Abstract
Rhodopsin, a prototypical G protein-coupled receptor, is a membrane protein that can sense dim light. This highly effective photoreceptor is known to be sensitive to the composition of its lipidic environment, but the molecular mechanisms underlying this fine-tuned modulation of the receptor’s function and structural stability are not fully understood. There are two competing hypotheses to explain how this occurs: 1) lipid modulation occurs via solvent-like interactions, where lipid composition controls membrane properties like hydrophobic thickness, which in turn modulate the protein’s conformational equilibrium; or 2) protein-lipid interactions are ligand-like, with specific hot spots and long-lived binding events. By analyzing an ensemble of all-atom molecular dynamics simulations of five different states of rhodopsin, we show that a local ordering effect takes place in the membrane upon receptor activation. Likewise, docosahexaenoic acid acyl tails and phosphatidylethanolamine headgroups behave like weak ligands, preferentially binding to the receptor in inactive-like conformations and inducing subtle but significant structural changes.
Introduction
Lipids serve both structural and functional roles in biological membranes. They act as diffusion barriers that allow the generation of electrochemical potentials, contribute to the selectivity of external cues in signaling and provide a diverse milieu for membrane proteins (1, 2). Many integral membrane proteins are sensitive to the composition of their lipid bilayers (3, 4, 5, 6, 7, 8). Thus, factors that modulate the relative abundance of membrane lipids, such as dietary intake and age, can also influence the activity and organization of proteins (2, 9). Alterations in the lipid composition of membranes have been linked to the onset of various pathologies, including cardiovascular diseases, obesity, cancer, and neural and retinal degeneration (10, 11). Therefore, understanding the molecular basis underlying lipid modulation of membrane proteins is key for developing new and more effective therapies, targeting both membrane proteins and membrane lipids (10, 12).
Lipid effects on membrane protein activity can be grouped into ligand-like and solvent-like effects (1, 12). Ligand-like effects are those where lipids make specific molecular interactions with the protein, including hydrogen bonding and charge-charge interactions (6, 13, 14). We define solvent-like effects as non-ligand-like effects that arise from intrinsic membrane properties, such as bilayer thickness (15, 16, 17), acyl chain order and packing (18), fluidity, curvature elastic stress (19, 20), and lateral pressure (8). Due to their different natures, these effects can often be distinguished by certain characteristics of the lipids from which they originate, namely, their exchange and diffusion rates, lifetimes at the protein surface, lateral and rotational mobility, and the degree to which they require structural specificity when interacting with the protein (1, 9, 21). Ultimately, the relative importance of ligand-like and solvent-like lipid effects on the structure and function of a membrane protein depends on the environment and the nature of the protein itself (9, 22).
G protein-coupled receptors (GPCRs) are ubiquitous seven-transmembrane (7TM) proteins whose primary function is to transduce information across lipid bilayers. They constitute the most numerous and diverse superfamily of proteins, with more than 825 distinct members identified in humans (23). Although GPCRs have a highly conserved topology, each receptor can specifically sense different external stimuli on one side of the membrane and start particular signaling responses on the other by binding a cytosolic partner, usually a G protein (24, 25). These responses mediate signaling pathways that play central roles in many physiological processes, including inflammation, vision, blood pressure, and pain (26, 27, 28, 29). Not surprisingly, one-third to one-half of all small-molecule drugs on the market target GPCRs (30).
Lipids have been shown to influence the function and activity of GPCRs at different levels. For instance, ligand-binding affinity was found to be modulated by cholesterol in the receptor (5-HR) and the chemokine receptor CXCR4; similarly, lipid headgroup composition can modulate the ligand affinity of the -adrenergic receptor (AR) (31, 32, 33). For the smoothened (Smo) receptor, cholesterol alone can act as an activating ligand and initiate signaling (34). In the cytosol, G protein recruitment and post-activation G protein subunit sorting have been observed to be sensitive to the non-lamellar-phase propensity and charge of phospholipid headgroups (35, 36). Receptor dimerization and oligomerization are also thought to be tuned by direct lipid binding or by lipid-composition-dependent bilayer properties. The presence of docosahexaenoic acid (DHA), for example, modifies the rate of association of the dopamine D2 receptor and the adenosine 2A receptor (), whereas palmitoylation and cholesterol also play a role in the formation of μ-opioid receptor oligomers (33, 37, 38).
Experimental and computational evidence have shown that GPCRs can sample ensembles of distinct conformations, which result in a broad range of signaling efficacies and functional differences (39, 40, 41, 42, 43, 44, 45). The equilibrium among these ensembles of conformations has also been proposed to be affected by the lipid composition of membranes through different mechanisms. In particular, putative cholesterol binding sites have been identified in several GPCRs, including the AR and the . These non-annular cholesterol molecules are hypothesized to have allosteric and structure-stabilizing roles (46, 47, 48). Likewise, specific receptor-lipid interactions with anionic phospholipid headgroups have been proposed to stabilize the active conformation of the AR (49). Solvent-like effects have also been shown to alter the activity of these proteins, as is the case with 5-HR, where cholesterol-induced membrane ordering favors receptor activation (18). Given the large variety of membranes that contain GPCRs, it is not unreasonable that the vast functional diversity and specificity of these receptors may result in part from membrane-composition-dependent lipid effects.
Rhodopsin is a prototypical GPCR that mediates scotopic vision. This highly efficient photoreceptor is located in the disk membranes of the outer segment of rod cells in the retina. In its ground state (dark state), rhodopsin has negligible activity, because its covalently bound ligand, retinal, is an exceptional inverse agonist when it is in the 11-cis form (50). Light absorption induces the cis-to-trans isomerization of the ligand (to an agonist) and starts a relaxation process that drives the receptor through a series of spectroscopically distinguishable intermediates (19). This relaxation culminates when rhodopsin reaches an equilibrium between the inactive metarhodopsin I (Meta I) and active metarhodopsin II (Meta II) states. Meta II is the G protein-activating state of the receptor wherein the outer rotation of helix 6 creates a cleft at the cytoplasmic end of the protein that allows the G protein transducin to bind (51, 52). The Meta I-Meta II equilibrium takes place on the millisecond timescale and has been shown to be influenced by temperature, pH, and lipidic environment (4, 53). The activation process ends when retinal is hydrolyzed from the binding pocket producing ligand-free opsin and the receptor is regenerated by binding 11-cis retinal to restart the photocycle (54).
Rhodopsin is a good example of protein-lipid co-evolution for optimal function. The receptor constitutes ∼90% of all proteins in rod outer segment (ROS) disk membranes, and the lipid composition of ROS disk membranes is highly specialized, containing a large fraction of polyunsaturated fatty acids, particularly DHA (55, 56). Studies where rhodopsin was reconstituted in model membranes show that increasing concentrations of DHA enhance rhodopsin’s activity (57, 58, 59) and that this effect is amplified by phosphatidylethanolamine (PE) headgroups (60). In turn, the presence of cholesterol drives the Meta I-Meta II equilibrium toward the inactive state (61). Both solvent-like and ligand-like lipid effects have been proposed to explain these phenomena (9, 62, 63, 64, 65, 66).
Rhodopsin is sensitive to bilayer thickness, membrane order, lipid packing, and curvature elastic stress (67). Accordingly, DHA, which is known to lower the packing density and order of lipid membranes, was proposed to facilitate the conformational transitions that lead to the Meta II state, whereas cholesterol was suggested to counteract these effects by ordering the membrane and increasing its thickness (57, 68). In addition, DHA and PE are non-lamellar-phase-forming lipid components and introduce negative-curvature elastic stress in membranes. Using flash photolysis experiments with different bilayer compositions, Botelho et al. (20) proposed that this so-called membrane frustration might be relieved by the transition to Meta II. Although sophisticated in its conception, this and other models arguing in favor of solvent-like lipid effects do not take into account the heterogeneity of the hydrophobic surface of the protein and the distinct characteristics of the lipids that are close or far from the receptor (69). For instance, PE has hydrogen-bonding capacity, whereas the multiple double bonds of DHA can form π-π stacking interactions with aromatic side chains and the remaining single bonds have low isomerization energy barriers that make them extremely flexible and could potentially facilitate protein-lipid interactions (60, 70, 71). Indeed, saturation-transfer NMR experiments have shown that lipids directly associate with rhodopsin and behave as weak ligands (65, 69). However, the exact molecular details of these putative protein-lipid interactions are still largely unknown.
Here, we investigate the role of lipid membranes in modulating rhodopsin’s structure and dynamics at atomic resolution. We present an ensemble of multi-microsecond all-atom simulations of rhodopsin along its photocycle embedded in 1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphatidylethanolanime (SDPE) membranes. Our analysis of the resulting trajectories suggests that membrane modulation of rhodopsin activity is due to both solvent-like and ligand-like properties of lipids and that these effects are state dependent.
Methods
System construction
Crystal structures have been solved for bovine rhodopsin in the dark state (72), Meta II (52), and opsin (73) (PDB: 1U19, 3PXO, and 3CAP, respectively). We used these structures as the starting points of four of our systems, including dark-state rhodopsin with retinal removed (hereafter referred to as dark opsin). Opsin exists in an equilibrium between inactive-like and active-like receptor conformations. It was crystallized in low-pH conditions and has a Cα root mean-square deviation of 0.51 and 2.8 Å with respect to the Meta II and dark-state structures, respectively (52, 72, 73). However, at pH ∼6.1, its Fourier transform infrared spectrum closely resembles that of the dark state, and the apo protein can activate transducin only very poorly (50). Therefore, we consider dark opsin a model for the hypothesized inactive-like opsin structure.
The starting structure of the Meta I state was obtained from previous simulation work in which the protein was initialized and equilibrated in the dark state and retinal isomerization was accomplished by applying an external potential on the C10-C11=C12-C13 torsion (74). This model of Meta I formation emulates the complex counterion hypothesis formulated by Lüdeke et al. (75) and was validated by direct comparison of the 2H NMR spectra of retinal’s methyl groups calculated from simulation and measured experimentally (74).
