Abstract
Although colistin’s clinical use is limited due to its nephrotoxicity, colistin is considered to be an antibiotic of last resort because it is used to treat patients infected with multidrug-resistant bacteria. In an effort to provide molecular details about colistin’s ability to kill Gram-negative (G(−)) but not Gram-positive (G(+)) bacteria, we investigated the biophysics of the interaction between colistin and lipid mixtures mimicking the cytoplasmic membrane of G(+), G(−) bacteria as well as eukaryotic cells. Two different models of the G(−) outer membrane (OM) were assayed: lipid A with two deoxy-manno-octulosonyl sugar residues, and Escherichia coli lipopolysaccharide mixed with dilaurylphosphatidylglycerol. We used circular dichroism and x-ray diffuse scattering at low and wide angle in stacked multilayered samples, and neutron reflectivity of single, tethered bilayers mixed with colistin. We found no differences in secondary structure when colistin was bound to G(−) versus G(+) membrane mimics, ruling out a protein conformational change as the cause of this difference. However, bending modulus KC perturbation was quite irregular for the G(−) inner membrane, where colistin produced a softening of the membranes at an intermediate lipid/peptide molar ratio but stiffening at lower and higher peptide concentrations, whereas in G(+) and eukaryotic mimics there was only a slight softening. Acyl chain order in G(−) was perturbed similarly to KC. In G(+), there was only a slight softening and disordering effect, whereas in OM mimics, there was a slight stiffening and ordering of both membranes with increasing colistin. X-ray and neutron reflectivity structural results reveal colistin partitions deepest to reach the hydrocarbon interior in G(−) membranes, but remains in the headgroup region in G(+), OM, and eukaryotic mimics. It is possible that domain formation is responsible for the erratic response of G(−) inner membranes to colistin and for its deeper penetration, which could increase membrane permeability.
Introduction
Colistin (i.e., polymyxin E) was first isolated by Koyama et al. (1) from the broth of Bacillus polymyxa in 1949; it is a linear trilipopeptide linked to a cyclic heptapeptide that is produced by nonribosomal peptide synthetase systems in Gram-positive (G(+)) bacteria (2). The fatty acid tail can consist of seven, eight, or nine carbons (3). It has a narrow antibacterial spectrum, mainly against Gram-negative bacteria (G(−)) (4, 5), but not G(+) bacteria. Clinical use of colistin decreased in the 1970s due to nephrotoxicity and neurotoxicity after intravenous administration. However, the world is now facing a growing threat from bacteria that are resistant to all available antibiotics (6, 7), and colistin has reemerged as an antibiotic of last resort (3), and is in use to treat cystic fibrosis patients. The incidence of resistance to colistin is relatively low (8), but resistance in Gram-negative pathogens can emerge both in vitro (9, 10) and clinically (11, 12, 13, 14).
Colistin interacts with high affinity with the lipopolysaccharide (LPS) of the outer membrane (OM) of G(−) bacteria (15), which docks the positively charged (+5 e) antibiotic to the cell surface. A critical interaction of colistin with the lipid A component of LPS is suggested by the charge screening mechanism of bacterial resistance whereby negative phosphate charges are neutralized by the addition of aminoarabinose and ethanolamine residues (16). However, membrane blebbing (17) and electrochemical transmembrane potential dissipation in cells treated with colistin have also been reported in previous works (18), which suggests that inner membrane (IM) permeabilization leads to bacterial cell death. What is the mechanism of colistin’s rapid, concentration-dependent bacterial killing with negligible postantibiotic effects (10)? Hancock and co-workers (19, 20, 21, 22, 23) have proposed a general model for several antimicrobial peptides called the “self-promoted uptake” model, where colistin’s aggregation promotes its own uptake across the OM, and subsequent pore formation of the IM. An alternative proposed mechanism is the vesicle-vesicle contact pathway (24, 25) where a colistin dimer can mediate the contacts between periplasmic leaflets of inner and outer membranes (24). A third possible mechanism is a generalized mechanism for bactericidal agents, in which an oxidative burst produces a reactive hydroxyl radical (•OH) that can induce rapid cell death (26).
This work aims to delineate the role of lipids in the interactions between colistin and bacterial lipid membrane mimics in an effort to understand the molecular details of colistin’s bactericidal mechanism. It is of interest to determine if there are structural or elastic properties that differ between G(+) and G(−) lipid membrane mimics that could be responsible for colistin’s preferential killing of G(−) bacteria. As a first approach, we studied the secondary structure of the peptide by means of circular dichroism in the absence and presence of lipid membranes mimicking the outer leaflet (OM) and IM of G(−), cytoplasmic membrane of G(+) bacteria, and eukaryotic cells. With the same membrane mimics, by means of x-ray diffuse scattering we measured both structure (membrane thickness, area/lipid, and peptide position) and the elastic parameter bending modulus (KC), which yields information about membrane softening. In addition, an order parameter that indicates lipid acyl chain ordering is obtained (Sxray). Finally, the third biophysical method, neutron scattering, was used for confirming colistin’s location in the membrane mimics. Because our experimental x-ray system is symmetric, we cannot probe the vesicle-vesicle contact model, and we are also not studying oxidized lipids, so we cannot comment on these other two models for bactericidal killing.
Materials and Methods
Reagents
The synthetic lyophilized lipids 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′rac-glycerol) sodium salt (POPG), 1′, 3′-bis[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol sodium salt (TOCL, i.e., cardiolipin), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dioleoyl-3-trimeathylammonium-propane chloride salt (DOTAP), di[3-deoxy-D-manno-octulosonyl]-lipid A ammonium salt (KDO2), and 1,2-dioleoyl-sn-glycero-3-[phospho-rac-(3-lysyl(1-glycerol))] (chloride salt) (Lysyl PG) were purchased from Avanti Polar Lipids (Alabaster, AL) and used as received. Cholesterol was from Nu-Chek-Prep (Waterville, MN). HPLC grade organic solvents, LPS from Escherichia coli 0111:B4, and colistin sulfate salt were purchased from Sigma-Aldrich (St. Louis, MO) and used as received.
