Abstract
Single-molecule methods provide direct measurements of macromolecular dynamics, but are limited by the number of degrees of freedom that can be followed at one time. High-resolution rotor bead tracking (RBT) measures DNA torque, twist, and extension, and can be used to characterize the structural dynamics of DNA and diverse nucleoprotein complexes. Here, we extend RBT to enable simultaneous monitoring of additional degrees of freedom. Fluorescence-RBT (FluoRBT) combines magnetic tweezers, infrared evanescent scattering, and single-molecule FRET imaging, providing real-time multiparameter measurements of complex molecular processes. We demonstrate the capabilities of FluoRBT by conducting simultaneous measurements of extension and FRET during opening and closing of a DNA hairpin under tension, and by observing simultaneous changes in FRET and torque during a transition between right-handed B-form and left-handed Z-form DNA under controlled supercoiling. We discover unanticipated continuous changes in FRET with applied torque, and also show how FluoRBT can facilitate high-resolution FRET measurements of molecular states, by using a mechanical signal as an independent temporal reference for aligning and averaging noisy fluorescence data. By combining mechanical measurements of global DNA deformations with FRET measurements of local conformational changes, FluoRBT will enable multidimensional investigations of systems ranging from DNA structures to large macromolecular machines.
Main Text
Dynamic observations of individual molecules have contributed unique insight into biological processes such as macromolecular folding and molecular motor function (1, 2). However, whereas the conformational dynamics of biological macromolecules are highly multidimensional, single-molecule measurements project these dynamics onto only a few degrees of freedom—sometimes a single dimension, such as the end-to-end extension of the molecule or the distance between two fluorescent dye labels. Combining different single-molecule techniques in the same assay can leverage their individual strengths and increase the number of observables (3). High-resolution mechanical measurements of extension from optical trapping and magnetic tweezers have been combined with single-molecule fluorescence and FRET to track local conformational transitions or observe changes in composition during binding and unbinding events (4, 5, 6, 7, 8, 9, 10). In recent applications, high-resolution optical trapping combined with fluorescence has been used to relate different DNA unwinding modes of the helicase UvrD to its conformation and multimeric composition (11), to observe diffusion of single-stranded DNA binding protein on DNA (12), and to relate the tertiary structure of the thiamine pyrophosphate riboswitch to affinity for its ligand (13). Simultaneous fluorescence and manipulation or particle tracking (5) has also been used to correlate mechanics with conformational changes or nucleotide turnover in both linear (14) and rotary (15, 16) molecular motors.
Rotor bead tracking (RBT) is a single-molecule method that can directly monitor two mechanical degrees of freedom at high spatiotemporal resolution (17, 18, 19). A micron-sized magnetic bead is typically used to apply forces and torques on a DNA molecule, whereas a separate nanoscale rotor bead attached to the side of the DNA is used as a reporter of DNA angle and extension. In the context of a dynamic nucleoprotein complex, changes in extension can arise from bending, stretching, or sequestering of DNA contour length, whereas changes in angle can result from topoisomerization or from trapped writhe (e.g., due to DNA wrapping) or twist (e.g., due to DNA unwinding) (19). Previously, RBT has been used to study DNA structural dynamics under torque, including DNA melting and the transition between B- and Z-DNA (18, 19, 20, 21), and to investigate the mechanochemical cycle of the supercoiling motor DNA gyrase (18, 19, 22). However, RBT is limited to observing only global DNA structural deformations. Local structural features, such as the locations or domain configurations of Z-DNA or melted bubble regions, cannot be directly distinguished. Enzymes bound to DNA cannot be detected unless they deform the DNA substrate, and protein domain movements leading to changes in DNA topology can only be postulated based on other structural or biochemical data.
Here we introduce Fluorescence-RBT (FluoRBT), which combines RBT and single-molecule fluorescence. We test the capabilities of FluoRBT in experiments that probe conformational changes in DNA, highlighting the ability to simultaneously apply mechanical perturbations, measure DNA extension, twist, and torque, and probe local structural transitions via FRET. FluoRBT is implemented by adapting an instrument configured for evanescent dark field imaging and magnetic tweezers manipulation (18), adding optical paths for simultaneous multiwavelength single-molecule fluorescence measurements (Fig. 1 a). For imaging the rotor bead, total internal reflection illumination is achieved by focusing an infrared laser beam at the periphery of the objective back focal plane using a pair of small metallic mirrors positioned under the objective (18, 23). For fluorescence measurements, visible lasers are similarly coupled via a second pair of small mirrors, creating evanescent illumination. The infrared light scattered by the rotor bead is separated from the fluorophore emission using a dichroic beamsplitter. Rotor bead tracking is performed on a fast CMOS camera, and fluorescence is imaged on a sensitive EMCCD camera, after separating light emitted from different fluorophores using an image splitter. Complete methods and technical characterization (Figs. S1–S4) can be found in the Supporting Material.