We built six independent replicates of each of the five protein structures, for a total of 30 trajectories and ∼163 μs of simulation time. In each system, the receptor was embedded in a lipid bilayer composed of 123 SDPE molecules. These phospholipids contain a PE headgroup, an 18-carbon saturated stearoyl (STEA) chain and an ω3 22-carbon DHA chain with six double bonds. ROS membranes are highly enriched in DHA (∼35–60% of all phospholipid acyl tails), whereas PE constitutes ∼40% of all headgroups. Hence, we chose SDPE as a simplified approximation of rhodopsin’s lipid environment. Experimental work with pure SDPE bilayers is challenging, since these lipids are not only susceptible to oxidation, but also tend to form an inverse hexagonal (HII) phase at physiological temperatures (293–323 K), as repeatedly shown by NMR spectroscopy and x-ray diffraction (76, 77). In practice, SDPE is often titrated in with other lipids like 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine, whose spontaneous monolayer curvatures are not as extreme (70). However, this phase transition does not occur on the timescale of our simulations. Our choice of membrane takes into consideration that the presence of a second lipid species necessarily introduces lateral membrane-reorganization degrees of freedom, which have timescales of hundreds of nanoseconds to microseconds (78, 79, 80). It also reflects that any lipid modulation effects are to be more cleanly discernible if the Meta I-Meta II equilibrium is shifted in a single direction, as is the case for DHA and PE, which favor the active state of the receptor (57). These membranes were constructed according to a procedure analogous to that described by Grossfield et al. (45, 79, 81), fully hydrated with ∼8000 waters, and embedded in 74 Å × 74 Å × 90 Å periodic boxes with 100 mM NaCl (in addition to neutralizing salt). Total system sizes were ∼46,000 atoms. Because the coordinates of the non-protein components of the system are randomized during construction, this approach ensures that every replicate is as independent as possible, with the only caveat being that the same initial protein structure must be used. This protocol has been recently implemented as an automated membrane-building tool in LOOS, a Python/C++ open-source library for molecular dynamics (MD) analysis (82).
Simulation details
We ran these simulations using the NAMD simulation package (version 2.8) (83) with the CHARMM22 force field with CMAP corrections for protein parameters (84, 85) and the CHARMM36 force field for lipid parameters (86). 11-cis and all-trans retinal parameters were kindly provided by Dr. Scott Feller (87). The systems were energy minimized and initially equilibrated for ∼10 ns in the NPAT ensemble. We have found that using a constant lateral surface area as part of energy minimization helps to stabilize the volume of the simulation box, allowing the water in the bulk region to equilibrate. Production was carried out in the NPγT ensemble, with γ = 30 dyn/cm (see Supporting Material, Section S1, for further details). The pressure (1 bar) was regulated using a Langevin piston with an oscillation period of 200 ns and a damping timescale of 100 ns, whereas the temperature (310.15 K) was controlled using a Langevin thermostat with a damping coefficient of 2 . We used the Velocity Verlet integrator with a 2 fs timestep and the SHAKE algorithm to constrain hydrogen-containing bonds (88). For the treatment of long-range electrostatics, we employed the smooth particle-mesh Ewald (PME) summation method (89) with a cutoff of 10 Å and a grid of 75 × 75 × 96 points (∼1 Å/grid point). All simulations were run on the BlueGene/Q supercomputer of the Center for Integrated Research Computing (CIRC) at the University of Rochester.
Simulation analysis
Analysis was performed with the trajectories sampled at 1 ns resolution (except as indicated), with the first 500 ns excluded to allow the protein and membrane to relax. Unless explicitly noted, we used either existing analysis tools included in the current LOOS distribution (version 2.3.1) (82) or in-house code generated using LOOS. The source code for LOOS is available for download at GitHub (http://github.com/GrossfieldLab/loos). Trajectory visualization, image rendering and data plotting were carried out using VMD (version 1.9.2) (90), PyMOL (version 1.8.0.0) (91), and gnuplot (version 5.0, www.gnuplot.info). We refer the reader to the Supporting Material for an extensive description of the analysis mentioned in the main text.
Results
We examined the possibility that lipids could affect rhodopsin’s function by differently modulating the receptor along its photocycle via solvent-like or ligand-like effects. We carried out extensive simulations of rhodopsin in SDPE membranes with starting structures corresponding to the dark state, Meta I, Meta II, opsin, and dark opsin (an apo dark-like state; see Methods for details). The aggregate data set (∼163 μs) included six independently constructed replicates of each protein state that we used to assess the statistical significance of our observations.
Changes in membrane structure during rhodopsin activation
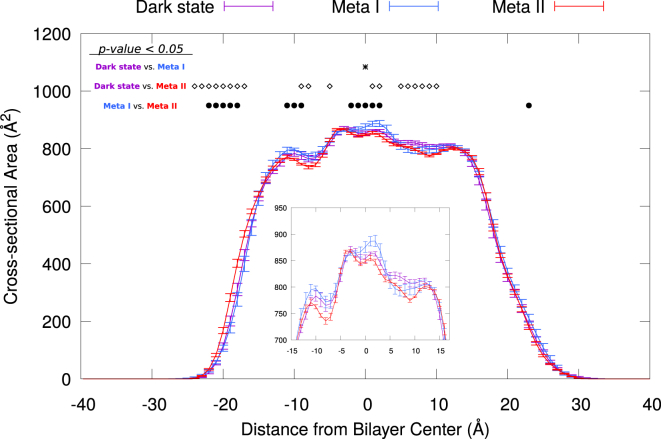
Reconstitution experiments of rhodopsin in different membrane compositions have shown that the presence of PE headgroups and DHA acyl chains favors rhodopsin activation (3, 57). This effect has been proposed to be the result of curvature elastic stress in the membrane coupled with changes in the shape of the receptor during activation (19, 20, 92). To examine any changes in the shape of the protein in the context of our simulations, we computed the cross-sectional area of the receptor for the dark, Meta I, and Meta II states, which are part of rhodopsin’s activation process. Briefly, we sliced the systems into thin sections along the membrane normal that were partitioned using a Voronoi decomposition in the plane of the membrane. The cross-sectional area of each thin slice was taken as the smallest convex hull delimiting the atoms of the protein (see Supporting Material, Section S2, for further details). Figs. 1 and S2 show the average cross-sectional areas of rhodopsin along the bilayer normal computed from the dark-state, Meta I, and Meta II systems. Our simulations reveal small changes in the transmembrane region of the protein, wherein the receptor undergoes a decrease in cross-sectional area (; Fig. 1, inset) and appears to be slightly (but significantly) more elongated in the Meta II state. Treating the individual trajectories as single measurements, we were able to calculate accurate error bars and also determine whether the noted changes are statistically significant (see Figs. 1 and S2, b–d).
Figure 1.
Cross-sectional area of rhodopsin during activation. Average cross-sectional area profiles of rhodopsin states are calculated from six trajectories per protein state. Error bars indicate the mean ± SE, computed by treating each simulation as a single data point. Significantly different cross-sectional areas among protein states (p < 0.05) were determined with a t-test for each pairwise comparison and are annotated at the top by stars (dark state versus Meta I), open diamonds (dark state versus Meta II), and dots (Meta I versus Meta II). The inset corresponds to the cross-sectional areas of the −15 < z < 15 region, with the y axis scaled up. To see this figure in color, go online.
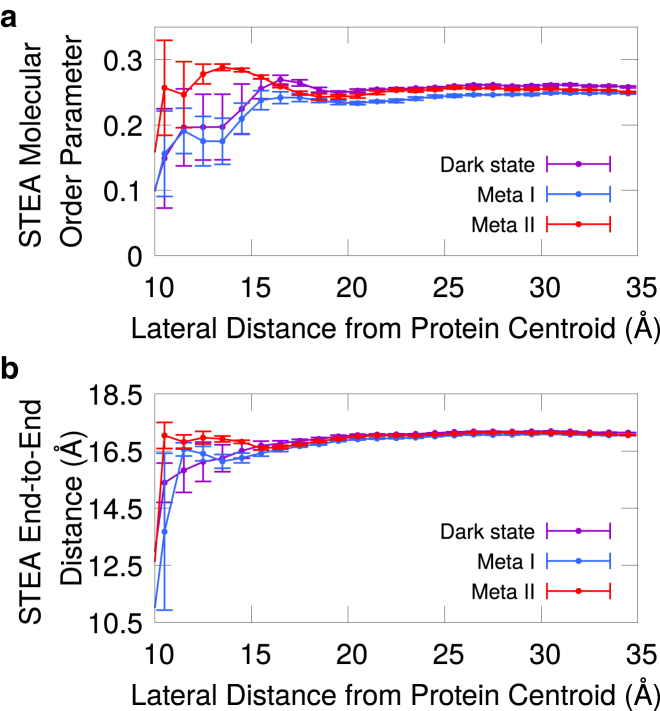
To understand the effects of these local changes in cross-sectional area, we studied the organization and structure of the lipids surrounding the receptor. Experimentally, 2H quadripolar splittings measured via NMR provide a way to quantify the average orientations sampled by C-D bonds with respect to the membrane normal along perdeuterated fatty acid tails (93). This observable is customarily referred to as C-D bond order parameters and is used to gauge membrane disorder and assess the average structure of lipid bilayers. We investigated activation-induced changes in local membrane disorder by computing a whole-chain analog of C-D bond order parameters (hereafter molecular order parameters), where the angle between the second or third principal axes of phospholipid acyl chains and the membrane normal is averaged instead (see Supporting Material, Section S3, for details).