Sample preparation
Membrane mimics were prepared by first dissolving lyophilized lipids in chloroform/methanol (8:2, v/v), or LPS in chloroform/methanol/H2O (2:1:1, v/v/v). Lipid stock solutions were mixed to create lipid mixtures in molar ratios mimicking bacterial membranes: G(−) IM, POPE/POPG/TOCL (7:2:1, molar ratio); G(+) membrane, POPG/DOTAP/POPE/TOCL (6:1.5:1.5:1); and eukaryotic membrane, POPC/POPE/cholesterol (5:1:1.8). For the outer membrane of G(−) bacteria, two different mimics were used: lipid A with two deoxy-manno-octulosonyl sugar residues of the inner core (KDO2), and a mixture of LPS/DLPG (1:3). Stock solutions of colistin (MW = 1400) were prepared in Milli-Q water (Millipore, Billerica, MA).
Multilamellar stacked samples for x-ray scattering were prepared by mixing 4 mg of the different lipid mixtures plus colistin into a glass test tube in various mole ratios from 750:1 to 50:1, lipid/peptide. Solvents were removed by evaporation under vacuum and samples were redissolved in appropriate HPLC-grade solvents for spreading: G(−), G(+), chloroform/trifluoroethanol (TFE) 7:3 (v/v); eukaryotic, chloroform/TFE 1:1 (v/v); LPS model, chloroform/TFE/H2O (5:5:1); and KDO2, chloroform/methanol/H2O (90:10:1). These mixtures were plated onto silicon wafers (15 × 30 × 1 mm) via the rock-and-roll method (27) to produce stacks of ∼1800 well-aligned bilayers. After solvent removal under vacuum for 2 h, hydration occurred through the vapor in a thick-walled x-ray hydration chamber (28).
Samples for circular dichroism (CD) spectroscopy were prepared by spreading a thin film of 0.7 mg of the same bacterial mimic mixtures as for x-ray with/without 0.3 mg colistin onto the inner wall of a 1-cm quartz cuvette. After solvent removal under vacuum, CD samples were fully hydrated at room temperature overnight with 100 μL Milli-Q water in the bottom of the sealed cuvette.
Samples for densimetry were prepared as in (29). A quantity of 10–50 mg dried lipid mixture was mixed with ∼1.2 mL water. This mixture was hydrated by temperature cycling three times from 60 to 0°C with vortexing to produce multilamellar vesicles.
Samples for neutron scattering were prepared by adding colistin to 8 mg lipid mixtures in a 50:1 lipid/peptide molar ratio. Organic solvent was removed by evaporation and samples were rehydrated in a 2 M NaCl aqueous solution to a final concentration of 5–6 mg/mL and bath-sonicated for 60–90 min until clarity. The self-assembled monolayers of HC18 tethers were formed on 3" diameter silicon wafers (30) and sparsely tethered bilayer lipid membranes were formed by exposing the self-assembled monolayer to the vesicle suspension for 60 min in a National Institute of Standards and Technology (NIST) reflectivity flow cell. This was followed by a rinse with 40 mL deionized water (31).
Methods
Low-angle x-ray scattering. Low-angle x-ray scattering (LAXS) data from oriented, fully hydrated samples were obtained at the G1 line at the Cornell High Energy Synchrotron Source (CHESS, Ithaca, NY) with previously described methods (32, 33, 34) on two separate trips using x-ray wavelengths 1.108 and 1.096 Å and sample-to-detector (S)-distances of 387.2 and 396.6 mm. In addition, a laboratory x-ray source RUH3R Rotating Anode X-Ray Generator (Rigaku, Tokyo, Japan) with a FOX 2D Focusing Collimator (Xenocs, Sassenage, France) and a Mercury CCD Detector (Rigaku) were used with an x-ray wavelength of 1.5418 Å and S-distance of 280.6 mm. Full hydration is judged by no further increase in lamellar D-spacing over time. Measurements were carried out in the fluid phase typically at 37°C, except for KDO2 samples, which were studied at 55°C due to the high melting temperature of the lipid. Details about determination of KC from diffuse LAXS images and electron density profiles are given in the Supporting Material.
Wide-angle x-ray scattering. Wide-angle x-ray scattering (WAXS) was obtained at CHESS (S-distances = 163.4 and 179.3 mm) and at Carnegie Mellon University (Pittsburgh, PA) (S-distance = 125.7 mm) as described in (29, 35). Hydrocarbon chain order parameters were estimated by measuring the angular dependence of the interchain WAXS signal according to the model developed by Mills et al. (35), which is also based upon liquid crystal theory. More details of the WAXS analysis are given in the Supporting Material.
Neutron reflectivity. Neutron reflectivity (NR) measurements were performed at the NGD-MAGIK reflectometer at the NIST Center for Neutron Research (Gaithersburg, MD) over a momentum transfer range 0–0.25 Å−1. More details about NR are given in the Supporting Material.
CD spectroscopy. CD spectra in the 200–240-nm range were collected with a model No. 715 Spectropolarimeter (Jasco, Oklahoma City, OK) by accumulating 15 or 20 spectra at 37°C, at 100 nm/min with a step resolution of 1 nm. Samples were performed in duplicate or triplicate. More details of the CD spectroscopy are given in the Supporting Material.
