Abstract
Background
Gibberellin (GA) treatments can induce parthenocarpy in the main crop of San Pedro-type figs, the native non-parthenocarpic fruit, however, the underlying mechanism is still largely unclear.
Results
In our study, GA3 was applied to San Pedro-type fig main crop at anthesis. Sharply increased GA3 content was detected in both female flowers and receptacle, along with significantly decreased indole-3-acetic acid (IAA), zeatin and abscisic acid (ABA) levels in female flowers, and increased zeatin peak intensity and earlier ABA peak in receptacles. Transcriptome comparison between control and treatment groups identified more differentially expressed genes (DEGs) in receptacles than in female flowers 2 and 4 days after treatment (DAT); 10 DAT, the number of DEGs became similar in the two tissues. Synchronized changing trends of phytohormone-associated DEGs were observed in female flowers and receptacles with fruit development. Modulation of ethylene and GA signaling and auxin metabolism by exogenous GA3 occurred mainly 2 DAT, whereas changes in auxin, cytokinin and ABA signaling occurred mainly 10 DAT. Auxin-, ethylene- and ABA-metabolism and response pathways were largely regulated in the two tissues, mostly 2 and 10 DAT. The major components altering fig phytohormone metabolic and response patterns included downregulated GA2ox, BAS1, NCED and ACO, and upregulated ABA 8′-h and AUX/IAA.
Conclusions
Thus GA-induced parthenocarpy in fig is co-modulated by the female flowers and receptacle, and repression of ABA and ethylene biosynthesis and GA catabolism might be the main forces deflecting abscission and producing fig parthenocarpy.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1318-1) contains supplementary material, which is available to authorized users.
Keywords: Ficus carica, Gibberellin treatment, Parthenocarpy, Transcriptome analysis, Plant hormone
Background
Gibberellins (GAs) are involved in almost every aspect of plant growth and development. In plant reproductive processes, GAs regulate floral initiation [1], fruit set and growth [2], and seed maturation and germination [3, 4]. Among the abundant GAs identified to date, only GA1, GA3, GA4 and GA7 have been recognized as bioactive [5], and only GA3, GA4 and/or GA7 have been successfully applied for parthenocarpic fruit induction [6].
Parthenocarpy, a highly valuable agronomic characteristic, especially in edible fruit crops, means that fruit set and development can be uncoupled from pollination and fertilization and thus the fruit is seedless, such as in fig, banana, persimmon, pineapple and pear [6]. Parthenocarpic fruit can also be triggered by application of exogenous plant hormones, termed artificial parthenocarpy. Auxin and GAs are the most frequently used hormones in parthenocarpic fruit induction [7, 8]. Other plant hormones, including cytokinin [9], ethylene [10] and abscisic acid (ABA) [11], also regulate or participate in parthenocarpic fruit set and growth.
Parthenocarpy requires the unique and coordinated action of different phytohormones, whereas non-parthenocarpic fruit will senescence and abscise if fertilization is not achieved. Auxin, GA and cytokinin are generally recognized as the major regulators of fruit set. Elevated levels of these hormones in tomato ovaries have been associated with fruit set and early fruit growth [12]. Auxin and GA, and the crosstalk between them, largely modulate pollination-dependent and parthenocarpic fruit set in tomato [13]. Transcriptome comparisons between pollinated and parthenocarpic cucumber fruit have shown crosstalk of auxin, GA and cytokinin at the transcriptional level during parthenocarpic fruit set [14]. Exogenous GA3 application significantly modulated the expression patterns of plant hormone metabolism and signaling genes in seedless grape, as determined by transcriptome analysis [15]. Previous studies in several plants have demonstrated that parthenocarpy induced by auxin and cytokinin requires downstream GA biosynthesis. Application of exogenous auxin and cytokinin in grape increased the expression of GA-biosynthesis genes, such as VvGA3ox1, and repressed GA-catabolism genes, such as VvGA2ox3 and VvGA2ox4, suggesting that auxin- and cytokinin-induced parthenocarpic fruit set requires enhanced GA biosynthesis [9]. Parthenocarpy induction in tomato by cytokinin depends in part on modulation of GA and indole-3-acetic acid (IAA) metabolism, as reflected by the upregulated expression levels of GA-biosynthesis genes such as GPS, GA20ox and GA3ox, and the IAA-biosynthesis gene ToFZY, and downregulation of the GA-inactivation gene GA2ox [16]. Auxin-induced parthenocarpic tomato fruit set is partially mediated by GAs, as indicated by the dependence of parthenocarpic fruit formation in the unfertilized entire tomato mutant with a defective SlIAA9 gene on regulation of GA metabolism [17]. SlIAA9, a member of the Aux/IAA family, is an auxin-signaling repressor that prevents tomato ovary development [18]. Moreover, fruit set in tomato induced by auxin analogue 2,4-D was significantly suppressed by GA-biosynthesis inhibitors [7]. Similar to the situation in Arabidopsis and tomato, auxin is the major inducer of fruit set in Capsicum annuum, depending in part on enhanced GA biosynthesis [19]. Ethylene is also considered to be a key regulator in coordinating fruit set, and ethylene reduces fruit set by suppressing GA metabolism [10].
Ficus carica L. (Moraceae) bears urn-shaped fruit with an enclosed inflorescence structure termed syconium. There are two major sex types in fig trees: caprifig (male fig) and female fig; the latter is further classified into three types, common, Smyrna and San Pedro, based on cropping characteristics [20]. Parthenocarpic or pollinated non-parthenocarpic fig fruit show a typical double-sigmoid growth curve, with two rapid fruit-size-increment phases, i.e., stages I and III, separated by a lag phase (stage II) [21]. Every year, San Pedro fig produces a parthenocarpic first crop called breba, and a non-parthenocarpic main crop which constitutes the main yield [20]. Studies on non-parthenocarpic Smyrna fig cv. Calimyrna revealed that parthenocarpic fruit set can be induced through auxin, GA and cytokinin treatments [22, 23]. Analysis of auxin and GA content in the two crops of San Pedro fig cv. King demonstrated higher auxin content in the parthenocarpic breba than the pollinated non-parthenocarpic main crop, whereas GA was higher in the latter than in the former [24]. Our previous studies revealed that compared with San Pedro parthenocarpic breba, expression of GA- and auxin-biosynthesis genes is repressed in its non-parthenocarpic main crop female flowers, whereas ABA- and ethylene-biosynthesis genes are enhanced [25]. Though exogenous hormone treatment can induce fruit set and development of non-parthenocarpic fig, our understanding of the underlying mechanisms is limited.
In this study, the effect of exogenous GA3 treatment on the main crop of San Pedro fig was studied, and hormone contents of the female flowers and receptacle were assayed. A corresponding global transcriptome comparison demonstrated a significant number of gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with spatiotemporal regulation characteristics. The expression patterns of annotated plant hormone metabolism and signal-transduction genes were analyzed in depth. GA treatment led mainly to the regulation of auxin, ethylene and ABA biosynthesis and signaling in fig. Differential expression of plant hormone genes revealed synchronized change trends with fruit development between female flowers and receptacles following GA treatment. Our work provides new information on GA-induced fruit set in San Pedro-type fig by unraveling key pathways and genes in the expression-regulation network, which could help elucidate the molecular mechanisms underlying the diversified parthenocarpic nature of fig species.
Methods
Plant materials and exogenous GA treatment
A San Pedro-type fig cultivar (Ficus carica L. ‘Asteran’) was used in this study. The trees were planted in a commercial orchard in Beijing with regular management. Main crop fruits at pre-anthesis (15–20 mm), anthesis (20–25 mm) and post-anthesis (> 25 mm) stages were selected to evaluate the time window of GA treatment; 200 μl of 25 mg/l GA3 solution was injected through the ostiole into the fruit cavity with a 1-ml syringe. Solution without GA3 was injected into the control group. For each treatment and control group, 100 fruits were injected and fruit-set rates were calculated.
Growth curves of control and GA3-treated fruit were established with 30 tagged fruits using a digital slide caliper. Three biological replicates were collected from pools of at least 30 fruits 2, 4 and 10 days after treatment (DAT). For transcriptome sequencing and plant hormone analysis, main crop fruits at anthesis, 22 ± 1 mm in transverse diameter, were used. Female flowers and receptacle were carefully separated, frozen in liquid nitrogen on site, pulverized in the lab, and stored at − 80 °C for further use.