We calculated the molecular order parameters of DHA and STEA acyl chains as a function of distance from the receptor to track down changes in the structure of the bilayer during rhodopsin activation. At 11–15 from the centroid of the protein, we observe that where STEA order is reduced compared to the distal region of the membrane (>25 ) in the dark and Meta I states, it is slightly increased in the case of Meta II (Fig. 2 a). These differences in STEA molecular order parameters between the inactive (dark and Meta I states) and active (Meta II state) distributions are statistically significant in this region of the membrane (Fig. S3). A lower molecular order parameter is indicative of either increased chain disorder or systematic tilting of the chain. Therefore, our analysis of the dark-state, Meta I, and Meta II trajectories suggests that there is a local ordering effect in the lipids surrounding the receptor upon activation, wherein STEA acyl chains become more ordered around the protein. This observation is further supported by the distribution of end-to-end distances of STEA acyl chains as a function of distance from the protein, which shows that these fatty acids are in more stretched conformations near the receptor in the Meta II state compared to the dark or Meta I states (Fig. 2 b). Taken together, these results suggest that when the transmembrane region of the protein gets elongated upon activation, the lipid tails in the vicinity of the protein (∼11–15 from the protein centroid) can compensate for the resulting hydrophobic mismatch by adopting more stretched conformations. This end-to-end elongation of the acyl chains in Meta II brings about a local ordering of the membrane that is not present in the dark state or Meta I (Fig. 2). The decrease in the receptor’s cross-sectional area in Meta II, which occurs mainly in the hydrocarbon region (Fig. S2), could also facilitate these changes in the structure of the membrane.
Figure 2.
Activation-induced changes in the structure of the membrane. (a) Average distributions of molecular order parameters of STEA acyl chains as a function of distance from the protein centroid are shown. (b) Average distributions of STEA end-to-end distances as a function of distance from the protein centroid computed from atoms C1 and C18 of the acyl chain are shown. These averages were computed from the distributions of six trajectories per protein state. Error bars indicate the mean ± SE, calculated by treating each trajectory as a single data point. To see this figure in color, go online.
State dependence of lipid-binding lifetimes
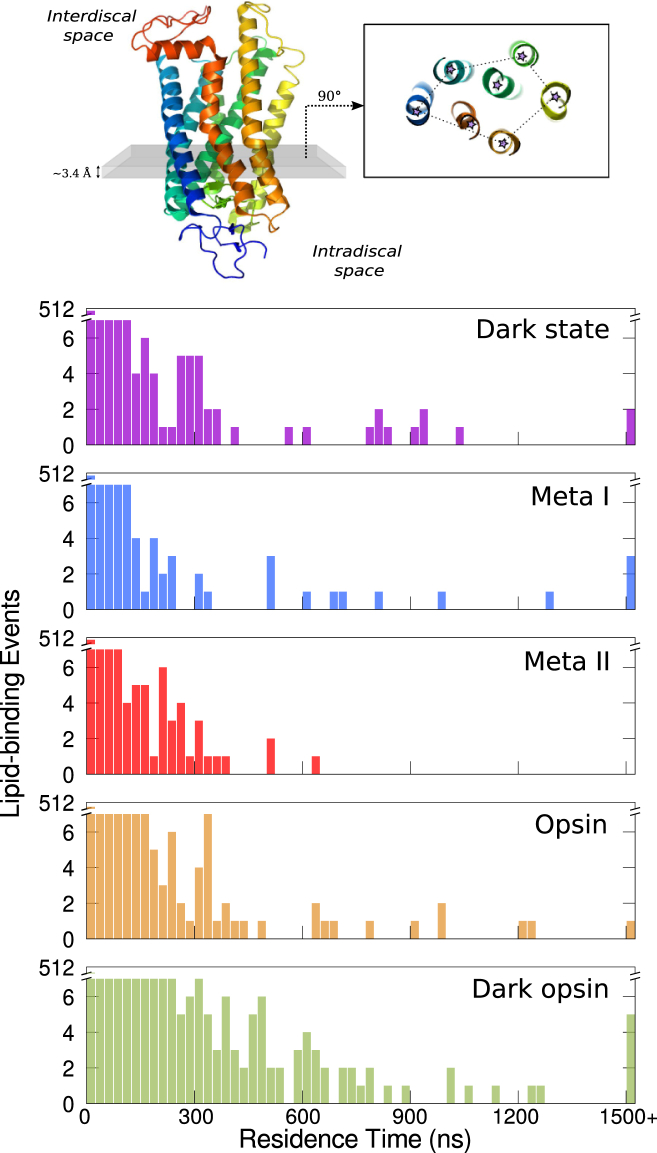
Differences in the structure and distribution of phospholipids surrounding the receptor during activation may also modulate direct interactions between rhodopsin and annular or non-annular lipids. In previous work, we had identified regions on the protein surface that are likely to be enriched in tight-packing lipid events (79). Here, with the benefit of far longer simulations in different functional states, we identified lipids that form long-lived interactions where the chains intercalate themselves between the helices. To determine whether these interactions are state dependent, we monitored the lifetimes of lipid-binding events in rhodopsin simulations starting from different functional states. For the purpose of our analysis, we reasoned that instances of fatty acid tails penetrating the helical bundle, as opposed to acyl chains just contacting the surface of the protein, would be more stable and more clearly discernible. For this reason, lipid-binding events were defined by the presence of one or more lipid heavy atoms inside the protein core at a given time point. To discriminate the interior from the exterior of the protein, we again took thin slices of the receptor along the membrane normal and applied a Voronoi analysis, using only the geometric centroids of the receptor’s transmembrane helices to define convex hulls (see Figs. 3, S4, and S5, and refer to Supporting Material, Section S5, for further details).
Figure 3.
Residence times of DHA acyl chains inside rhodopsin. Top: Scheme illustrating the approach we employed to quantify DHA-binding events in our simulations is shown. Briefly, we took thin slices of the system along the membrane normal and used a Voronoi analysis to find the smallest convex hull containing all transmembrane helices in a slice. We counted a DHA tail as being inside the protein if one or more of its heavy atoms was inside any of these convex hulls. Bottom: Distributions of the residence times of DHA-binding events by protein state computed from the full trajectories (including the first 500 ns of the simulation). Binding events >1500 ns are grouped together. To see this figure in color, go online.
Fig. S4 shows the distributions of residence times of DHA and STEA binding events computed from the full trajectories. The total number of DHA penetration events was larger than the total number of STEA penetration events for every protein state. This result is consistent with previous work suggesting that DHA is enriched at the surface of the receptor and can penetrate deeper than STEA into rhodopsin’s core (63, 79, 94). Since long-lived binding events are more likely to have ligand-like effects and induce changes in the protein, we examined the possibility that the observed preference for DHA translated to longer residence times for this fatty acid inside the receptor. There were at least 1.7 times more long-lived DHA binding events (those lasting >500 ns) than STEA binding events in each protein state, suggesting that DHA-rhodopsin interactions are more stable. The vast majority of the long-lived DHA penetration events occurred when the protein was in inactive-like states—dark, Meta I, or dark opsin—as well as in one opsin trajectory that spontaneously deactivated ∼550 ns into the simulation (Fig. 3). To assess the statistical significance of these observations, we performed a t-test to compare the number of total, short-lived (<20 ns), and long-lived DHA penetration events per microsecond of simulation time among protein states. Interestingly, although the number of total and short-lived events per microsecond was not significantly different between active-like and inactive-like protein states (p > 0.05), the number of long-lived events in the dark-state, Meta I, and dark-opsin trajectories were significantly different (p < 0.05) from the ones found in the Meta II simulations. We repeated these calculations excluding the first 500 ns and the first microsecond from the simulations (see Fig. S5). Although the number of lipid penetrations varied in each case, the differences between the inactive and active states in terms of their observed long-lived binding events are robust. Overall, these results suggest that DHA interacts with rhodopsin in a state- and conformation-dependent manner.
Ligand-like effects of DHA on protein structure and ligand behavior
The premise that DHA-rhodopsin interactions might be state dependent prompted us to investigate the possible origins of this preference. It has been previously proposed that the intrinsic geometry of a given protein state might be better suited to accommodate particular lipid species, such as DHA (70). Another possibility is that specific regions or specific residues on the protein surface differentially associate with lipids in active-like and inactive-like conformations of the receptor, depending on their accessibility and availability for binding (63, 67).
