Abstract
Monomers of amyloid-β (Aβ) protein are known to be disordered, but there is considerable controversy over the existence of residual or transient conformations that can potentially promote oligomerization and fibril formation. We employed single-molecule Förster resonance energy transfer (FRET) spectroscopy with site-specific dye labeling using an unnatural amino acid and molecular dynamics simulations to investigate conformations and dynamics of Aβ isoforms with 40 (Aβ40) and 42 residues (Aβ42). The FRET efficiency distributions of both proteins measured in phosphate-buffered saline at room temperature show a single peak with very similar FRET efficiencies, indicating there is apparently only one state. 2D FRET efficiency-donor lifetime analysis reveals, however, that there is a broad distribution of rapidly interconverting conformations. Using nanosecond fluorescence correlation spectroscopy, we measured the timescale of the fluctuations between these conformations to be ∼35 ns, similar to that of disordered proteins. These results suggest that both Aβ40 and Aβ42 populate an ensemble of rapidly reconfiguring unfolded states, with no long-lived conformational state distinguishable from that of the disordered ensemble. To gain molecular-level insights into these observations, we performed molecular dynamics simulations with a force field optimized to describe disordered proteins. We find, as in experiments, that both peptides populate configurations consistent with random polymer chains, with the vast majority of conformations lacking significant secondary structure, giving rise to very similar ensemble-averaged FRET efficiencies.
Introduction
Amyloid-β (Aβ) protein is a fragment comprising between 39 and 43 residues of amyloid precursor protein found in amyloid plaques in the brains of Alzheimer’s disease patients (1). X-ray fiber diffraction and solid-state NMR have shown that the Aβ fibrils found in these plaques contain ordered cross-β structures (2, 3, 4). There is considerable complexity, however, at the molecular scale: solid-state NMR structures of fibrils formed by the 40-residue (Aβ40) (5, 6, 7) and 42-residue (Aβ42) proteins (8, 9, 10) are quite different and structural polymorphism is found even in the fibrils of the same protein isoform depending on aggregation conditions (11, 12, 13). In addition, recent studies have shown that the structure of fibrils seeded from material derived from patients is different from those grown in vitro from the monomer state (7, 14). Beyond this structural heterogeneity, the mechanism of fibril formation is also nontrivial and the hardest to study experimentally due to the heterogeneity and transient nature of the initial oligomers. Aggregation of Aβ is characterized by a rapid fibril growth phase preceded by a long lag period, which is required for the formation of aggregation seeds (15). Once an aggregation seed is formed, it acts as a template for fibril elongation. The fibrils themselves can also accelerate the formation of seeds for new fibrils, a process known as “secondary nucleation” (16). Although the same overall kinetic mechanism appears to describe both proteins, aggregation of Aβ42 is much faster than that of Aβ40 (17) and the relative importance of primary versus secondary nucleation differs (18, 19). In addition to seeding aggregation, recent studies have suggested that oligomers may be more toxic than fibrils and interact with various cellular components and synaptic receptors (20, 21, 22, 23). Therefore, characterization of the seed formation from monomers should be helpful for understanding the complexity and heterogeneity of the process and possibly the disease mechanism.
As a first step to understanding the assembly, monomers of Aβ have been extensively studied, mostly using solution NMR and molecular dynamics (MD) simulations. A consensus of conclusions of these studies is that Aβ is largely disordered and does not have a large fraction of stable conformations. Nonetheless, many studies have suggested that local secondary structures or transient conformations with tertiary contacts exist, although the structures and populations of these conformations vary among studies (24, 25, 26, 27, 28) despite similarities in experimental conditions (neutral pH and 4–8°C). Because the only difference between Aβ40 and Aβ42 is the two additional hydrophobic residues at the C-terminus of Aβ42, the interpretation of different behaviors of the two peptides has been focused on the effect of these two residues. For example, Yan et al. (26) have used NMR relaxation data to suggest that the C-terminal end of Aβ42 is more rigid and shows a higher β-strand propensity than that of Aβ40, which may have implications in explaining the increased aggregation rates of Aβ42. In a study of the decapeptide Aβ21–30 identified by limited proteolysis, Lazo et al. (29) raised a possibility of the formation of a β-turn in the hydrophilic region (residue 23–30) that resembles the solid-state NMR structure of Aβ40 fibril (30); long-range contacts required for this hairpin structure were not, however, observed in the MD simulation and NMR study by Fawzi et al. (31) for the same peptide. In the most recent and extensive NMR study, at 4°C, Roche et al. (32) have shown that the chemical shifts and J couplings of both Aβ40 and Aβ42 minimally deviate from those of a random coil, and, more strikingly, there is virtually no difference in these values between the two proteins from the N-terminus to residue 34. In addition, there are no long-range nuclear Overhauser effects (NOEs) in either protein, contrary to earlier studies (28, 33, 34). This result strongly suggests that both Aβ40 and Aβ42 are predominantly disordered, very similar to each other, and that the population of compact structures, such as β-hairpins, in the hydrophilic region is very low or negligible.
MD simulations are a valuable addition to experiment for characterizing disordered proteins, as determining structural features of a heterogeneous distribution of conformations from ensemble-averaged experimental data is clearly very challenging. The results of past simulation studies, however, vary largely in various measures such as the secondary structure content and the nature of representative conformations. Garcia and co-workers (35, 36, 37, 38) have observed a more structured C-terminus for Aβ42 than for Aβ40, a strong propensity for turn formation in the hydrophilic region (residues 24–28), and relatively high secondary structure contents. In contrast, Lin and co-workers (39, 40) have reported significantly reduced β-contents in this region. Head-Gordon and co-workers (33, 34) have performed a number of studies of Aβ40 and Aβ42 as well as derived fragments, with careful comparison to NMR data. They found that a broad ensemble of local secondary structure was formed in both Aβ40 and Aβ42. They also suggested significant differences in where local β-structure was formed in the two peptides. One likely reason for the discrepancies between the various studies is residual inaccuracies of the force fields, to which disordered proteins are particularly sensitive, as well as the challenge of adequately sampling the disordered ensemble. The first of these systematic errors is the relative propensity for forming different types of secondary structure, a deficiency that has largely been corrected in newer force fields by empirical corrections using solution NMR data (41). In addition, until recently, unfolded proteins were more compact than observed experimentally; in the past 2 years, this has also been addressed by modifying the simulation water models (42, 43, 44, 45, 46). Somewhat remarkably, given this difficulty, the simulation results have always been found to be consistent with experimental observations, usually solution NMR data such as chemical shifts, J-couplings, and short-range NOEs (33, 34, 35, 36). Part of the explanation for this may be that those parameters are primarily sensitive to local structure in a disordered chain. Complementary information on the global chain dimensions should better help to discriminate between different models. In principle, one source of such information could be NMR data obtained from paramagnetic relaxation enhancement and residual dipolar couplings (47, 48).
In this work, we compare the behaviors of Aβ40 and Aβ42 using a close combination of single-molecule Förster resonance energy transfer (FRET) spectroscopy and MD simulations. Single-molecule spectroscopy can virtually eliminate one of the key difficulties in quantitative experimental characterizations of the monomeric state of Aβ: namely the formation of oligomers and larger aggregates at high micromolar concentrations. This is because the experimental concentration is so low (100 pM or below) and the detection of a monomer can be ensured by observation of a single photobleaching step of the dyes. This avoids the need to perform experiments at low temperatures to slow down the aggregation, as often done in NMR studies. A second advantage of FRET is that it probes global, long-range intramolecular distances, thus complementing the more local information provided by many NMR observables. We labeled the N- and C-termini of Aβ with Alexa 488 and Alexa 647 site-specifically by incorporating an unnatural amino acid, 4-acetylphenylalanine (49, 50) at the N-terminus and cysteine at the C-terminus to exclude any potential ambiguity caused by the two species with different donor and acceptor positions. We compared FRET efficiencies of the two proteins in solution and after immobilization on a glass surface at room temperature. We found that each protein apparently populates a single state, with Aβ42 being slightly more compact than Aβ40. However, 2D FRET efficiency-donor lifetime analysis shows that there is a distribution of different conformations, which are rapidly interconverting, similar to unfolded proteins. We did not observe any dynamics on the timescale longer than ∼1 μs except acceptor photoblinking, whereas nanosecond fluorescence correlation spectroscopy (nsFCS) experiments measured an end-to-end distance relaxation time of ∼35 ns, which agrees well with those measured for other intrinsically disordered proteins (IDPs) (51, 52). Nanosecond-timescale conformational fluctuation has also been observed for an amyloid forming protein, the N-terminal region of the yeast prion protein Sup35 (53). These results suggest that both proteins are disordered and there is no significant population of a stable ordered structure, which is consistent with the most recent NMR result (32). However, the observed FRET efficiency values of both isoforms are much lower than those expected from previous MD simulation studies, which have reported disordered but compact conformational ensembles.
Our MD simulation results give a consistent picture of the peptide structure and dynamics. We ran both conventional and temperature replica-exchange MD (REMD) of both isoforms in explicit solvent using two recently developed force fields that do not suffer from the systematic global drive toward collapse and artificial stabilization of secondary structure in IDPs observed using earlier energy functions (44, 54). The simulations are consistent with previously published NMR data, reflecting primarily local structure formation. In addition, we find that both peptides have virtually indistinguishable ensemble-averaged radii of gyration Rg, end-to-end distances Ree, and FRET efficiencies E, as in the single-molecule experiments. Distributions of these parameters, however, show a very small population at low Ree for Aβ42 that is absent for Aβ40. Performing a machine learning cluster analysis of the peptides’ contact maps allows us to resolve the conformational ensemble into groups of structures with similar properties. As expected, these analyses show that the vast majority of populated states are random coil. However, we do find a very small subpopulation of states in the ensemble of Aβ42 that differs significantly from other states of Aβ42 and all states of Aβ40 in that it exhibits long-range terminal contacts that result in hairpin formation. This local β-structure is responsible for the low Ree probability shoulder and is consistent with the marginally higher FRET efficiency observed for freely diffusing Aβ42 compared with that of Aβ40.
