAP1;2 (AT2G19480); AtNAP1;3 (AT5G56950); AtNAP1;4 (AT3G13782); AtNRP1 (AT1G74560) and AtNRP2 (AT1G18800). b. Sequence alignment of AtNAP1s and AtNRPs. Black represents conserved amino acids (consensus), pink for 75% identity, blue for 50% and yellow for 33% identity. c. Comparison of the transcriptional expression pattern of AtNAP1 paralog genes in flower from Arabidopsis eFP Browser (http://bar.utoronto.ca/efp_arabidopsis). (JPG 3822 kb)
Table S1. Analysis of protein identity among AtNRP1s and AtNRPs. (PDF 98 kb)
Acknowledgements
We thank Prof. Peter Doerner (University of Edinburg, UK) for providing the seeds of CYCLIN B1::GUS expressing plant and Prof. Changbin Chen (University of Minnesota, Saint Paul, USA) for critically reading the manuscript.
Funding
This work was financially supported by NSF grants (MCB 0548525 to ZH) and by the Agricultural Science and Technology Innovation Program from China (ASTIP-IAS14 to QY). The funding bodies did not play a role in the design of the study and collection, analysis, or interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on request.
Abbreviations
- DAG
Days after germination
- DAI
Days after initiation
- GUS
β-glucuronidase
- MDM2
Murine Double Minute 2
- NAP1
NUCLEOSOME ASSEMBLY PROTEINS1
- NRPs
NAP1-RELATED PROTEINs
- NS
Nucleostemin
- NSN
Nucleostemin-like
Authors’ contributions
ZH, XW, ZW and QCY designed the research; XW, ZW and BX performed experiments, and XW and ZW wrote the article. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1289-2) contains supplementary material, which is available to authorized users.
Zhen Wang and Xiaomin Wang contributed equally to this work.
Contributor Information
Zhen Wang, Email: wangzhen@caas.cn.
Xiaomin Wang, Email: wxm7516@gmail.com.
Bo Xie, Email: xiebo@mail.ccnu.edu.cn.
Zonglie Hong, Email: zhong@uidaho.edu.
Qingchuan Yang, Email: qchyang66@163.com.
References
- 1.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16(23):2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Q, Yasumoto H, Tsai RY. Nucleostemin delays cellular senescence and negatively regulates TRF1 protein stability. Mol Cell Biol. 2006;26(24):9279–9290. doi: 10.1128/MCB.00724-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng L, Lin T, Tsai RY. Nucleoplasmic mobilization of nucleostemin stabilizes MDM2 and promotes G2-M progression and cell survival. J Cell Sci. 2008;121(Pt 24):4037–4046. doi: 10.1242/jcs.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng L, Yasumoto H, Tsai RY. Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J Cell Sci. 2006;119(Pt 24):5124–5136. doi: 10.1242/jcs.03292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol. 2008;28(13):4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma H, Pederson T. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol Biol Cell. 2007;18(7):2630–2635. doi: 10.1091/mbc.e07-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romanova L, Grand A, Zhang L, Rayner S, Katoku-Kikyo N, Kellner S, Kikyo N. Critical role of nucleostemin in pre-rRNA processing. J Biol Chem. 2009;284(8):4968–4977. doi: 10.1074/jbc.M804594200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 9.Britton RA. Role of GTPases in bacterial ribosome assembly. Annu Rev Microbiol. 2009;63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- 10.Meng L, Zhu Q, Tsai RY. Nucleolar trafficking of nucleostemin family proteins: common versus protein-specific mechanisms. Mol Cell Biol. 2007;27(24):8670–8682. doi: 10.1128/MCB.00635-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168(2):179–184. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Politz JC, Polena I, Trask I, Bazett-Jones DP, Pederson T. A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol Biol Cell. 2005;16(7):3401–3410. doi: 10.1091/mbc.e05-02-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Q, Meng L, Hsu JK, Lin T, Teishima J, Tsai RY. GNL3L stabilizes the TRF1 complex and promotes mitotic transition. J Cell Biol. 2009;185(5):827–839. doi: 10.1083/jcb.200812121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeon Y, Park YJ, Cho HK, Jung HJ, Ahn TK, Kang H, Pai HS. The nucleolar GTPase nucleostemin-like 1 plays a role in plant growth and senescence by modulating ribosome biogenesis. J Exp Bot. 2015;66(20):6297–6310. doi: 10.1093/jxb/erv337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Gingrich DK, Deng Y, Hong Z. A nucleostemin-like GTPase required for normal apical and floral meristem development in Arabidopsis. Mol Biol Cell. 2012;23(8):1446–1456. doi: 10.1091/mbc.e11-09-0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Xie B, Zhu M, Zhang Z, Hong Z. Nucleostemin-like 1 is required for embryogenesis and leaf development in Arabidopsis. Plant Mol Biol. 2012;78(1–2):31–44. doi: 10.1007/s11103-011-9840-7. [DOI] [PubMed] [Google Scholar]
- 17.Ohkuni K, Shirahige K, Kikuchi A. Genome-wide expression analysis of NAP1 in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2003;306(1):5–9. doi: 10.1016/S0006-291X(03)00907-0. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Zhu Y, Gao J, Yu F, Dong A, Shen WH. Molecular and reverse genetic characterization of NUCLEOSOME ASSEMBLY PROTEIN1 (NAP1) genes unravels their function in transcription and nucleotide excision repair in Arabidopsis thaliana. Plant J. 2009;59(1):27–38. doi: 10.1111/j.1365-313X.2009.03844.x. [DOI] [PubMed] [Google Scholar]
- 19.Yoon HW, Kim MC, Lee SY, Hwang I, Bahk JD, Hong JC, Ishimi Y, Cho MJ. Molecular cloning and functional characterization of a cDNA encoding nucleosome assembly protein 1 (NAP-1) from soybean. Mol Gen Genet. 1995;249(5):465–473. doi: 10.1007/BF00290572. [DOI] [PubMed] [Google Scholar]
- 20.Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci U S A. 2006;103(5):1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lankenau S, Barnickel T, Marhold J, Lyko F, Mechler BM, Lankenau DH. Knockout targeting of the Drosophila nap1 gene and examination of DNA repair tracts in the recombination products. Genetics. 2003;163(2):611–623. doi: 10.1093/genetics/163.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong A, Zhu Y, Yu Y, Cao K, Sun C, Shen WH. Regulation of biosynthesis and intracellular localization of rice and tobacco homologues of nucleosome assembly protein 1. Planta. 2003;216(4):561–570. doi: 10.1007/s00425-002-0910-6. [DOI] [PubMed] [Google Scholar]
- 23.Liu ZQ, Gao J, Dong AW, Shen WH. A truncated Arabidopsis NUCLEOSOME ASSEMBLY PROTEIN 1, AtNAP1;3T, alters plant growth responses to abscisic acid and salt in the Atnap1;3-2 mutant. Mol Plant. 2009;2(4):688–699. doi: 10.1093/mp/ssp026. [DOI] [PubMed] [Google Scholar]
- 24.Galichet A, Gruissem W. Developmentally controlled farnesylation modulates AtNAP1;1 function in cell proliferation and cell expansion during Arabidopsis leaf development. Plant Physiol. 2006;142(4):1412–1426. doi: 10.1104/pp.106.088344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Son O, Kim S, Shin YJ, Kim WY, Koh HJ, Cheon CI. Identification of nucleosome assembly protein 1 (NAP1) as an interacting partner of plant ribosomal protein S6 (RPS6) and a positive regulator of rDNA transcription. Biochem Biophys Res Commun. 2015;465(2):200–205. doi: 10.1016/j.bbrc.2015.07.150. [DOI] [PubMed] [Google Scholar]