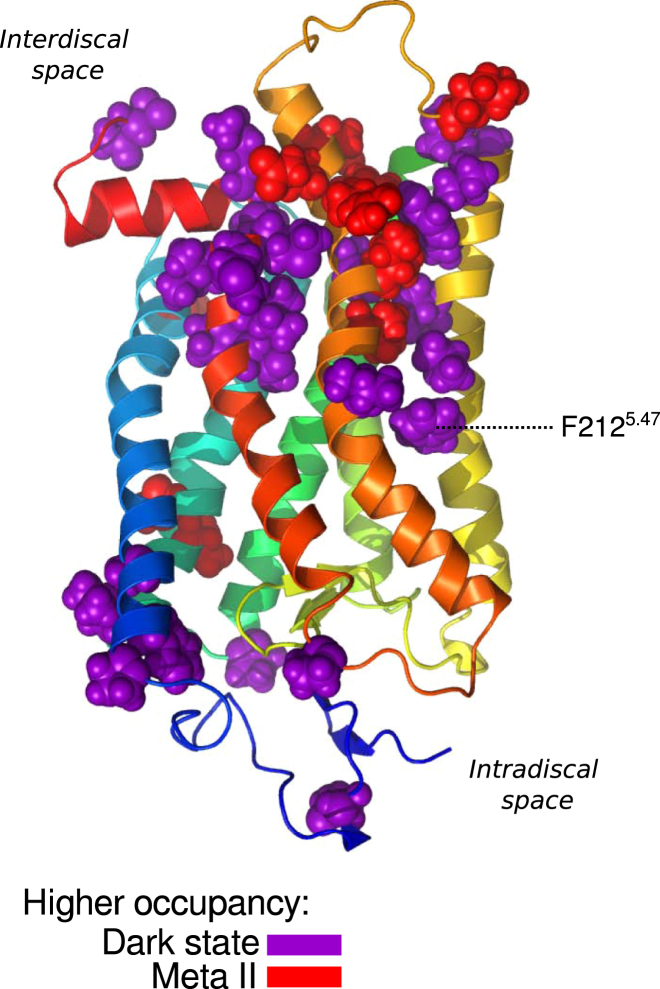
To test the latter hypothesis, we investigated whether there were also significant differences among states regarding protein residues involved in DHA-binding events. Specifically, we computed the likelihood for a given residue to make contact with DHA (see Supporting Material, Section S6, for further details). For our six replicates per protein state, we calculated the fraction of simulation time that each protein residue was in contact with any given DHA acyl chain and normalized this quantity by the length of the trajectory. Using these results, we carried out a t-test to find which residues were occupied by DHA at significantly differently rates in the five protein states, comparing two states at a time. A table with these comparisons is shown in Fig. S6. For the dark state and Meta II (which can be thought of as the end points of the rhodopsin activity spectrum), these residues are mapped in sphere representation onto the dark-state crystal structure (PDB: 1U19 (72)) in Fig. 4. Residues with higher occupancies (i.e., normalized fraction of simulation time in contact with DHA) in the dark state are colored purple, whereas residues with higher occupancies in Meta II are colored red. For clarity, residues with small overall occupancies are not shown, even if the differences between protein states are statistically significant. Unsurprisingly, most of the residues are located in the interface of helices 5 and 6 and the interdiscal end of helix 7, where the largest conformational changes occur during activation.
Figure 4.
Protein residues with significantly different DHA occupancies in the dark state and Meta II. Amino acids that spent significantly different simulation times (on average) in contact with DHA in either the dark state or the Meta II trajectories are shown in sphere representation. Significance was judged as p < 0.05, using six trajectories per protein state as single measurements for the calculation. To see this figure in color, go online.
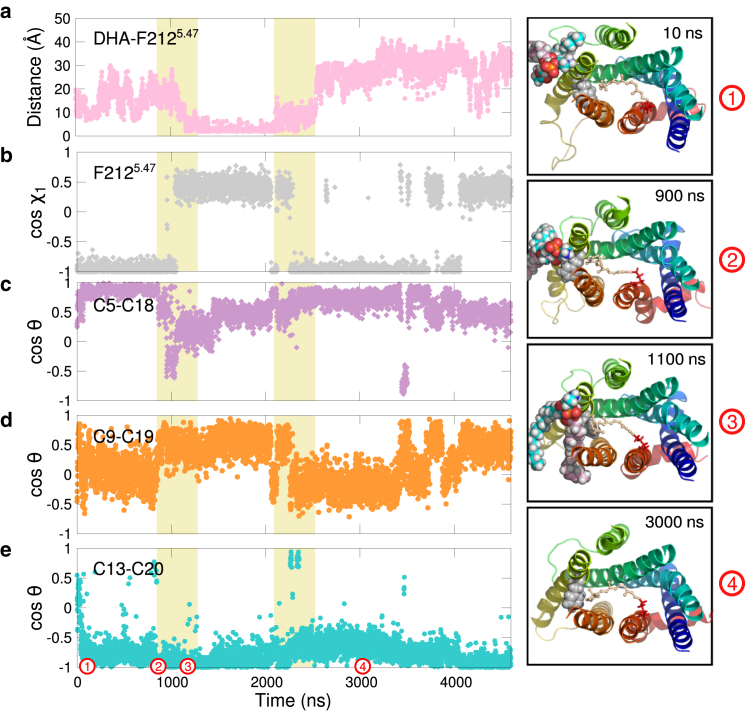
Residue F212, or 5.47 in Ballesteros-Weinstein notation (95), is found in the middle section of helix 5 and spends significantly more time in contact with DHA when the protein is in the dark state (65% of the simulation time) than when it is in Meta II (21% of the simulation time). Fig. 5 a shows a direct interaction between a DHA acyl chain and this residue in one of our dark-state trajectories, as defined by the minimum interatomic distance between the two. In the simulation, undergoes a rotameric switch when the DHA tail of the phospholipid binds between helices 5 and 6 (Fig. 5, a and b, 950 ns). This is not an isolated event; indeed, the rate of isomerization was at least 1.5 times higher in the dark-state trajectories than in Meta II.
Figure 5.
Direct DHA-rhodopsin interactions can alter the orientations of aromatic side chains and retinal methyl groups. (a) Time series of the minimum heavy-atom-to-heavy-atom distance between a DHA acyl chain and protein residue F2125.47 in one of the dark-state trajectories. (b) Time series of the χ1 torsion angle of F2125.47 computed from the same dark-state trajectory as in (a). (c–e) Retinal methyl orientations as a function of simulation time computed from the same trajectory as in (a) and (b): (c) C5-methyl (C5-C18), (d) C9-methyl (C9-C19), and (e) C13-methyl (C13-C20). Right column: Time stills showing rhodopsin viewed from the intradiscal side of the membrane in cartoon representation (only TM segments are shown for clarity). K2967.43 and retinal are shown in stick representation. An SDPE phospholipid is drawn in sphere representation. To see this figure in color, go online.
Evidence from previous simulation work and NMR experiments shows that polyunsaturated acyl chains have a strong preference for aromatic amino acid residues due to interactions between the double bonds in the hydrocarbon tails and aromatic side chains, and that these interactions seem to be largely non-specific (63, 67, 71). Notably, rhodopsin’s ligand-binding site is lined by several aromatic residues that directly interact with the hydrophobic ligand (96). For instance, is positioned within ∼5–7 of retinal’s β-ionone ring (97). Therefore, we hypothesized that the isomerizations of some of these aromatic residues induced by DHA binding could propagate to the ligand, altering its orientation. To test this, we monitored the orientations of the three methyl groups of retinal, as described by Leioatts et al. (45). Briefly, we tracked the vector orientations between retinal atom pairs C5-C18, C9-C19, and C13-C20 with respect to the membrane normal as a function of simulation time (see Supporting Material, Section S7, for further details). Fig. 5, c–e, shows time series of retinal methyl-group orientations computed from the same dark-state trajectory as in Fig. 5, a and b. There, retinal orientation is concomitantly altered with the torsion angle of F2125.47.
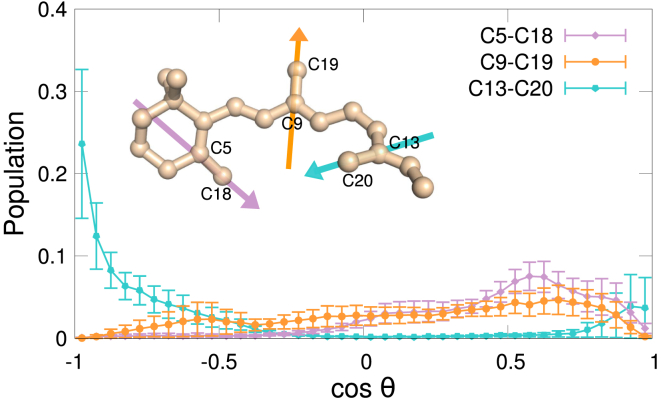
Although Fig. 5 focuses on a single event in one trajectory, we observed other instances of coupling between changes in the orientation of retinal’s methyl groups, the torsion angles of aromatic amino acids and the formation/breakage of interactions between these residues and DHA acyl chains. This was especially the case for F2125.47, W2656.46, and Y2686.51, which account for ∼30% of the ligand’s binding pocket in the ground state (24). When we histogrammed the observed retinal methyl orientations in the dark state and averaged the resulting distributions of our six trajectories, we found that the three methyl groups sampled a wide range of orientations (Fig. 6), suggesting that long-lived interactions between DHA and aromatic residues are at least partially responsible for alterations in the orientation of the ligand.
Figure 6.
Retinal methyl orientations in dark-state rhodopsin. Average distributions of the orientations sampled by retinal methyl groups: C5-methyl (C5-C18), C9-methyl (C9-C19), and C13-methyl (C13-C20) are shown. cos θ = 1 means that the vector is parallel to the membrane normal (i.e., pointing toward the interdiscal/intracellular side); and cos θ = −1 indicates that it is anti-parallel to the membrane normal (i.e., pointing toward the intradiscal/extracellular side). Error bars indicate the mean ± SE of six trajectories. To see this figure in color, go online.
PE headgroups alter the conformation of intracellular loop 3
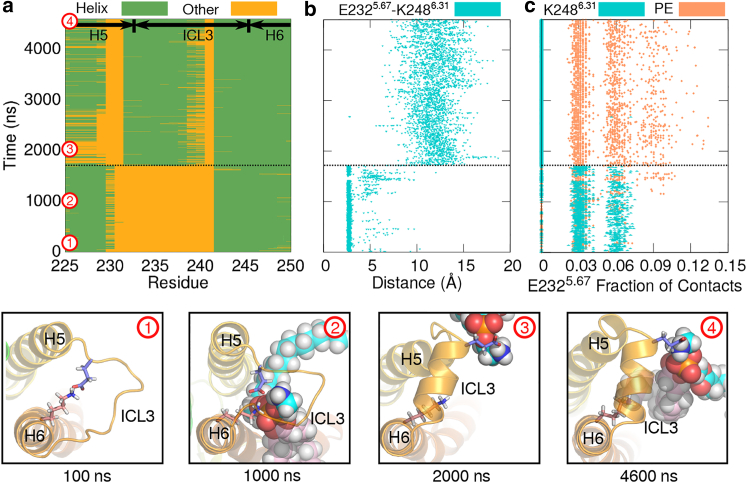
Intracellular loop 3 (ICL3) of rhodopsin connects helices 5 and 6 at the cytoplasmic side of the receptor and has been implicated in protein-protein interactions with transducin, rhodopsin kinase, and arrestin (98, 99, 100, 101, 102). This region possesses intrinsic structural plasticity, as evidenced by its high B-factors and lack of electron density in some dark-state crystal structures; in other GPCRs, it must be truncated or artificially stabilized by a fusion protein for crystalization to occur (103, 104). Our simulations suggest that this protein region is also altered by direct protein-lipid interactions. From crystal structures of the active state, ICL3 is known to become partially structured upon activation (52, 73). However, we observed that the loop can acquire some degree of partial structuring even when the receptor is in inactive-like protein states (dark state, Meta I, and dark opsin).
