Abstract
Single-molecule tracking (SMT) offers rich information on the dynamics of underlying biological processes, but multicolor SMT has been challenging due to spectral cross talk and a need for multiple laser excitations. Here, we describe a single-molecule spectral imaging approach for live-cell tracking of multiple fluorescent species at once using a single-laser excitation. Fluorescence signals from all the molecules in the field of view are collected using a single objective and split between positional and spectral channels. Images of the same molecule in the two channels are then combined to determine both the location and the identity of the molecule. The single-objective configuration of our approach allows for flexible sample geometry and the use of a live-cell incubation chamber required for live-cell SMT. Despite a lower photon yield, we achieve excellent spatial (20–40 nm) and spectral (10–15 nm) resolutions comparable to those obtained with dual-objective, spectrally resolved Stochastic Optical Reconstruction Microscopy. Furthermore, motions of the fluorescent molecules did not cause loss of spectral resolution owing to the dual-channel spectral calibration. We demonstrate SMT in three (and potentially more) colors using spectrally proximal fluorophores and single-laser excitation, and show that trajectories of each species can be reliably extracted with minimal cross talk.
Introduction
Single-molecule tracking (SMT) is an effective tool for probing the diffusional states of target molecules and detecting spatial partitioning events (1, 2, 3). When a large number of diffusion trajectories are obtained (4), robust statistical analysis such as variational Bayes single-particle tracking (vbSPT) can be applied to derive critical information on the underlying biological processes, including the occurrence of transient interactions and state conversion kinetics (5, 6). Large numbers of trajectories are often obtained via combining SMT with single-molecule localization techniques such as fluorescence photoactivated localization microscopy (7, 8) and direct Stochastic Optical Reconstruction Microscopy (STORM) (9, 10) and alternatively with various labeling strategies that achieve sparse fluorescence localization events (11, 12).
SMT becomes especially powerful if multiple molecular species can be studied simultaneously (11, 13, 14, 15, 16, 17, 18, 19). It can be used, for example, to probe whether multiple species are sequestered to the same cellular compartments, which can have important implications for understanding their biological functions. However, to date, multicolor SMT has been challenging for at least two reasons. First, multicolor fluorescence detection is implemented using bandpass filters to discriminate between fluorophores, which requires that emission spectra are separated by 50–100 nm for minimal signal bleed-through. Although SMT in three–four colors has been demonstrated (13, 18, 19), for each color a separate acquisition channel is needed, an approach not easily scalable to tracking in even more colors. Second, each of the multiple fluorophores also requires a distinct excitation wavelength, which both complicates the optical setup and adds to the overall dose of illumination on the cells. In particular, cells are more sensitive to blue and green than to far red and near infrared laser excitations, and cellular response to the short wavelength excitations potentially causes complications to the acquired trajectories (20). Hence, there is an immediate need for better strategies in multicolor SMT.
Recent progress on multiplexed single-molecule imaging has offered potential solutions to this challenge. A particularly powerful approach distinguishes between individual fluorophores based on their spectral signatures instead of using emission filters, which is achieved by recording the position and emission spectrum simultaneously for each fluorophore. This has been achieved using confocal (21) or line-scanning (18) schemes, but both are slow for some applications, including live-cell imaging. Sonehara et al. (22) introduced a prism-based, wide field, single-molecule spectral imaging scheme to obtain the emission spectra of all fluorescent molecules in the field of view at once. However, fluorophore positions had to be obtained by imaging gold nanoparticles, to which the fluorophores were attached, at a separate time point. Although the authors were able to distinguish four fluorophores (with emission maxima 540–620 nm), this approach is not practical for SMT in live cells. Broeken and colleagues (23) used a spatial light modulator to disperse the fluorescence signal from single fluorophores to simultaneously record the positions and spectra, where the zeroth-order diffraction recorded the position, and the distance between zeroth- and first-order spots corresponds to the emission wavelength. This eliminated the need for separate positional markers at the same time as achieving a spectral resolution of ∼50 nm.
A more recent approach to multicolor single-molecule imaging is spectrally resolved superresolution microscopy (24, 25). In this approach, each fluorescent molecule generates two images, a positional image and a spectral image, which can be achieved using either a dual-objective (24) or a single-objective configuration (25). For each detected fluorescent molecule, the positional image is used to determine its precise location, and the spectral image is used to determine its identity. This approach has been used to acquire four-color images of fixed cells using four fluorophores with highly overlapping emission spectra with a nominal spectral resolution of ∼10 nm (24), and to analyze the spectral fluctuations of fluorescent proteins (25).
This is a significant improvement in spectral resolution over previously reported single-molecule localization techniques. However, the use of this approach for multicolor SMT has not been demonstrated. Although the approach offers much improved spectral resolution in single-molecule imaging, it is unclear whether similar spectral and spatial resolutions could be achieved in imaging diffusing molecules in cells. Here, we describe a single-objective, single-molecule spectral imaging system for simultaneous three (or more)-color SMT in live cells at ∼20 ms time resolution, during which cells are kept under physiological conditions using an on-stage incubator. Tens of thousands of SMT trajectories were obtained using spectrally overlapping fluorophores. Trajectories of each species were extracted reliably without cross talk, each exhibiting its unique diffusion characteristics, thus allowing parallel analysis and direct comparison of their diffusion properties in the same cellular space.