Materials and Methods
Materials
Details of the expression, purification, and dye-labeling of proteins are described in the Supporting Material.
Single-molecule spectroscopy
Details of free-diffusion, immobilization, and nsFCS experiments are described in the Supporting Material. For the accurate determination of the FRET efficiency and donor lifetime, we performed various corrections for background, donor leak into the acceptor channel, ratios of the detection efficiencies, and quantum yields of the donor and acceptor (γ-factor) and acceptor blinking (55, 56). See the Supporting Material for the details.
MD simulation
Details of the REMD simulations are described in the Supporting Material. Briefly, we ran long isobaric-isothermal simulations on the order of 750 ns for each peptide using a temperature ladder from T = 277 K to T = 355 K in steps of 2 K, using both the Amber ff99SBws and Amber ff03ws force fields. Both optimized force fields give consistent results, demonstrating that our observations are not systematic artifacts. We report in the main text the results from Amber ff99SBws at 299 K with 20 mM NaCl, with analogous results for Amber ff03ws at 299 K included in the Supporting Material. The results from both force fields at 277 K are also consistent with experimental NMR signals. In addition, we ran multiple long simulations with chromophores attached to obtain dynamical properties. Further details are given in the Supporting Material.
Results and Discussion
Single-molecule free-diffusion experiment shows that Aβ is disordered
We performed two types of single-molecule experiments. First, by immobilizing molecules on a polyethylene glycol (PEG)-coated glass surface, it is possible to collect long fluorescence trajectories, which allows for accurate determination of FRET efficiencies and fluorescence lifetimes. However, there is a possibility that protein-surface interactions can affect the observed protein conformations and dynamics even though the surface is coated with PEG. In addition, the linker amino acid sequence required for immobilization may also affect protein conformations and dynamics. Therefore, we employed a second experiment, in which molecules freely diffuse in solution and a burst of fluorescence is detected when a molecule crosses the confocal volume. There is no protein-surface interaction in this experiment as long as the focus is sufficiently far from any surface. However, the residence time in the focal volume is relatively short (∼1 ms), which leads to a shot-noise broadening of the FRET efficiency distribution due to the limited photons collected, and limits the measurable timescale of dynamics to a few milliseconds (57). Our experimental strategy was to perform both experiments and compare the mean FRET efficiencies obtained from the two experiments. If these values are similar, this would lend confidence to more detailed analyses of the data obtained from immobilization experiments.
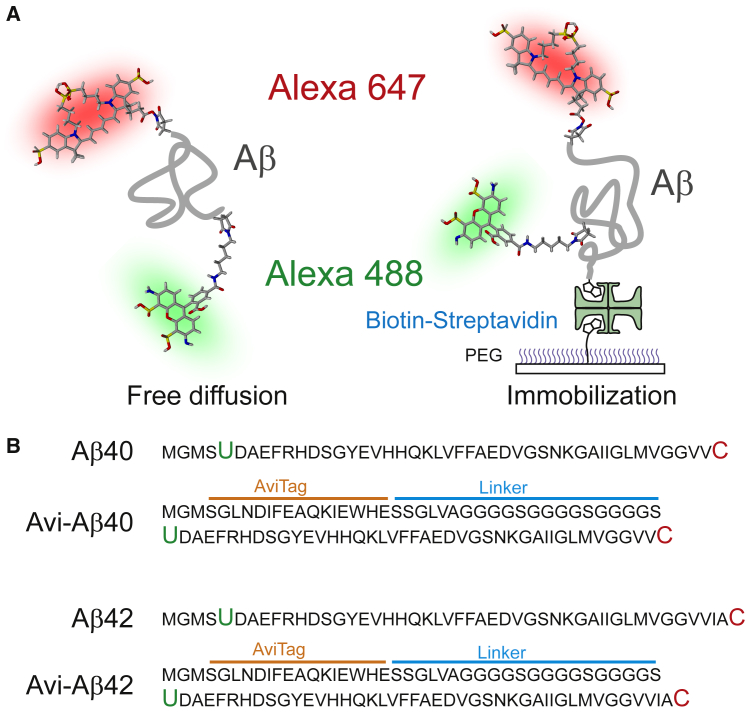
Fig. 1 shows four protein constructs used in the free-diffusion experiment: Aβ40 and Aβ42, with and without an immobilization tag (AviTag) sequence. Details of the gene construction and incorporation of an unnatural amino acid are described in Protein Expression and Incorporation of an Unnatural Amino Acid in the Supporting Material. Alexa 488 (donor) and Alexa 647 (acceptor) were attached to the N- and C-termini site-specifically. Two shorter sequences of Aβ40 and Aβ42 without the immobilization tag were designed as the least perturbed constructs for the free-diffusion experiment. These constructs serve as controls for those with AviTag (Avi-Aβ) to characterize the potential interference of AviTag and the linker in Aβ protein conformations and dynamics.
Figure 1.
Schematic diagram of four amyloid β-protein constructs. (A) Aβ40 and Aβ42 (thick gray lines) are labeled with Alexa 488 (donor) and Alexa 647 (acceptor) site-specifically at the N- and C-termini, respectively. Experiments were carried out in the free-diffusion mode (left) or with molecules immobilized on a PEG-coated glass surface via a biotin-streptavidin linkage (right). (B) Amino acid sequence of the four proteins. Alexa 488 hydroxylamine is attached to the unnatural amino acid (4-acetylphenylalanine, green U) and Alexa 647 maleimide is attached to the cysteine residue (red C). For immobilization, a biotin acceptor sequence (AviTag) is added to the N-terminus of Aβ (Avi-Aβ). The glycine-rich flexible linker is inserted between AviTag and the protein to prevent potential protein-surface interactions. To see this figure in color, go online.
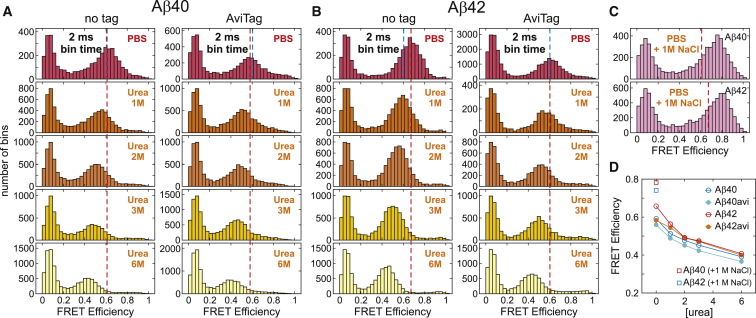
FRET efficiency histograms (E = nA/(nA + nD), where nA and nD are respectively the numbers of detected acceptor and donor photons) of these four constructs are compared in Fig. 2. FRET efficiency distributions measured (40–100 pM protein concentration) in phosphate-buffered saline (PBS) at room temperature (22°C) (top row in Fig. 2) show only one peak at E ∼ 0.6. The other peak near E = 0 results from the molecules without an active acceptor (either unlabeled or photobleached). The single peak at E ∼ 0.6 can be explained by the protein having a single state on the millisecond timescale (the bin time is 2 ms). This state could be a disordered (unfolded) state as the FRET efficiency is similar to that of a 42-residue IDP, the tetramerization domain of p53 (56). It is also possible, however, that this state could be well structured. To rule out this second possibility, we investigated the denaturant dependence of the FRET efficiency distributions. As the urea concentration is increased, the single peak gradually moves to the lower FRET efficiency side (Fig. 2, A and B). This result strongly suggests that the proteins are already unfolded under native conditions (PBS). The gradual decrease in the FRET efficiency is consistent with gradual expansion of polypeptide chains observed in proteins unfolded by chemical denaturant and IDPs (53, 58, 59, 60, 61, 62, 63, 64, 65, 66). Even for two-state proteins for which separate folded and unfolded peaks are expected, however, it is possible to observe a single peak whose FRET efficiency shifts with denaturant concentration. This occurs when the exchange between the folded and unfolded states is much faster than the bin time (2 ms), as seen in fast-folding proteins such as the 35-residue subdomain of villin headpiece and WW domain (67, 68, 69). This possibility will be discussed in the immobilization experiment section below.
Figure 2.
FRET efficiency histograms obtained from free diffusion experiments for Aβ40 (A) and Aβ42 (B). (A and B) Apparent (uncorrected) FRET efficiencies were calculated using the photons collected in 2 ms bins (≥30 photons). “No tag” stands for the protein samples without AviTag and linker and AviTag represents Avi-Aβ. Blue dashed lines indicate the center of the FRET efficiency peak (Gaussian fit) of Avi-Aβ42 measured in PBS and are drawn for the comparison of the FRET efficiencies between different constructs. Red dashed lines indicate the center of the peaks of individual constructs measured in PBS. The FRET efficiency decreases gradually as the urea concentration is increased, indicating the expansion of disordered proteins. (C) Apparent FRET efficiency histograms for Aβ40 (top) and Aβ42 (bottom) without AviTag in PBS with the addition of 1 M NaCl. Red dashed lines indicate the center of the peaks of the histograms measured in PBS. (D) FRET efficiency changes as a function of urea concentration. FRET efficiencies were corrected for background, donor leak, and γ-factor. Square symbols are the data collected in PBS with an additional 1 M NaCl. To see this figure in color, go online.