Fig. 7 a shows the secondary structure assignments of residues Q2255.60–V2506.33 computed from one of the dark-state trajectories (see Supporting Material, Section S9, for details). A salt bridge between E2325.67 and K2486.31 breaks concomitantly with ICL3 gaining helical structure (Fig. 7 b). Surprisingly, we found that this interaction is bridged and then disrupted by PE headgroups (Fig. 7, bottom row). The amino group of PE forms a hydrogen bond with the side chain of E2325.67, displacing K2486.31, whereas K2486.31 hydrogen-bonds with PE’s phosphate group (Fig. 7 c); similar behavior was observed in five of the six dark-state trajectories. Although Fig. 7 shows a single trajectory, we consistently saw ICL3 undergoing a conformational change and PE bridging the interaction between E2325.67 and K2486.31 in the dark-state (Fig. S7), Meta I, and dark-opsin simulations. The resulting helix varied in length and starting position. Its average length was 5.2 ± 0.5 residues over the three inactive-like protein states, with residues 233–235 forming the central part of the resulting secondary structure. These observations suggest that PE headgroups favor the open state of the salt bridge and stabilize the partial structuring of ICL3 in inactive-like protein states, and they further support the view that rhodopsin structure can be modulated by direct protein-lipid interactions in a conformation-dependent manner.
Figure 7.
ICL3 samples multiple conformations in inactive-like rhodopsin. (a) Time course of secondary structure assignments of protein residues Q2255.60–V2506.33 in one of the dark-state trajectories is shown. ICL3 partially turns into a helix ∼1700 ns into the simulation. (b) Time course of the minimum distance between E2325.67 and K2486.31 computed from the same dark-state trajectory as in (a). (c) Fraction of contacts between E2325.67 and K2486.31 or PE headgroups computed from the same trajectory are shown. The fraction of contacts between PE and E2325.67 increases concomitantly with the breakage of the E2325.67–K2486.31 salt bridge and ICL3 structuring. Bottom row: Conformation of ICL3 at different time points of the same trajectory, seen from the cytoplasmic side of the receptor. E2325.67 and K2486.31 are shown in stick representation. SDPE phospholipids located within 3.2 Å of E2325.67 are shown in sphere representation. To see this figure in color, go online.
Discussion
MD simulations have been used in the past to examine protein-lipid interactions in rhodopsin systems (63, 64, 79). However, most of this work predated the crystal structure of Meta II (52) and, due to the computational power available at the time, was limited to the nanosecond timescale. The work presented here extends previous analysis to include multiple protein states and longer simulation times. Generally, GPCR state transitions have remained elusive for unbiased simulations with standard force fields, given their intrinsic timescales (105). We take advantage of the existing number of rhodopsin structures in multiple states and prior simulations to study the effects of lipids along its photocycle.
Our analysis of the dark, Meta I, and Meta II states suggests that activation-related conformational changes in the receptor are coupled with changes in the structure of the bilayer. We note that there is an elongation of the transmembrane region of the protein in the Meta II state (Fig. 1) that seemingly increases the hydrophobic surface of the protein, particularly on the intradiscal side of the membrane. We also note that STEA acyl chains are more ordered around the protein (∼11–15 from the centroid of the receptor) in the Meta II state than in the dark or Meta I states (Fig. 2 a). Our results suggest that this local ordering effect is likely to be associated with the stretching of these fatty acid tails that occurs in that area of the membrane in the active state (Fig. 2 b), perhaps in an effort to compensate for the changes in the conformation of the protein. These observations support the view that rhodopsin activation induces hydrophobic mismatch at the membrane-water interface (16, 106), and they agree with the model proposed by Botelho et al. (20), which suggests that this activation-induced mismatch promotes stretching in the membrane, which in turn could reduce the curvature stress of the bilayer. Moreover, in the case of the dark state and Meta I systems, where the membrane is not as ordered around the protein, it is likely that DHA acyl chains may move more freely, conceivably facilitating direct interactions with the protein, as suggested by Fig. 3.
Because of their extreme flexibility, DHA acyl chains increase the fluidity of lipid membranes (68). Thus, bilayers containing high concentrations of DHA tend to be more loosely packed and less ordered. This effect, which is particularly enhanced at the annular region of certain proteins, such as the 5′-nucleotidase and the Mg2+-ATPase, is thought to be responsible for increasing membrane protein activity by allowing proteins greater mobility (5). In the case of rhodopsin, evidence from ultraviolet-visible spectroscopy indicates that the presence of these polyunsaturated lipids shifts the Meta I-Meta II equilibrium toward Meta II (4). Several lines of investigation, including solid-state NMR, FRET, and MD simulations, have shown that there is a strong preference for DHA, as compared to other lipid species, to accumulate at the lipid-protein interface of rhodopsin (63, 64, 69, 79, 107, 108). Based on this work, it was previously suggested that the ability of DHA to more deeply penetrate the receptor’s core may also have functionally relevant implications in rhodopsin activation (63, 94). Our results suggest that DHA may behave as a weak ligand with multiple low-affinity binding sites (Figs. 3 and 4). This view is consistent with saturation-transfer NMR experiments, where the effect of titrating DHA in rhodopsin-containing reconstituted membranes was fit to a ligand-binding model with ∼16 binding sites (69).
Our simulations also suggest that DHA interacts with rhodopsin in a state-dependent manner and that it associates differently with inactive-like and active-like protein states (Fig. 3), which is a necessary feature of allosteric modulation of function. Specifically, we found that DHA has a larger number of long-lived binding events in inactive-like protein states (dark, Meta I, and dark opsin). Botelho et al. (20) suggested that non-lamellar-phase-forming lipids, such as DHA, stabilize the Meta II state by introducing curvature elastic stress in the membrane (a solvent-like lipid effect). Our data support a complementary destabilizing role for DHA, where these polyunsaturated fatty acids preferentially bind inactive-like protein states and potentially disrupt stabilizing interactions within the receptor (a ligand-like lipid effect) (66). Indeed, the presence of these polyunsaturated acyl chains has been shown to decrease the stability of the receptor (61, 107), although separating the role of lipid-protein effects from those due to the lower Tm of polyunsaturated lipid membranes is problematic.
Spontaneous cis-to-trans isomerization of rhodopsin-bound retinal is on the order of 10−8 s−1, owing to a photoactivation energy of ∼48 kcal/mol (109, 110). For isolated retinal, however, this energy barrier is significantly lower (∼14.9 kcal/mol) (111). This discrepancy suggests that rhodopsin dramatically reduces the rate of retinal isomerization caused by thermal noise, which is important for achieving a high signal/noise ratio in dim-light conditions. Within the binding pocket, retinal’s Schiff base and its counterion (E1133.28) form a stabilizing salt-bridge interaction, but the ligand is also tightly packed and its mobility is hindered by steric constraints (96). Unexpectedly, we observed that 11-cis retinal sampled a broad distribution of orientations in the dark-state simulations (Fig. 6). Our analysis of these trajectories and the long-lived DHA-binding events that we observed in them (e.g., Fig. 5) suggests that direct protein-lipid interactions are at least partially responsible for these broad distributions of orientations and supports the possibility of a destabilizing role for DHA in inactive-like protein conformations.
Other direct protein-lipid interactions can also modulate rhodopsin’s conformational ensemble (12, 112). Protein residues at the ends of transmembrane helices can often satisfy their own hydrogen-bond requirements with lipid headgroups (12). Our simulations suggest that the hydrogen-bonding ability of PE headgroups can favor the formation of secondary structure in rhodopsin’s ICL3. These findings illustrate the structural plasticity of this region and help reconcile the different ICL3 conformations observed in existing dark-state crystal structures (72, 113, 114). Our dark-state trajectories were initialized from the tetragonal P41 crystal structure solved by Okada et al. (72) (PDB: 1U19), in which ICL3 is entirely unstructured. During the simulations, ICL3 sampled an ensemble of conformations that included an additional helical turn at the end of helix 5 (Figs. 7 and S7), as observed in the trigonal P31 crystal structure solved by Li et al. (113) (PDB: 1GZM). PDB: 1GZM has been suggested to adopt a more native-like conformation than PDB: 1U19 in this region (115, 116). Interestingly, ICL3 stays unstructured in simulations of the dark state embedded in 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine (DMPC) bilayers (117). DMPC phospholipids contain two short, saturated myristoyl acyl chains and a phosphatidylcholine headgroup, which cannot hydrogen bond. In these membranes and in the absence of a Gα-derived peptide, the loop swiftly becomes disordered (<200 ns) in multiple trajectories even when started from the active state, where helix 5 is elongated (117). This result is consistent with experimental evidence showing that rhodopsin is largely inactive in DMPC membranes (19, 57, 118). Conversely, ICL3 remains very close to its initial conformation when active-like (Meta II and opsin) in our simulations with SDPE membranes, suggesting that the lipid environment of the receptor contributes to modulating the conformations accessible to this protein region.