Phytohormone content assays
The levels of IAA, GA3, GA4, zeatin and ABA were monitored by reverse-phase HPLC–MS/MS [26]. In brief, about 50 mg of the frozen and pulverized plant material was placed in 500 μl extraction solvent (isopropanol:H2O:concentrated HCl, 2:1:0.002, v/v), then shaken (100 rpm) at 4 °C for 30 min. Dichloromethane (1 ml) was added, followed by shaking (100 rpm) at 4 °C for 30 min. After centrifugation (10,000 × g) at 4 °C for 5 min, the solution was divided into two phases, and the lower phase (approximately 1 ml) was collected. The solvent mixture was concentrated to dryness using a Termovap sample concentrator (N-EVAP, Organomation, USA), then redissolved in 100 μl methanol. The extracted hormone solutions were run in an AB SCIEX (USA) Triple Quad™ 5500 LC–MS/MS system. For each sample, three independent and parallel extractions were carried out. Presented values are means of all replicates ± SD. Significance analysis was performed by SPSS 17.0.
RNA extraction
Total RNA from female flowers and receptacles taken 2, 4 and 10 DAT and the corresponding controls was isolated using the CTAB method [27] and treated with RNase-free DNase I (TaKaRa, China) according to the manufacturer’s instructions. RNA quality and quantity were assessed by Nanodrop 2000 (Thermo Scientific, USA) and electrophoresis in a 1% agarose gel. RNA integrity number, analyzed by Agilent 2100 Bioanalyzer, was > 8.0.
Library preparation and paired-end transcriptome sequencing
The mRNA of each sample’s three biological replicates were respectively enriched using cellulose oligo (dT) magnetic beads (Invitrogen, USA), then fragmented into ca. 200-bp fragments. The fragments were transcribed and double-stranded cDNA was synthesized, then end repair, 3′-end single-nucleotide A (adenine) addition and ligation of adaptors were performed according to the manufacturer’s instructions. The resultant fragments were enriched by PCR and purified using 2% Certified Low Range Ultra Agarose (Bio-Rad) to create the final cDNA library. The quantitative assay was conducted using Picogreen fluorescent dye (Invitrogen, USA) with a TBS-380 fluorimeter (Invitrogen, USA). After bridge PCR amplification on Illumina cBot using Truseq PE Cluster Kit v3-cBot-HS, paired-end (2 × 151 bp) sequencing of the cDNA library products was carried out using the Illumina HiSeq4000 platform.
Illumina read processing and functional annotation
Clean reads were generated by removing reads with adaptors and more than 10% unknown nucleotides, and low-quality reads (the rate of reads with quality value ≤10 was more than 20%) from the raw data. Then clean reads were mapped to our reported reference sequences deposited at DDBJ/EMBL/GenBank under the accession number GDKC00000000 [25] using Bowtiealigner (http://bowtie-bio.sourceforge.net/index.shtml) [28] with no more than two mismatches allowed. The number of mapped clean reads for each transcript was normalized to FPKM (fragments per kilobase of exon model per million mapped reads) by RSEM (http://deweylab.biostat.wisc.edu/rsem/). Genes that were differentially expressed between GA3-treated samples and their corresponding controls were analyzed by edgeR (http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html) [29], and “false discovery rate (FDR) < 0.01 and absolute value of log2 (FPKMtreatment/FPKMcontrol) ≥ 1” were set as the thresholds to determine significant differences in gene expression. For GO annotation of all transcripts (http://www.geneontology.org/), Goatools (https://github.com/tanghaibao/Goatools) was used. Pathway-enrichment analysis was carried out based on the KEGG pathway database by KOBAS (http://kobas.cbi.pku.edu.cn/) [30]. Both GO and KEGG enrichment analyses were based on Fisher’s Exact Test [31] with multiple testing correction of FDR [32], and corrected P-value ≤0.05 was selected as the threshold for significance.
Quantitative real-time PCR (qRT-PCR) verification
Thirteen phytohormone-related genes were selected for validation by qRT-PCR and the specific primers are shown in Additional file 1: Table S1. Isolation of total RNA was as described previously and RNase-free DNase I (D2270A, TaKaRa) treatment was performed to digest DNA in the sample. For the first-strand cDNA synthesis, 1000 ng of total RNA was reverse-transcribed with a synthesis kit (D6210A, TaKaRa). Ten-fold-diluted cDNA template was used for the qRT-PCR assay performed by the SYBR Premix Ex Taq Kit (DRR420A, TaKaRa) on a 7500 Real-Time PCR System (Applied Biosystems, USA) according to the manufacturer’s protocol. Relative quantitative method (2−ΔΔCt) was used to calculate the level of expression of the tested genes [33]. All of the reactions were performed in three replicates using actin as the internal gene [34].
Results
Developmental characteristics and endogenous hormone changes upon GA3 treatment
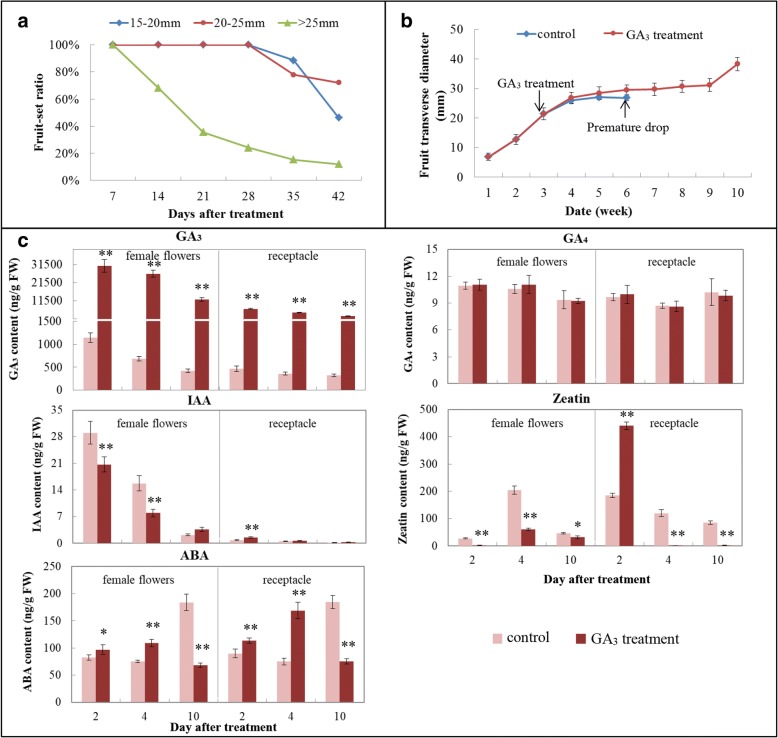
To optimize the GA3 treatment time window at stage I, fruits at pre-anthesis (15–20 mm in diameter), anthesis (20–25 mm) and post-anthesis (> 25 mm) stages were grouped and treated with GA3. GA3 treatment at anthesis gave a fruit-set rate of about 72% 42 DAT, much higher than the pre- and post-anthesis stage treatments (40 and 10%, respectively) (Fig. 1a). Anthesis-stage fruit with transverse diameter of 22 ± 1 mm were therefore selected for further plant hormone and transcriptome experiments. The growth curve of GA3-induced parthenocarpic fruit is shown in Fig. 1b; the typical double-sigmoid curve pattern was in agreement with previous reports [35]. However, fruit abscission was happened in control group at the beginning of stage II, which was called “premature drop”.
Fig. 1.
Physiological features of San Pedro-type fig main crop fruit after gibberellin (GA3) application. a Effect of application time window on fruit-set ratio. b Growth curves of control and GA3-treated fruits (GA3 was applied at anthesis). c GA3, GA4, indole acetic acid (IAA), zeatin and ABA concentration in control and GA3-treated female flowers and receptacle. Error bars indicate standard deviation. Significance analysis was performed by SPSS 17.0. *P ≤ 0.05, **P ≤ 0.01; FW, fresh weight
Exogenous GA3 treatment directly increased GA concentration in the female flowers, yielding about 26.8-, 38.8- and 28.7-fold its amount in controls 2, 4 and 10 DAT, respectively (Fig. 1c). Exogenous GA3 was also immediately transported into the receptacle, with 15-fold control GA3 levels in the treatment group 2 DAT. Decreasing trends of GA3 content were seen from 2 to 10 DAT in both control and GA3-treated female flowers and receptacles, indicating the fig tissues’ strong capacity to deplete this hormone (Fig. 1c). GA4 was present in fig tissues at a low and stable level, and was insensitive to GA3 treatment. The declining phase of native IAA in stage I female flowers was greatly accelerated by GA3 treatment 2 and 4 DAT, but 10 DAT, IAA degradation ceased in the treated flowers and its concentration accumulated to almost 2-fold that in the controls. At this stage, the receptacle contained much less IAA than the female flowers, and GA3 treatment only slightly changed its level 2 DAT (Fig. 1c); thereafter, IAA decreased to very low levels in both control and treated receptacles. Exogenous GA3 treatment dramatically decreased zeatin peak intensity in the female flowers 4 DAT, whereas it specifically enhanced the zeatin peak to 2.4-fold that in control receptacles 2 DAT (Fig. 1c); this action was opposite that in female flowers. ABA content was higher in GA3-treated female flowers and receptacles than in their control counterparts 2 and 4 DAT, whereas its level was repressed in both tissues 10 DAT, to a level that was less than half of the control. Thus, GA3 caused a shift in the ABA peak to 6 days earlier than its natural peak in female flowers and receptacle (Fig. 1c).