Materials and Methods
Microscopy
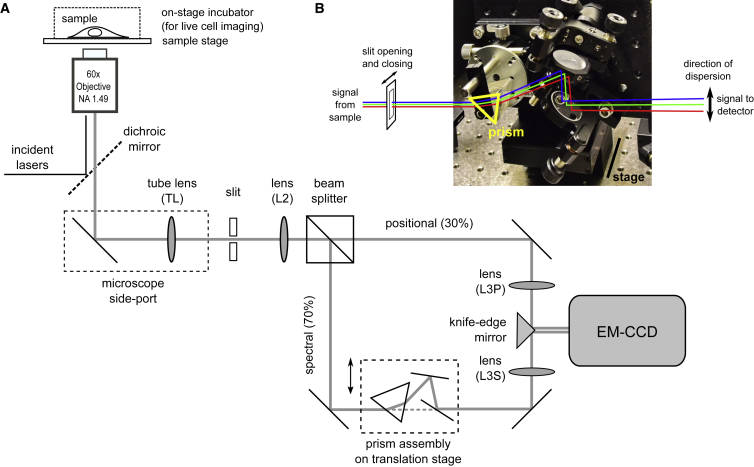
The custom single-molecule spectral imaging system was constructed with a similar illumination path to that described previously (26). Briefly, two lasers emitting at 405 (Coherent OBIS 405, 100 mW) and 637 nm (Coherent OBIS 637, 140 mW) were combined and introduced into the back of a Nikon Ti-U microscope equipped with a 60× total internal reflection fluorescence objective (Nikon, NA 1.49). The illumination was continuously tuned between epi-fluorescence and strict total internal reflection fluorescence modes by shifting the incident laser horizontally with a translational stage before entering the microscope. A single-edge dichroic mirror (LPD02-633RU) was used to reflect the laser into the objective and clean up fluorescence signals from the sample; these dichroic mirrors also reflect 405 nm light needed for photoactivation and photoswitching. A short-pass filter (BLP-633R; Semrock) was placed in front of the 637 nm laser to clean up the output. The OBIS 637 laser was set to operate at 11°C to bring the center wavelength down to ∼635 nm in order to work with these filters.
Fluorescence signals were collected at the side-port of the microscope, with a slit (VA100; Thorlabs, Newton, NJ) at the intermediate image plane to narrow the field of view. An infinity space was created by placing a triplet lens (PAC076; Newport, f = 125 mm) at the Fourier plane of the intermediate image. A filter wheel was mounted in the infinity space right after the triplet lens to position emission and/or notch filters for further clean-up of the signals and specification of detection wavelength range. A nonpolarizing beam splitter (BS022; Thorlabs) was inserted after the filter mount to divide the signal into positional (30%, transmitted) and spectral (70%, reflected) channels. Signal in each channel was refocused with another triplet lens (f = 125 mm) before the two channels were combined using a knife-edge mirror (MRAK25-E02; Thorlabs) and projected onto the left and right halves of the same electron multiplied charge-coupled device (EM-CCD) (iXon Ultra 897; Andor). To disperse fluorescence signals in the spectral channel, an equilateral prism (PS863; Thorlabs) was placed after the beam splitter and at the Fourier plane of the last focusing (triplet) lens. Two steering mirrors were used to bring the light path back to the original direction (i.e., in the absence of the prism). The prism and two steering mirrors were mounted vertically on a translational stage, and the whole assembly could be moved in and out of the light path to facilitate alignment. An effective pixel size of 178 nm was used in both channels.
Spectral calibration
Spectral calibration of the single-molecule spectral imaging system was performed by imaging 40 nm fluorescent beads (F8793; Life Technologies, Carlsbad, CA). Narrow bandpass filters were inserted in the infinity space below the objective to specify the wavelength range of signals to reach the detectors. This resulted in a relatively narrow image in the spectral channel to allow for calculation of the distance between the precise centroid positions of each molecule in the two channels. The bandpass filters used for this purpose were all from Semrock, with part numbers FF01-572/15, FF01-605/15, FF01-635/18, FF01-661/11, FF01-673/11, FF01-711/25, and LD01-785/10. Of these, the FF01-661/11 filter was used to overlap the positional and spectral images; the same filter was also used before and after each imaging session to ensure alignment between the two channels and to obtain a registration matrix specific for the imaging session.
Tissue culture
U2OS cells (human osteosarcoma, American Type Culture Collection, HTB-96) were maintained at 37°C and under 5% CO2 in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Life Technologies, 11995 and 10082, respectively). For imaging, LabTek (155409; Thermo Fisher Scientific) or μ-Slide (80827; ibidi) chambers with a #1.5 coverglass bottom were first cleaned by incubating with 1 M NaOH for 2 h at room temperature, followed by washing five times with MilliQ water and incubation with MilliQ water or PBS overnight. Cells were plated in the chambers 36–48 h before imaging in phenol red-free DMEM (21063; Life Technologies) supplemented with 10% FBS.
Live-cell SMT
Cells were cultured using the previously described procedure first. 2 h before imaging, the growth medium was replaced with Fluorobrite DMEM (A18967; Thermo Fisher Scientific) supplemented with 10% FBS and 1:100 (v/v) ProLong Live Antifade Reagent (P36975; Thermo Fisher Scientific). Live-cell SMT experiments were performed at 37°C and 5% CO2 using a temperature- and CO2-controlled sample stage (Tokai Hit, INUG2F-SSI-W and UNIV2-CSG) connected to a house CO2 line. After the temperature and CO2 level stabilized, fluorescent labeling reagents were added to the well of interest. Wheat germ agglutinin (WGA)-CF633 (29024; Biotium) and human transferrin (HT)-CF680R (00086; Biotium) were added to a final concentration of 0.25–1 nM, whereas CellMask Deep Red (CellMask DR) (C10046; Life Technologies) was added to a final concentration of 1−4 × 10−4 [X], where [X] was the concentration recommended by the manufacturer for sample staining. The sample was incubated for 5 min to allow the fluorescent molecules to bind to the cell membrane, and the temperature and CO2 level to stabilize. Sample illumination and signal detection were performed in the same manner as described for fixed cells. Typically, 10,000–30,000 frames were recorded at 20–50 frames per second.
Data acquisition and analysis
Acquisition of raw images was performed using the open source micromanager software suite (https://micro-manager.org/). Image analyses for extracting single-molecule localization, spectra, and trajectories were all performed with custom MATLAB (The MathWorks, Natick, MA) scripts as described previously (26, 27).
For spectral analysis, individual molecules were first identified in the positional channel. A corresponding center position for each molecule in the spectral channel was computed based on a transformation matrix, which was determined by registering positional and spectral images of fluorescent beads on the coverslip taken with the 661 ± 5.5 nm bandpass filter. For each molecule, a 51 (rounded center position ± 25 along the dispersion direction) pixel ×3 (orthogonal direction) pixel region in the spectral image was used as its raw spectrum. The raw spectrum was first smoothed by moving average to locate the pixel with maximum intensity. Next, the precise pixel position of maximum emission was calculated by using a second-order polynomial fitting of pixels surrounding the one with maximum intensity; this was used to calculate the spectral shift distance (ssd). For multicolor data, coordinates of each fluorophore were first separated based on ssd values as described in the text; each channel was then rendered separately and recombined in ImageJ (https://imagej.nih.gov/ij/) (28) into a composite image.