Conclusions
Is the lipid control of rhodopsin’s function a ligand-like effect or a solvent-like effect? Our analysis suggests that the answer is Yes. These are often presented as mutually exclusive hypotheses, but this work demonstrates that both phenomena are present in rhodopsin systems. In our simulations, activation-induced changes in the structure of the receptor were coupled to changes in the structure of the membrane. Specifically, we observed an increase in the local order and effective length of STEA acyl chains in the vicinity of the protein. On the other hand, our data also suggest that direct protein-lipid interactions may play important roles in modulating rhodopsin structure and that these contributions are state dependent. For instance, we found that DHA acyl chains can bind to the receptor preferentially to inactive-like protein conformations, acting as a weak ligand; likewise, direct interactions between rhodopsin and PE headgroups seemed to stabilize the partial structuring of ICL3 in inactive-like protein states.
Given the current difficulties in capturing GPCR state transitions using all-atom unbiased MD, simulating five different functional states along rhodopsin’s photocycle allowed us to study different mechanisms by which the receptor interacts with its surrounding membrane in each protein state. Multiple replicates corresponding to each of these states were required to assess the significance of our observations. Our work emphasizes the value of the high level of detail provided by all-atom simulations in hypothesis testing and in the formulation of experimentally verifiable queries.
Author Contributions
A.G., L.A.S.-E., and N.L. designed the research. L.A.S.-E., N.L., and T.D.R. performed the research. T.D.R., A.G., and L.A.S.-E. contributed analytic tools. L.A.S.-E. analyzed the data and wrote the manuscript. All authors copy-edited the manuscript.
Acknowledgments
The authors thank the Center for Integrated Research Computing at the University of Rochester for providing computational resources and technical support. L.A.S.-E. also thanks The American Association of University Women (AAUW) for awarding her an International Fellowship during the 2016–2017 cycle.
This work was supported by grant R01GM095496 (to A.G.) from the National Institutes of Health.
Editor: Emad Tajkhorshid.
Footnotes
Supporting Materials and Methods and seven figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31253-5.
Supporting Citations
References (119, 120, 121, 122, 123, 124, 125, 126, 127, 128, 129, 130) appear in the Supporting Material.
Supporting Material
References
- 1.Palsdottir H., Hunte C. Lipids in membrane protein structures. Biochim. Biophys. Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Vigh L., Escribá P.V., Harwood J.L. The significance of lipid composition for membrane activity: new concepts and ways of assessing function. Prog. Lipid Res. 2005;44:303–344. doi: 10.1016/j.plipres.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Gibson N.J., Brown M.F. Lipid headgroup and acyl chain composition modulate the MI-MII equilibrium of rhodopsin in recombinant membranes. Biochemistry. 1993;32:2438–2454. doi: 10.1021/bi00060a040. [DOI] [PubMed] [Google Scholar]
- 4.Litman B.J., Mitchell D.C. A role for phospholipid polyunsaturation in modulating membrane protein function. Lipids. 1996;31:S193–S197. doi: 10.1007/BF02637075. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto M., Hossain M.S., Shido O. Effects of docosahexaenoic acid on annular lipid fluidity of the rat bile canalicular plasma membrane. J. Lipid Res. 2001;42:1160–1168. [PubMed] [Google Scholar]
- 6.Valiyaveetil F.I., Zhou Y., MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 7.Williamson I.M., Alvis S.J., Lee A.G. The potassium channel KcsA and its interaction with the lipid bilayer. Cell. Mol. Life Sci. 2003;60:1581–1590. doi: 10.1007/s00018-003-3172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gullingsrud J., Schulten K. Lipid bilayer pressure profiles and mechanosensitive channel gating. Biophys. J. 2004;86:3496–3509. doi: 10.1529/biophysj.103.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee A.G. Lipid-protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 10.Escribá P.V. Membrane-lipid therapy: a new approach in molecular medicine. Trends Mol. Med. 2006;12:34–43. doi: 10.1016/j.molmed.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 11.de Velasco P.C., Mendonça H.R., Serfaty C.A. Nutritional restriction of omega-3 fatty acids alters topographical fine tuning and leads to a delay in the critical period in the rodent visual system. Exp. Neurol. 2012;234:220–229. doi: 10.1016/j.expneurol.2011.12.032. [DOI] [PubMed] [Google Scholar]
- 12.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Nogi T., Fathir I., Miki K. Crystal structures of photosynthetic reaction center and high-potential iron-sulfur protein from Thermochromatium tepidum: thermostability and electron transfer. Proc. Natl. Acad. Sci. USA. 2000;97:13561–13566. doi: 10.1073/pnas.240224997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlame M., Rua D., Greenberg M.L. The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 2000;39:257–288. doi: 10.1016/s0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- 15.Mouritsen O.G., Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys. J. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fattal D.R., Ben-Shaul A. A molecular model for lipid-protein interaction in membranes: the role of hydrophobic mismatch. Biophys. J. 1993;65:1795–1809. doi: 10.1016/S0006-3495(93)81249-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pignataro M.F., Dodes-Traian M.M., Rossi J.P.F.C. Modulation of plasma membrane Ca2+-ATPase by neutral phospholipids: effect of the micelle-vesicle transition and the bilayer thickness. J. Biol. Chem. 2015;290:6179–6190. doi: 10.1074/jbc.M114.585828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutierrez M.G., Mansfield K.S., Malmstadt N. The functional activity of the human serotonin 5-HT1A receptor is controlled by lipid bilayer composition. Biophys. J. 2016;110:2486–2495. doi: 10.1016/j.bpj.2016.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown M.F. Modulation of rhodopsin function by properties of the membrane bilayer. Chem. Phys. Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 20.Botelho A.V., Gibson N.J., Brown M.F. Conformational energetics of rhodopsin modulated by nonlamellar-forming lipids. Biochemistry. 2002;41:6354–6368. doi: 10.1021/bi011995g. [DOI] [PubMed] [Google Scholar]
- 21.Contreras F.-X., Ernst A.M., Brügger B. Specificity of intramembrane protein-lipid interactions. Cold Spring Harb. Perspect. Biol. 2011;3:a004705. doi: 10.1101/cshperspect.a004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeagle P.L., Young J., Rice D. Effects of cholesterol on (Na+,K+)-ATPase ATP hydrolyzing activity in bovine kidney. Biochemistry. 1988;27:6449–6452. doi: 10.1021/bi00417a037. [DOI] [PubMed] [Google Scholar]
- 23.Chan W.K.B., Zhang H., Zhang Y. GLASS: a comprehensive database for experimentally validated GPCR-ligand associations. Bioinformatics. 2015;31:3035–3042. doi: 10.1093/bioinformatics/btv302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deupi X., Standfuss J. Structural insights into agonist-induced activation of G-protein-coupled receptors. Curr. Opin. Struct. Biol. 2011;21:541–551. doi: 10.1016/j.sbi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Deupi X., Standfuss J., Schertler G. Conserved activation pathways in G-protein-coupled receptors. Biochem. Soc. Trans. 2012;40:383–388. doi: 10.1042/BST20120001. [DOI] [PubMed] [Google Scholar]
- 26.Sun L., Ye R.D. Role of G protein-coupled receptors in inflammation. Acta Pharmacol. Sin. 2012;33:342–350. doi: 10.1038/aps.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garriga P., Manyosa J. The eye photoreceptor protein rhodopsin. Structural implications for retinal disease. FEBS Lett. 2002;528:17–22. doi: 10.1016/s0014-5793(02)03241-6. [DOI] [PubMed] [Google Scholar]
- 28.Stone L.S., Molliver D.C. In search of analgesia: emerging roles of GPCRs in pain. Mol. Interv. 2009;9:234–251. doi: 10.1124/mi.9.5.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brinks H.L., Eckhart A.D. Regulation of GPCR signaling in hypertension. Biochim. Biophys. Acta. 2010;1802:1268–1275. doi: 10.1016/j.bbadis.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overington J.P., Al-Lazikani B., Hopkins A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 31.Pucadyil T.J., Chattopadhyay A. Cholesterol modulates ligand binding and G-protein coupling to serotonin1A receptors from bovine hippocampus. Biochim. Biophys. Acta. 2004;1663:188–200. doi: 10.1016/j.bbamem.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Dawaliby R., Trubbia C., Govaerts C. Allosteric regulation of G protein-coupled receptor activity by phospholipids. Nat. Chem. Biol. 2016;12:35–39. doi: 10.1038/nchembio.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gahbauer S., Böckmann R.A. Membrane-mediated oligomerization of G protein coupled receptors and its implications for GPCR function. Front. Physiol. 2016;7:494. doi: 10.3389/fphys.2016.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luchetti G., Sircar R., Rohatgi R. Cholesterol activates the G-protein coupled receptor smoothened to promote Hedgehog signaling. eLife. 2016;5:e20304. doi: 10.7554/eLife.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vögler O., Casas J., Escribá P.V. The Gbetagamma dimer drives the interaction of heterotrimeric Gi proteins with nonlamellar membrane structures. J. Biol. Chem. 2004;279:36540–36545. doi: 10.1074/jbc.M402061200. [DOI] [PubMed] [Google Scholar]
- 36.Inagaki S., Ghirlando R., Grisshammer R. Modulation of the interaction between neurotensin receptor NTS1 and Gq protein by lipid. J. Mol. Biol. 2012;417:95–111. doi: 10.1016/j.jmb.2012.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palczewski K. Oligomeric forms of G protein-coupled receptors (GPCRs) Trends Biochem. Sci. 2010;35:595–600. doi: 10.1016/j.tibs.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goddard A.D., Dijkman P.M., Watts A. Lipid-dependent GPCR dimerization. Methods Cell Biol. 2013;117:341–357. doi: 10.1016/B978-0-12-408143-7.00018-9. [DOI] [PubMed] [Google Scholar]