Another notable feature is that the FRET efficiency of Avi-Aβ is slightly lower than that of Aβ without the immobilization tag for both Aβ40 and Aβ42 (Fig. 2 D). This difference may result from the slightly higher net negative charge of Avi-Aβ compared to that without a tag (−7 vs. −3), which may increase electrostatic repulsion, or from the excluded volume effect of the additional amino acid sequence at the N-terminus of Avi-Aβ, which prohibits certain conformations with two dyes close to each other. The increase of the FRET efficiency (collapse of unfolded molecules) by the addition of 1 M NaCl supports the former (Fig. 2, C and D). In any case, the similar FRET efficiency values indicate that the effect of the immobilization tag on the conformational dynamics of Aβ is small.
Although the FRET efficiencies of Aβ40 and Aβ42 in PBS are very similar, the FRET efficiency of Aβ42 is slightly higher than that of Aβ40, despite the two extra residues. This small difference is not an artifact, because the result is consistent in the three different cases: Aβ without AviTag and Avi-Aβ in free-diffusion experiments, and the immobilization experiment in the next section (see Table S1 for the comparison). The higher FRET efficiency of Aβ42 may result from greater hydrophobic collapse induced by two additional C-terminal residues, or from differences in local structure. We have performed molecular simulations to address this issue, as discussed below.
Immobilized Aβ shows no conformational transitions on the μs to ms timescale
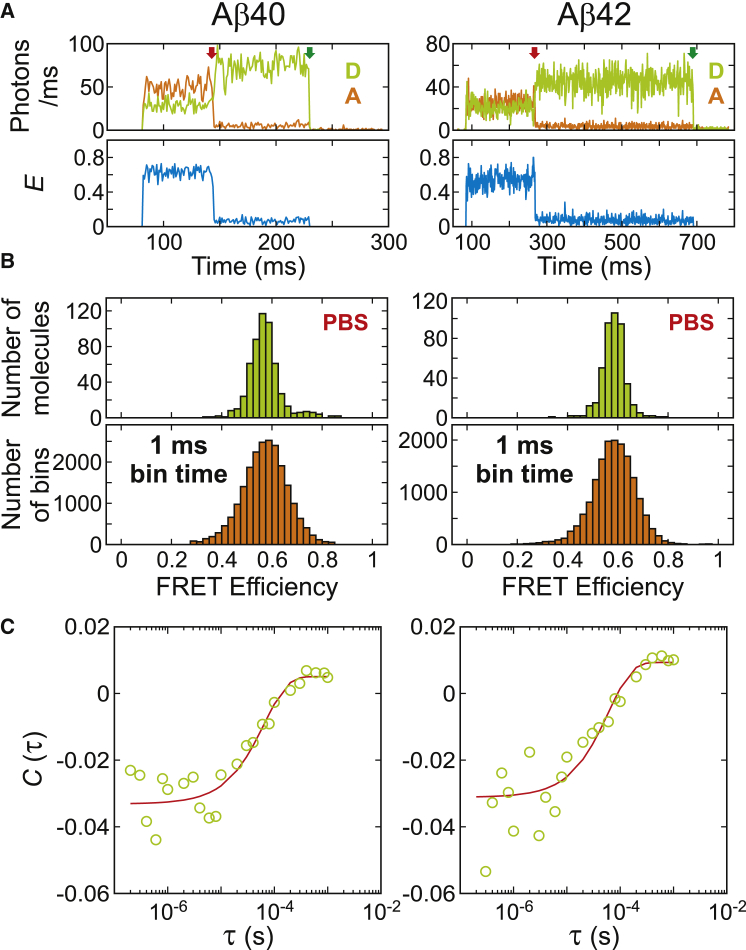
To collect single-molecule trajectories, long enough to measure the submillisecond dynamics, we immobilized Aβ molecules on a glass surface and acquired >600 trajectories for both Aβ40 and Aβ42 in PBS. The representative fluorescence and FRET efficiency trajectories obtained by continuous-wave excitation are shown in Fig. 3 A. Most of the trajectories show a constant FRET efficiency level near E = 0.6 for both Aβ40 and Aβ42 without any transition before photobleaching of the donor or acceptor. The FRET efficiency histograms constructed from the mean values of 1-ms bins show a single peak (lower panels in Fig. 3 B) similar to the free-diffusion data in Fig. 2. The mean FRET efficiencies are similar to those in the free-diffusion data, and the FRET efficiency of Aβ42 is slightly higher than that of Aβ40, consistent with the free-diffusion data in Fig. 2. The width of the peak in the immobilization data is narrower than that of the free-diffusion data because of higher photon count rates (50–100 ms−1) and consequently smaller shot noise. The similarity of the results in the two types of experiments suggests that the effect of immobilization is very small.
Figure 3.
Immobilization experiment to probe the conformational dynamics of Aβ40 (left) and Aβ42 (right) on the timescale of microseconds to milliseconds. Experiments were performed at the native condition (PBS, 22°C). (A) Representative fluorescence (D, donor; A, acceptor) and FRET efficiency (E) trajectories of Aβ40 and Aβ42 (1 ms bin time) obtained by continuous-wave excitation. Photon count rates were 50–100 ms−1. Both trajectories show a constant FRET efficiency (E = 0.5–0.6) followed by successive acceptor (red arrow) and donor (green arrow) photobleaching. (B) The FRET efficiency histograms in the upper panels are constructed using the FRET efficiency values calculated from the initial segment of the trajectory of each molecule. The histograms in the lower panels are constructed using the FRET efficiency values of all 1 ms bins in the initial segment of each trajectory. FRET efficiencies were corrected for background photons. (C) Donor-acceptor cross correlation averaged over the initial segments of trajectories in (B). Experimental data (green circles) are fitted to a single exponential function (red). The relaxation rates are 15.3 and 16.5 ms−1 for Aβ40 and Aβ42, respectively. To see this figure in color, go online.
As mentioned above, even though there is only one peak in the FRET efficiency distribution, it is possible that there are multiple states that interconvert much more rapidly than the bin time of 1 ms. In fact, numerous simulation and NMR studies have raised the possibility that stable conformations such as a turn in the hydrophilic region (residue 22–30) (27, 29, 36) could exist transiently. This structure has been of particular interest because it resembles the structure of the fibril of Aβ40 (30), which suggests this transient structure can be a template for fibril formation. Lazo et al. (29) found that this region was resistant to enzymatic cleavage and the stability of the 10-residue peptide (residue 21–30) was correlated with the aggregation propensity of the mutant Aβ proteins (70). Yamaguchi et al. (27) observed that the NMR signal of monomer Aβ became weaker and eventually disappeared as the temperature was raised, which was attributed to the fast chemical exchange between unstructured and a hairpin structure in the hydrophilic region. If this structure exists, we anticipate that its FRET efficiency is higher than 0.6 because the average end-to-end distance will be shorter. In fact, there is a small fraction of molecules exhibiting high FRET efficiencies (E = 0.7–0.8) in Fig. 3 B (upper panels) and Fig. 4 B. However, >50% of these molecules show slow and irreversible FRET efficiency changes, which we attribute to the unusual photophysics of Alexa 647 (56) (see Fig. S1, A and B).
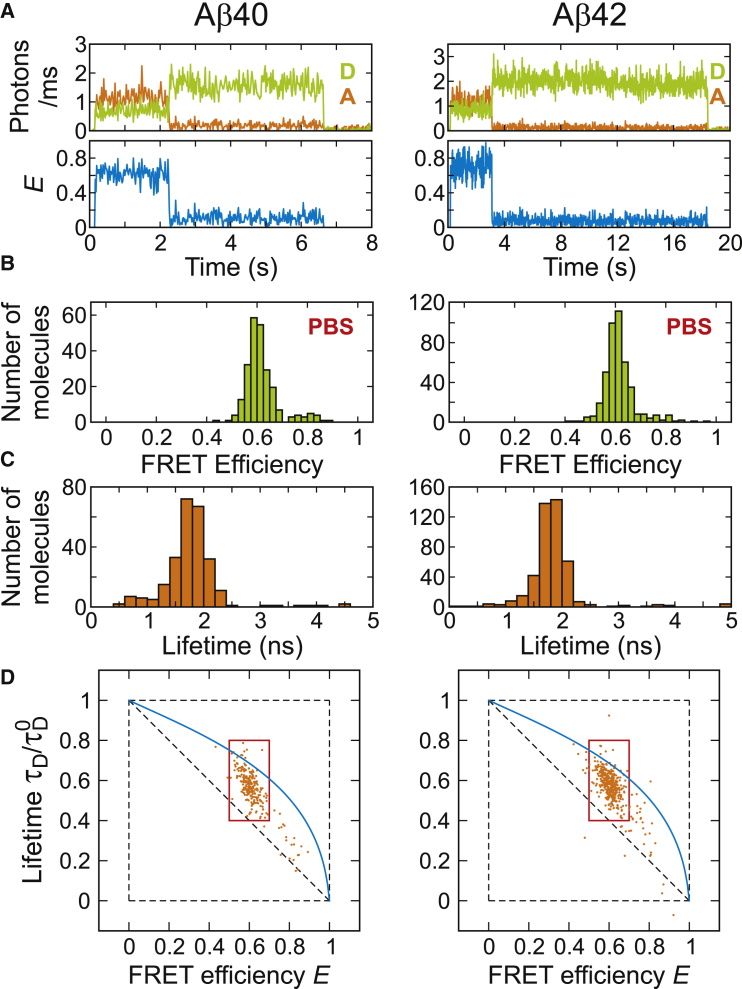
Figure 4.