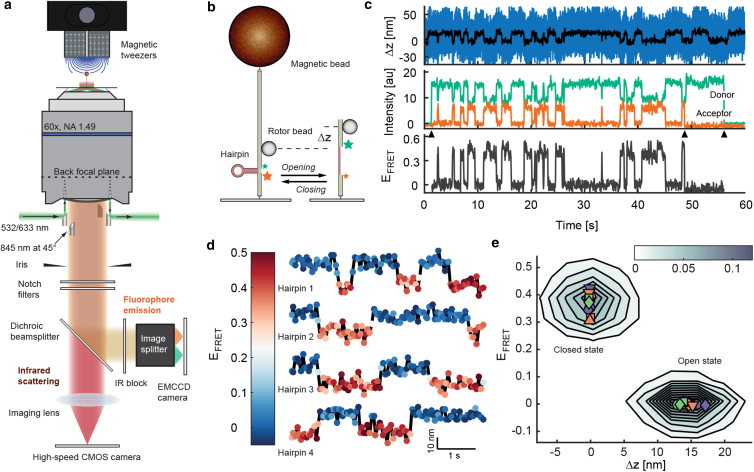
Figure 1.
Design and demonstration of combined extension and FRET measurements with FluoRBT. (a) A simplified schematic of the experimental setup is shown. Evanescent excitation with 845- and 532-nm light is achieved using a set of small mirrors positioned under the objective. Emitted light is spectrally separated via a dichroic beamsplitter; fluorophore emission is further isolated with a set of optical filters and an IR block. A pair of magnets above the sample is used to apply forces and torques on DNA. See Fig. S1. (b–e) Simultaneous measurements of FRET and extension (Δz) were collected during opening and closing of a hairpin under tension. (b) A DNA tether was stretched using a magnetic bead, a separate rotor bead was used as an evanescent scattering probe of Δz, and a FRET dye pair (green star, Quasar 570; orange star, Quasar 670) reported locally on the state of an incorporated hairpin, consisting of 15 basepair stem region and eight nucleotide loop. (c) Simultaneously acquired Δz (top, 2000 Hz raw and 20 Hz averaged), donor and acceptor intensities (middle, 20 Hz), and calculated FRET efficiency (bottom) are shown for a single DNA hairpin fluctuating between closed and open states under constant force. Arrowheads show start of donor excitation (∼2 s), acceptor bleaching (∼48 s), and donor bleaching (∼57 s). (d) Correspondence between Δz (vertical axis) and FRET efficiency (color scale) is shown for four separate hairpin tethers. (e) Data from 13 hairpin tethers were pooled together to generate a 2D histogram of 6180 instantaneous measurements, presented as a contour plot. Contours are equally spaced; color bar indicates the relative number of observations for each area. The mean Δz extension and FRET efficiency of both states for each molecule are shown with colored symbols. The mean Δz position of the closed state and the mean FRET efficiency of the open state were set to zero in our analysis procedure, as described in Supporting Material.
We first tested the ability of FluoRBT to simultaneously measure extension and FRET during reversible unfolding of a DNA hairpin, a model system that has been employed in prior demonstrations of methods combining the application of force with FRET measurements (9, 10). We incorporated a previously characterized hairpin construct (9, 24) in the DNA tether, and used both the rotor bead height and a local FRET dye pair to monitor its unfolding (Fig. 1 b). Under constant force (∼8 pN; see Supporting Materials and Methods in the Supporting Material), we observed concurrent changes in Δz and changes in intensity of the donor and acceptor dyes (Fig. 1, c and d), reflecting equilibrium fluctuations between the open and closed states of the hairpin. A histogram of data collected from 13 different molecules demonstrates that the instrument resolves the two hairpin states along both extension and FRET efficiency coordinates (Fig. 1 e). The measured extension change upon opening of the hairpin is 14.5 ± 1.1 nm, and the FRET efficiency of the closed state is 0.37 ± 0.04 (mean ± SD, n = 13 molecules), in agreement with the expected values of ∼14 nm and ∼0.4, respectively (see Supporting Materials and Methods in the Supporting Material).