Transcriptome sequencing analysis and qRT-PCR validation
The transcriptomes of female flowers and receptacles from control and GA3-treated fruit were sequenced 2, 4 and 10 DAT. After removing adaptors and low-quality sequences, each library generated 29.7 to 40.5 million clean reads (Additional file 1: Table S2). Sequencing quality compliance was indicated by an error ratio of single bases of less than 0.0097%; Q20 and Q30 (the percentage of bases with base quality greater than 20 or 30 among the total bases) were around 98.6 and 96.0%, respectively. The number of G and C bases was about 47.0% for each gene-expression library. Alignment of the clean reads to the reference sequence showed over 24.7 million total mapped reads, with the ratio varying from 82.11 to 83.89% of the total clean reads (Additional file 1: Table S2).
Thirteen genes related to phytohormone metabolism or signal transduction were selected for validation. Positive correlations were found between the RNA sequencing (RNA-Seq) and qRT-PCR data in all pairwise comparison groups (Additional file 1: Figure S1 and S2), confirming the consistency, validity and representativeness of our RNA-Seq data.
Differences in gene expression pattern between control and GA3-treated fruit
Comparative transcriptome analysis was performed to identify the differentially expressed genes (DEGs) in GA3-treated samples compared to controls. Filtered by a minimum 2-fold FPKM difference with FDR < 0.01, for female flower samples, pairwise comparisons revealed 685, 294 and 1811 DEGs at 2, 4 and 10 DAT, respectively, with 400 (58.4%), 103 (35.0%) and 701 (38.7%) upregulated and 255 (41.6%), 191 (65.0%) and 1110 (61.3%) downregulated in the GA3-treated samples relative to controls; for receptacle samples, 908, 911 and 1672 genes were differentially expressed at 2, 4 and 10 DAT, respectively, with 436 (48.0%), 509 (55.9%) and 755 (45.2%) upregulated and 472 (52.0%), 402 (44.1%) and 917 (54.8%) downregulated in the treated samples relative to controls (Additional file 1: Figure S3).
Significantly enriched terms in the GO and KEGG databases were analyzed to identify the principle biological functions of the DEGs. In female flowers, 38, 37 and 45 significant GO terms of biological process, cellular component and molecular function were enriched 2, 4 and 10 DAT, respectively, and in the receptacle, 40, 39 and 41 GO terms were enriched (Additional file 1: Table S3). In general, the enrichment patterns of up- and downregulated genes were similar in female flowers and receptacles; the shared enriched terms in the two different tissues 2, 4 and 10 DAT were: binding (GO:0005488) and catalytic activity (GO:0003824) terms in the molecular function category, cell part (GO:0044464) and cell (GO:0005623) terms in the cellular component group, and metabolic process (GO:0008152), cellular process (GO:0009987) and single-organism process (GO:0044699) terms in the biological process group.
DEGs identified 2, 4 and 10 DAT were assigned to 4, 1 and 13 significantly different KEGG pathways in female flowers and 9, 4 and 12 pathways in the receptacle, respectively (Additional file 1: Table S4). The largest number of significantly changed pathways was found 10 DAT in both tissues. Among the hormone-related pathways, tryptophan metabolism (ko00380) was significantly enriched in female flowers 10 DAT, and plant hormone-signal transduction (ko04075) was significantly enriched in receptacles 2 and 10 DAT.
Expression pattern of genes involved in plant hormone metabolism and signal-transduction pathways
Plant hormone-related genes differentially expressed in female flowers and/or receptacles are summarized in Additional file 1: Tables S5 and S6. Briefly, most of the genes belonged to the ethylene-metabolism and response pathways, followed by auxin, ABA and GA; relatively fewer DEGs were related to cytokinin. A FPKM value > 10 for at least one sample was set as the threshold to select genes for further analysis. The expression profiles of the rest of the hormone-related DEGs, with FPKM values of all samples < 10, are shown in Additional file 1: Figure S4.
Gibberellin
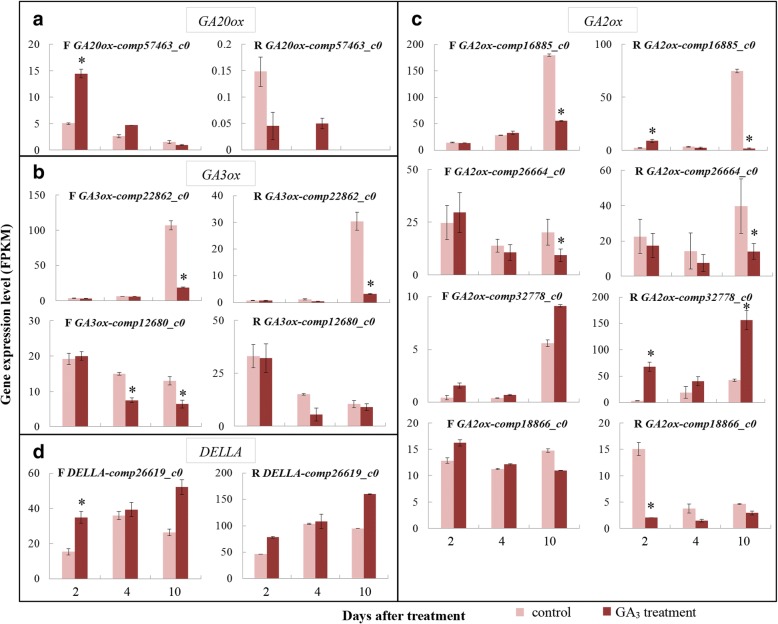
We identified 10 GA biosynthesis and catabolism genes encoding GA 20-oxidase (GA20ox), GA 3-oxidase (GA3ox) and GA 2-oxidase (GA2ox) (Additional file 1: Table S5). The expression level of the only GA20ox identified in this study was dramatically higher in female flowers than receptacles, and it was significantly upregulated in GA3-treated female flowers 2 DAT compared to controls, then downregulated until 10 DAT in both control and treated samples (Fig. 2a). Two GA3ox genes showed different expression patterns: comp22862_c0 was weakly expressed in all studied samples 2 and 4 DAT and showed sharp upregulation in control female flowers and receptacle 10 DAT, to significantly higher levels compared to their GA3-treated counterparts; comp12680_c0 showed the highest expression level 2 DAT and then a decrease during development, and was generally downregulated in the two different tissues after GA3 treatment (Fig. 2b).
Fig. 2.
Expression patterns of GA-metabolism and signaling genes. a GA20ox. b GA3ox. c GA2ox. d DELLA. F, female flowers; R, receptacle. *Fold change of FPKM ≥2 and FDR ≤ 0.001
Among all GA2ox genes, comp16885_c0 was most highly expressed, but its expression was repressed 3.3- and 41.1-fold in GA3-treated female flowers and receptacle compared to controls at 10 DAT, respectively. Another GA2ox gene (comp26664_c0) also exhibited significantly decreased expression in GA3-treated vs. control samples at 10 DAT. However, comp32778_c0 expression was upregulated after treatment at all sampling stages in both female flowers and receptacle, and remarkably higher expression was found in the receptacle vs. female flowers. In addition, comp18866_c0 only showed a significant difference in expression between control and treated receptacles 2 DAT, with the latter being 7.5-fold lower than the former (Fig. 2c).
With respect to GA-signal transduction, only one DEG encoding DELLA protein was identified, which was more highly expressed in GA3-treated fruit than in controls; its upregulation was significant in GA3-treated female flowers 2 DAT (Fig. 2d). These results suggest that exogenous GA3 treatment mainly influences GA-biosynthesis and catabolism pathways in both female flowers and receptacle, but not the signal-transduction process.