Densimetry. Approximately one milliliter of multilamellar vesicles were loaded into the model No. 5000M Densimeter (Anton-Paar, Graz, Austria). Density measurements were recorded as in (36), by first measuring Milli-Q water at 37°C and then measuring the sample density at 37°C. The sample was withdrawn and remeasured several times to ensure reproducibility. The molecular volume was calculated from density using Avogadro’s number and the combined molecular weight of the lipids assuming additivity. Colistin’s volume was measured separately as an aqueous solution at 37°C (7.9 mg/mL).
Results
CD
Colistin adopted a secondary structure in water that contained primarily β-sheet, random coil, and β-turn (see Table 1). When associated with lipids in thin films, however, β-turn and β-sheet structure were reduced, and a signal resembling α-helix could be fitted. However, it must be recalled that in peptides, some turns can show the same negative band at 222 nm as a result of n-π∗ transitions (37, 38). What is striking is that colistin’s secondary structure is quite similar in all of the lipid models, with slightly higher α-helix-type signal in LPS and eukaryotic membranes. In particular, there is little difference between colistin in G(−) and G(+) membrane mimics. Light microscopy pictures of the thin film samples in the dried state that were then hydrated for CD are shown in Fig. S1. CD ellipticity data that produced the results in Table 1 can be found in Fig. S2.
Table 1.
CD Results of Colistin in Water and Membrane Mimics
| Colistin Sample | α-Helix-Type % | β-Sheet % | β-Turn % | Random Coil % | R2 |
|---|---|---|---|---|---|
| Water | 0 ± 1 | 48 ± 6 | 16 ± 5 | 36 ± 8 | 0.88 |
| G(−) | 24 ± 6 | 18 ± 8 | 0 ± 1 | 59 ± 7 | 0.99 |
| G(+) | 17 ± 3 | 17 ± 4 | 2 ± 1 | 64 ± 2 | 0.99 |
| LPS | 31 ± 11 | 3 ± 3 | 0 ± 1 | 67 ± 3 | 0.99 |
| Eukaryotic | 30 ± 10 | 9 ± 8 | 0 ± 1 | 62 ± 12 | 0.99 |
Percent composition was obtained by normalizing to 100% the unitless coefficients in the linear least squares fit of the structural motifs to the data. R2 values indicate goodness of fit, with 1 indicating a perfect fit to the data.
Diffuse scattering
Fig. S4 shows typical LAXS data from oriented, fully hydrated stacks of membrane mimics containing colistin, 100:1 lipid/colistin. The results of the LAXS fitting are shown in Figs. 1, A and B, 2, and 3.
Figure 1.
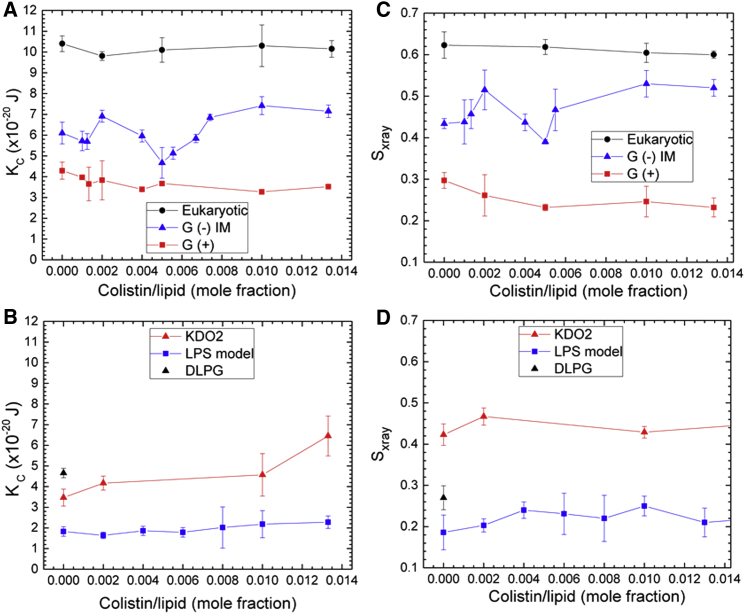
Elasticity (KC) results for (A) eukaryotic, G(−) and G(+) membrane mimics, (B) KDO2 and LPS model, and DLPG control. Sxray order parameter results for (C) eukaryotic, G(−) and G(+) membrane mimics, (D) KDO2 and LPS model, and DLPG control. To see this figure in color, go online.
Figure 2.
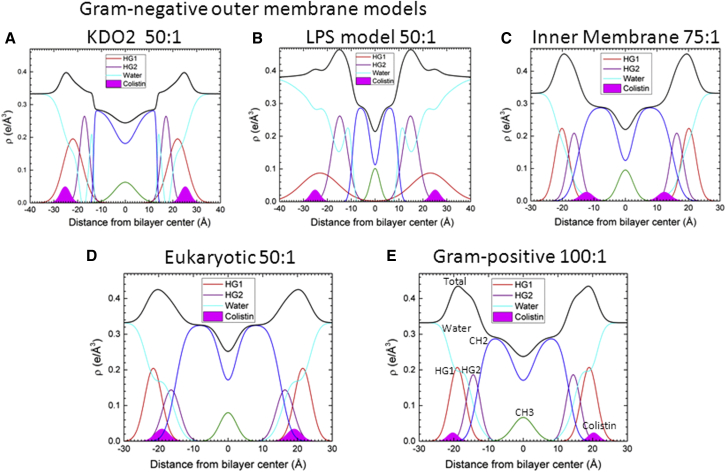
EDPs for (A) KDO2, 50:1 lipid/peptide molar ratio; (B) LPS model, 50:1; (C) IM, 75:1; (D) Eukaryotic, 50:1; (E) G(+), 100:1. Component groups are headgroup HG1 (red), headgroup HG2 (violet), methylene CH2 region (blue), methyl CH3 region (green), colistin (solid magenta), water (cyan), and total (black). To see this figure in color, go online.
Figure 3.