FRET efficiency and donor lifetime analysis of immobilized Aβ. (A) Representative fluorescence and FRET efficiency trajectories of Aβ40 (left) and Aβ42 (right) obtained by pulsed-mode laser excitation with 20 MHz repetition rate. The illumination intensity is lowered by 10 times compared to that in Fig. 3 to collect longer trajectories for the accurate determination of donor lifetimes. The bin time is 20 ms. The experiments were performed in PBS (22°C). (B and C) FRET efficiency and donor lifetime histograms were constructed using the values calculated from the initial segment of each trajectory. Segments containing >1000 photons were analyzed. The FRET efficiency was corrected for background, donor leak into the acceptor channel, γ-factor, direct acceptor excitation, and acceptor blinking. The donor lifetime was corrected for background and acceptor blinking. (D) 2D FRET efficiency-donor lifetime plots constructed using the values in (B) and (C). The shift of the distribution above the diagonal indicates rapid conformational fluctuations of disordered Aβ. The data inside the red rectangles were used to calculate the variance (σc2) of the FRET efficiency distribution caused by these fluctuations. σc2 = 0.07 (±0.02) for both Aβ40 and Aβ42. Errors are standard deviations calculated by error propagation using the standard deviation and covariance values of the FRET efficiencies and donor lifetimes in the 2D distributions. The blue solid curve indicates the lifetime dependence on the FRET efficiency of a Gaussian chain. To see this figure in color, go online.
To probe any fast dynamics, we first calculated the average donor-acceptor cross-correlation function (see Eq. S17 in the Supporting Material) of the segments containing both donor and acceptor fluorescence of the immobilized trajectories (Fig. 3 C). There are low-amplitude decays for both proteins, and single-exponential fitting yields relaxation times of 65 and 61 μs for Aβ40 and for Aβ42, respectively (see Table S2). One explanation for this relaxation could be conformational exchange such as the hairpin formation dynamics mentioned above (27).
To obtain more detailed information, we used a maximum likelihood method that analyzes photon trajectories directly without binning and extracts FRET efficiencies, relative populations of states, and rate coefficients between them (69, 71). We first used the simplest two-state model (see Maximum Likelihood Analysis of Acceptor Blinking in the Supporting Material). The extracted apparent (uncorrected) FRET efficiencies of the two states are 0.60 (0.61) and 0.13 (0.13), the population of the high-FRET state is 0.93 (0.94), and the relaxation rate is 12.2 ms−1 (14.1 ms−1) for Aβ40 (Aβ42) (Table S2). These relaxation rates are similar to those obtained from the correlation analysis, indicating that the two-state model is a reasonably good model. Because the major species with the high-E value (E ∼0.6, 94%) is the disordered state, the low-FRET state should be a potentially structured state if it exists. However, the FRET efficiencies of the low-FRET states, corrected for background, donor-leak (0.05), and γ-factor (ratio of the detection efficiencies and quantum yields of the donor and acceptor) are 0.07 and 0.08 for Aβ40 and Aβ42, respectively. E = 0.07 correspond to 8 nm in distance between the two fluorophores (with Förster radius of 5.2 nm), and it is very unlikely that both Aβ40 and Aβ42, which are very short proteins, take this extended conformation with similar stability (lifetime of ∼60 μs) and similar relative populations (see Simulation Results in the Supporting Material). Instead, we suspect that this additional state results from acceptor blinking. When the acceptor is in the dark state, there is no energy transfer from the donor, and acceptor intensity decreases whereas donor intensity increases. This appears as anticorrelation in the cross-correlation function. The apparent FRET efficiency of 0.13 is slightly higher than the expected FRET efficiency of the acceptor dark state, which is the same as that of the donor-only (acceptor-bleached) state, 0.06. However, the maximum likelihood analysis with the fixed FRET efficiency of 0.06 for the second state results in similar parameters, including the relaxation rates and relative populations (see Table S2).
The results so far show that both Aβ40 and Aβ42 are disordered, exhibit apparently one state, and there are no conformational dynamics on a timescale longer than ∼1 μs. If there is an additional stable state, its population should be much lower than the population of the acceptor dark state, ∼5%, or its FRET efficiency should be so similar to that of the disordered Aβ, 0.6, that these are indistinguishable.
2D FRET efficiency-lifetime analysis and nsFCS reveal nanosecond dynamics of disordered Aβ
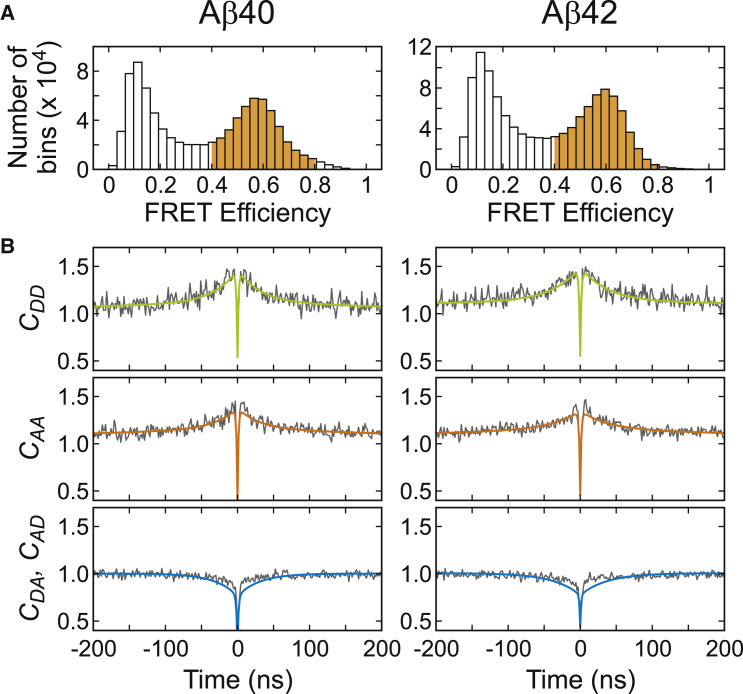
2D FRET efficiency-lifetime analysis (51, 56, 61, 72, 73, 74, 75), which visualizes the correlation between the FRET efficiency and the donor fluorescence lifetime, provides further evidence that Aβ is disordered. For this analysis, we collected trajectories of immobilized molecules illuminated by a picosecond-pulsed laser. In this experiment, in addition to the photon count rates (intensity) of the donor and acceptor, the delay times between the laser pulse and photon arrivals are measured. Representative trajectories and the histograms of the mean FRET efficiency and donor lifetime values calculated for individual trajectories (i.e., individual molecules) are shown in Fig. 4, A–C.
The 2D FRET efficiency-lifetime plots in Fig. 4 D were constructed using these values. When the distance between the donor and acceptor is fixed (i.e., a single conformation), the FRET efficiency and donor lifetime are related as τD/τD0 = 1 – E, where τD and τD0 are the donor lifetimes in the presence and absence of the acceptor, respectively. In this case, the distribution should be located on the diagonal of the 2D plot. On the other hand, when there is a distribution of conformations with different donor-acceptor distances, which are so rapidly interconverting that the FRET efficiency histogram shows a single peak, the relationship between the FRET efficiency and donor lifetime is different, and given by τD/τD0 = 1 – E + σc2/(1 – E). The value σc2 is the variance of the FRET efficiency of the underlying conformational distribution (74). (Note that this is not the variance of the peak of the FRET efficiency histograms in Fig. 4 B.) In this case, the 2D distribution is positively shifted from the diagonal. This shift results from the fact that the donor lifetime is determined by the donor photons and more donor photons are emitted from the conformations with low FRET efficiency values. In other words, the donor lifetime is the average lifetime weighted by 1 – E of the conformations. This shift has been observed in unfolded proteins and IDPs (51, 52, 56, 61, 75). As shown in Fig. 4 D, both Aβ40 and Aβ42 exhibit a positive shift. This shift can be compared with that of a random polymer model (Gaussian chain), which has been widely used to describe the dynamics of chemically unfolded proteins and intrinsically disordered proteins (51, 58, 59, 61, 63, 76). In this model, the distance distribution can be obtained from the mean experimental FRET efficiency, and therefore, σc2 can be calculated. The expected donor lifetime is calculated as a function of E in Fig. 4 D. The σc2 values are 0.07 (±0.02) for both proteins, indicating that they are largely disordered although the values are slightly smaller than 0.11 expected from a Gaussian chain.
As mentioned above, we have not detected any conformational dynamics on a timescale longer than ∼1 μs. Therefore, the conformational fluctuations of disordered Aβ should occur on the nanosecond timescale. To determine this timescale, we performed nsFCS experiments (77, 78). Fig. 5 shows the correlation functions. All three correlation functions (donor and acceptor autocorrelations and donor-acceptor cross correlation) have three components: antibunching, conformational dynamics, and triplet blinking. The three correlations were fitted to a triexponential function (see Eq. S16 in the Supporting Material), which results in the timescale of the conformational dynamics of 30 and 37 ns for Aβ40 and Aβ42, respectively, similar to those of other IDPs (51, 52, 53). The σc2 value calculated from the correlation amplitudes (79, 80) (see Eq. S16 in the Supporting Material) and the FRET efficiencies of Avi-Aβ measured in the free-diffusion experiment (Table S1) is 0.06 for both Aβ40 and Aβ42, which agrees very well with the values obtained from the 2D FRET efficiency-lifetime analysis above.
Figure 5.
nsFCS measurement of Avi-Aβ40 and Avi-Aβ42. (A) Fluorescence bursts with FRET efficiencies between 0.4 and 0.8 (shaded in orange) were analyzed. (B) Global fitting of three correlation data (autocorrelation of the donor and acceptor and cross correlation) results in the conformational fluctuation timescale (τCD) and σc2 values of 30 (±2) ns and 0.059 (±0.003) for Aβ40 and 37 (±2) ns and 0.057 (±0.002) for Aβ42. σc2 values are consistent with the value 0.07 obtained from the 2D FRET efficiency-lifetime analysis. To see this figure in color, go online.