For trajectory analyses, two molecules in adjacent frames were connected into a trajectory if they were located within 500 nm (i.e., around three pixels on the EM-CCD) of each other for data taken at 50 fps, and adjusted accordingly for data taken at slower rates. This could be performed before or after separating the coordinates into individual colors. Of note, molecules often fluctuated in brightness during diffusion, hence some frames within a trajectory contained noisier spectra than others. To avoid having to discard the entire trajectory due to these noisy spectra, spectral data from two–three successive frames could be combined to obtain an average spectrum. This procedure resulted in a much better retention rate of trajectories without adversely affecting the spectral resolution than solely using single-frame spectra for color separation. Diffusion state analysis with vbSPT was performed using the MATLAB scripts provided by the authors (http://sourceforge.net/projects/vbspt/) with minor modifications (26).
Results
The single-molecule spectral imaging system was constructed on a commercial, inverted microscope frame (Fig. 1 A). The same, high-numerical aperture objective was used to illuminate the sample and to collect the fluorescence signal. The signal was subsequently split at an ∼30:70 ratio between a positional channel and a spectral channel, respectively. The resulting signals in the two channels were projected onto the left and right halves of the same EM-CCD to yield two side-by-side images. A prism assembly was then inserted in the Fourier plane in the spectral channel to disperse the signal from each fluorophore into its spectral components. A slit was placed at the intermediate image plane to clip the image to a size matching half of the CCD chip.
Figure 1.
The single-molecule spectral imaging setup. (A) Optical scheme of the single-molecule spectral imaging system, constructed based on a standard, single-objective, single-molecule localization microscope is shown. To achieve simultaneous recording of the positions and emission spectra of all fluorescent molecules in the field of view, signals from the sample were split into two channels at ∼30:70 for the positional and spectral channels, respectively. The signals were then projected to the left and right sides of the detector after passing through the lenses L3S (spectral) and L3P (positional), respectively, and combining on a knife-edge mirror. In the spectral channel, a prism assembly consisting of an equilateral prism and a pair of mirrors was inserted in the infinity space between lenses L2 and L3S to disperse the signal. The prism assembly was mounted on a translational stage so that it could be moved in and out of the beam path, and the pair of mirrors after the prism was used to align the light path so that the overall direction of light propagation did not change in this channel. (B) A picture of the prism assembly in the microscope setup. The prism was mounted on its side so that the signals came in at the minimum dispersion angle and induced dispersion in the direction perpendicular to the closing direction (horizontal in this case) of the slit. To see this figure in color, go online.
We constructed the prism assembly on a translational stage to conveniently move it in and out of the light path, similarly to that described by Zhang et al. (24). With the assembly out, the two channels were aligned to yield nearly identical images with subpixel precision. The assembly, which comprises an equilateral prism and a pair of mirrors, was then put into the light path of the spectral channel to disperse the signal. The prism also diverts the signal beam, but the diversion is corrected by aligning the pair of mirrors behind the prism to steer the spectral image back to the same location on the EM-CCD as without the prism assembly (Fig. 1 B). Fine adjustment of the prism assembly is further discussed in the next section. Of note, we chose to orient the prism to disperse light in the direction parallel to the left and right edges of the slit, so that the slit edges do not cast shadows in the spectral image (Fig. S1).
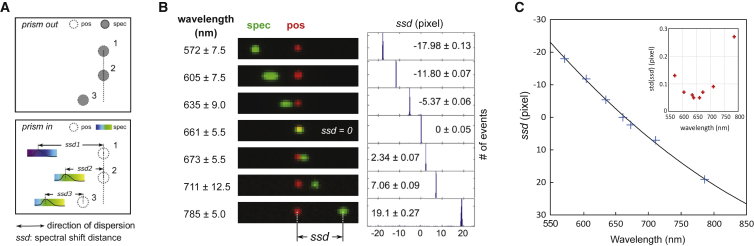
The single-molecule spectral imaging system generates a positional image and a spectral image for each fluorescent object in the field of view. With the prism out, the two images are nearly identical, and all the spectral components are focused to the same diffraction-limited spot on the detector (Fig. 2 A, top). With the prism in, the spectral image of each object becomes elongated along the direction of dispersion and the intensity profile of this image encodes the emission spectrum of the object. Each wavelength component in the signal from an object is deflected at a distinct angle by the prism and projected onto the detector at a specific distance away from the original, undeflected spectral image. The distance between the centroids of the positional and spectral images of a fluorophore in the overlaid image is defined as the spectral ssd for the fluorophore (Fig. 2 A, bottom).
Figure 2.
Calibration of the single-molecule spectral imaging system. (A) Principles of spectral measurement on the single-molecule spectral imaging system are shown. The positional (empty circle) and spectral (filled circle) images were first aligned with high precision before the prism was inserted (top panel). With the prism inserted, the spectral images of all fluorescent objects became elongated, and the intensity profile of each elongated image represented the emission spectrum of the corresponding object (bottom panel). For each object in the overlaid image, its ssd was defined as the distance between the centroid of its positional image and the (sub-) pixel position of maximum emission intensity in the spectral image. In this case, objects two and three are the same type of fluorophore and they exhibit the same ssd (i.e., ssd2 = ssd3) values, whereas object one is in a different color exhibiting a different ssd value (ssd1); (B) Overlaid positional (pos) (red) and spectral (spec) (green) images of fluorescent beads (broad emission between 500 and 800 nm when excited at 488 or 561 nm) after passing through a series of narrow bandpass filters as indicated. The data point at ∼572 nm was taken with 488 nm excitation and the remaining data were taken with 561 nm excitation. The positional and spectral images were intentionally overlapped with subpixel precision at 661 ± 5.5 nm (i.e., ssd = 0.0). Thus, images acquired at shorter wavelengths had negative ssd with the spectral image to the left of the positional image, and the opposite in images taken at longer wavelengths (left). Shown on the right, histograms of ssd at each wavelength with the means and SD indicated. (C) The calibration curve showing the relationship between the center wavelength and the measured ssd values is shown. Inset is the SD of ssd at each wavelength. To see this figure in color, go online.