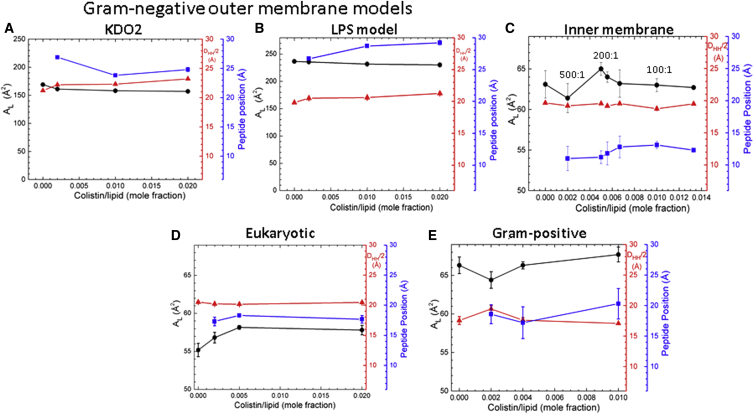
Structural parameters, area/lipid AL (black lines), DHH/2 (red lines) and colistin position (blue lines), as a function of increasing colistin mole fraction in five lipid membrane mimics: (A) KDO2, (B) LPS model, (C) G(−) IM, (D) Eukaryotic, and (E) G(+). Left axis shows AL (black); right axes show DHH/2 (red) and peptide position (blue). DHH/2 and peptide position are in Å units from the bilayer center, and AL is in units of Å2. To see this figure in color, go online.
Fig. S7 shows WAXS data obtained from the same sample concentrations as in Fig. S4. The results of the WAXS fitting are shown in Fig. 1, C and D.
Bending moduli and Sxrayorder parameters
Fig. 1 shows the results of fitting the LAXS (Fig. 1, A and B) and WAXS (Fig. 1, C and D) diffuse data for the five lipid membrane mimics containing colistin. Not surprisingly, KC is highest (stiffest membrane) for the eukaryotic membrane mimic, in which cholesterol interacts favorably with the saturated palmitoyl chain (39) in POPC and POPE (Fig. 1 A). Colistin caused only a slight softening of the eukaryotic mimic with increasing concentration. G(+) membrane mimics had a much lower KC, indicating a more bendable membrane, which was also only slightly softened by colistin. G(−), on the other hand, showed a complex behavior of KC with colistin concentration. At low (500:1) and high peptide concentration (>100:1 lipid/peptide molar ratio), colistin induced stiffening, but at intermediate ratios (∼200:1), a significant membrane softening was obtained (Fig. 1 A). For studying the interaction of colistin with the outer membrane of G(−) bacteria, a model mixture consisting of LPS/DLPG (1:3 molar ratio) was employed, because multilayered stacks of pure LPS showed a lamellar x-ray pattern with D-spacing of ∼290 Å but no diffuse signal, indicating nonfluctuating bilayers and impairing the analysis of mechanical and structural properties of the membrane within the theoretical framework of liquid crystals. The KC values in Fig. 1 B for LPS/DLPG model were lower than those for the G(+) membrane mimic, indicating that the model used for OM is a relatively soft membrane, whereas DLPG alone (black triangle) had a higher KC, indicating that the additional sugar residues on LPS cause a softening. KDO2 stacked bilayers in fluid phase at 55°C had a higher KC than LPS/DLPG at 37°C and showed stiffening with increasing colistin content (Fig. 1 B). In Fig. 1, C and D, the complementary Sxray results for the KC results in Fig. 1, A and B are shown. Sxray, which indicates acyl chain order, paralleled membrane bending in all of the membrane mimics. As shown in Fig. 1 C, eukaryotic membrane, containing cholesterol, had the most ordered chains, and colistin only slightly disordered them. G(+) had the most disordered chains and colistin slightly disordered them. G(−) showed the same irregular behavior for Sxray as for KC, first increasing chain order at 500:1 lipid/peptide molar ratio, then decreasing chain order at 200:1, then increasing chain order to a maximum at 100:1. In the OM mimics in Fig. 1 D, the chain order increased slightly with increasing colistin concentration, similar to the slight membrane stiffening shown in Fig. 1 B. Similar to its KC effect, KDO2 had a higher chain order compared to LPS model. LPS model also has more disordered chains than DLPG, suggesting a chain disordering effect of the sugar residues.
Structural results
Fig. 2 shows the electron density profiles (EDPs) of the five membrane mimics containing the highest concentration in our study of colistin for each mimic. The measured volumes required for determining the EDPs appear in Table S1. Colistin (solid magenta, Gaussian) showed a deeper partitioning into the G(−) IM membrane mimics (Figs. 2 C and 3 C) when compared to both G(+) and eukaryotic mimics at the different lipid/peptide mole ratios assayed. In G(−) IM, colistin is located in the hydrocarbon region just within the interfacial region, 11–14 Å from the bilayer center, at all concentrations. In eukaryotic and G(+) membrane, colistin locates in the headgroup region, just within DHH/2. Colistin in G(−) mimics locates in the headgroup region in KDO2 (Fig. 2 A) and outside of the headgroup maximum (DHH/2) in the mixture LPS/DLPG (1:3) (Fig. 2 B). The EDP of the LPS/DLPG mixture was calculated taking into account the electrons from the Re variant of the LPS molecule (lipid A plus the inner sugar core, consisting of three residues of deoxy-manno-octulosonyl and 2 phospho-heptose), whereas the remainder sugar moieties in the core and O-antigen regions were included in the water electron density.