Auxin
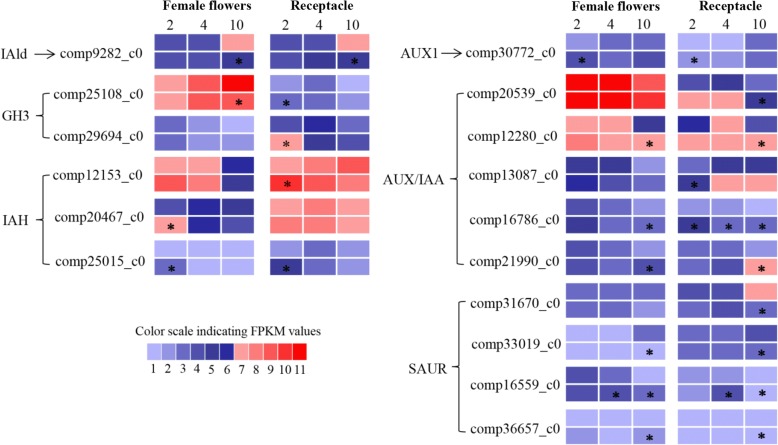
In GA3-treated fig, auxin-pathway genes constituted the largest group of DEGs among growth- and development-promoting phytohormones (Additional file 1: Table S5). The expression level of indole-3-acetaldehyde dehydrogenase (IAld) remained stable in female flowers and receptacle 2 and 4 DAT, whereas 10 DAT, it was upregulated in the two control tissues, to significantly higher levels than in their GA3-treated counterparts (Fig. 3). Two auxin-responsive Gretchen Hagen 3 (GH3) family genes revealed different expression patterns in the female flowers and similar patterns in the receptacles. In female flowers, comp25108_c0 showed continuously increasing expression during development in control tissues, more than 2-fold higher than in the treated tissues 10 DAT; in contrast, the other GH3 (comp29694_c0) decreased from 2 to 10 DAT in female flowers, and showed no significant difference between control and treated flowers. In receptacles, both GH3 genes exhibited over 2-fold upregulation 2 DAT (Fig. 3). Three IAA-amino acid hydrolase (IAH) genes demonstrated overall moderate changes in expression during development—increasing from 2 to 4 DAT and then decreasing in control female flowers and receptacle; however, GA3 treatment significantly elevated IAH expression 2 DAT, followed by a continuous decrease, resulting in similar expression levels between control and treated tissues 4 and 10 DAT. This implies that GA only transiently modulates IAH expression in female flowers and receptacles (Fig. 3).
Fig. 3.
Expression patterns of auxin-metabolism and signaling genes. Grids with 11 different colors represent FPKM values of 0–10, 10–20, 20–40, 40–60, 60–80, 80–100, 100–200, 200–300, 300–400, 400–500, and over 500, respectively. For each gene, the upper and lower rows refer to the results of control and GA3 treatment, respectively. 2, 4 and 10 = days after treatment. *Fold change of FPKM ≥2 and FDR ≤ 0.001
With respect to the auxin-response pathway (Additional file 1: Table S5), auxin-influx carrier (AUX1) demonstrated higher expression in GA3-treated fruit female flowers than in controls on all sampling days, and in the treated receptacle, upregulation was observed 2 DAT, whereas downregulation was identified 10 DAT (Fig. 3). Expression patterns of four small auxin-up RNA (SAUR) genes could be separated into two groups: DEGs comp31670_c0 and comp33019_c0 were significantly repressed in GA3-treated fruit female flowers and receptacle 10 DAT, whereas comp16559_c0 and comp36657_c0 were generally upregulated in the treatment group on all three sampling days relative to their control counterparts (Fig. 3). The negative auxin-response regulator AUX/IAA revealed divergent expression patterns between female flowers and receptacle and between control and treatment groups. In general, AUX/IAA presented higher expression in GA3-treated samples than in controls (Fig. 3).
Cytokinin
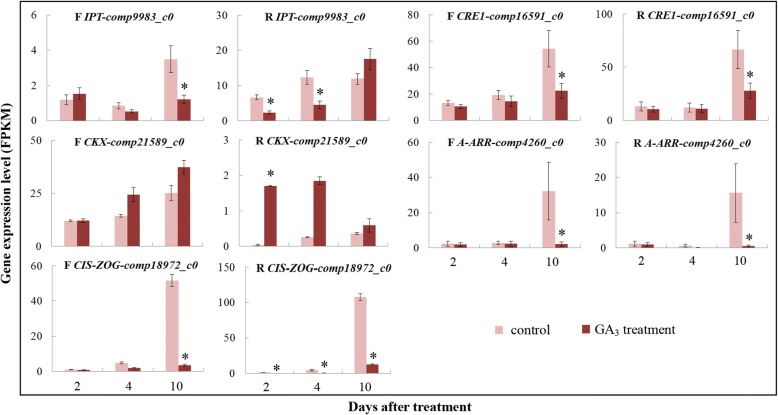
Differential expression of cytokinin metabolism and signal-transduction genes was identified (Fig. 4, Additional file 1: Table S5). Isopentenyl transferase (IPT) expression was higher in GA3-treated female flowers than controls 2 DAT, but lower 4 and 10 DAT. On the other hand, in the receptacle, IPT was downregulated 2 and 4 DAT, and moderately upregulated 10 DAT. Compared to controls, cytokinin oxidase/dehydrogenase (CKX) was upregulated in GA3-treated samples, and its expression was remarkably higher in female flowers vs. receptacle. Cis-zeatin O-glucosyltransferase (CIS-ZOG) showed very low expression in all samples 2 and 4 DAT, and maintained this low expression in GA3-treated fruit, whereas 10 DAT, it was 14.7- and 8.6-fold higher in control vs. GA3-treated female flowers and receptacle, respectively (Fig. 4). Note that in rice, the activity of cis-zeatin is comparable to that of trans-zeatin and can upregulate cytokinin-inducible genes, whereas overexpression of cis-ZOG delays rice leaf senescence [36].
Fig. 4.
Expression patterns of cytokinin-metabolism and signaling genes. F, female flowers; R, receptacle. *Fold change of FPKM ≥2 and FDR ≤ 0.001
In cytokinin-signal transduction, the cytokinin receptor cytokinin response1 (CRE1) was downregulated in both female flowers and receptacle after GA3 treatment, especially 10 DAT (Fig. 4). The negative A-type Arabidopsis response regulators (ARRs) remained stable with a low expression level in GA3-treated fruit at all tested stages, whereas a very significant increase was identified in controls 10 DAT, presenting 16- and 31-fold upregulation in female flowers and receptacle, respectively, compared to the corresponding GA3-treated tissues (Fig. 4).