We calibrated the instrument spectrally by imaging 40 nm fluorescent beads that emit broadly between 550 and 800 nm when excited at 488 or 561 nm. Specific wavelength ranges were selected by inserting filters with 10–20 nm transmission bandwidths, yielding much less elongated and symmetric spectral images to facilitate both alignment and centroid determination (Fig. 2 B). We adjusted the system using the pair of mirrors in the prism assembly so that the positional and spectral images acquired using a 661 ± 5.5 nm bandpass filter coincided (Fig. 2 B, middle panel). We noted that the overlap gradually deteriorated toward the periphery, because light from peripheral objects enters the prism at different angles than that from a central object. We corrected for this by computing a registration matrix between the two images. This reduced the registration error to ∼0.05 pixels across the whole field of view. The same matrix was used to register paired positional and spectral images taken at other wavelengths. In this way, the ssd was set to be 0 (± 0.05) pixels for objects emitting at 661 nm on the instrument (Fig. 2 B, right panel). We chose 661 nm as the central wavelength because it is roughly at the midpoint of the emission wavelength range for commonly used fluorophores (∼520 to ∼800 nm).
We obtained ssd values for other wavelengths using six different bandpass filters with center transmission wavelengths ranging from 572 to 785 nm (Fig. 2 B). Thousands of beads were measured at each wavelength to determine the mean and SD for that ssd (Fig. 2 B, right panel). We designated the ssd values at wavelengths shorter than 661 nm to have a negative sign and those at longer wavelengths to be positive. We fitted a second-order polynomial to the calibration curve and used the resulting analytic formula to convert ssd values from pixels to wavelength units (Fig. 2 C). ssd values referenced in wavelength or pixel units will be used interchangeably for the rest of this manuscript. Notably, the SD of ssd values typically were ∼0.1 pixel or lower, except for the last data point at 785 nm, where emission from the beads was weak (Fig. 2 B and C, inset in the latter). As one pixel corresponds to ∼6 nm in spectral space according to our calibration procedure, this indicates that the inherent spectral precision of this instrument, in terms of identifying emission peak positions of bright emitters, should be better than 0.6 nm.
To demonstrate the spectral and spatial resolutions of our single-molecule spectral imaging system in comparison with spectrally-resolved stochastic optical reconstruction microscopy (SR-STORM), we performed four-color superresolution imaging of fixed U2OS cells using four photoswitchable fluorophores—Dyomics 634 (DY634), DyLight 650 (DL650), CF660C, and CF680—which have been used previously for four-color SR-STORM. As expected, the fluorophores also performed well on our instrument, showing efficient photoswitching and good single-molecule photon yields, with ∼ 1000 median photons in the positional channel alone. The ssd values measured on our instrument were 662.8 ± 3.5, 680.5 ± 4.0, 696.8 ± 4.2, and 711.9 ± 2.9 nm (mean ± SD), respectively. Thus, these fluorophores were easily distinguished (Fig. S2 A), even though their emission maxima were separated by only 10–15 nm.
Images of U2OS cells simultaneously stained with four fluorophores labeling mitochondria (DY634), vimentin (DL650), microtubule (CF660C), and the plasma membrane (CF680) clearly showed four distinct structures with excellent spatial resolution and spectral separation. Clearly resolved single-molecule images with robust photoswitching were recorded for these samples, and the four fluorophores were visually distinguished from each other in overlaid spectral and positional images (Fig. S2 B). Histograms of ssd values for all single molecules detected showed four well separated peaks corresponding to the four fluorophores (Fig. S3). The histograms were fitted with four Gaussian distributions, each of which had a mean and SD that largely agreed with the single-color results. Based on the fitting results, we defined an empirical ssd range for each fluorophore that included ∼80% of all events. Events outside of the defined ranges were discarded. We estimate that the false assignment rate between neighboring fluorophores is < 2% with these cutoff criteria (Fig. S3). This leaves sufficient localization events to reconstruct a high-resolution image in each of the four channels with very low spectral cross talk (Fig. S2 C). Even lower false assignment rates could be achieved by further narrowing the acceptable ssd ranges at the cost of more discarded localization events and potentially less continuous structures.
We also tested whether using only ∼30% of the photons in the positional channel would significantly impact the spatial resolution by imaging microtubules in U2OS cells labeled with AF647 via indirect immunofluorescence. We imaged the sample on our instrument using standard STORM imaging buffer and acquisition settings, and used similar settings in data processing (in particular, the same threshold for picking up single particles) as in conventional STORM. The reconstructed image (Fig. S4) clearly shows the hollow structure of microtubules, a commonly used standard for assessing imaging resolution of STORM (29). We measured the widths of the microtubules to be ∼39 nm. This is comparable to other reported measurements and is consistent with an average spatial resolution of ∼26 nm (29, 30).
With the demonstrated spectral and spatial resolutions and the single-lens architecture, our single-molecule spectral imaging system is potentially well suited for multicolor live-cell superresolution imaging and SMT, the latter of which is demonstrated herein. By freeing up the space above the sample, live cells can be maintained in an on-stage incubator with temperature and CO2 control. For SMT, we used conventional instead of photoswitchable fluorophores, which eliminates the need for potentially toxic STORM imaging buffers and high-power illumination needed to induce photoswitching. We enabled single-molecule imaging by using a sparse labeling strategy, in which low concentrations (0.5–5 nM) of fluorescently labeled affinity reagents were kept in the growth medium throughout the imaging experiment. In this strategy, binding of new fluorescent probes to cellular targets replenished probes lost to photobleaching. We sought to keep the particle density at no higher than ∼200 molecules within a 200 × 256 pixel field of view (∼0.1 molecules/μm2) to prevent excessive occurrences of overlapping spectra from nearby molecules. Particles with overlapping spectra were discarded.