Fig. 3 summarizes the structural results for all five membrane mimics with increasing colistin concentration. AL is the area/unit cell, which includes the lipid and the colistin, but not the cholesterol (for eukaryotic membrane). In G(−) IM, AL (black lines) decreased at 500:1 lipid/peptide molar ratio but increased at 200:1. These changes are in the expected direction according to the Sxray results in Fig. 1 C, because increasing chain order decreases AL, as the chains straighten. At peptide concentrations higher than 100:1, AL showed no significant differences when compared to control. The overall bilayer thickness (DHH/2, red lines) did not change appreciably. In G(+) membranes, AL first decreases slightly then increases with colistin concentration, whereas in eukaryotic membranes, there is an increase in AL with colistin concentration. AL is fairly constant as colistin is added to KDO2 and LPS model, as is the bilayer thickness, DHH/2. Table S4 compares our KDO2 control area with literature values.
NR
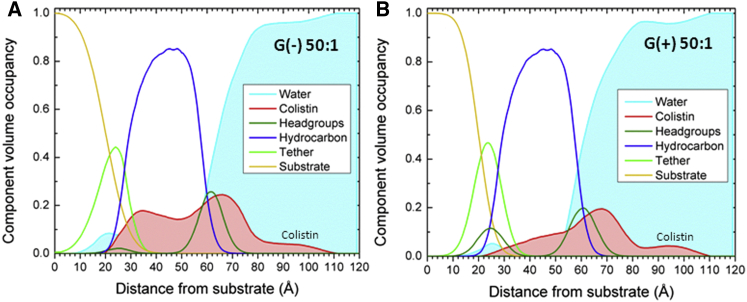
Fig. 4 shows the NR results as component volume occupancy versus distance from the gold substrate. At this concentration of lipid/peptide (50:1), colistin (red line) enters more deeply into the G(−) membrane (Fig. 4 A) than into the G(+) membrane (Fig. 4 B). However, compared to the x-ray result, there is considerable colistin outside of the outer leaflet headgroup Gaussian (cyan) for both G(−) and G(+) membrane mimics. This could be due to the different sample preparation using vesicle fusion to form a single tethered bilayer, compared to the stacked system of 1800 bilayers for x-ray. The significance of the NR result is that there is a deeper penetration of colistin into the hydrocarbon core in G(−) membrane mimics, similar to the x-ray result. Additional details of the NR results are shown in the Supporting Material.
Figure 4.
Component volume occupancy versus distance from substrate. (A) G(−) IM mimic; (B) G(+) mimic. Lipid/peptide 50:1 molar ratio. Component groups are shown: water (cyan), colistin (red), headgroups (olive), hydrocarbons (blue), tether (green), and substrate (gold). To see this figure in color, go online.
Discussion
Colistin secondary structure is similar in different membrane mimics
A shape analysis procedure (38) for fitting the experimental CD spectra was undertaken for estimating the secondary structure of colistin, using a set of spectra of typical motifs obtained from standard proteins/polypeptides (40). Although these reference spectra correspond to proteins formed by the naturally occurring L isomer of amino acids, we employed them for this colistin analysis, whose primary structure is formed of nine L-amino acids plus one D-amino acid. This method was appropriate because several previous reports indicated that CD is a sensitive measurement of the secondary structural changes caused by single or double D-amino acid substitution in short peptides (41, 42, 43). On the other hand, complete inverse CD structure is obtained in an all-D model α-helixes (44, 45). Our study indicates that, in agreement with previous NMR results (46, 47), the colistin backbone in the aqueous phase has a significant degree of freedom, with a high random coil or disordered structure plus β-turn and β-sheet. When bound to lipids, both β-turn and β-sheet content decreased whereas disordered structure increased, which agrees with the weakening of the β-turn NMR signal observed previously (46). However, the α-helix-type feature was also found, which could be ascribed to distorted turns, as previously observed for other cyclic peptides and D-amino acid-containing peptides (43).
The further result that there were no significant differences in secondary structure of colistin when bound to the different membrane mimics, suggests that a change in colistin secondary structure cannot be the reason for differences in bactericidal mechanism in G(−) versus G(+) bacteria.
Colistin’s effects on G(−) IM membrane mechanical properties and order parameters may involve lateral heterogeneity
Contrary to the CD results, dramatic changes were observed in the elastic behavior of colistin interacting with G(−) IM mimics compared to G(+) mimics. As shown in Fig. 1 A, colistin caused alternating stiffening, softening and stiffening, as its concentration increased in G(−) IM mimics. This erratic behavior was mirrored by the Sxray order parameter (Fig. 1 C), where colistin alternately ordered, disordered and ordered the acyl chains at the same concentrations as for KC. These changes are unlike any seen in our previous investigations: increasing concentration of HIV-1 fusion peptide (48) or alamethicin (49) decreased the bending modulus KC in DOPC and diC22:1PC in an exponential fashion, and increasing HIV-1 matrix31 peptide caused a gradual increase in KC and Sxray then a gradual decrease in both in PS-containing membranes (29). The fact that this sharply irregular behavior occurred only in G(−) IM mimics may offer a clue to colistin’s bactericidal mechanism in G(−) bacteria. Compared to G(+) mimics, which displayed only a slight softening and disordering of lipid chains, G(−) IM mimics were greatly perturbed, in both directions, by colistin.
The G(+) mimic used in this work consisted of the lipid mixture POPG/DOTAP/POPE/TOCL (6:1.5:1.5:1 molar ratio), as many Gram-positive bacteria, such as Staphylococcus aureus, contain a high amount of the negatively charged lipids PG and cardiolipin, but also a positively charged derivative of PG, lysyl-PG, in which a lysine residue is bonded to the PG headgroup (50). Due to the high amounts of lipids needed for preparing samples in a wide range of peptide concentrations, a more affordable lipid, DOTAP, was used instead of lysyl-PG, which is also positively charged. A control sample prepared with lysyl-PG showed no significant differences in AL or KC compared to mixtures prepared with DOTAP, indicating that the latter is a good surrogate for lysyl-PG. Unlike G(+) membranes, the inner membranes of G(−) bacteria are highly enriched in PE but also contain a significant amount of negatively charged lipids (51, 52), so this mimic contained POPE/POPG/TOCL (7:2:1).