Acharya et al. (81) have recently investigated the chain dynamics of Aβ by monitoring quenching of the triplet state of tryptophan by cysteine. The inferred timescale of the contact formation between residue 4 and residue 35 was several microseconds. The contact formation time is generally much longer than the end-to-end distance correlation time. Although the measured chain diffusion of Aβ42 was ∼5 times slower than that of Aβ40, they observed Aβ42 was slightly more compact than Aβ40, similar to our result that the FRET efficiency of Aβ42 is marginally higher than that of Aβ40.
Simulation results
The experimental results above show that both Aβ40 and Aβ42 are largely disordered and their reconfiguration time is ∼35 ns. The slightly higher FRET efficiency value of Aβ42 indicates that it is slightly more compact than Aβ40 despite having two more residues at the C-terminus. It is very difficult, however, to investigate the origin of this difference experimentally. To obtain molecular insight into Aβ conformations, we performed all-atom MD simulations in explicit solvent, as described in the Materials and Methods. Most notably, we have used the Amber ff99SBws force field, which has been optimized to reproduce the dimensions of disordered proteins.
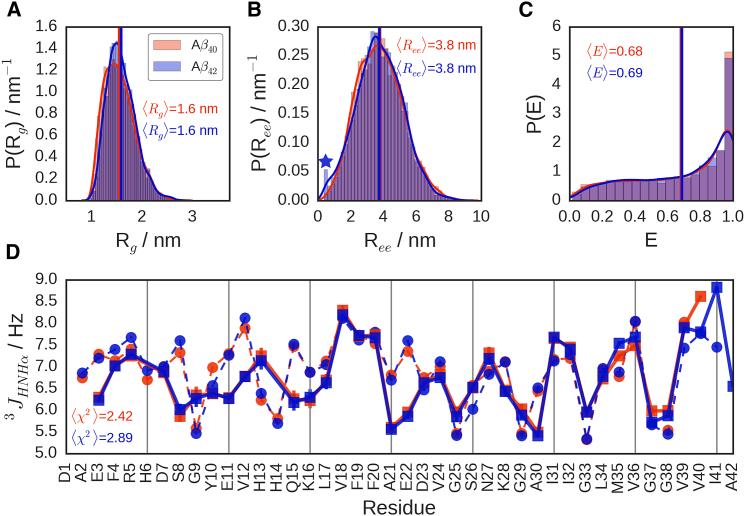
We first calculated average configurational quantities for both peptides from REMD simulation. The distributions of radius of gyration (Rg), end-to-end distance (Ree), and E at 299 K, shown in Fig. 6, demonstrate that both peptides populate closely overlapping ensembles for all quantities, with nearly indistinguishable ensemble averages: 〈Rg〉 = 1.56 ± 0.01 nm (1.59 ± 0.01 nm), 〈Ree〉 = 3.80 ± 0.05 nm (3.77 ± 0.04 nm), and 〈E〉 = 0.68 ± 0.01 (0.69 ± 0.01), with a distribution variance of σc2 = 0.09 (0.08) for Aβ40 (Aβ42). Although the FRET efficiency variances agree quantitatively with experimental results, the mean FRET efficiency values are slightly higher than those obtained experimentally. We note, however, that because we are close to the range where efficiency varies most sharply with distance, 〈E〉 can be altered by relatively small errors in the simulation distance distribution. We illustrate in Fig. S2 the small scaling that would need to be applied to the distance distributions for 〈E〉 to match experiment. Although the results above were obtained from simulations of unlabeled proteins, we have also performed long equilibrium MD simulations with explicit fluorophores attached, as well as separate REMD simulations with a different force field, with all methods yielding consistent results (Table S1). Although we cannot obtain dynamical properties from the REMD simulations, we have used our long simulations with attached fluorophores to determine the FRET efficiency autocorrelation function. We find an approximately biexponential relaxation for both peptides, with relaxation times of ∼4 and ∼40 ns (Fig. S3). The faster component would not be visible in the experiment due to overlap with the antibunching contribution to the correlation function, but the slow components are in excellent agreement with the experimental relaxation times. There is the possibility that the difference between the FRET efficiencies derived from simulation and experiment could be due to the quenching of photoactivated donors by the acceptors for short interdye separations. We find, however, that this effect is unlikely to be a significant factor in our simulations because the simulated transfer efficiencies are barely affected by excluding frames in which the dyes are in close proximity (Table S3).
Figure 6.
Ensemble observables from simulations of Aβ40 (red) and Aβ42 (blue). (A–C) Probability densities for peptide radii of gyration (Rg), end-to-end distances (Ree), and FRET efficiencies (E), showing nearly overlapping distributions and ensemble averages (shown as vertical lines and as text annotations) for both isoforms. The major difference is the low end-to-end distance probability shoulder observed only for Aβ42, denoted by a blue star in (B). (D) J couplings from simulation (dashed lines and circles), compared with those derived from experiment (solid thick lines and squares, data from (32)), showing good agreement, as demonstrated by the low values of 〈χ2〉, where the average is taken over all residues. To see this figure in color, go online.
Whereas the ensemble averages of Rg and Ree are very similar, the distribution of Ree for Aβ42 shows a significant shoulder at very short end-to-end distances, which is not seen for Aβ40. The distributions of Rg do not show such a feature for either peptide and behave as expected, with the longer peptide having a slightly larger size. The simplest interpretation of these effects is the presence of a small population of long-range contacts within the Aβ42 peptide without a global collapse that would affect the radius of gyration (see below). Our simulations are also in overall good agreement with NMR J-couplings and chemical shifts, which have been used to validate earlier MD studies. In Fig. 6 D, we show the 3JHNHα scalar couplings computed from the simulations with the Karplus equation (82) using a recent set of parameters (83), whereas Fig. S4 shows predicted chemical shifts. Although there are some localized discrepancies for certain residue types, probably reflecting residual force-field inaccuracies, both J-couplings and chemical shifts are generally in good agreement with experimental values from (32) (note that the RMSDs between the predicted and experimental chemical shifts are smaller than the prediction error of SPARTA+, ∼1 ppm for both carbon shifts (84)). Importantly, our simulations are also consistent with estimates of the hydrodynamic radius of Aβ40 from diffusion measurements by NMR (1.73 ± 0.01 nm, as calculated using Hydropro (85), compared with the experimental value of 1.6 ± 0.1 nm (86)), which confirms that the overall dimensions of the protein are reasonable.
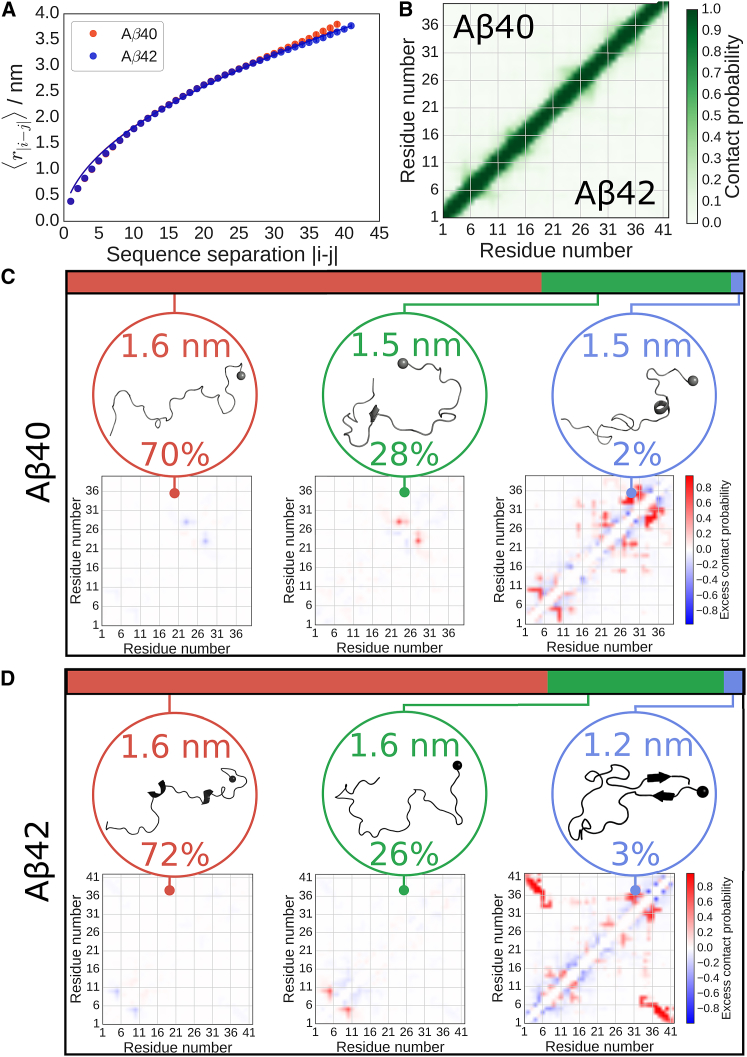
Having shown that the simulations are able to recapitulate experimental observations, we sought to explain the differences in the Ree distributions at a residue-level by performing a contact analysis. The ensemble-averaged intramolecular contact maps (Fig. 7 B) reveal a nearly complete absence of long-range contacts, suggesting that these peptides are almost entirely disordered. We have characterized this disorder by fitting an approximate scaling exponent to the mean intramolecular distances r|i – j| (Fig. 7 A) as a function of their sequence separation |i – j|. Indeed, the fitted scaling exponents of ν = 0.5197 ± 0.0006 and 0.5180 ± 0.0004, respectively, for Aβ40 and Aβ42, indicate that both peptides are close to the Θ-state in which attractive interactions approximately balance repulsive excluded volume interactions. A similar conclusion was reached from analysis of FRET data for a number of unfolded and intrinsically disordered proteins under folding conditions (87), suggesting that Aβ has global properties typical of other disordered proteins. Because structured species with low population may still be lost in such an average, we also examined in more detail the contact maps of individual structures. We partitioned the conformational ensemble of each peptide into clusters of structures with similar features in the contact map, using the k-means machine learning algorithm, which we optimized to avoid overfitting and validated as being robust in cluster assignment despite its stochastic mechanism (see Fig. S5; Table S4). As expected, and most significantly, we find that the vast majority of the ensemble populated by both peptides is completely devoid of any long-range contacts or appreciable structure (Fig. 7, C and D); the major clusters for both peptides show featureless average intramolecular contact maps, and the most representative structures are largely random coil, save for short stretches of helix in the Aβ42 cluster center that are unlikely to be significant (and definitely do not account for the shoulder in the distribution of Ree in Fig. 6 B). This random coil subpopulation accounts for the majority (70.1 and 71.6%, respectively, for Aβ40 and Aβ42) of the sampled ensembles, indicating that neither peptide is subject to a strong drive toward folding, as expected for an IDP and in agreement with the average contact maps. The secondmost populated clusters observed are still largely disordered, as evident from the contact maps, but include some short-range contacts between residues, with the largest sequence separation of contacting residues being ∼5. For Aβ40, these contacts are between residues located in the loop of the fibril structures, specifically between D23 and K28 (this same pair of residues interact as an intermolecular salt bridge in solid-state NMR fibril structures of Aβ40 (5, 13), and was also observed in earlier MD studies), whereas for Aβ42, the main contacts are in the N-terminus, between the two aromatic residues Y10 and F4. These locally structured states account, respectively, for 27.6 and 25.6% of the Aβ40 and Aβ42 ensembles, but have FRET efficiencies (0.70 and 0.72, respectively) very similar to the ensemble averages.