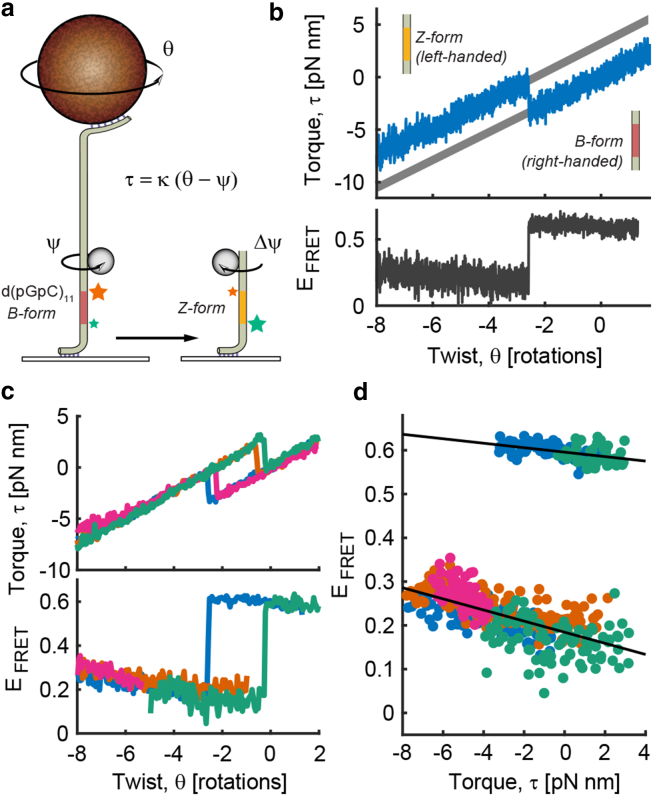
An advantage of FluoRBT over other tools that combine extension measurements with fluorescence is that it can also simultaneously measure twist and torque at high resolution (18). RBT has been previously used to observe torque-induced structural transitions in DNA such as duplex melting and Z-DNA formation (18, 20, 21). In the “static RBT” assay (19), the total twist of the DNA is changed by rotating the magnetic bead, and torque is measured based on the calibrated angular deflection of the DNA segment between the magnetic bead and the rotor bead. Structural transitions in DNA produce characteristic signatures in the torque-twist relationship. As a test of combined FRET and torque spectroscopy, we observed the transition of a GC-repeat sequence of interest (SOI) between right-handed B-form and left-handed Z-form DNA under controlled supercoiling. Torque spectroscopy has previously shown that the B-Z transition of a short GC-repeat sequence displays two-state behavior with a jump in torque at the transition event (21). The B-Z transition has also been observed via single-molecule FRET by incorporating a dye pair in the DNA backbone (7); high salt concentration or negative supercoiling (applied using conventional magnetic tweezers without torque measurement) were found to drive this sequence from a high-FRET state in B-DNA to a low-FRET state in Z-DNA. For FluoRBT measurements, we used a FRET-labeled d(pGpC)11 insert, designed based on the construct reported in Lee et al. (7) (Fig. 2 a). When we twisted the DNA under 2 pN of tension, we detected the B-Z transition as a jump in torque concurrent with a change in the FRET efficiency of the dye pair incorporated in the DNA backbone (Fig. 2 b), consistent with the previous results. Unexpectedly, we also discovered a continuous change in FRET as a function of twist (Fig. 2, c and d; Figs. S5 and S6), which likely reflects compliance in the DNA duplex, particularly in the torsionally soft Z-DNA state. This previously unreported effect (7) may arise from changes in both orientation and separation of the dyes (25) (Fig. S6), and might be exploited to develop molecular torque sensors, analogous to previously described molecular force sensors (26).
Figure 2.