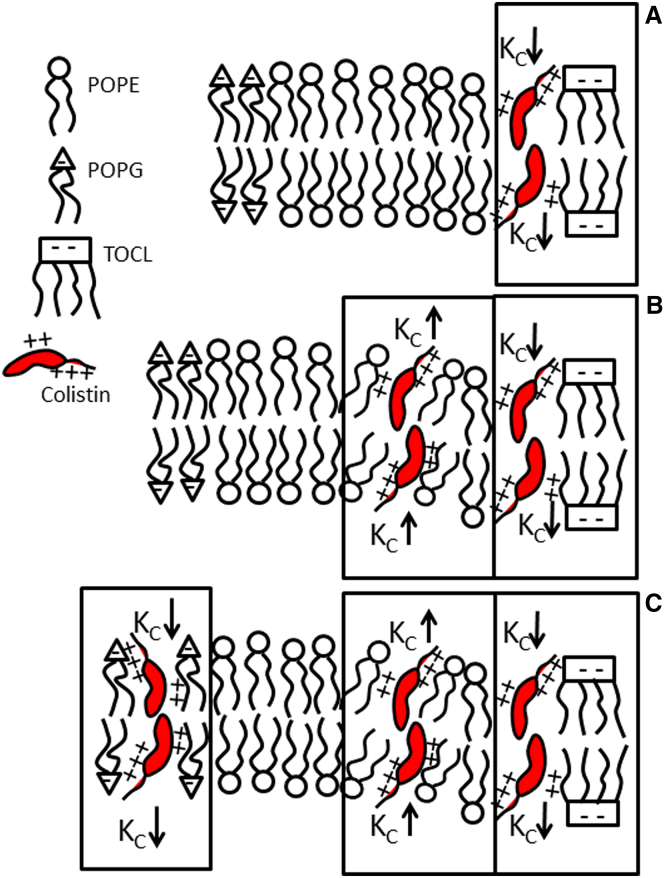
It has been suggested that cardiolipin-rich domains exist in G(−) membranes (53) and similar cationic antimicrobial peptides can perturb bilayer permeability by inducing phase separation in membranes containing PE and cardiolipin (54). Thus, we propose that colistin may induce membrane lateral heterogeneity by clustering anionic lipids due to Coulombic interactions in the presence of high content of PE, as in IM G(−) membranes but not in a low content of PE, as in G(+) membranes. Formation of lipid domains can lead to interfacial curvature stress in bilayers, due to a nonbilayer tendency that some of them have, like PE (55) or cardiolipin (56). A previous work found that the increase in current due to permeability perturbation in asymmetric model membranes occurred above a threshold colistin concentration, which was also postulated to be a consequence of preferential binding of the peptide with negatively charged lipids and phase separation (57). The presence of lipid domains and interfacial curvature stress would explain transient membrane permeabilization along the defects at domain boundaries (54). In a recent molecular dynamics (MD) simulation (58), it was shown that the bending modulus was altered as a cationic AMP was added to a binary lipid membrane containing the zwitterionic DPPC and the negatively charged DPPS. The authors suggested that when cationic AMPs encounter bacterial membranes, domains with different material moduli are formed, which could lead to a destabilization at the boundaries of these domains. To investigate this possibility further, we have studied the KC values of the individual components of G(−) IM, with and without 0.01 mol fraction colistin, with results shown in Table 2. KC of POPG is reduced by about half when colistin is added, whereas KC of TOCL is only slightly reduced. POPE with 0.01 colistin, or even 0.001 colistin, did not fluctuate, indicating that KC has been greatly increased by colistin (i.e., membrane is stiffer). Thus, even though PE is the lipid at highest proportion in G(−) IM, the softening of the mixture at intermediate colistin mole fractions should be produced by a preferential interaction of colistin with PG or TOCL due to Coulombic forces, whereas the stiffening caused by colistin-PE interaction is evidenced at intermediate and higher concentrations. When these results are compared to Fig. 1 A, we might interpret the initial slight lowering of KC as due to colistin binding to TOCL. As concentration increases, colistin could bind to POPE, thus increasing KC. When colistin encounters POPG, a larger decrease in KC is observed. At the highest concentrations, colistin interacts with the remaining POPE, causing a final increase in KC. A cartoon summarizing this scenario is presented in Fig. 5.
Table 2.
Bending Moduli, KC Values, of Components of G(−) Inner Membranes
| Colistin Mole Fraction | POPE | POPG | TOCL |
|---|---|---|---|
| 0 | 9.7 ± 1.0 | 7.5 ± 0.3 | 4.4 ± 0.4 |
| 0.01 | ND | 3.7 ± 0.4 | 3.3 ± 1.0 |
Kc units (×10−20 J). ND, none detected.
Figure 5.
Cartoon showing colistin interacting with individual lipid types in G(−) IM as its concentration increases. (A) By comparing to Fig. 1A, colistin may first interact with TOCL, slightly lowering KC. (B) Colistin may then interact with POPE, increasing KC. (C) Colistin may then interact with POPG, decreasing KC substantially. Finally, colistin may interact with the remaining POPE (data not shown), causing an increase in KC. To see this figure in color, go online.
A second reason for differences between G(+) and G(−) IM is that colistin reaches the deepest location in the hydrocarbon region in G(−) IM when compared to the other membrane mimics. The EDPs and summary of structural results show that colistin is located in the headgroup region at all concentrations for all mimics except for G(−) IM. NR supports this location, although the contrast between G(−) and G(+) is not as dramatic as in x-ray, presumably due to the different sample preparations. This deeper location of colistin in G(−) IM could cause a defect that would allow for a deeper penetration of water into the bilayer, leading to permeabilization and bacterial cell death.