Figure 7.
Conformational ensembles of Aβ40 and Aβ42. (A) Internal Cα atom distance scalings for each peptide, with data shown as circles, and fits to a power law as described in the Supporting Material as solid lines, giving scaling exponents of ν = 0.5197 ± 0.0006 and 0.5180 ± 0.0004, respectively, for Aβ40 and Aβ42. Note that the similarity in peptide behaviors means that the fitted curve for Aβ40 overlaps with that of Aβ42. (B) Simulation-averaged intramolecular contact maps show no significant secondary structure for either peptide, with Aβ40 shown in the upper half and Aβ42 in the lower. (C) Major clusters for the structural ensemble of Aβ40. The top colored bar denotes the observed statistical weights of each subpopulation, for which the most representative structure (with the N-terminus denoted by a sphere) and average intramolecular contact map relative to the ensemble-averaged contact map are shown. Blue contacts are those populated less frequently than the average, whereas red are those populated more frequently. The average radius of gyration and population for each cluster are shown accompanying the structure. The first cluster is random coil, the second populates very local contacts, and the last shows structure within the termini. (D) Major clusters for the structural ensemble of Aβ42. Similarly to the Aβ40 ensemble, the first cluster is random coil and the second populates very local contacts, whereas the last shows the structure formation between the termini. To see this figure in color, go online.
The smallest clusters (2.3 and 2.8%, respectively, for Aβ40 and Aβ42) of both peptides show more extensive contact formation. The central structure of this cluster for Aβ40 shows the formation of several locally structured regions, with the maximum sequence-separation of contacts being ∼15 residues. In contrast, Aβ42 forms long-range β-sheet between N- and C-termini involving hydrogen bonding of backbone atoms of G38, V39, V40, and I41 with F4, E3, A2, and D1, with the C-terminal A42 excluded from the motif perhaps due to electrostatic clashes between the charged carboxyl terminus and the side chains of the N-terminal residues D1 and E3 (see Fig. S6 B). This N- to C-terminal strand formation results in a very low end-to-end distance, thereby explaining the low-Ree probability shoulder of the Aβ42 distribution in Fig. 6 B, and may help to explain the marginally higher experimentally observed FRET efficiency of freely diffusing Aβ42. This interterminal contact was observed independently in several different REMD windows, and was reversible on the timescale of the simulations (Fig. S7). Nevertheless, because of the low population of this structure, and limitations of current force fields and conformational sampling, we conducted a similar analysis on a completely separate set of REMD simulations performed with a different force field (Amber ff03ws). This force field differs from Amber ff99SBws in both the atomic partial charges and the backbone and side-chain torsion angle potentials, although both force fields have been shown to yield reasonable properties for disordered proteins (43). This force field yielded values of the peptide FRET efficiencies and NMR signals consistent with those obtained using ff99SBws (Fig. S8). Remarkably, we find a very similar picture (Fig. S9), with the same β-structure forming between the termini of Aβ42, but not Aβ40, lending confidence to our interpretation. We also note that there are no contacts between these residues in the structures used to initialize the simulations (contact maps shown in Fig. S10, A–D), confirming that they were formed independently during the course of the two simulations. Why, then, is the short β-sheet formed between the N- and C-termini in Aβ42 not observed in Aβ40, given the extremely high sequence similarity? One possible reason is that, in the shorter isoform, the terminal V40 would carry the C-terminal charge, which would interact unfavorably with residues D1 and E3. It would also be difficult to form a sheet with a shift of register due to the presence of glycine residues at positions 37 and 38, as this residue disfavors formation of secondary structure.
Conclusions
A large body of experimental and simulation studies has considered the structure of monomeric Aβ, as this is the starting point for the formation of all Aβ amyloid fibrils, with many of these studies inferring a substantial population of structured species. Although our experimental results cannot rule out such structured species, the timescale of their formation or breaking would have to be <1 μs, which is shorter than the folding time of the fastest-folding β-hairpins (88), making it unlikely. An alternative possibility is that these stable states have a similar FRET efficiency as the average of the unstructured states. The most probable interpretation of the data, including urea denaturation, 2D FRET efficiency-lifetime analysis, and FCS measurements, however, is that the peptides populate a broad ensemble of structures interconverting on a timescale of tens of nanoseconds, as is typical for disordered proteins. Our simulations, which are in excellent agreement with experiment, provide a consistent picture in which the vast majority of conformations are disordered, or form only local structure. Intriguingly, we do observe a very small population (∼3%) of β-structure formed between the termini of Aβ42 only, which may explain its slightly higher experimental FRET efficiency. The main difference from earlier simulations, which also predicted a population of disordered states, is that the population of collapsed and structured states is extremely small in our simulations; earlier results also suggested a smaller average radius of gyration of 1.0–1.2 nm (38, 86). It is unlikely that structures with such a low Rg could be consistent with the FRET experiments reported here. Those simulations were nonetheless in agreement with NMR data such as NMR J couplings, NOEs, and chemical shifts that primarily reflect local structure formation. The new FRET data, although not as detailed as from NMR, therefore provide us with a more powerful discriminating factor between different simulation ensembles through constraints on global peptide behavior.
What is the relation, if any, between monomer structure and the different rates of primary nucleation for these two peptides? One may speculate that the formation of β-structure between the termini of Aβ42 reduces the entropic cost for formation of subsequent ordered species, leading to the experimentally observed higher nucleation rate (17). Overall, however, the similarity in properties between the two peptides, and lack of stable structure in either, suggests that monomer structure alone is unlikely to play the deciding role in the nucleation differences, consistent with the nucleated conformational conversion model of amyloid fibril formation (53, 89, 90, 91, 92), but that, instead, the effect possibly arises from enhanced intermolecular interactions due to the additional two hydrophobic C-terminal residues.
Author Contributions
F.M., M.M.J.B., R.B.B., and H.S.C. designed research and wrote the manuscript. F.M. performed experiments and analyzed experimental data. M.M.J.B. performed MD simulations and analyzed simulation data. J.-Y.K. contributed analytic tools. G.H.Z. analyzed simulation data.
Acknowledgments
We thank I. V. Gopich and B. Schuler for very helpful suggestions and comments on the nsFCS experiment and analysis; R. Tycko, W. A. Eaton, and A. Szabo, for numerous helpful discussions and comments; P. G. Schultz for sharing the plasmid for the expression and incorporation of an unnatural amino acid 4-acetyl phenylalanine; J. Roche for providing carbon chemical shift data; and J. M. Louis for advice and suggestions on protein expression and purification. This work utilized the high performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (NIH), Bethesda, MD (http://biowulf.nih.gov).
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institutes of Health (NIH). M.M.J.B. was supported by the NIH-Oxford/Cambridge Scholars Program and the Cambridge Commonwealth, European and International Trust.
Editor: Elizabeth Rhoades.
Footnotes
Fanjie Meng and Mathias M. J. Bellaiche contributed equally to this work.
Supporting Materials and Methods, ten figures, and four tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)35128-7.
Contributor Information
Robert B. Best, Email: robert.best2@nih.gov.
Hoi Sung Chung, Email: chunghoi@niddk.nih.gov.
Supporting Citations
References (93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 103, 104) appear in the Supporting Material.