- 39.Ghanouni P., Gryczynski Z., Kobilka B.K. Functionally different agonists induce distinct conformations in the G protein coupling domain of the β2 adrenergic receptor. J. Biol. Chem. 2001;276:24433–24436. doi: 10.1074/jbc.C100162200. [DOI] [PubMed] [Google Scholar]
- 40.Peleg G., Ghanouni P., Zare R.N. Single-molecule spectroscopy of the β2 adrenergic receptor: observation of conformational substates in a membrane protein. Proc. Natl. Acad. Sci. USA. 2001;98:8469–8474. doi: 10.1073/pnas.151239698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kofuku Y., Ueda T., Shimada I. Efficacy of the β2-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat. Commun. 2012;3:1045. doi: 10.1038/ncomms2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nygaard R., Zou Y., Kobilka B.K. The dynamic process of β2-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maeda R., Hiroshima M., Imamoto Y. Single-molecule observation of the ligand-induced population shift of rhodopsin, a G-protein-coupled receptor. Biophys. J. 2014;106:915–924. doi: 10.1016/j.bpj.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manglik A., Kobilka B. The role of protein dynamics in GPCR function: insights from the β2AR and rhodopsin. Curr. Opin. Cell Biol. 2014;27:136–143. doi: 10.1016/j.ceb.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leioatts N., Romo T.D., Grossfield A. Retinal conformation changes rhodopsin’s dynamic ensemble. Biophys. J. 2015;109:608–617. doi: 10.1016/j.bpj.2015.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanson M.A., Cherezov V., Stevens R.C. A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J.Y., Lyman E. Predictions for cholesterol interaction sites on the A2A adenosine receptor. J. Am. Chem. Soc. 2012;134:16512–16515. doi: 10.1021/ja307532d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manna M., Niemelä M., Vattulainen I. Mechanism of allosteric regulation of β2-adrenergic receptor by cholesterol. eLife. 2016;5:e18432. doi: 10.7554/eLife.18432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neale C., Herce H.D., García A.E. Can specific protein-lipid interactions stabilize an active state of the β2 adrenergic receptor? Biophys. J. 2015;109:1652–1662. doi: 10.1016/j.bpj.2015.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han M., Smith S.O., Sakmar T.P. Constitutive activation of opsin by mutation of methionine 257 on transmembrane helix 6. Biochemistry. 1998;37:8253–8261. doi: 10.1021/bi980147r. [DOI] [PubMed] [Google Scholar]
- 51.Altenbach C., Yang K., Hubbell W.L. Structural features and light-dependent changes in the cytoplasmic interhelical E-F loop region of rhodopsin: a site-directed spin-labeling study. Biochemistry. 1996;35:12470–12478. doi: 10.1021/bi960849l. [DOI] [PubMed] [Google Scholar]
- 52.Choe H.-W., Kim Y.J., Ernst O.P. Crystal structure of metarhodopsin II. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 53.Parkes J.H., Gibson S.K., Liebman P.A. Temperature and pH dependence of the metarhodopsin I-metarhodopsin II equilibrium and the binding of metarhodopsin II to G protein in rod disk membranes. Biochemistry. 1999;38:6862–6878. doi: 10.1021/bi9827666. [DOI] [PubMed] [Google Scholar]
- 54.Cooper A., Dixon S.F., Robb J.L. Mechanism of retinal Schiff base formation and hydrolisis in relation to visual pigment photolysis and regeneration: resonance raman spectroscopy of a tetrahedral carbinolamine intermediate and oxygen-18 labeling of retinal at the metarhodopsin state of photoreceptor membranes. J. Am. Chem. Soc. 1987;109:7254–7263. [Google Scholar]
- 55.Mason W.T., Fager R.S., Abrahamson E.W. Lipid and fatty acid composition of frog photoreceptor outer segments. Biochemistry. 1973;12:2147–2150. doi: 10.1021/bi00735a021. [DOI] [PubMed] [Google Scholar]
- 56.Albert A.D., Young J.E., Paw Z. Phospholipid fatty acyl spatial distribution in bovine rod outer segment disk membranes. Biochim. Biophys. Acta. 1998;1368:52–60. doi: 10.1016/s0005-2736(97)00200-9. [DOI] [PubMed] [Google Scholar]
- 57.Mitchell D.C., Straume M., Litman B.J. Role of sn-1-saturated,sn-2-polyunsaturated phospholipids in control of membrane receptor conformational equilibrium: effects of cholesterol and acyl chain unsaturation on the metarhodopsin I in equilibrium with metarhodopsin II equilibrium. Biochemistry. 1992;31:662–670. doi: 10.1021/bi00118a005. [DOI] [PubMed] [Google Scholar]
- 58.Mitchell D.C., Lawrence J.T., Litman B.J. Primary alcohols modulate the activation of the G protein-coupled receptor rhodopsin by a lipid-mediated mechanism. J. Biol. Chem. 1996;271:19033–19036. doi: 10.1074/jbc.271.32.19033. [DOI] [PubMed] [Google Scholar]
- 59.Mitchell D.C., Niu S.-L., Litman B.J. DHA-rich phospholipids optimize G-protein-coupled signaling. J. Pediatr. 2003;143:S80–S86. doi: 10.1067/s0022-3476(03)00405-0. [DOI] [PubMed] [Google Scholar]
- 60.Teague W.E., Jr., Soubias O., Gawrisch K. Elastic properties of polyunsaturated phosphatidylethanolamines influence rhodopsin function. Faraday Discuss. 2013;161:383–395. doi: 10.1039/c2fd20095c. discussion 419–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett M.P., Mitchell D.C. Regulation of membrane proteins by dietary lipids: effects of cholesterol and docosahexaenoic acid acyl chain-containing phospholipids on rhodopsin stability and function. Biophys. J. 2008;95:1206–1216. doi: 10.1529/biophysj.107.122788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huber T., Rajamoorthi K., Brown M.F. Structure of docosahexaenoic acid-containing phospholipid bilayers as studied by 2H NMR and molecular dynamics simulations. J. Am. Chem. Soc. 2002;124:298–309. doi: 10.1021/ja011383j. [DOI] [PubMed] [Google Scholar]
- 63.Feller S.E., Gawrisch K., Woolf T.B. Rhodopsin exhibits a preference for solvation by polyunsaturated docosohexaenoic acid. J. Am. Chem. Soc. 2003;125:4434–4435. doi: 10.1021/ja0345874. [DOI] [PubMed] [Google Scholar]
- 64.Pitman M.C., Grossfield A., Feller S.E. Role of cholesterol and polyunsaturated chains in lipid-protein interactions: molecular dynamics simulation of rhodopsin in a realistic membrane environment. J. Am. Chem. Soc. 2005;127:4576–4577. doi: 10.1021/ja042715y. [DOI] [PubMed] [Google Scholar]
- 65.Soubias O., Gawrisch K. Probing specific lipid-protein interaction by saturation transfer difference NMR spectroscopy. J. Am. Chem. Soc. 2005;127:13110–13111. doi: 10.1021/ja0538942. [DOI] [PubMed] [Google Scholar]
- 66.Grossfield A., Feller S.E., Pitman M.C. Contribution of omega-3 fatty acids to the thermodynamics of membrane protein solvation. J. Phys. Chem. B. 2006;110:8907–8909. doi: 10.1021/jp060405r. [DOI] [PubMed] [Google Scholar]
- 67.Soubias O., Gawrisch K. The role of the lipid matrix for structure and function of the GPCR rhodopsin. Biochim. Biophys. Acta. 2012;1818:234–240. doi: 10.1016/j.bbamem.2011.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soubias O., Gawrisch K. Docosahexaenoyl chains isomerize on the sub-nanosecond time scale. J. Am. Chem. Soc. 2007;129:6678–6679. doi: 10.1021/ja068856c. [DOI] [PubMed] [Google Scholar]
- 69.Soubias O., Teague W.E., Gawrisch K. Evidence for specificity in lipid-rhodopsin interactions. J. Biol. Chem. 2006;281:33233–33241. doi: 10.1074/jbc.M603059200. [DOI] [PubMed] [Google Scholar]
- 70.Soubias O., Teague W.E., Jr., Gawrisch K. Contribution of membrane elastic energy to rhodopsin function. Biophys. J. 2010;99:817–824. doi: 10.1016/j.bpj.2010.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yau W.M., Wimley W.C., White S.H. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 72.Okada T., Sugihara M., Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 73.Park J.H., Scheerer P., Ernst O.P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 74.Grossfield A., Pitman M.C., Gawrisch K. Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. J. Mol. Biol. 2008;381:478–486. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lüdeke S., Beck M., Vogel R. The role of Glu181 in the photoactivation of rhodopsin. J. Mol. Biol. 2005;353:345–356. doi: 10.1016/j.jmb.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 76.Shaikh S.R., Brzustowicz M.R., Wassall S.R. Formation of inverted hexagonal phase in SDPE as observed by solid-state 31P NMR. Biochem. Biophys. Res. Commun. 2001;286:758–763. doi: 10.1006/bbrc.2001.5454. [DOI] [PubMed] [Google Scholar]
- 77.Teague W.E., Fuller N.L., Gawrisch K. Polyunsaturated lipids in membrane fusion events. Cell. Mol. Biol. Lett. 2002;7:262–264. [PubMed] [Google Scholar]
- 78.Katsaras J., Gutberlet T. Springer-Verlag; Berlin, Germany: 2001. Lipid Bilayers: Structure and Interactions. [Google Scholar]
- 79.Grossfield A., Feller S.E., Pitman M.C. A role for direct interactions in the modulation of rhodopsin by omega-3 polyunsaturated lipids. Proc. Natl. Acad. Sci. USA. 2006;103:4888–4893. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hong C., Tieleman D.P., Wang Y. Microsecond molecular dynamics simulations of lipid mixing. Langmuir. 2014;30:11993–12001. doi: 10.1021/la502363b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Woolf T.B., Roux B. Structure, energetics, and dynamics of lipid-protein interactions: a molecular dynamics study of the gramicidin A channel in a DMPC bilayer. Proteins. 1996;24:92–114. doi: 10.1002/(SICI)1097-0134(199601)24:1<92::AID-PROT7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 82.Romo T.D., Leioatts N., Grossfield A. Lightweight object oriented structure analysis: tools for building tools to analyze molecular dynamics simulations. J. Comput. Chem. 2014;35:2305–2318. doi: 10.1002/jcc.23753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 85.Mackerell A.D., Jr., Feig M., Brooks C.L., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 86.Klauda J.B., Venable R.M., Pastor R.W. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu S., Brown M.F., Feller S.E. Retinal conformation governs pKa of protonated Schiff base in rhodopsin activation. J. Am. Chem. Soc. 2013;135:9391–9398. doi: 10.1021/ja4002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ryckaert J.-P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 89.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 90.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 91.Schrödinger, L. L. C., 2015. The PyMOL Molecular Graphics System, Version 1.8.