Colistin locates in the headgroup region in G(−) OM mimics
As for the G(−) OM mimics, we observe that the LPS model (LPS/DLPG 1:3) is softer and less ordered than DLPG or KDO2, indicating that the LPS component with its core and O-antigen sugar residues induces a softening and disordering effect. This contradicts the idea that the OM has low fluidity (59, 60), but is in agreement with order parameters calculated by MD simulation that showed |SCD| decreases with increasing sugar residues on lipid A (61). The hydrocarbon half-thickness for the two OM control mimics (EDPs not shown) are both thinner than usual (DC = 12.2 Å for KDO2 and 9.2 Å for LPS model), whereas the other control mimics have more normal thicknesses (DC = 14.4 Å for G(+), 14.8 Å for G(−), and 15.4 Å for eukaryotic). We find that colistin slightly orders both KDO2 and the LPS model, contradicting the Fourier transform infrared spectroscopy results of (62), which showed a fluidizing effect of the related polymyxin B on KDO2. However, in that study, the smallest concentration of lipid/peptide was 8:1, a much larger amount of colistin than in our study. The small effect of colistin in our model OM systems agrees with previous results with polymyxin B, in which lower charge screening and permeabilization are observed in LPS- compared to PG-containing lipid vesicles (63).
Colistin’s location in the headgroup of KDO2 and the LPS model suggests that the outer membrane destabilization observed in previous biophysical and microbiological works must occur through divalent cation displacement by colistin (18, 64). Although our study did not add Ca2+ or Mg2+ as variables, our main conclusion, that colistin remains in the headgroup region of G(−) OM models, is consistent with the idea that it could displace divalent cations. Future experiments could explore the dependence of colistin’s location on type of divalent cation. The divalent cations play an important role in the assembly of the LPS molecules in the outer membrane by screening the repulsive Coulombic forces between the phosphate residues at the lipid A and sugar inner core levels (59). It was previously reported that colistin binding to lipid A is inhibited by divalent cations and high ionic strength (64). By weakening the outer membrane, colistin could induce its self-promoted uptake toward the periplasmic space and reach its final target, the inner membrane of G(−) bacteria. A recent MD simulation confirmed our headgroup location of colistin in KDO2 and the LPS model (65).
Conclusions
Using a structural and materials approach, this work attempted to clarify molecular steps in colistin’s bactericidal mechanism. We found no difference in secondary structure of colistin in G(−) IM and G(+) membranes, thus ruling out a protein conformational change as the cause of colistin’s ability to kill G(−) but not G(+) bacteria. However, dramatic differences between colistin’s effect on G(−) IM versus G(+) membrane mimics were observed in the elasticity results; whereas G(−) IM mimics were softened at a critical lipid/peptide molar ratio (200:1), they were stiffened above and below that ratio. G(+) membranes, on the other hand, were only slightly softened, a very small perturbation. We suggest that colistin induces domains in G(−) IM with different adjacent elasticity, which could lead to permeation through the domain boundaries. Chain ordering paralleled membrane elasticity to indicate that lipid acyl chains are significantly perturbed only in the case of G(−) IM. In addition, colistin located at a deepest position in the interior of the hydrocarbon region in G(−) membranes. Therefore, the elasticity, chain ordering, and peptide membrane location point to colistin’s ability to enter into and perturb the G(−) inner membrane, which could lead to an increase in permeability. As for the G(−) OM, colistin remains in the headgroup region at all the concentrations that we studied, and increases slightly the OM stiffness and chain order. Therefore, colistin is in position to displace divalent cations, leading to OM perturbation, and eventually, to its self-promoted uptake.
Author Contributions
F.G.D. and S.T.-N. designed the experimental research, performed experiments, analyzed data, and wrote the manuscript. I.P., K.A., and F.H. performed experiments and analyzed data. M.F.P. and Y.E. performed experiments.
Acknowledgments
The authors thank Prof. Emeritus John F. Nagle for help with the data collection at CHESS and for discussions concerning the use of the SDP program, and Dr. Yohei Doi for critical reading of the manuscript and Dr. Arthur Woll for his help at the G1 CHESS beamline.
Parts of this research were performed at the NIST Center for Nanoscale Science. Certain commercial materials, equipment, and instruments are identified in this work to describe the experimental procedure as completely as possible. In no case does such an identification imply a recommendation or endorsement by NIST, nor does it imply that the materials, equipment, or instrument identified are necessarily the best available for the purpose.
Support for this work was from the Samuel and Emma Winters Foundation (STN), The National Scientific and Technical Research Council, Argentina (CONICET) under Programa de Becas Externas (to F.G.D.), The National Scientific and Technical Research Council, Argentina (CONICET) (MFP), and National Institutes of Health (NIH) R01GM101647 (to F.H.). This work is based upon research conducted at Carnegie Mellon University and at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation (NSF) under award No. DMR-1332208. The neutron work was supported by the Department of Commerce through its Measurement Science and Engineering Program (70NANB13H009), the National Institute of Standards and Technology (NIST) Center for Neutron Research Comprehensive Grant Program (70NANB17H299), and the NIST IMS Program “Precision Measurement for Integral Membrane Proteins”.
Editor: Georg Pabst.
Footnotes
Supporting Materials and Methods, ten figures, and four tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)35132-9.
Supporting Citations
References (66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79) appear in the Supporting Material.