Supporting Material
References
- 1.Selkoe D.J. Alzheimer’s disease. Cold Spring Harb. Perspect. Biol. 2011;3:a004457. doi: 10.1101/cshperspect.a004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kirschner D.A., Abraham C., Selkoe D.J. X-ray diffraction from intraneuronal paired helical filaments and extraneuronal amyloid fibers in Alzheimer disease indicates cross-β conformation. Proc. Natl. Acad. Sci. USA. 1986;83:503–507. doi: 10.1073/pnas.83.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malinchik S.B., Inouye H., Kirschner D.A. Structural analysis of Alzheimer’s β(1–40) amyloid: protofilament assembly of tubular fibrils. Biophys. J. 1998;74:537–545. doi: 10.1016/S0006-3495(98)77812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu. Rev. Phys. Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petkova A.T., Yau W.-M., Tycko R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paravastu A.K., Leapman R.D., Tycko R. Molecular structural basis for polymorphism in Alzheimer’s β-amyloid fibrils. Proc. Natl. Acad. Sci. USA. 2008;105:18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J.-X., Qiang W., Tycko R. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao Y., Ma B., Ishii Y. Aβ(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat. Struct. Mol. Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wälti M.A., Ravotti F., Riek R. Atomic-resolution structure of a disease-relevant Aβ(1–42) amyloid fibril. Proc. Natl. Acad. Sci. USA. 2016;113:E4976–E4984. doi: 10.1073/pnas.1600749113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colvin M.T., Silvers R., Griffin R.G. Atomic resolution structure of monomorphic Aβ42 amyloid fibrils. J. Am. Chem. Soc. 2016;138:9663–9674. doi: 10.1021/jacs.6b05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petkova A.T., Leapman R.D., Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer’s β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 12.Tycko R. Amyloid polymorphism: structural basis and neurobiological relevance. Neuron. 2015;86:632–645. doi: 10.1016/j.neuron.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tycko R., Wickner R.B. Molecular structures of amyloid and prion fibrils: consensus versus controversy. Acc. Chem. Res. 2013;46:1487–1496. doi: 10.1021/ar300282r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiang W., Yau W.-M., Tycko R. Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature. 2017;541:217–221. doi: 10.1038/nature20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knowles T.P.J., Vendruscolo M., Dobson C.M. The amyloid state and its association with protein misfolding diseases. Nat. Rev. Mol. Cell Biol. 2014;15:384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 16.Cohen S.I.A., Linse S., Knowles T.P.J. Proliferation of amyloid-β42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. USA. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meisl G., Yang X., Knowles T.P.J. Differences in nucleation behavior underlie the contrasting aggregation kinetics of the Aβ40 and Aβ42 peptides. Proc. Natl. Acad. Sci. USA. 2014;111:9384–9389. doi: 10.1073/pnas.1401564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bitan G., Kirkitadze M.D., Teplow D.B. Amyloid β-protein (Aβ assembly: Aβ 40 and Aβ 42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. USA. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernstein S.L., Dupuis N.F., Bowers M.T. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat. Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haass C., Selkoe D.J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 21.Selkoe D.J. Soluble oligomers of the amyloid β-protein impair synaptic plasticity and behavior. Behav. Brain Res. 2008;192:106–113. doi: 10.1016/j.bbr.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson M.E., Lesné S.E. Soluble Aβ oligomer production and toxicity. J. Neurochem. 2012;120(Suppl 1):125–139. doi: 10.1111/j.1471-4159.2011.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uversky V.N. Mysterious oligomerization of the amyloidogenic proteins. FEBS J. 2010;277:2940–2953. doi: 10.1111/j.1742-4658.2010.07721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riek R., Güntert P., Wüthrich K. NMR studies in aqueous solution fail to identify significant conformational differences between the monomeric forms of two Alzheimer peptides with widely different plaque-competence, Aβ(1–40)(ox) and Aβ(1–42)(ox) Eur. J. Biochem. 2001;268:5930–5936. doi: 10.1046/j.0014-2956.2001.02537.x. [DOI] [PubMed] [Google Scholar]
- 25.Hou L., Shao H., Zagorski M.G. Solution NMR studies of the Aβ(1-40) and Aβ(1-42) peptides establish that the Met35 oxidation state affects the mechanism of amyloid formation. J. Am. Chem. Soc. 2004;126:1992–2005. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- 26.Yan Y., Wang C. Aβ42 is more rigid than Aβ40 at the C terminus: implications for Aβ aggregation and toxicity. J. Mol. Biol. 2006;364:853–862. doi: 10.1016/j.jmb.2006.09.046. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi T., Matsuzaki K., Hoshino M. Transient formation of intermediate conformational states of amyloid-β peptide revealed by heteronuclear magnetic resonance spectroscopy. FEBS Lett. 2011;585:1097–1102. doi: 10.1016/j.febslet.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Vivekanandan S., Brender J.R., Ramamoorthy A. A partially folded structure of amyloid-β(1–40) in an aqueous environment. Biochem. Biophys. Res. Commun. 2011;411:312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazo N.D., Grant M.A., Teplow D.B. On the nucleation of amyloid β-protein monomer folding. Protein Sci. 2005;14:1581–1596. doi: 10.1110/ps.041292205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petkova A.T., Ishii Y., Tycko R. A structural model for Alzheimer’s β-amyloid fibrils based on experimental constraints from solid state NMR. Proc. Natl. Acad. Sci. USA. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fawzi N.L., Phillips A.H., Head-Gordon T. Structure and dynamics of the Aβ(21–30) peptide from the interplay of NMR experiments and molecular simulations. J. Am. Chem. Soc. 2008;130:6145–6158. doi: 10.1021/ja710366c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roche J., Shen Y., Bax A. Monomeric Aβ1–40 and Aβ1–42 peptides in solution adopt very similar Ramachandran map distributions that closely resemble random coil. Biochemistry. 2016;55:762–775. doi: 10.1021/acs.biochem.5b01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ball K.A., Phillips A.H., Head-Gordon T. Homogeneous and heterogeneous tertiary structure ensembles of amyloid-β peptides. Biochemistry. 2011;50:7612–7628. doi: 10.1021/bi200732x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ball K.A., Phillips A.H., Head-Gordon T. Differences in β-strand populations of monomeric Aβ40 and Aβ42. Biophys. J. 2013;104:2714–2724. doi: 10.1016/j.bpj.2013.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sgourakis N.G., Yan Y., Garcia A.E. The Alzheimer’s peptides Aβ40 and 42 adopt distinct conformations in water: a combined MD/NMR study. J. Mol. Biol. 2007;368:1448–1457. doi: 10.1016/j.jmb.2007.02.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sgourakis N.G., Merced-Serrano M., Garcia A.E. Atomic-level characterization of the ensemble of the Aβ(1–42) monomer in water using unbiased molecular dynamics simulations and spectral algorithms. J. Mol. Biol. 2011;405:570–583. doi: 10.1016/j.jmb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenman D.J., Connors C.R., García A.E. Aβ monomers transiently sample oligomer and fibril-like configurations: ensemble characterization using a combined MD/NMR approach. J. Mol. Biol. 2013;425:3338–3359. doi: 10.1016/j.jmb.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenman D.J., Wang C., García A.E. Characterization of Aβ monomers through the convergence of ensemble properties among simulations with multiple force fields. J. Phys. Chem. B. 2016;120:259–277. doi: 10.1021/acs.jpcb.5b09379. [DOI] [PubMed] [Google Scholar]
- 39.Lin Y.-S., Bowman G.R., Pande V.S. Investigating how peptide length and a pathogenic mutation modify the structural ensemble of amyloid-β monomer. Biophys. J. 2012;102:315–324. doi: 10.1016/j.bpj.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin Y.-S., Pande V.S. Effects of familial mutations on the monomer structure of Aβ42. Biophys. J. 2012;103:L47–L49. doi: 10.1016/j.bpj.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Best R.B. Atomistic molecular simulations of protein folding. Curr. Opin. Struct. Biol. 2012;22:52–61. doi: 10.1016/j.sbi.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Nettels D., Müller-Späth S., Schuler B. Single-molecule spectroscopy of the temperature-induced collapse of unfolded proteins. Proc. Natl. Acad. Sci. USA. 2009;106:20740–20745. doi: 10.1073/pnas.0900622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Best R.B., Zheng W., Mittal J. Balanced protein-water interactions improve properties of disordered proteins and non-specific protein association. J. Chem. Theory Comput. 2014;10:5113–5124. doi: 10.1021/ct500569b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piana S., Klepeis J.L., Shaw D.E. Assessing the accuracy of physical models used in protein-folding simulations: quantitative evidence from long molecular dynamics simulations. Curr. Opin. Struct. Biol. 2014;24:98–105. doi: 10.1016/j.sbi.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 45.Piana S., Donchev A.G., Shaw D.E. Water dispersion interactions strongly influence simulated structural properties of disordered protein states. J. Phys. Chem. B. 2015;119:5113–5123. doi: 10.1021/jp508971m. [DOI] [PubMed] [Google Scholar]
- 46.Huang J., Rauscher S., MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dedmon M.M., Lindorff-Larsen K., Dobson C.M. Mapping long-range interactions in α-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J. Am. Chem. Soc. 2005;127:476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- 48.Salmon L., Nodet G., Blackledge M. NMR characterization of long-range order in intrinsically disordered proteins. J. Am. Chem. Soc. 2010;132:8407–8418. doi: 10.1021/ja101645g. [DOI] [PubMed] [Google Scholar]
- 49.Brustad E.M., Lemke E.A., Deniz A.A. A general and efficient method for the site-specific dual-labeling of proteins for single molecule fluorescence resonance energy transfer. J. Am. Chem. Soc. 2008;130:17664–17665. doi: 10.1021/ja807430h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young T.S., Ahmad I., Schultz P.G. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 51.Soranno A., Buchli B., Schuler B. Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy. Proc. Natl. Acad. Sci. USA. 2012;109:17800–17806. doi: 10.1073/pnas.1117368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuler B., Soranno A., Nettels D. Single-molecule FRET spectroscopy and the polymer physics of unfolded and intrinsically disordered proteins. Annu. Rev. Biophys. 2016;45:207–231. doi: 10.1146/annurev-biophys-062215-010915. [DOI] [PubMed] [Google Scholar]