- 92.Carrillo-Tripp M., Feller S.E. Evidence for a mechanism by which omega-3 polyunsaturated lipids may affect membrane protein function. Biochemistry. 2005;44:10164–10169. doi: 10.1021/bi050822e. [DOI] [PubMed] [Google Scholar]
- 93.Vermeer L.S., de Groot B.L., Czaplicki J. Acyl chain order parameter profiles in phospholipid bilayers: computation from molecular dynamics simulations and comparison with 2H NMR experiments. Eur. Biophys. J. 2007;36:919–931. doi: 10.1007/s00249-007-0192-9. [DOI] [PubMed] [Google Scholar]
- 94.Horn J.N., Kao T.-C., Grossfield A. Coarse-grained molecular dynamics provides insight into the interactions of lipids and cholesterol with rhodopsin. Adv. Exp. Med. Biol. 2014;796:75–94. doi: 10.1007/978-94-007-7423-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ballesteros J.A., Weinstein H. Receptor Molecular Biology. Volume 25. Elsevier; 1995. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors; pp. 366–428. [Google Scholar]
- 96.Pope A., Eilers M., Smith S.O. Amino acid conservation and interactions in rhodopsin: probing receptor activation by NMR spectroscopy. Biochim. Biophys. Acta. 2014;1837:683–693. doi: 10.1016/j.bbabio.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ahuja S., Crocker E., Smith S.O. Location of the retinal chromophore in the activated state of rhodopsin∗. J. Biol. Chem. 2009;284:10190–10201. doi: 10.1074/jbc.M805725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Franke R.R., König B., Hofmann K.P. Rhodopsin mutants that bind but fail to activate transducin. Science. 1990;250:123–125. doi: 10.1126/science.2218504. [DOI] [PubMed] [Google Scholar]
- 99.Shi W., Osawa S., Weiss E.R. Rhodopsin mutants discriminate sites important for the activation of rhodopsin kinase and Gt. J. Biol. Chem. 1995;270:2112–2119. doi: 10.1074/jbc.270.5.2112. [DOI] [PubMed] [Google Scholar]
- 100.Acharya S., Saad Y., Karnik S.S. Transducin-α C-terminal peptide binding site consists of C-D and E-F loops of rhodopsin. J. Biol. Chem. 1997;272:6519–6524. doi: 10.1074/jbc.272.10.6519. [DOI] [PubMed] [Google Scholar]
- 101.Kisselev O.G., Downs M.A., Hargrave P.A. Conformational changes in the phosphorylated C-terminal domain of rhodopsin during rhodopsin arrestin interactions. J. Biol. Chem. 2004;279:51203–51207. doi: 10.1074/jbc.M407341200. [DOI] [PubMed] [Google Scholar]
- 102.Jones Brunette A.M., Sinha A., Farrens D.L. Evidence that the rhodopsin kinase (GRK1) N-terminus and the transducin Gα C-terminus interact with the same “hydrophobic patch” on rhodopsin TM5. Biochemistry. 2016;55:3123–3135. doi: 10.1021/acs.biochem.6b00328. [DOI] [PubMed] [Google Scholar]
- 103.Cherezov V., Rosenbaum D.M., Stevens R.C. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rosenbaum D.M., Zhang C., Kobilka B.K. Structure and function of an irreversible agonist-β2 adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dror R.O., Arlow D.H., Shaw D.E. Activation mechanism of the β2-adrenergic receptor. Proc. Natl. Acad. Sci. USA. 2011;108:18684–18689. doi: 10.1073/pnas.1110499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Soubias O., Niu S.-L., Gawrisch K. Lipid-rhodopsin hydrophobic mismatch alters rhodopsin helical content. J. Am. Chem. Soc. 2008;130:12465–12471. doi: 10.1021/ja803599x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Polozova A., Litman B.J. Cholesterol dependent recruitment of di22:6-PC by a G protein-coupled receptor into lateral domains. Biophys. J. 2000;79:2632–2643. doi: 10.1016/S0006-3495(00)76502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gawrisch K., Soubias O., Mihailescu M. Insights from biophysical studies on the role of polyunsaturated fatty acids for function of G-protein coupled membrane receptors. Prostaglandins Leukot. Essent. Fatty Acids. 2008;79:131–134. doi: 10.1016/j.plefa.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cooper A. Energy uptake in the first step of visual excitation. Nature. 1979;282:531–533. doi: 10.1038/282531a0. [DOI] [PubMed] [Google Scholar]
- 110.Flyvbjerg H., Jülicher F., David F. Springer-Verlag; Berlin, Germany: 2002. Physics of Bio-Molecules and Cells: Les Houches Session LXXV, 2–27 July 2001, [Google Scholar]
- 111.Dilger J., Musbat L., Toker Y. Direct measurement of the isomerization barrier of the isolated retinal chromophore. Angew. Chem. Int. Ed. Engl. 2015;54:4748–4752. doi: 10.1002/anie.201411894. [DOI] [PubMed] [Google Scholar]
- 112.Krishna A.G., Menon S.T., Sakmar T.P. Evidence that helix 8 of rhodopsin acts as a membrane-dependent conformational switch. Biochemistry. 2002;41:8298–8309. doi: 10.1021/bi025534m. [DOI] [PubMed] [Google Scholar]
- 113.Li J., Edwards P.C., Schertler G.F.X. Structure of bovine rhodopsin in a trigonal crystal form. J. Mol. Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 114.Piscitelli C.L., Angel T.E., Lawrence C.M. Equilibrium between metarhodopsin-I and metarhodopsin-II is dependent on the conformation of the third cytoplasmic loop. J. Biol. Chem. 2006;281:6813–6825. doi: 10.1074/jbc.M510175200. [DOI] [PubMed] [Google Scholar]
- 115.Schertler G.F.X. Structure of rhodopsin and the metarhodopsin I photointermediate. Curr. Opin. Struct. Biol. 2005;15:408–415. doi: 10.1016/j.sbi.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 116.Kobilka B.K. G protein coupled receptor structure and activation. Biochim. Biophys. Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Elgeti M., Rose A.S., Heck M. Precision vs flexibility in GPCR signaling. J. Am. Chem. Soc. 2013;135:12305–12312. doi: 10.1021/ja405133k. [DOI] [PubMed] [Google Scholar]
- 118.Baldwin P.A., Hubbell W.L. Effects of lipid environment on the light-induced conformational changes of rhodopsin. 1. Absence of metarhodopsin II production in dimyristoylphosphatidylcholine recombinant membranes. Biochemistry. 1985;24:2624–2632. doi: 10.1021/bi00332a006. [DOI] [PubMed] [Google Scholar]
- 119.Klauda J.B., Monje V., Im W. Improving the CHARMM force field for polyunsaturated fatty acid chains. J. Phys. Chem. B. 2012;116:9424–9431. doi: 10.1021/jp304056p. [DOI] [PubMed] [Google Scholar]
- 120.Venable R.M., Brown F.L.H., Pastor R.W. Mechanical properties of lipid bilayers from molecular dynamics simulation. Chem. Phys. Lipids. 2015;192:60–74. doi: 10.1016/j.chemphyslip.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Flyvbjerg H., Petersen H.G. Error estimates on averages of correlated data. J. Chem. Phys. 1989;91:461–466. [Google Scholar]
- 122.Grossfield A., Zuckerman D.M. Quantifying uncertainty and sampling quality in biomolecular simulations. Annu. Rep. Comput. Chem. 2009;5:23–48. doi: 10.1016/S1574-1400(09)00502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barber C.B., Dobkin D.P., Huhdanpaa H. The Quickhull algorithm for convex hulls. ACM Trans. Math. Softw. 1996;22:469–483. [Google Scholar]
- 124.Oliphant T.E. Python for scientific computing. Comput. Sci. Eng. 2007;9:10–20. [Google Scholar]
- 125.Lindahl E., Edholm O. Mesoscopic undulations and thickness fluctuations in lipid bilayers from molecular dynamics simulations. Biophys. J. 2000;79:426–433. doi: 10.1016/S0006-3495(00)76304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sonne J., Hansen F.Y., Peters G.H. Methodological problems in pressure profile calculations for lipid bilayers. J. Chem. Phys. 2005;122:124903. doi: 10.1063/1.1862624. [DOI] [PubMed] [Google Scholar]
- 127.Horn J.N., Romo T.D., Grossfield A. Simulating the mechanism of antimicrobial lipopeptides with all-atom molecular dynamics. Biochemistry. 2013;52:5604–5610. doi: 10.1021/bi400773q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Leioatts N., Mertz B., Brown M.F. Retinal ligand mobility explains internal hydration and reconciles active rhodopsin structures. Biochemistry. 2014;53:376–385. doi: 10.1021/bi4013947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 130.Touw W.G., Baakman C., Vriend G. A series of PDB-related databanks for everyday needs. Nucleic Acids Res. 2015;43:D364–D368. doi: 10.1093/nar/gku1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.