Simultaneous measurements of FRET and torque. (a) A GC-repeat sequence was incorporated in a DNA tether. The magnetic bead was rotated to change the total twist (θ) in the DNA while using a rotor bead to measure the torque (τ) and a local FRET pair (green star, Cy3; orange star, Cy5) to monitor the B-Z structural transition. (b) The B-Z transition is observed via simultaneous jumps in torque (top, 250 Hz) and FRET efficiency (bottom, 5 Hz). Only the rewinding portion of the torque-twist curve is shown. Thick gray lines are fits to torque-twist data before and after the B-Z transition. (c) Combined torque and FRET measurements are shown for four different molecules. In the orange and magenta traces, one of the fluorophores bleaches before the structural transition in the SOI is observed. Data are block-averaged in 0.05 rotation (2 s) windows. (d) The FRET efficiency shows continuous variation with torque in both the B- and Z-DNA states of the SOI. Trend lines (black) were fit to the data from (c), after replotting on the torque-FRET plane. FRET changes at a rate of −0.5% (pN nm)−1 when the SOI is in B-form (top) and at a rate of −1.3% (pN nm)−1 when the SOI is in Z-form (bottom).
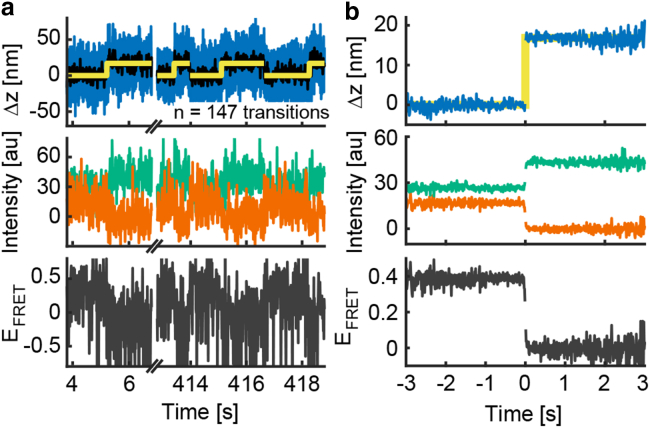
Fluorescence is a powerful complement to mechanical measurements as it can report on local structural transitions. However, due to limited photon budgets, single-molecule fluorescence imaging often requires tradeoffs between high signal-to-noise ratio and long observation times (27). In a combined assay, a mechanical signal can in principle provide an independent temporal reference for interpreting fluorescence data, addressing challenges associated with detecting state transitions in noisy FRET traces. Using the DNA hairpin system, we obtained high-resolution FRET measurements of molecular states by averaging fluorescence intensities over many transitions, identified and aligned in time via the extension signal (Fig. 3). We recorded 147 opening transitions in a single molecule, using low fluorescence excitation conditions to extend the observation window at the expense of signal-to-noise ratio (Fig. 3 a; Fig. S8). After postsynchronization (28) to transitions in extension, the averaged FRET data (Fig. 3 b) show two well-resolved states of the hairpin, as expected (9, 24). When applied to systems that visit multiple states during a single mechanical step, this approach may also reveal short-lived structural intermediates as transients in the averaged FRET data.
Figure 3.
High-resolution FRET measurements of molecular states by postsynchronization of fluorescence data to mechanical measurements. DNA hairpin state transitions (yellow step function) were assigned based on the Δz signal; data from (a) were excised, aligned in time to the opening event, and averaged to generate (b). Fluorescence intensity is acquired at 100 Hz. See Fig. S8 for full data trace and further analysis.
FluoRBT expands the toolbox of fluorescence-enabled force spectroscopy techniques by providing the capability to measure FRET simultaneously with DNA torque, twist, and extension. This capability complements tools that apply forces and exclusively measure FRET (9, 29), or combine the measurement of fluorescence intensity (6, 8, 30) or FRET (7, 10, 11, 12, 13) with extension alone. The use of magnetic tweezers provides constant force without the need for feedback, and may allow the technique to be extended for parallel measurements on multiple molecules. Alternative methods for measuring torque and twist (19, 31, 32, 33) could in principle also be combined with single-molecule fluorescence, but do not provide the high spatiotemporal resolution in torque and twist achieved by RBT (18, 19, 31). FluoRBT may further be extended for imaging of more than two fluorophores and for multicolor FRET, facilitated by the spatial separation of the fluorescence excitation and emission paths (23). The combination of high-speed evanescent dark field tracking (34) with FRET implemented here can also be adapted for investigations of diverse molecular machines, including cytoskeletal motors and rotary ATPases (16). In future applications, FluoRBT may be used to conduct detailed multiparameter studies of DNA structures under torque, and to map the structural conformations of nucleoprotein machines such as gyrase (18, 22) or RNA polymerase (35, 36, 37) during active cycling through functional states, where well-defined mechanical transitions could be used to temporally align FRET signals repeated using multiple dye pairs, placing constraints on modeling (38) dynamically interconverting molecular architectures.