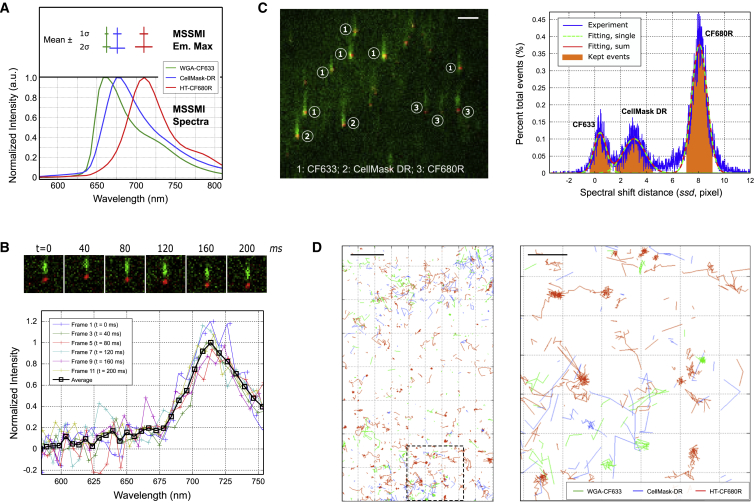
We tested the performance of our instrument for SMT in live U2OS cells stained for glycoproteins using WGA conjugated to CF633, transferrin receptors using HT conjugated to CF680R, and the CellMask DR reagent comprised of a fluorophore coupled to a lipid-soluble membrane anchor. CF633, Cellmask DR, and CF680R were efficiently excited with a single 637 nm laser at a low excitation dose, and all exhibited adequate photostability to allow effective SMT in single-color tests (Fig. S5). We used a relaxed total internal reflection illumination scheme to excite fluorophores on the cell surface above the glass substrate on which the cells were grown. We assessed the single-molecule spectral characteristics of these fluorophores, including the average emission spectra, the mean, and SD of ssd in single-color live-cell imaging experiments (Fig. 3 A). The measured ssd values for CF633, Cellmask DR, and CF680R in the conjugated forms were 662.3 ± 2.8, 677.6 ± 5.2, and 708.9 ± 5.5 nm (mean ± SD), respectively.
Figure 3.
Multicolor single-molecule tracking with the single-molecule spectral imaging system in living cells. (A) The lower panel shows the average single-molecule spectra of CF633, CellMask DR, and CF680R molecules measured in live U2OS cells. The upper panel shows the mean (vertical line) and spread (horizontal line) of emission maxima for each fluorophore. (B) Series of single-molecule positional (pos) (red) and spectral (spec) (green) images of CF680R recorded at 20 ms per frame and shown in 40 ms (every two frames) intervals are shown. Here, the spectra have not been corrected by image registration; (C) Left: a representative raw image frame taken on a live U2OS cell simultaneously labeled with WGA-CF633, CellMask DR, and HT-CF680R, showing three distinct populations of molecules based on the separation between the positional (red) and spectral (green) images of the single molecules. Markers 1, 2, and 3 indicate CF633, CellMask DR, and CF680R molecules, respectively. Frame acquisition time was 20 ms. All three dyes were excited with the same 637 nm laser. Right: a histogram of ssd values from a three-color SMT experiment and the results of fitting with three Gaussian distributions, where the green curves represent individual fittings and the red line is the sum of all three fittings. Areas with orange shades indicate ranges of ssd values of which the associated localization events were kept and assigned to specific fluorophores. Other localization events were discarded. (D) Example diffusion trajectories of the three molecular species in a live U2OS cell, obtained simultaneously on the single-molecule spectral imaging system but separated into the three channels during sample processing are shown. Also shown on the left is an overview of a part of a cell, and on the right is a zoom-in view of the boxed region in the left plot. The scale bars represent 2 μm (C, left and D, left) and 500 nm (D, right), respectively. MSSMI, multispectral single-molecule imaging.
Importantly, we found that single-molecule spectra (measured as the ssd histogram) for these fluorophores were not broadened by molecular motions (Fig. 3 B), in large part because motions led to the same displacement in the positional and spectral images. To further investigate the potential impact of molecular motions on the measured emission spectra, we performed a systematic simulation study, where molecules were allowed to diffuse at specific rates, and the resulting smearing of emission spectra was calculated based on the coordinates sampled within the frame acquisition time (see Figs. S6–S11 and the related text in the Supporting Material). From these results, it became apparent that at frame acquisition times < 50 ms, significant broadening of the ssd histograms by > 2 nm (SD) did not happen until very high diffusion coefficients (> 5 μm2/s). When photon flux is taken into consideration, longer acquisition times actually present a slight advantage in yielding a slightly narrower distribution of ssd (hence slightly better spectral resolution), presumably at a cost of spatial and temporal resolution.
With these considerations, we used an acquisition time of 20–30 ms per frame and a laser power density of 0.5–1 kW/cm2 for a good compromise between trajectory length, temporal resolution, and spectral resolution. This power density is significantly lower than that typically used for photoswitchable dyes and comparable to that typically used in single-color SMT. By exciting all three (potentially more) fluorophores with a single-laser emitting at a far-red wavelength, our approach significantly reduced light induced cell damage in multicolor SMT experiments. We also added the Prolong Live Antifade reagent, specially formulated for live-cell imaging, to further improve fluorophore photostability and trajectory length (see Materials and Methods).
We first measured the movements of the three fluorophore-labeled cell surface targets individually in single-color labeling experiments. Visual inspection of the movies (Movie S1) showed that both protein-coupled fluorophores displayed more than one movement pattern. A small population of molecules moved rapidly along linear tracks indicative of directed transport, others displayed a seemingly random pattern, whereas others appeared to be constrained to move within a small region. The lipid-anchored CellMask DR made much larger, random excursions on average than the other two fluorophores (Fig. S5), and we saw no evidence for directed or constrained movement.