- 26.Zhou W, Gao J, Ma J, Cao L, Zhang C, Zhu Y, Dong A, Shen WH. Distinct roles of the histone chaperones NAP1 and NRP and the chromatin-remodeling factor INO80 in somatic homologous recombination in Arabidopsis thaliana. Plant J. 2016;88(3):397–410. doi: 10.1111/tpj.13256. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg GR, Hilfinger JM. Regulation of synthesis of ribonucleotide reductase and relationship to DNA replication in various systems. Prog Nucleic Acid Res Mol Biol. 1996;53:345–395. doi: 10.1016/S0079-6603(08)60150-6. [DOI] [PubMed] [Google Scholar]
- 28.Cockcroft CE, den Boer BG, Healy JM, Murray JA. Cyclin D control of growth rate in plants. Nature. 2000;405(6786):575–579. doi: 10.1038/35014621. [DOI] [PubMed] [Google Scholar]
- 29.Menges M, de Jager SM, Gruissem W, Murray JA. Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J. 2005;41(4):546–566. doi: 10.1111/j.1365-313X.2004.02319.x. [DOI] [PubMed] [Google Scholar]
- 30.Doerner P, Jorgensen JE, You R, Steppuhn J, Lamb C. Control of root growth and development by cyclin expression. Nature. 1996;380(6574):520–523. doi: 10.1038/380520a0. [DOI] [PubMed] [Google Scholar]
- 31.Colon-Carmona A, You R, Haimovitch-Gal T, Doerner P. Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 1999;20(4):503–508. doi: 10.1046/j.1365-313x.1999.00620.x. [DOI] [PubMed] [Google Scholar]
- 32.Criqui MC, Weingartner M, Capron A, Parmentier Y, Shen WH, Heberle-Bors E, Bogre L, Genschik P. Sub-cellular localisation of GFP-tagged tobacco mitotic cyclins during the cell cycle and after spindle checkpoint activation. Plant J. 2001;28(5):569–581. doi: 10.1046/j.1365-313X.2001.01180.x. [DOI] [PubMed] [Google Scholar]
- 33.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412(6846):553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 34.Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006;48(6):947–961. doi: 10.1111/j.1365-313X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- 35.Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 2008;56(3):505–516. doi: 10.1111/j.1365-313X.2008.03612.x. [DOI] [PubMed] [Google Scholar]
- 36.Inze D, De Veylder L. Cell cycle regulation in plant development. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 37.Gutierrez C. Coupling cell proliferation and development in plants. Nat Cell Biol. 2005;7(6):535–541. doi: 10.1038/ncb0605-535. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Dong A, Meyer D, Pichon O, Renou JP, Cao K, Shen WH. Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell. 2006;18(11):2879–2892. doi: 10.1105/tpc.106.046490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37(5):501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 40.Granier C, Massonnet C, Turc O, Muller B, Chenu K, Tardieu F. Individual leaf development in Arabidopsis thaliana: a stable thermal-time-based programme. Ann Bot. 2002;89(5):595–604. doi: 10.1093/aob/mcf085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220(4601):1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- 42.Marchant A, Kargul J, May ST, Muller P, Delbarre A, Perrot-Rechenmann C, Bennett MJ. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999;18(8):2066–2073. doi: 10.1093/emboj/18.8.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence analysis and expression profiles of NAP1 family members. a. Homology analysis of AtNAP1 and AtNRP proteins using DNAMAN version 7. Sequence accession number: AtNAP1;1 (AT4G26110.2); AtNAP1;2 (AT2G19480); AtNAP1;3 (AT5G56950); AtNAP1;4 (AT3G13782); AtNRP1 (AT1G74560) and AtNRP2 (AT1G18800). b. Sequence alignment of AtNAP1s and AtNRPs. Black represents conserved amino acids (consensus), pink for 75% identity, blue for 50% and yellow for 33% identity. c. Comparison of the transcriptional expression pattern of AtNAP1 paralog genes in flower from Arabidopsis eFP Browser (http://bar.utoronto.ca/efp_arabidopsis). (JPG 3822 kb)
Table S1. Analysis of protein identity among AtNRP1s and AtNRPs. (PDF 98 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on request.