Author Contributions
I.E.I., P.L., F.C.O., and Z.B. designed research. I.E.I., P.L., F.C.O, and A.I. performed research. C.H.S. and A.C.P. contributed modeling and analytic tools. I.E.I. analyzed data. Z.B. supervised research. I.E.I. and Z.B. wrote the manuscript with input from all authors.
Acknowledgments
This work was supported by a Tusher Family Stanford Interdisciplinary Graduate Fellowship to I.E.I. and by a National Institutes of Health grant GM106159 to Z.B.
Editor: Enrique De La Cruz.
Footnotes
Paul Lebel’s present address is Chan Zuckerberg Biohub, San Francisco, California.
Florian C. Oberstrass’s present address is Ultima Genomics, Inc., Fremont, California.
Supporting Materials and Methods, eight figures, two tables, one movie, and one data file are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31246-8.
Supporting Citations
References (39, 40, 41, 42, 43, 44, 45, 46, 47) appear in the Supporting Material.
Supporting Material
Movie showing simultaneous imaging of the rotor bead (left), and the donor (middle) and acceptor (right) dyes on a DNA hairpin under tension. Opening and closing of the hairpin leads to concurrent changes in rotor bead extension and donor and acceptor intensity, shown as moving traces at the top of each channel. The state of the hairpin is indicated in the bottom left corner, and time is shown in the bottom right corner. Messages indicating donor and acceptor photobleaching also appear in the respective channels. The rotor bead was imaged at 2000 Hz, but only every 100th frame is shown in this movie, and the extension trace is block-averaged to 20 Hz. Fluorescence was imaged at 20 Hz. The movie plays in real time. Data and frames from this trace are also shown in Figure S2.
MATLAB Figure. FluoRBT trace showing many transitions between the open and closed states of a DNA hairpin. Low fluorescence excitation is used to extend the lifetime of the fluorophores. This trace is used for making high-resolution FRET measurements of the hairpin states by post-synchronization of fluorescence intensity to extension data (Fig. 3 and Fig. S8). Top: Raw (2000 Hz, blue) and block-averaged (100 Hz, black) δz, and state assignment by HMM fit (yellow). Middle: Donor (100 Hz, green) and acceptor (orange) fluorescence intensity. Bottom: Instantaneous FRET efficiency.
References
- 1.Dulin D., Lipfert J., Dekker N.H. Studying genomic processes at the single-molecule level: introducing the tools and applications. Nat. Rev. Genet. 2013;14:9–22. doi: 10.1038/nrg3316. [DOI] [PubMed] [Google Scholar]
- 2.Fazal F.M., Block S.M. Optical tweezers study life under tension. Nat. Photonics. 2011;5:318–321. doi: 10.1038/nphoton.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ha T. Single-molecule methods leap ahead. Nat. Methods. 2014;11:1015–1018. doi: 10.1038/nmeth.3107. [DOI] [PubMed] [Google Scholar]
- 4.Gross P., Farge G., Wuite G.J.L. Combining optical tweezers, single-molecule fluorescence microscopy, and microfluidics for studies of DNA-protein interactions. In: Walter N.G., editor. Methods in Enzymology. Academic Press; Cambridge, MA: 2010. pp. 427–453. [DOI] [PubMed] [Google Scholar]
- 5.Cordova J.C., Das D.K., Lang M.J. Combining single-molecule manipulation and single-molecule detection. Curr. Opin. Struct. Biol. 2014;28:142–148. doi: 10.1016/j.sbi.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Lang M.J., Fordyce P.M., Block S.M. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat. Methods. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee M., Kim S.H., Hong S.C. Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension. Proc. Natl. Acad. Sci. USA. 2010;107:4985–4990. doi: 10.1073/pnas.0911528107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comstock M.J., Ha T., Chemla Y.R. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat. Methods. 2011;8:335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long X., Parks J.W., Stone M.D. Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 2013;41:2746–2755. doi: 10.1093/nar/gks1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemmerich F.E., Swoboda M., Schlierf M. Simultaneous single-molecule force and fluorescence sampling of DNA nanostructure conformations using magnetic tweezers. Nano Lett. 2016;16:381–386. doi: 10.1021/acs.nanolett.5b03956. [DOI] [PubMed] [Google Scholar]