ABA
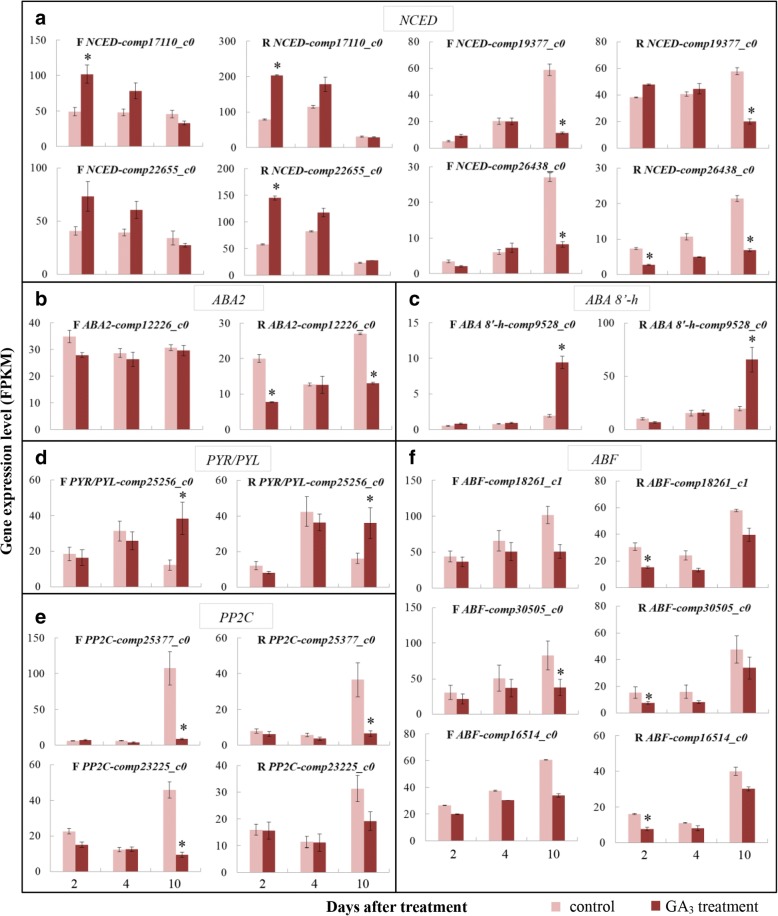
All identified ABA-synthesis, catabolism and response DEGs are shown in Additional file 1: Table S6. NECD (9-cisepoxycarotenoid dioxygenase) is the first committed and rate-limiting enzyme in ABA biosynthesis [37]. Two NCED genes—comp17110_c0 and comp22655_c0—were enhanced in the both tissues 2 and 4 DAT with GA3 (Fig. 5a). In controls, their expression was relatively stable during development in female flowers, and rose first and then fell at the 10 DAT in the receptacle; however, decreasing trends were found in both female flowers and receptacle of GA3-treated fruit. The other two NCED—comp19377_c0 and comp26438_c0—generally increased in the two tissues of control fruit during development, especially from 4 to 10 DAT, whereas their levels changed moderately in GA3-treated tissues; ultimately, their expression was significantly lower in treatment vs. control tissues 10 DAT (Fig. 5a). ABA2, encoding another ABA-biosynthesis enzyme, was downregulated in GA3-treated vs. control tissues, but with no significant difference in the female flowers (Fig. 5b). The process catalyzed by ABA 8′-hydroxylase (ABA 8′-h) is considered to be the main pathway for ABA catabolism [37]. ABA 8′-h expression remained relatively stable in the controls, whereas remarkable upregulation was found in the GA3-treated samples 10 DAT—4.9- and 3.3-fold higher in female flowers and receptacle, respectively, than in controls (Fig. 5c).
Fig. 5.
Expression patterns of ABA-metabolism and signaling genes. a NCED. b ABA2. c ABA 8′-h. d PYR/PYL. e PP2C. f ABF. F, female flowers; R, receptacle. *Fold change of FPKM ≥2 and FDR ≤ 0.001
In the ABA-signaling pathway, PYR/PYL expression was slightly lower in GA3-treated samples than in controls 2 and 4 DAT, but significantly higher 10 DAT (Fig. 5d). Protein phosphatase 2C (PP2C) usually represses ABA responses. PP2C expression was not significantly different between the control and treated fruit 2 and 4 DAT, whereas it was dramatically downregulated in GA3-treated female flowers and receptacle 10 DAT (Fig. 5e). ABRE binding factor (ABF) functions as a positive regulator of ABA signaling. The ABF genes’ expression was consistent with an increasing tendency from 2 to 10 DAT in both control and GA3-treated samples, and their expression was more or less downregulated in all studied samples after GA3 treatment (Fig. 5f).
Ethylene
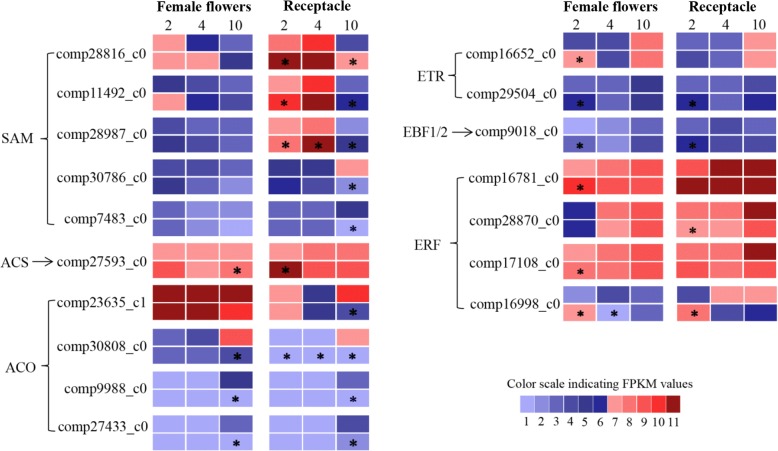
A large number of ethylene biosynthesis- and signaling-related DEGs were found in female flowers and/or receptacles after GA3 treatment (Additional file 1: Table S6). S-adenosylmethionine synthetase (SAM) displayed the highest expression 2 DAT in control and GA3-treated female flowers, followed by a gradual decrease, with no significant differences between control and treatment (Fig. 6). Three SAM genes—comp28816_c0, comp11492_c0 and comp28987_c0—peaked 4 DAT and showed their lowest expression 10 DAT in control and GA3-treated receptacles; they demonstrated higher expression after treatment than in controls. The other two SAM genes—comp30786_c0 and comp7483_c0—increased during the control samples’ development, but showed decreasing trends after GA3 treatment, leading to 6.8- and 7.7-fold downregulation 10 DAT, respectively, in the GA3-treated receptacle (Fig. 6). One gene annotated as 1-aminocyclopropane-1-carboxylate (ACC) synthase (ACS) presented higher expression in GA3-treated samples than in controls. ACC oxidase (ACO) genes, except comp23635_c1, had low expression in all tested control and GA3-treated samples 2 and 4 DAT, whereas dramatic upregulation was detected in control fruit female flowers and receptacle 10 DAT, resulting in significantly higher expression in the two control vs. treated tissues (2.5- to 42-fold) (Fig. 6).
Fig. 6.
Expression patterns of ethylene-biosynthesis and signaling genes. Grids with 11 different colors represent FPKM values 0–10, 10–20, 20–40, 40–60, 60–80, 80–100, 100–200, 200–300, 300–400, 400–500 and over 500, respectively. For each gene, upper and lower rows refer to the results of control and GA3 treatment, respectively. 2, 4 and 10 = days after treatment. *Fold change of FPKM ≥2 and FDR ≤ 0.001
In the ethylene-response pathway, ETR transcripts increased during development, showing the highest expression levels 10 DAT in controls. For GA3-treated samples, their expression levels fell first and then rose, with a peak at 2 or 10 DAT (Fig. 6). The expression levels of ETR and EBF1/2 were significantly upregulated in GA3-treated female flowers and receptacle 2 DAT. In general, ERF transcripts revealed an increasing trend in control fruit female flowers and receptacle, whereas they presented divergent expression patterns after GA3 treatment: comp16781_c0 and comp16998_c0 had the highest expression 2 DAT in female flowers and receptacle; comp28870_c0 and comp17108_c0 displayed upward trends during development similar to their expression in the controls (Fig. 6). ERFs were generally upregulated in the two treatment tissues 2 DAT, whereas 10 DAT, their expression levels were slightly lower in GA3-treated vs. control receptacles.
Discussion
Our aim was to investigate the role of GA in parthenocarpy induction in fig by comparing control and GA3-treated fruit’s female flowers and receptacles. We focused our study on 2, 4 and 10 days after exogenous GA3 application in a San Pedro-type fig cultivar’s main crop, which is genetically non-parthenocarpic. By Illumina RNA-Seq transcriptome analysis, 1.3- and 3.1-fold more DEGs were identified in the receptacle vs. female flowers at 2 and 4 DAT, respectively, whereas the number of DEGs was a little higher in female flowers than in the receptacle 10 DAT. This implied that aside from the reproductive female flowers, which mainly influence fruit development [38], the vegetative receptacle tissue might also have some regulatory effect on parthenocarpic fruit set and growth. GA, auxin, cytokinin, ABA and ethylene metabolism- and response-related genes showed differential expression between control and GA3-treated fruit, and phytohormone content assay revealed a new balance of endogenous phytohormone levels in both female flowers and receptacle following exogenous GA treatment (Fig. 1c). This indicated that parthenocarpy induction by GA in fig is the result of the coordinated action of different phytohormones.