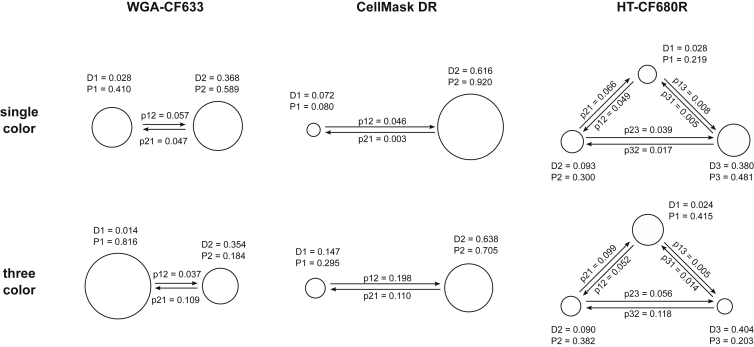
To analyze these complex movement patterns, we used vbSPT (5), which was designed to extract diffusional models and model parameters from a large number of single-molecule trajectories, even if the trajectories are relatively short (∼20 steps or less). In vbSPT, the number of diffusion states in the mode is varied and, for each model, the diffusion constants associated with each state and the state conversion rates are inferred based on the trajectories using an approximate Bayesian approach. The model that best fits the data is chosen as the output (5). Analysis with vbSPT suggested that WGA-CF633 exhibited two distinct movement patterns: one characterized by a diffusion constant (D) of ∼0.36 μm2/s, and the other by D ∼ 0.01 μm2/s. HT-680R displayed three movement patterns characterized by Ds of 0.02, 0.09, and 0.39 μm2/s. We recognize that the presence of directed movements makes trajectory analysis using vbSPT not entirely appropriate, but it is presented here as an expedient to demonstrate the existence of multiple movement patterns (6). As expected for a small molecule probe, most CellMask DR fluorophores moved in a seemingly random pattern associated with D ∼ 0.62 μm2/s, although a small fraction exhibited a slower diffusion rate (D ∼ 0.07 μm2/s) (Fig. 4, top).
Figure 4.
Single-molecule diffusion properties measured with single-molecule spectral imaging and analyzed with vbSPT (5). The diffusion state graphs obtained from single-color (top) and three-color (bottom) experiments are shown for WGA-CF633 (left), CellMask DR (middle), and HT-CF680R (right) as connected circles. Size of a circle indicates probabilities of the molecules residing in that specific state. Diffusion constants of state i are labeled as Di (units: μm2/s), probability of molecules in state i is Pi, and the probability of transitioning from state i to state j within one frame is labeled as pij.
We then performed simultaneous imaging of all three fluorophore-tagged probes using the single-molecule spectral imaging. Cells were labeled and imaged under the same conditions used for single-color experiments, except that the culture medium contained all three labeling reagents. Despite the increased density and coexistence of all three labeling reagents, clear images of single molecules were recorded at 20–30 ms/frame (Movie S1). The three molecular species were assigned based on ssd values (Fig. 3 C, left). Histograms of ssd values for all detected localization events showed three distinct peaks, and each peak was assigned to a fluorophore as shown in Fig. 3 C (right panel). From the histogram, it is evident that the spectral cross talk in identifying the three fluorescent species is essentially negligible; furthermore, there is clearly additional spectral space to be filled with appropriate fluorophores to achieve SMT in even more colors.
After fluorophore assignment, single-molecule localization events were separated into three channels based on ssd value, and the localizations were connected into trajectories based on spatial proximity in successive frames (Fig. 3 D). This allowed diffusion properties of all three species to be extracted from a single experiment. By comparing the vbSPT outputs from single-color (Fig. 4, top) and three-color (Fig. 4, bottom) experiments, we found that both the number of diffusion states and the respective diffusion constants agreed very well, although the relative populations of each diffusion state vary to some extent, likely due to variations between individual cells. This consistency is also reflected in the histograms of frame-to-frame displacement of the three species in the single- and three-color SMT (Fig. S5). These data show that our instrument and the associated algorithms were able to correctly identify individual fluorophores in the data and extract the unique movement properties of each species. These results validate the use of single-molecule spectral imaging for simultaneous tracking of multiple targets in live cells at the single-molecule level.
Discussion
In this article, we show that multicolor spatial and spectral imaging using single-molecule localization principles can be achieved using a relatively simple, single-objective imaging system that is well suited to both fixed and live-cell imaging. Using fluorophores tested herein, single-molecule spectral imaging achieves far better spectral resolution at 10–15 nm compared to that achieved with conventional fluorescence detection schemes based on emission filters (50–100 nm), without compromising spatial resolution. By including additional excitation lasers (e.g., 561 or 488 nm), and provided there is an optimal set of fluorophores with emission wavelengths spanning the ∼500 to ∼850 nm range, single-molecule spectral imaging has the potential to image samples labeled with 6–10 different colors. If implemented, this would offer a transformative tool for analyzing complex, multicomponent interactions in intact biological specimens.
A unique advantage of single-molecule spectral imaging, as already demonstrated with SR-STORM, is the essential elimination of spectral cross talk at the same time as using fluorophores with broad emission and significant spectral overlap, provided that the ssd values for each fluorophore do not vary substantially. The fluorophores used in this study for both fixed and live-cell experiments satisfy these criteria; their ssd values typically fluctuate within just 3–5 nm, and the shapes of the single-molecule spectra are mostly invariant across the sample (this work and Zhang et al. (24)). We show that this is true even when the molecules are moving (Fig. 4 B). This allowed us to use a panel of fluorophores whose emission spectra maxima differed by only ∼15 nm.
We established fluorophore classification criteria that allowed ∼80% of the fluorophore events to be localized at the same time as misclassifying only 1–2%. The misclassification rate could be further reduced by adopting a more stringent assignment strategy, but at the expense of discarding more localization events. Although here we only used ssd values to distinguish fluorophores, other spectral properties such as the shape of the emission spectra could also be used to further improve fluorophore identification. Additionally, synthesis and screening of fluorophores that exhibit even narrower ssd distributions than those used in this study would be beneficial.
The ability to use spectrally proximal fluorophores in single-molecule spectral imaging offers a number of advantages. First, all the fluorophores can be efficiently excited at once with a single-laser, which greatly simplifies experimentation and reduces the total laser power and the associated phototoxic effects during live-cell imaging (15, 31). This is particularly beneficial when a single red or near infrared laser is used for SMT in more than three colors. Multicolor excitation would traditionally require multiple lasers, including those emitting at short wavelengths (blue or green), and would be more detrimental to cell health. Thus, for live-cell imaging, the benefit of using single-laser excitation may well offset the somewhat lower spatial resolution as a result of using only 30% of the photons emitted from each fluorophore for molecular localization. Second, registration errors between the different color channels are essentially eliminated by using fluorophores with closely spaced emission maxima (e.g., ∼15 nm apart).