- 11.Comstock M.J., Whitley K.D., Chemla Y.R. Protein structure. Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science. 2015;348:352–354. doi: 10.1126/science.aaa0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suksombat S., Khafizov R., Chemla Y.R. Structural dynamics of E. coli single-stranded DNA binding protein reveal DNA wrapping and unwrapping pathways. eLife. 2015;4:e08193. doi: 10.7554/eLife.08193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duesterberg V.K., Fischer-Hwang I.T., Block S.M. Observation of long-range tertiary interactions during ligand binding by the TPP riboswitch aptamer. eLife. 2015;4:e12362. doi: 10.7554/eLife.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishijima A., Kojima H., Yanagida T. Simultaneous observation of individual ATPase and mechanical events by a single myosin molecule during interaction with actin. Cell. 1998;92:161–171. doi: 10.1016/s0092-8674(00)80911-3. [DOI] [PubMed] [Google Scholar]
- 15.Adachi K., Oiwa K., Kinosita K., Jr. Coupling of rotation and catalysis in F(1)-ATPase revealed by single-molecule imaging and manipulation. Cell. 2007;130:309–321. doi: 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 16.Sugawa M., Okazaki K., Nishizaka T. F1-ATPase conformational cycle from simultaneous single-molecule FRET and rotation measurements. Proc. Natl. Acad. Sci. USA. 2016;113:E2916–E2924. doi: 10.1073/pnas.1524720113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryant Z., Stone M.D., Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 18.Lebel P., Basu A., Bryant Z. Gold rotor bead tracking for high-speed measurements of DNA twist, torque and extension. Nat. Methods. 2014;11:456–462. doi: 10.1038/nmeth.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bryant Z., Oberstrass F.C., Basu A. Recent developments in single-molecule DNA mechanics. Curr. Opin. Struct. Biol. 2012;22:304–312. doi: 10.1016/j.sbi.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oberstrass F.C., Fernandes L.E., Bryant Z. Torque spectroscopy of DNA: base-pair stability, boundary effects, backbending, and breathing dynamics. Phys. Rev. Lett. 2013;110:178103. doi: 10.1103/PhysRevLett.110.178103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberstrass F.C., Fernandes L.E., Bryant Z. Torque measurements reveal sequence-specific cooperative transitions in supercoiled DNA. Proc. Natl. Acad. Sci. USA. 2012;109:6106–6111. doi: 10.1073/pnas.1113532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basu A., Parente A.C., Bryant Z. Structural dynamics and mechanochemical coupling in DNA gyrase. J. Mol. Biol. 2016;428:1833–1845. doi: 10.1016/j.jmb.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman L.J., Chung J., Gelles J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys. J. 2006;91:1023–1031. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodside M.T., Behnke-Parks W.M., Block S.M. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. USA. 2006;103:6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sindbert S., Kalinin S., Seidel C.A.M. Accurate distance determination of nucleic acids via Förster resonance energy transfer: implications of dye linker length and rigidity. J. Am. Chem. Soc. 2011;133:2463–2480. doi: 10.1021/ja105725e. [DOI] [PubMed] [Google Scholar]
- 26.Grashoff C., Hoffman B.D., Schwartz M.A. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy R., Hohng S., Ha T. A practical guide to single-molecule FRET. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrov A., Chen J., Puglisi J.D. Single-molecule analysis of translational dynamics. Cold Spring Harb. Perspect. Biol. 2012;4:a011551. doi: 10.1101/cshperspect.a011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hohng S., Zhou R., Ha T. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the Holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitley K.D., Comstock M.J., Chemla Y.R. Elasticity of the transition state for oligonucleotide hybridization. Nucleic Acids Res. 2017;45:547–555. doi: 10.1093/nar/gkw1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipfert J., Kerssemakers J.W.J., Dekker N.H. Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments. Nat. Methods. 2010;7:977–980. doi: 10.1038/nmeth.1520. [DOI] [PubMed] [Google Scholar]