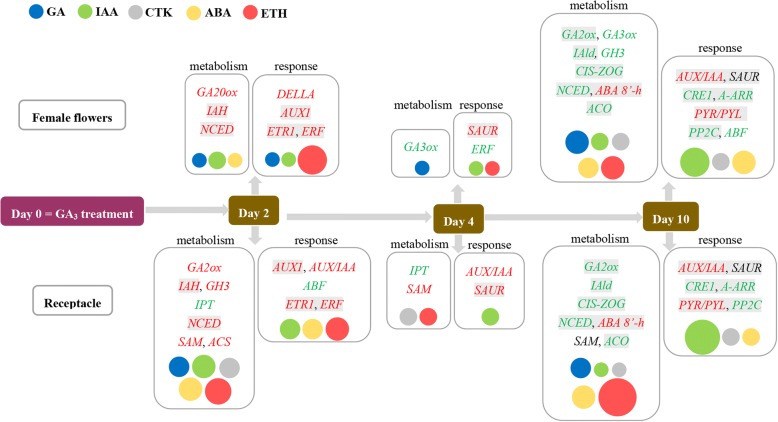
Exogenous GA3 application reset the transcriptional expression of plant hormone-related genes. In female flowers, 4, 1 and 15 phytohormone-metabolism genes and 8, 2 and 12 response-related genes were significantly differently expressed 2, 4 and 10 DAT, respectively. In the receptacle, 19, 4 and 18 phytohormone-metabolism and 10, 2 and 12 signal-transduction DEGs were identified 2, 4 and 10 DAT, respectively. The higher representation of plant hormone transcripts’ differential expression in receptacles compared to female flowers may further suggest an indelible role for receptacle tissue in controlling fruit set and development. Concrete spatiotemporal transcriptional changes in phytohormone-related genes induced by GA3 are summarized in Fig. 7. There were more common DEGs with the same changing trends in female flowers and receptacles 10 DAT than 2 and 4 DAT, indicating that GA3 treatment induces synchronized patterns of changing gene expression between the two tissues as fruit development progresses. In addition, in both female flowers and receptacles 10 DAT, the number of DEGs related to ethylene metabolism was highest, followed by those related to GA and ABA metabolism; for phytohormone response, the highest number of DEGs were related to auxin.
Fig. 7.
Summary of main differentially expressed phytohormone-related genes identified in female flowers and receptacle after GA3 treatment. Different colored circles represent different phytohormones and circle size indicates relative number of DEGs—the higher the number, the bigger the circle. Genes in red (green) color are upregulated (downregulated). Gray highlight represents genes with same changing trend in both female flowers and receptacle following GA3 application
Phytohormone-metabolism response to exogenous GA3
Previous studies have shown that transcription of GA-metabolism genes is regulated by the content of bioactive GA [2, 39]. GA oxidases modulate the GA-biosynthetic pathway mainly through feedback-loop mechanisms [40] and expression regulation that depends on the tissue and developmental stage [39, 41]. Exogenous GA application before or after bloom in grapevine leads to downregulation of the GA-biosynthetic genes GA20ox and GA3ox and upregulation of the GA-catabolic gene GA2ox [15, 42, 43]; this has also been observed in Arabidopsis [39] and tobacco [44]. Conversely, in the present study, GA20ox was upregulated in female flowers 2 DAT and GA2ox was repressed in both female flowers and receptacle 10 DAT. This difference could be explained by the change in endogenous GA content: in both fig and grapevine, GA content increases significantly and immediately after treatment and then decreases; however, for grapevine, GA content drops to levels equal to or significantly less than the control 3 DAT [43, 45], whereas in fig, it is still significantly higher than the control 10 DAT (Fig. 1c).
Auxin is one of the main regulators of fruit set and development. Similar to [42], auxin-biosynthesis genes, e.g. YUCCA, did not show significant regulation after GA3 treatment in fig. The main differentially expressed auxin-metabolism genes identified in the present study were IAH and GH3. The former encodes enzymes that produce free IAA from IAA–amino acid conjugates, whereas the latter catalyzes auxin conjugation [46]. IAH and GH3 were mainly significantly upregulated 2 DAT; upregulation of GH3 has also been reported previously [42], indicating that bioactive auxin levels are co-mediated by IAH and GH3 in fig. Cytokinin induces parthenocarpy by altering the expression of GA-biosynthesis genes [9, 16]. Only a few metabolic genes’ transcriptional levels were regulated by GA3 treatment in our dataset. For female flowers, cytokinin-metabolism transcripts were only mediated by GA3 treatment 10 DAT, whereas for the receptacle, exogenous GA3 also regulated cytokinin metabolism 2 and 4 DAT (Fig. 7). Zeatin concentration increased dramatically in the receptacle 2 DAT, then decreased to a markedly lower level than controls, suggesting that cytokinin-biosynthesis genes respond more rapidly (in under 2 days) in the receptacle following GA3 application.
Aside from growth-promoting hormones, GA3 treatment also modulated the biosynthesis of ABA and ethylene, which mainly play GA-antagonistic roles in the control of many plant developmental processes [47]. Enhancement of ABA and ethylene biosynthesis 2 DAT might reflect ‘feedback’ regulation of the highly significantly increased GA3 content in fig fruit following exogenous GA3 application. This ‘feedback’ response then declines, with almost no ABA- or ethylene-related DEGs being identified 4 DAT. Along with fruit development, both female flowers and receptacle revealed suppression of ABA and ethylene biosynthesis 10 DAT, with downregulation of NCED and ACO and upregulation of ABA 8′-h, in accordance with the lower ABA level in GA3-treated fruits. Previous reports have shown that expression of the ethylene-biosynthesis genes ACOs decreases during parthenocarpic fruit set and development or after GA3 treatment [13].
Phytohormone signaling in response to exogenous GA3
DELLA protein has been characterized as a negative regulator in GA signaling [48]. One DELLA gene exhibited upregulation after GA3 application, most markedly in female flowers 2 DAT, in accordance with previously reported results [7, 13, 43]. This indicates GA-perception feedback regulation in female flowers and receptacle.
Studies of the crosstalk between GA and auxin have revealed that GA-induced parthenocarpy arises from the interaction of GA with auxin signaling [42], and that auxin-induced fruit set is mediated in part by GA [9]. In the present study, AUX/IAA gene expression was significantly upregulated in the receptacle on all sampling days and in female flowers 10 DAT, suggesting feedback regulation of the auxin response following GA3 treatment, in agreement with results in grape [15]. In addition to the AUX/IAAs, differential expression of SAUR transcript levels was seen in both female flowers and receptacles 10 DAT (Fig. 3). Further study is needed to elucidate their roles in fig fruit development. DEGs related to cytokinin signaling were only identified 10 DAT in the female flowers and receptacle (Fig. 7).
GA3 treatment also changed the signaling of ABA and ethylene. Enhancement of ethylene signaling was identified 2 DAT. Crosstalk internodes between GA and the ethylene-signaling pathway were elucidated by the role of group VII ERFs as DELLA partners [49], and ERF11 activates GA biosynthesis and signaling [50]. Genes encoding ABA receptor PYR/PYL and PP2C, a negative regulator in the ABA-response pathway, displayed upregulation and downregulation, respectively, in both tissues 10 DAT, leading to enhancement of ABA-signal transduction in both female flowers and receptacle. A recent study on rice revealed the central role of PYR/PYL in the antagonistic action of GA and ABA [51].
Conclusions
GA3 treatment, performed to induce parthenocarpy in San Pedro-type fig main crop, resulted in a highly significant GA3 content increment in both female flowers and receptacle tissues of fig fruit. Changes in the expression of key genes in plant hormone-synthesis and metabolism pathways—as reflected by modulated endogenous plant hormone levels—and in the hormone-signaling pathway, converted the pre-dropping fruit pattern to an artificial parthenocarpic developmental pattern. Spatiotemporal characteristics of the changes in plant hormone levels and gene-expression patterns in GA3-induced fig parthenocarpy were reflected by differentially reacting female flowers and receptacle tissue, and temporal waves in major plant hormone and relevant genes’ expression were illustrated. Further study on fig fruit development with the application of other exogenous plant hormones, e.g., auxin and cytokinin, will serve to further understand the mechanisms of GA-induced parthenocarpy and the key plant hormones in fig fruit set and development.
Additional file
Table S1. Primer sequences of genes used for validation of RNA-Seq results by quantitative real-time PCR, Table S2. Summary of sequencing results for control and GA3-treated female flowers and receptacles, Table S3. Significant GO terms (corrected P-value ≤0.05) identified between control and GA3-treated female flowers and receptacles, Table S4. Significant KEGG pathways (corrected P-value ≤0.05) identified between control and GA3-treated female flowers and receptacles, Table S5. All gibberellin-, auxin- and cytokinin-synthesis, catabolism and response transcripts identified in this study which were differentially expressed (FDR < 0.01 and the absolute value of log2 (FPKMtreatment/FPKMcontrol) ≥ 1) in at least one pairwise comparison group, Table S6. All abscisic acid- and ethylene-synthesis, catabolism and response transcripts identified in this study which were differentially expressed (FDR < 0.01 and the absolute value of log2 (FPKMtreatment/FPKMcontrol) ≥ 1) in at least one pairwise comparison group, Figure S1. Verification of RNA-Seq results by qRT-PCR. Bars represent standard deviation. F, female flowers; R, receptacle, Figure S2. Correlation of fold changes in gene expression between RNA-Seq and qRT-PCR. Equation of linear regression and correlation coefficient (R2) are shown. DAT, days after treatment, Figure S3. Number of differentially expressed genes (FDR ≤ 0.001 and log2 (FPKMtreatment/FPKMcontrol) ≥ 1 or ≤ − 1) between control and GA3-treated female flowers and receptacles. DAT, days after treatment, Figure S4. Heat maps of hormone-related genes with low expression. FPKM of all samples was < 10. (PDF 1241 kb)
Acknowledgments
Funding
This work was financially supported by the grants from National Natural Science Foundation of China project NSFC [31372007].