We used a single-objective configuration for single-molecule spectral imaging, which has significant implications in the practical use of the technique. Among others, this design can be constructed on most commercial, inverted microscope frames, and existing single-molecule fluorescence setups can be easily modified to perform single-molecule spectral imaging. More importantly, this configuration is fully compatible with live-cell imaging, as demonstrated in this work. Importantly, this benefit did not come with a noticeable cost in either spectral or spatial (Fig. S4) resolutions, in part owing to the good photon yields of the fluorophores such that the resolutions were mostly not photon-limited. Recently, Mlodzianoski et al. (25) also reported single-objective spectral photoactivated localization microscopy; however, proof of its superior spectral and spatial resolution, and its utility in multicolor SMT, has not been demonstrated.
With single-molecule spectral imaging, we demonstrate three-color SMT in live cells using spectrally similar dyes (CF633, CellMask DR, and CF680R), which emit at wavelengths only 15–30 nm apart and can be excited by a single 637 nm laser. The diffusion properties derived from a simultaneous three-color tracking experiment agreed very well with the single-color controls, confirming accurate assignment of fluorophores in a sample with mixed staining. This greatly extends the ability of using SMT to probe dynamic biological processes. In particular, the presence of molecular states that are almost completely immobile (such as those of WGA-CF633 and HT-CF680R shown in Fig. 4) often indicates localization to nanodomains (such as “lipid rafts”) or interaction with cellular scaffolds (32, 33), an increasingly appreciated mechanism of biological regulation. However, previous SMT data were mostly single- or dual-color. With simultaneous multicolor SMT, we can address more complex questions such as dynamic colocalization of multiple signaling molecules in the same membrane domains that would be difficult to capture in snapshots of static images. Recent works by English and colleagues (19, 33) demonstrated three-color SMT using three lasers and three cameras. This should be readily achievable using single-molecule spectral imaging with a single-laser line and a single camera. As alluded to earlier, SMT in even more colors should be possible with an appropriate panel of fluorophores that fill in the remaining spectral space (Fig. 3 C). Multicolor three-dimensional SMT should also be possible with this method by including a cylindrical lens in the positional channel. This should be feasible because it is an extension of the three-dimensional SR-STORM imaging demonstrated by Zhang et al. (24). Lastly, although here we only demonstrated live-cell SMT, we anticipate that live-cell multicolor single-molecule localization microscopy would be feasible with the same system.
Imaging spectroscopy has become an increasingly attractive approach to studying biology given the rich information contained in molecular spectra. Besides the aforementioned reports (24, 25) and this work, a number of recent studies have used spectral information in superresolution microscopy for multicolor imaging (34) or improved spatial resolution (35); spectral imaging has also been used to greatly increase the multiplexity of fluorescence life-time microscopy (36). In addition to multicolor imaging, we also recognize the possibility of using single-molecule spectral imaging with environment-sensitive fluorophores (37) to assess heterogeneities in the micro- and nanoenvironment, an inherent and important property of biological systems. Work in this direction is currently underway.
In summary, we have shown that a single-objective, single-molecule spectral imaging system provides a flexible and powerful platform for multiplexed single-molecule localization and tracking with rich spatial and spectroscopic information. We realized nanoscopic imaging of multiple cellular structures and tracking of multiple molecular species simultaneously in fixed or live cells using a single-laser excitation and a panel of spectrally similar dyes with minimal spectral cross talk.
Author Contributions
J.W.G, S.J., S.L.G., and X.N. conceived the project. S.J. designed the initial line-scanning microscope configuration (not discussed here). X.N. and T.H. designed the dual (not discussed here) and single-objective microscope configuration. T.H., J.W., and X.N. constructed the microscope. T.H. developed software for data acquisition and analysis. T.H., J.W., L.-J.L., and A.B. tested fluorophores and optimized antibody labeling conditions. T.H. and J.W. performed multicolor labeling and imaging in fixed cells and analyzed data. C.P. and X.N. designed live-cell single-molecule tracking experiments, and C.P. performed the experiments and analyzed data. C.P. performed the simulations. Z.S. assisted with data analysis. T.H., X.N., J.W.G., and C.P. wrote the manuscript. J.W.G. and X.N. supervised the research.
Acknowledgments
We are grateful to Dr. Don Kania (formerly FEI) and many other colleagues for their support and helpful discussions. S.L.G. thanks Thermo Fisher Scientific for gifts of fluorescent agents.
X.N., S.L.G., and J.W.G. acknowledge financial support from the Damon Runyon Cancer Research Foundation, the FEI company, the M.J. Murdock Charitable Trust, and the Oregon Health and Science University Knight Cancer Institute. X.N. also acknowledges support by a supplement to NIH U54CA163123 (PI Lisa Coussens). J.W.G. was supported by the Office of Science and the Office of Biological and Environmental Research, both part of the U.S. Department of Energy, under contract DE-AC02-05CH11231, the W. M. Keck Foundation, and by a Department of the Army award (W81XWH-07-1-0663).
Editor: Antoine van Oijen.
Footnotes
Tao Huang and Carey Phelps contributed equally to this work.
Supporting Materials and Methods, eleven figures and one movie are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)31242-0.
Supporting Material
Raw data frames showing single-molecule images in the spectral (left) and positional (right) channels from a live U2OS cell simultaneously labeled with CF633 (WGA), CellMask DR, and HT-CF680R.