- 53.Mukhopadhyay S., Krishnan R., Deniz A.A. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc. Natl. Acad. Sci. USA. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Best R.B. Computational and theoretical advances in studies of intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2017;42:147–154. doi: 10.1016/j.sbi.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 55.Gopich I.V., Szabo A. Theory of single-molecule FRET efficiency histograms. Adv. Chem. Phys. 2012;146:245–297. [Google Scholar]
- 56.Chung H.S., Meng F., Louis J.M. Oligomerization of the tetramerization domain of p53 probed by two- and three-color single-molecule FRET. Proc. Natl. Acad. Sci. USA. 2017;114:E6812–E6821. doi: 10.1073/pnas.1700357114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoffmann A., Nettels D., Schuler B. Quantifying heterogeneity and conformational dynamics from single molecule FRET of diffusing molecules: recurrence analysis of single particles (RASP) Phys. Chem. Chem. Phys. 2011;13:1857–1871. doi: 10.1039/c0cp01911a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schuler B., Lipman E.A., Eaton W.A. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. doi: 10.1038/nature01060. [DOI] [PubMed] [Google Scholar]
- 59.Sherman E., Haran G. Coil-globule transition in the denatured state of a small protein. Proc. Natl. Acad. Sci. USA. 2006;103:11539–11543. doi: 10.1073/pnas.0601395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merchant K.A., Best R.B., Eaton W.A. Characterizing the unfolded states of proteins using single-molecule FRET spectroscopy and molecular simulations. Proc. Natl. Acad. Sci. USA. 2007;104:1528–1533. doi: 10.1073/pnas.0607097104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hoffmann A., Kane A., Schuler B. Mapping protein collapse with single-molecule fluorescence and kinetic synchrotron radiation circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA. 2007;104:105–110. doi: 10.1073/pnas.0604353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ziv G., Thirumalai D., Haran G. Collapse transition in proteins. Phys. Chem. Chem. Phys. 2009;11:83–93. doi: 10.1039/b813961j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Müller-Späth S., Soranno A., Schuler B. From the cover: charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. USA. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aznauryan M., Delgado L., Schuler B. Comprehensive structural and dynamical view of an unfolded protein from the combination of single-molecule FRET, NMR, and SAXS. Proc. Natl. Acad. Sci. USA. 2016;113:E5389–E5398. doi: 10.1073/pnas.1607193113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng W., Borgia A., Best R.B. Probing the action of chemical denaturant on an intrinsically disordered protein by simulation and experiment. J. Am. Chem. Soc. 2016;138:11702–11713. doi: 10.1021/jacs.6b05443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Borgia A., Zheng W., Schuler B. Consistent view of polypeptide chain expansion in chemical denaturants from multiple experimental methods. J. Am. Chem. Soc. 2016;138:11714–11726. doi: 10.1021/jacs.6b05917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chung H.S., McHale K., Eaton W.A. Single-molecule fluorescence experiments determine protein folding transition path times. Science. 2012;335:981–984. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chung H.S., Cellmer T., Eaton W.A. Measuring ultrafast protein folding rates from photon-by-photon analysis of single molecule fluorescence trajectories. Chem. Phys. 2013;422:229–237. doi: 10.1016/j.chemphys.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chung H.S., Gopich I.V. Fast single-molecule FRET spectroscopy: theory and experiment. Phys. Chem. Chem. Phys. 2014;16:18644–18657. doi: 10.1039/c4cp02489c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grant M.A., Lazo N.D., Teplow D.B. Familial Alzheimer’s disease mutations alter the stability of the amyloid β-protein monomer folding nucleus. Proc. Natl. Acad. Sci. USA. 2007;104:16522–16527. doi: 10.1073/pnas.0705197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gopich I.V., Szabo A. Decoding the pattern of photon colors in single-molecule FRET. J. Phys. Chem. B. 2009;113:10965–10973. doi: 10.1021/jp903671p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sisamakis E., Valeri A., Seidel C.A.M. Accurate single-molecule FRET studies using multiparameter fluorescence detection. Methods Enzymol. 2010;475:455–514. doi: 10.1016/S0076-6879(10)75018-7. [DOI] [PubMed] [Google Scholar]
- 73.Kalinin S., Valeri A., Seidel C.A.M. Detection of structural dynamics by FRET: a photon distribution and fluorescence lifetime analysis of systems with multiple states. J. Phys. Chem. B. 2010;114:7983–7995. doi: 10.1021/jp102156t. [DOI] [PubMed] [Google Scholar]
- 74.Gopich I.V., Szabo A. Theory of the energy transfer efficiency and fluorescence lifetime distribution in single-molecule FRET. Proc. Natl. Acad. Sci. USA. 2012;109:7747–7752. doi: 10.1073/pnas.1205120109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chung H.S., Louis J.M., Gopich I.V. Analysis of fluorescence lifetime and energy transfer efficiency in single-molecule photon trajectories of fast-folding proteins. J. Phys. Chem. B. 2016;120:680–699. doi: 10.1021/acs.jpcb.5b11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schuler B., Eaton W.A. Protein folding studied by single-molecule FRET. Curr. Opin. Struct. Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nettels D., Gopich I.V., Schuler B. Ultrafast dynamics of protein collapse from single-molecule photon statistics. Proc. Natl. Acad. Sci. USA. 2007;104:2655–2660. doi: 10.1073/pnas.0611093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nettels D., Hoffmann A., Schuler B. Unfolded protein and peptide dynamics investigated with single-molecule FRET and correlation spectroscopy from picoseconds to seconds. J. Phys. Chem. B. 2008;112:6137–6146. doi: 10.1021/jp076971j. [DOI] [PubMed] [Google Scholar]
- 79.Gopich I.V., Nettels D., Szabo A. Protein dynamics from single-molecule fluorescence intensity correlation functions. J. Chem. Phys. 2009;131:095102. doi: 10.1063/1.3212597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chung H.S., Louis J.M., Eaton W.A. Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. Proc. Natl. Acad. Sci. USA. 2009;106:11837–11844. doi: 10.1073/pnas.0901178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Acharya S., Srivastava K.R., Lapidus L.J. Monomer dynamics of Alzheimer peptides and kinetic control of early aggregation in Alzheimer’s disease. ChemPhysChem. 2016;17:3470–3479. doi: 10.1002/cphc.201600706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karplus M. Contact electron-spin coupling of nuclear magnetic moments. J. Chem. Phys. 1959;30:11–15. [Google Scholar]
- 83.Vögeli B., Ying J., Bax A. Limits on variations in protein backbone dynamics from precise measurements of scalar couplings. J. Am. Chem. Soc. 2007;129:9377–9385. doi: 10.1021/ja070324o. [DOI] [PubMed] [Google Scholar]
- 84.Shen Y., Bax A. SPARTA+: a modest improvement in empirical NMR chemical shift prediction by means of an artificial neural network. J. Biomol. NMR. 2010;48:13–22. doi: 10.1007/s10858-010-9433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ortega A., Amorós D., García de la Torre J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 2011;101:892–898. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Granata D., Baftizadeh F., Vendruscolo M. The inverted free energy landscape of an intrinsically disordered peptide by simulations and experiments. Sci. Rep. 2015;5:15449. doi: 10.1038/srep15449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hofmann H., Soranno A., Schuler B. Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc. Natl. Acad. Sci. USA. 2012;109:16155–16160. doi: 10.1073/pnas.1207719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kubelka J., Hofrichter J., Eaton W.A. The protein folding ‘speed limit’. Curr. Opin. Struct. Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 89.Serio T.R., Cashikar A.G., Lindquist S.L. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 90.Vitalis A., Pappu R.V. Assessing the contribution of heterogeneous distributions of oligomers to aggregation mechanisms of polyglutamine peptides. Biophys. Chem. 2011;159:14–23. doi: 10.1016/j.bpc.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee J., Culyba E.K., Kelly J.W. Amyloid-β forms fibrils by nucleated conformational conversion of oligomers. Nat. Chem. Biol. 2011;7:602–609. doi: 10.1038/nchembio.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Šarić A., Chebaro Y.C., Frenkel D. Crucial role of nonspecific interactions in amyloid nucleation. Proc. Natl. Acad. Sci. USA. 2014;111:17869–17874. doi: 10.1073/pnas.1410159111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng Q., Jockusch S., Blanchard S.C. The contribution of reactive oxygen species to the photobleaching of organic fluorophores. Photochem. Photobiol. 2014;90:448–454. doi: 10.1111/php.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng Q., Juette M.F., Blanchard S.C. Ultra-stable organic fluorophores for single-molecule research. Chem. Soc. Rev. 2014;43:1044–1056. doi: 10.1039/c3cs60237k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Páll S., Abraham M.J., Lindahl E. Lecture Notes in Computer Science. Springer; Cham, Switzerland: 2015. Tackling exascale software challenges in molecular dynamics simulations with GROMACS; pp. 3–27. [Google Scholar]
- 96.Abascal J.L.F., Vega C. A general purpose model for the condensed phases of water: TIP4P/2005. J. Chem. Phys. 2005;123:234505. doi: 10.1063/1.2121687. [DOI] [PubMed] [Google Scholar]
- 97.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 98.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 99.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 100.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 101.Graen T., Hoefling M., Grubmüller H. AMBER-DYES: characterization of charge fluctuations and force field parameterization of fluorescent dyes for molecular dynamics simulations. J. Chem. Theory Comput. 2014;10:5505–5512. doi: 10.1021/ct500869p. [DOI] [PubMed] [Google Scholar]
- 102.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 103.Kjaergaard M., Brander S., Poulsen F.M. Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J. Biomol. NMR. 2011;49:139–149. doi: 10.1007/s10858-011-9472-x. [DOI] [PubMed] [Google Scholar]
- 104.Kjaergaard M., Poulsen F.M. Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR. 2011;50:157–165. doi: 10.1007/s10858-011-9508-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.