Availability of data and materials
Raw data of all sample-sequencing results have been submitted to the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra) with accession number SRP113799.
Abbreviations
- ABA 8′-h
ABA 8′-hydroxylase
- ABA
Abscisic acid
- ABF
ABRE binding factor
- ACC
1-aminocyclopropane-1-carboxylate
- ACS
ACC synthase; ACO: ACC oxidase
- ARRs
Arabidopsis response regulators
- AUX1
auxin-influx carrier
- CIS-ZOG
cis-zeatin O-glucosyltransferase
- CKX
Cytokinin oxidase/dehydrogenase
- CRE1
Cytokinin response1
- DAT
Days after treatment
- DEGs
Differentially expressed genes
- FDR
False discovery rate
- FPKM
Fragments per kilobase of exon model per million mapped reads
- GA20ox
GA 20-oxidase
- GA2ox
GA 2-oxidase
- GA3ox
GA 3-oxidase
- GAs
Gibberellins
- GH3
Gretchen Hagen 3
- GO
Gene Ontology
- IAA
Indole-3-acetic acid
- IAH
IAA-amino acid hydrolase
- IAld
Indole-3-acetaldehyde dehydrogenase
- IPT
Isopentenyl transferase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NECD
9-cisepoxycarotenoid dioxygenase
- PP2C
Protein phosphatase 2C
- qRT-PCR
Quantitative real-time PCR
- RNA-Seq
RNA sequencing
- SAM
S-adenosylmethionine synthetase
- SAUR
Small auxin-up RNA
Authors’ contributions
HM and SC designed the experiments. LC and PC conducted the experiments and analyzed the results. LC and HM prepared the manuscript. SC and MF assisted in drafting the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The experiments did not involve endangered or protected species. No specific permits were required for these activities because the figs used in this study were obtained from an orchard in Beijing, which was a demonstration base of China Agricultural University.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Lijuan Chai and Peng Chai contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1318-1) contains supplementary material, which is available to authorized users.
Contributor Information
Lijuan Chai, Email: clj2010@cau.edu.cn.
Peng Chai, Email: chaipeng@cau.edu.cn.
Shangwu Chen, Email: swchen@cau.edu.cn.
Moshe A. Flaishman, Email: vhmoshea@volcani.agri.gov.il
Huiqin Ma, Email: hqma@cau.edu.cn.
References
- 1.Blazquez MR, Nilsson O, Sussman MR, Weigel D. Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell. 1998;10(5):791. doi: 10.1105/tpc.10.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serrani JC, Sanjuan R, Ruizrivero O, Fos M, Garciamartinez JL. Gibberellin regulation of fruit set and growth in tomato. Plant Physiol. 2007;145(1):246–257. doi: 10.1104/pp.107.098335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayele BT, Ozga JA, Reinecke DM. Regulation of GA biosynthesis genes during germination and young seedling growth of pea (Pisum sativum L.) J Plant Growth Regul. 2006;25(3):219–232. doi: 10.1007/s00344-006-0007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun T. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004;135(2):1008–1019. doi: 10.1104/pp.104.039578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 2000;5(12):523–530. doi: 10.1016/S1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- 6.Yarushnykov VV, Blanke MM. Alleviation of frost damage to pear flowers by application of gibberellin. Plant Growth Regul. 2005;45(1):21–27. doi: 10.1007/s10725-004-6893-5. [DOI] [Google Scholar]
- 7.Serrani JC, Ruizrivero O, Fos M, Garciamartinez JL. Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J. 2008;56(6):922–934. doi: 10.1111/j.1365-313X.2008.03654.x. [DOI] [PubMed] [Google Scholar]
- 8.Mesejo C, Yuste R, Reig C, Martínez-Fuentes A, Iglesias DJ, Muñoz-Fambuena N, Bermejo A, Germanà MA, Primo-Millo E, Agustí M. Gibberellin reactivates and maintains ovary-wall cell division causing fruit set in parthenocarpic Citrus species. Plant Sci. 2016;247:13–24. doi: 10.1016/j.plantsci.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 9.Lu L, Liang J, Zhu X, Xiao K, Li T, Hu J. Auxin- and Cytokinin-induced berries set in grapevine partly rely on enhanced gibberellin biosynthesis. Tree Genet Genomes. 2016;12(3):41. doi: 10.1007/s11295-016-0980-4. [DOI] [Google Scholar]
- 10.Shinozaki Y, Hao S, Kojima M, Sakakibara H, Ozekiiida Y, Zheng Y, Fei Z, Zhong S, Giovannoni JJ, Rose JKC. Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J. 2015;83(2):237–251. doi: 10.1111/tpj.12882. [DOI] [PubMed] [Google Scholar]
- 11.Nitsch L, Kohlen W, Oplaat C, Charnikhova T, Cristescu S, Michieli P, Wolters-Arts M, Bouwmeester H, Mariani C, Vriezen WH. ABA-deficiency results in reduced plant and fruit size in tomato. J Plant Physiol. 2012;169(9):878–883. doi: 10.1016/j.jplph.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Mariotti L, Picciarelli P, Lombardi L, Ceccarelli N. Fruit-set and early fruit growth in tomato are associated with increases in Indoleacetic acid, Cytokinin, and bioactive gibberellin contents. J Plant Growth Regul. 2011;30(4):405. doi: 10.1007/s00344-011-9204-1. [DOI] [Google Scholar]
- 13.Tang N, Deng W, Hu G, Hu N, Li Z. Transcriptome profiling reveals the regulatory mechanism underlying pollination dependent and parthenocarpic fruit set mainly mediated by auxin and gibberellin. PLoS One. 2015;10(4) [DOI] [PMC free article] [PubMed]
- 14.Ji L, Zhe W, Li C, Tinglin Z, Qinwei G, Jian X, Li J, Qunfeng L, Sanwen H, Zhengguo L. Transcriptome comparison of global distinctive features between pollination and parthenocarpic fruit set reveals transcriptional phytohormone cross-talk in cucumber (Cucumis sativus L.) Plant Cell Physiol. 2014;55(7):1325. doi: 10.1093/pcp/pcu051. [DOI] [PubMed] [Google Scholar]
- 15.Chai L, Li Y, Chen S, Perl A, Zhao F, Ma H. RNA sequencing reveals high resolution expression change of major plant hormone pathway genes after young seedless grape berries treated with gibberellin. Plant Sci. 2014;229:215–224. doi: 10.1016/j.plantsci.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 16.Ding J, Chen B, Xia X, Mao W, Shi K, Zhou Y, Yu J. Cytokinin-induced parthenocarpic fruit development in tomato is partly dependent on enhanced gibberellin and auxin biosynthesis. PLoS One. 2013;8(7):e70080. doi: 10.1371/journal.pone.0070080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mignolli F, Vidoz ML, Mariotti L, Lombardi L, Picciarelli P. Induction of gibberellin 20-oxidases and repression of gibberellin 2β-oxidases in unfertilized ovaries of entire tomato mutant, leads to accumulation of active gibberellins and parthenocarpic fruit formation. Plant Growth Regul. 2015;75(2):415–425. doi: 10.1007/s10725-014-0002-1. [DOI] [Google Scholar]
- 18.Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech J, Bouzayen M. The tomato aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell. 2005;17(10):2676–2692. doi: 10.1105/tpc.105.033415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiwari A, Offringa R, Heuvelink E. Auxin-induced fruit set in Capsicum annuum L. requires downstream gibberellin biosynthesis. J Plant Growth Regul. 2012;31(4):570–578. doi: 10.1007/s00344-012-9267-7. [DOI] [Google Scholar]
- 20.Flaishman MA, Rodov V, Stover E. The fig: botany, horticulture, and breeding. Horticultural Rev-Westport Then N Y. 2008;34:113. [Google Scholar]
- 21.Beck NG, Lord EM. Breeding system in Ficus carica, the common fig. II. Pollination events. Am J Bot. 1988;75(12):1913–1922. doi: 10.1002/j.1537-2197.1988.tb11272.x. [DOI] [Google Scholar]
- 22.Crane J, Campbell R. Breaking rest and inducing parthenocarpy in the Calimyrna fig with gibberellin: Proc 15th Int Cong Hort, Nice; 1958.