References
- 1.Manzo C., Garcia-Parajo M.F. A review of progress in single particle tracking: from methods to biophysical insights. Rep. Prog. Phys. 2015;78:124601. doi: 10.1088/0034-4885/78/12/124601. [DOI] [PubMed] [Google Scholar]
- 2.Kusumi A., Tsunoyama T.A., Fujiwara T.K. Tracking single molecules at work in living cells. Nat. Chem. Biol. 2014;10:524–532. doi: 10.1038/nchembio.1558. [DOI] [PubMed] [Google Scholar]
- 3.Yu J. Single-molecule studies in live cells. Annu. Rev. Phys. Chem. 2016;67:565–585. doi: 10.1146/annurev-physchem-040215-112451. [DOI] [PubMed] [Google Scholar]
- 4.Manley S., Gillette J.M., Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 5.Persson F., Lindén M., Elf J. Extracting intracellular diffusive states and transition rates from single-molecule tracking data. Nat. Methods. 2013;10:265–269. doi: 10.1038/nmeth.2367. [DOI] [PubMed] [Google Scholar]
- 6.Monnier N., Barry Z., Bathe M. Inferring transient particle transport dynamics in live cells. Nat. Methods. 2015;12:838–840. doi: 10.1038/nmeth.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 8.Hess S.T., Girirajan T.P., Mason M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heilemann M., van de Linde S., Sauer M. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 11.Bosch P.J., Corrêa I.R., Jr., Subramaniam V. Evaluation of fluorophores to label SNAP-tag fused proteins for multicolor single-molecule tracking microscopy in live cells. Biophys. J. 2014;107:803–814. doi: 10.1016/j.bpj.2014.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang H., English B.P., Ovryn B. Tracking surface glycans on live cancer cells with single-molecule sensitivity. Angew. Chem. Int. Ed. Engl. 2015;54:1765–1769. doi: 10.1002/anie.201407976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benke A., Olivier N., Manley S. Multicolor single molecule tracking of stochastically active synthetic dyes. Nano Lett. 2012;12:2619–2624. doi: 10.1021/nl301018r. [DOI] [PubMed] [Google Scholar]
- 14.Dunne P.D., Fernandes R.A., Klenerman D. DySCo: quantitating associations of membrane proteins using two-color single-molecule tracking. Biophys. J. 2009;97:L5–L7. doi: 10.1016/j.bpj.2009.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frigault M.M., Lacoste J., Brown C.M. Live-cell microscopy - tips and tools. J. Cell Sci. 2009;122:753–767. doi: 10.1242/jcs.033837. [DOI] [PubMed] [Google Scholar]
- 16.Subach F.V., Patterson G.H., Verkhusha V.V. Bright monomeric photoactivatable red fluorescent protein for two-color super-resolution sptPALM of live cells. J. Am. Chem. Soc. 2010;132:6481–6491. doi: 10.1021/ja100906g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnspang E.C., Brewer J.R., Lagerholm B.C. Multi-color single particle tracking with quantum dots. PLoS One. 2012;7:e48521. doi: 10.1371/journal.pone.0048521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cutler P.J., Malik M.D., Lidke K.A. Multi-color quantum dot tracking using a high-speed hyperspectral line-scanning microscope. PLoS One. 2013;8:e64320. doi: 10.1371/journal.pone.0064320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.English B.P., Singer R.H. A three-camera imaging microscope for high-speed single-molecule tracking and super-resolution imaging in living cells. Proc. SPIE Int. Soc. Opt. Eng. 2015;9550:955008. doi: 10.1117/12.2190246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wäldchen S., Lehmann J., Sauer M. Light-induced cell damage in live-cell super-resolution microscopy. Sci. Rep. 2015;5:15348. doi: 10.1038/srep15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundquist P.M., Zhong C.F., Zaccarin D. Parallel confocal detection of single molecules in real time. Opt. Lett. 2008;33:1026–1028. doi: 10.1364/ol.33.001026. [DOI] [PubMed] [Google Scholar]
- 22.Haga T., Sonehara T., Takahashi S. Prism-based spectral imaging of four species of single-molecule fluorophores by using one excitation laser. J. Fluoresc. 2013;23:591–597. doi: 10.1007/s10895-013-1208-8. [DOI] [PubMed] [Google Scholar]
- 23.Broeken J., Rieger B., Stallinga S. Simultaneous measurement of position and color of single fluorescent emitters using diffractive optics. Opt. Lett. 2014;39:3352–3355. doi: 10.1364/OL.39.003352. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z., Kenny S.J., Xu K. Ultrahigh-throughput single-molecule spectroscopy and spectrally resolved super-resolution microscopy. Nat. Methods. 2015;12:935–938. doi: 10.1038/nmeth.3528. [DOI] [PubMed] [Google Scholar]
- 25.Mlodzianoski M.J., Curthoys N.M., Hess S.T. Super-resolution imaging of molecular emission spectra and single molecule spectral fluctuations. PLoS One. 2016;11:e0147506. doi: 10.1371/journal.pone.0147506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nickerson A., Huang T., Nan X. Photoactivated localization microscopy with bimolecular fluorescence complementation (BiFC-PALM) for nanoscale imaging of protein-protein interactions in cells. PLoS One. 2014;9:e100589. doi: 10.1371/journal.pone.0100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nickerson A., Huang T., Nan X. Photoactivated localization microscopy with bimolecular fluorescence complementation (BiFC-PALM) J. Vis. Exp. 2015;106:e53154. doi: 10.3791/53154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schindelin J., Arganda-Carreras I., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dempsey G.T., Vaughan J.C., Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ries J., Kaplan C., Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods. 2012;9:582–584. doi: 10.1038/nmeth.1991. [DOI] [PubMed] [Google Scholar]
- 31.Laissue P.P., Alghamdi R.A., Shroff H. Assessing phototoxicity in live fluorescence imaging. Nat. Methods. 2017;14:657–661. doi: 10.1038/nmeth.4344. [DOI] [PubMed] [Google Scholar]
- 32.Dietrich C., Yang B., Jacobson K. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys. J. 2002;82:274–284. doi: 10.1016/S0006-3495(02)75393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz Z.B., English B.P., Singer R.H. Mapping translation ‘hot-spots’ in live cells by tracking single molecules of mRNA and ribosomes. Elife. 2016;5:e10415. doi: 10.7554/eLife.10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shechtman Y., Weiss L.E., Moerner W.E. Multicolour localization microscopy by point-spread-function engineering. Nat. Photonics. 2016;10:590–594. doi: 10.1038/nphoton.2016.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong B., Almassalha L., Zhang H.F. Super-resolution spectroscopic microscopy via photon localization. Nat. Commun. 2016;7:12290. doi: 10.1038/ncomms12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niehörster T., Löschberger A., Sauer M. Multi-target spectrally resolved fluorescence lifetime imaging microscopy. Nat. Methods. 2016;13:257–262. doi: 10.1038/nmeth.3740. [DOI] [PubMed] [Google Scholar]
- 37.Klymchenko A.S., Mely Y. Fluorescent environment-sensitive dyes as reporters of biomolecular interactions. Prog. Mol. Biol. Transl. Sci. 2013;113:35–58. doi: 10.1016/B978-0-12-386932-6.00002-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw data frames showing single-molecule images in the spectral (left) and positional (right) channels from a live U2OS cell simultaneously labeled with CF633 (WGA), CellMask DR, and HT-CF680R.