- 32.Lipfert J., Wiggin M., Dekker N.H. Freely orbiting magnetic tweezers to directly monitor changes in the twist of nucleic acids. Nat. Commun. 2011;2:439. doi: 10.1038/ncomms1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.La Porta A., Wang M.D. Optical torque wrench: angular trapping, rotation, and torque detection of quartz microparticles. Phys. Rev. Lett. 2004;92:190801. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- 34.Isojima H., Iino R., Tomishige M. Direct observation of intermediate states during the stepping motion of kinesin-1. Nat. Chem. Biol. 2016;12:290–297. doi: 10.1038/nchembio.2028. [DOI] [PubMed] [Google Scholar]
- 35.Abbondanzieri E.A., Greenleaf W.J., Block S.M. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma J., Bai L., Wang M.D. Transcription under torsion. Science. 2013;340:1580–1583. doi: 10.1126/science.1235441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakraborty A., Wang D., Ebright R.H. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi U.B., Strop P., Weninger K.R. Single-molecule FRET-derived model of the synaptotagmin 1-SNARE fusion complex. Nat. Struct. Mol. Biol. 2010;17:318–324. doi: 10.1038/nsmb.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalinin S., Peulen T., Seidel C.A.M. A toolkit and benchmark study for FRET-restrained high-precision structural modeling. Nat. Methods. 2012;9:1218–1225. doi: 10.1038/nmeth.2222. [DOI] [PubMed] [Google Scholar]
- 40.Nishizaka T., Hasimoto Y., Masaike T. Simultaneous observation of chemomechanical coupling of a molecular motor. In: Mashanov G.I., Batters C., editors. Single Molecule Enzymology: Methods and Protocols. Humana Press; Totowa, NJ: 2011. pp. 259–271. [DOI] [PubMed] [Google Scholar]
- 41.Sarkar A., Robertson R.B., Fernandez J.M. Simultaneous atomic force microscope and fluorescence measurements of protein unfolding using a calibrated evanescent wave. Proc. Natl. Acad. Sci. USA. 2004;101:12882–12886. doi: 10.1073/pnas.0403534101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe T.M., Sato T., Higuchi H. Three-dimensional nanometry of vesicle transport in living cells using dual-focus imaging optics. Biochem. Biophys. Res. Commun. 2007;359:1–7. doi: 10.1016/j.bbrc.2007.04.168. [DOI] [PubMed] [Google Scholar]
- 43.Milescu, L. S., C. Nicolai, and J. Bannen. 2000–2013. QuB Software.
- 44.Huguet J.M., Bizarro C.V., Ritort F. Single-molecule derivation of salt dependent base-pair free energies in DNA. Proc. Natl. Acad. Sci. USA. 2010;107:15431–15436. doi: 10.1073/pnas.1001454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng G., Lu X.-J., Olson W.K. Web 3DNA—a web server for the analysis, reconstruction, and visualization of three-dimensional nucleic-acid structures. Nucleic Acids Res. 2009;37:W240–W246. doi: 10.1093/nar/gkp358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stein I.H., Schüller V., Liedl T. Single-molecule FRET ruler based on rigid DNA origami blocks. ChemPhysChem. 2011;12:689–695. doi: 10.1002/cphc.201000781. [DOI] [PubMed] [Google Scholar]
- 47.Woźniak A.K., Schröder G.F., Oesterhelt F. Single-molecule FRET measures bends and kinks in DNA. Proc. Natl. Acad. Sci. USA. 2008;105:18337–18342. doi: 10.1073/pnas.0800977105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie showing simultaneous imaging of the rotor bead (left), and the donor (middle) and acceptor (right) dyes on a DNA hairpin under tension. Opening and closing of the hairpin leads to concurrent changes in rotor bead extension and donor and acceptor intensity, shown as moving traces at the top of each channel. The state of the hairpin is indicated in the bottom left corner, and time is shown in the bottom right corner. Messages indicating donor and acceptor photobleaching also appear in the respective channels. The rotor bead was imaged at 2000 Hz, but only every 100th frame is shown in this movie, and the extension trace is block-averaged to 20 Hz. Fluorescence was imaged at 20 Hz. The movie plays in real time. Data and frames from this trace are also shown in Figure S2.
MATLAB Figure. FluoRBT trace showing many transitions between the open and closed states of a DNA hairpin. Low fluorescence excitation is used to extend the lifetime of the fluorophores. This trace is used for making high-resolution FRET measurements of the hairpin states by post-synchronization of fluorescence intensity to extension data (Fig. 3 and Fig. S8). Top: Raw (2000 Hz, blue) and block-averaged (100 Hz, black) δz, and state assignment by HMM fit (yellow). Middle: Donor (100 Hz, green) and acceptor (orange) fluorescence intensity. Bottom: Instantaneous FRET efficiency.