- 23.Crane JC, Van Overbeek J. Kinin-induced parthenocarpy in the fig, Ficus carica L. Science. 1965;147(3664):1468–1469. doi: 10.1126/science.147.3664.1468. [DOI] [PubMed] [Google Scholar]
- 24.Lodhi F, Bradley MV, Crane JC. Auxins and gibberellin-like substances in parthenocarpic and non-parthenocarpic syconia of Ficus carica L., cv. King. Plant Physiol. 1969;44(4):555–561. doi: 10.1104/pp.44.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chai L, Wang Z, Chai P, Chen S, Ma H. Transcriptome analysis of San Pedro-type fig (Ficus carica L.) parthenocarpic breba and non-parthenocarpic main crop reveals divergent phytohormone-related gene expression. Tree Genet Genomes. 2017;13(4):83. doi: 10.1007/s11295-017-1166-4. [DOI] [Google Scholar]
- 26.Pan X, Welti R, Wang X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat Protoc. 2010;5(6):986. doi: 10.1038/nprot.2010.37. [DOI] [PubMed] [Google Scholar]
- 27.Reid KE, Olsson N, Schlosser J, Peng FY, Lund ST. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 2006;6(1):27. doi: 10.1186/1471-2229-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MD, Mccarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39 [DOI] [PMC free article] [PubMed]
- 31.Upton G. Fisher's exact test. J R Stat Soc A Sta. 1992;155:395–402. doi: 10.2307/2982890. [DOI] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57(1):289–300. [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Freiman ZE, Rosianskey Y, Dasmohapatra R, Kamara I, Flaishman MA. The ambiguous ripening nature of the fig (Ficus carica L.) fruit: a gene-expression study of potential ripening regulators and ethylene-related genes. J Exp Bot. 2015;66(11):3309. doi: 10.1093/jxb/erv140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marei N, Crane JC. Growth and respiratory response of fig (Ficus carica L. cv. Mission) fruits to ethylene. Plant Physiol. 1971;48(3):249–254. doi: 10.1104/pp.48.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kudo T, Makita N, Kojima M, Tokunaga H, Sakakibara H. Cytokinin activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-Zeatin-O-glucosyltransferase in rice. Plant Physiol. 2012;160(1):319. doi: 10.1104/pp.112.196733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nambara E, Marionpoll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol. 2005;56(1):165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 38.Rosianski Y, Doronfaigenboim A, Freiman ZE, Lama K, Milocochavi S, Dahan Y, Kerem Z, Flaishman MA. Tissue-specific transcriptome and hormonal regulation of pollinated and Parthenocarpic fig (Ficus carica L.) fruit suggest that fruit ripening is coordinated by the reproductive part of the Syconium. Front Plant Sci. 2016;7 [DOI] [PMC free article] [PubMed]
- 39.Rieu I, Ruizrivero O, Fernandezgarcia N, Griffiths J, Powers SJ, Gong F, Linhartova T, Eriksson S, Nilsson O, Thomas SG. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008;53(3):488. doi: 10.1111/j.1365-313X.2007.03356.x. [DOI] [PubMed] [Google Scholar]
- 40.Middleton AM, Ubedatomas S, Griffiths J, Holman T, Hedden P, Thomas SG, Phillips AL, Holdsworth MJ, Bennett MJ, King JR. Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling. Proc Natl Acad Sci U S A. 2012;109(19):7571–7576. doi: 10.1073/pnas.1113666109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi S, Kamiya Y, Sun T. Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 2001;28(4):443–453. doi: 10.1046/j.1365-313X.2001.01168.x. [DOI] [PubMed] [Google Scholar]
- 42.Jung CJ, Hur YY, Jung S-M, Noh J-H, Do G-R, Park S-J, Nam J-C, Park K-S, Hwang H-S, Choi D, et al. Transcriptional changes of gibberellin oxidase genes in grapevines with or without gibberellin application during inflorescence development. J Plant Res. 2014;127(2):359–371. doi: 10.1007/s10265-013-0623-x. [DOI] [PubMed] [Google Scholar]
- 43.Cheng C, Jiao C, Singer SD, Gao M, Xu X, Zhou Y, Li Z, Fei Z, Wang Y, Wang X. Gibberellin-induced changes in the transcriptome of grapevine ( Vitis labrusca × V. Vinifera ) cv. Kyoho flowers. BMC Genomics. 2015;16(1):128. doi: 10.1186/s12864-015-1324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallegogiraldo L, Ubedatomás S, Gisbert C, Garcíamartínez JL, Moritz T, Lópezdíaz I. Gibberellin homeostasis in tobacco is regulated by gibberellin metabolism genes with different gibberellin sensitivity. Plant Cell Physiol. 2008;49(5):679. doi: 10.1093/pcp/pcn042. [DOI] [PubMed] [Google Scholar]
- 45.Wang ZR, Zhao F, Zhao XB, Ge H, Chai L, Chen S, Perl A, Ma H. Proteomic analysis of berry-sizing effect of GA3 on seedless Vitis vinifera L. Proteomics. 2012;12(1):86–94. doi: 10.1002/pmic.201000668. [DOI] [PubMed] [Google Scholar]
- 46.Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004;135(2):978–988. doi: 10.1104/pp.104.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss D, Ori N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 2007;144(3):1240–1246. doi: 10.1104/pp.107.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun T. Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol. 2010;154(2):567–570. doi: 10.1104/pp.110.161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marínde lRN, Sotillo B, Miskolczi P, Gibbs DJ, Vicente J, Carbonero P, Oñatesánchez L, Holdsworth MJ, Bhalerao R, Alabadí D. Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol. 2014;166(2):1022–1032. doi: 10.1104/pp.114.244723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X, Zhang Z, Park J, Tyler L, Yusuke J, Qiu KQ, Nam E, Lumba S, Desveaux D, Mccourt P. The ERF11 transcription factor promotes internode elongation by activating gibberellin biosynthesis and signaling. Plant Physiol. 2016;171(4):2760–2770. doi: 10.1104/pp.16.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Q, Wu F, Sheng P, Zhang Z, Zhang X, Guo X, Wang J, Cheng Z, Wang J, Wang H. The SnRK2-APC/CTE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat Commun. 2015;6:7981. doi: 10.1038/ncomms8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences of genes used for validation of RNA-Seq results by quantitative real-time PCR, Table S2. Summary of sequencing results for control and GA3-treated female flowers and receptacles, Table S3. Significant GO terms (corrected P-value ≤0.05) identified between control and GA3-treated female flowers and receptacles, Table S4. Significant KEGG pathways (corrected P-value ≤0.05) identified between control and GA3-treated female flowers and receptacles, Table S5. All gibberellin-, auxin- and cytokinin-synthesis, catabolism and response transcripts identified in this study which were differentially expressed (FDR < 0.01 and the absolute value of log2 (FPKMtreatment/FPKMcontrol) ≥ 1) in at least one pairwise comparison group, Table S6. All abscisic acid- and ethylene-synthesis, catabolism and response transcripts identified in this study which were differentially expressed (FDR < 0.01 and the absolute value of log2 (FPKMtreatment/FPKMcontrol) ≥ 1) in at least one pairwise comparison group, Figure S1. Verification of RNA-Seq results by qRT-PCR. Bars represent standard deviation. F, female flowers; R, receptacle, Figure S2. Correlation of fold changes in gene expression between RNA-Seq and qRT-PCR. Equation of linear regression and correlation coefficient (R2) are shown. DAT, days after treatment, Figure S3. Number of differentially expressed genes (FDR ≤ 0.001 and log2 (FPKMtreatment/FPKMcontrol) ≥ 1 or ≤ − 1) between control and GA3-treated female flowers and receptacles. DAT, days after treatment, Figure S4. Heat maps of hormone-related genes with low expression. FPKM of all samples was < 10. (PDF 1241 kb)
Data Availability Statement
Raw data of all sample-sequencing results have been submitted to the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra) with accession number SRP113799.