Supporting Material
References
- 1.Koyama Y., Kurosawa A., Takakuta K. A new antibiotic ‘colistin’ produced by spore-forming soil bacteria. J. Antibiotics (Japan) 1950;3:457–458. [Google Scholar]
- 2.Stein T., Vater J., Morris H.R. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. J. Biol. Chem. 1996;271:15428–15435. doi: 10.1074/jbc.271.26.15428. [DOI] [PubMed] [Google Scholar]
- 3.Velkov T., Roberts K.D., Li J. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol. 2013;8:711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J., Nation R.L., Paterson D.L. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 2006;6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 5.Bergen P.J., Landersdorfer C.B., Li J. Pharmacokinetics and pharmacodynamics of ‘old’ polymyxins: what is new? Diagn. Microbiol. Infect. Dis. 2012;74:213–223. doi: 10.1016/j.diagmicrobio.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Infectious Diseases Society of America. The 10 × 20 initiative: pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin. Infect. Dis. 2010;50:1081–1083. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- 7.Talbot G.H., Bradley J., Bartlett J.G., Antimicrobial Availability Task Force of the Infectious Diseases Society of America Bad bugs need drugs: an update on the development pipeline from the Antimicrobial Availability Task Force of the Infectious Diseases Society of America. Clin. Infect. Dis. 2006;42:657–668. doi: 10.1086/499819. [DOI] [PubMed] [Google Scholar]
- 8.Gales A.C., Jones R.N., Sader H.S. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY antimicrobial surveillance program (2006–09) J. Antimicrob. Chemother. 2011;66:2070–2074. doi: 10.1093/jac/dkr239. [DOI] [PubMed] [Google Scholar]
- 9.Bergen P.J., Li J., Milne R.W. Comparison of once-, twice-, and thrice-daily dosing of colistin on antibacterial effect and emergence of resistance: studies with Pseudomonas aeruginosa in an in vitro pharmacodynamic model. J. Antimicrob. Chemother. 2008;61:636–642. doi: 10.1093/jac/dkm511. [DOI] [PubMed] [Google Scholar]
- 10.Poudyal A., Howden B.P., Li J. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 2008;62:1311–1318. doi: 10.1093/jac/dkn425. [DOI] [PubMed] [Google Scholar]
- 11.McGann P., Snesrud E., Schaecher K.E. Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of MCR-1 in the United States. Antimicrob. Agents Chemother. 2016;60:4420–4421. doi: 10.1128/AAC.01103-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qureshi Z.A., Hittle L.E., Doi Y. Colistin-resistant Acinetobacter baumannii: beyond carbapenem resistance. Clin. Infect. Dis. 2015;60:1295–1303. doi: 10.1093/cid/civ048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthaiou D.K., Michalopoulos A., Falagas M.E. Risk factors associated with the isolation of colistin-resistant Gram-negative bacteria: a matched case-control study. Crit. Care Med. 2008;36:807–811. doi: 10.1097/CCM.0B013E3181652FAE. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y.Y., Wang Y., Shen J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 15.Morrison D.C., Jacobs D.M. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry. 1976;13:813–818. doi: 10.1016/0019-2791(76)90181-6. [DOI] [PubMed] [Google Scholar]
- 16.Cai Y., Chai D., Bai N. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 2012;67:1607–1615. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- 17.Koike M., Iida K., Matsuo T. Electron microscopic studies on mode of action of polymyxin. J. Bacteriol. 1969;97:448–452. doi: 10.1128/jb.97.1.448-452.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L., Dhillon P., Hancock R.E. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000;44:3317–3321. doi: 10.1128/aac.44.12.3317-3321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock R.E.W. Alterations in outer membrane permeability. Annu. Rev. Microbiol. 1984;38:237–264. doi: 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- 20.Hancock R.E., Raffle V.J., Nicas T.I. Involvement of the outer membrane in gentamicin and streptomycin uptake and killing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1981;19:777–785. doi: 10.1128/aac.19.5.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock R.E.W., Chapple D.S. Peptide antibiotics. Antimicrob. Agents Chemother. 1999;43:1317–1323. doi: 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock R.E.W. Peptide antibiotics. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- 23.Hancock R.E.W., Falla T., Brown M. Cationic bactericidal peptides. Adv. Microb. Physiol. 1995;37:135–175. doi: 10.1016/s0065-2911(08)60145-9. [DOI] [PubMed] [Google Scholar]
- 24.Clausell A., Garcia-Subirats M., Cajal Y. Gram-negative outer and inner membrane models: insertion of cyclic cationic lipopeptides. J. Phys. Chem. B. 2007;111:551–563. doi: 10.1021/jp064757+. [DOI] [PubMed] [Google Scholar]
- 25.Cajal Y., Rogers J., Jain M.K. Intermembrane molecular contacts by polymyxin B mediate exchange of phospholipids. Biochemistry. 1996;35:299–308. doi: 10.1021/bi9512408. [DOI] [PubMed] [Google Scholar]
- 26.Kohanski M.A., Dwyer D.J., Collins J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 27.Tristram-Nagle S. Humana Press; Totowa, NJ: 2007. Preparation of Oriented, Fully Hydrated Lipid Samples for Structure Determination Using X-Ray Scattering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kucerka N., Liu Y., Nagle J.F. Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using x-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys. J. 2005;88:2626–2637. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Neil L., Andenoro K., Tristram-Nagle S. HIV-1 matrix-31 membrane binding peptide interacts differently with membranes containing PS vs. PI(4,5)P2. Biochim. Biophys. Acta. 2016;1858:3071–3081. doi: 10.1016/j.bbamem.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budvytyte R., Valincius G., Vanderah D.J. Structure and properties of tethered bilayer lipid membranes with unsaturated anchor molecules. Langmuir. 2013;29:8645–8656. doi: 10.1021/la401132c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barros M., Heinrich F., Lösche M. Membrane binding of HIV-1 matrix protein: dependence on bilayer composition and protein lipidation. J. Virol. 2016;90:4544–4555. doi: 10.1128/JVI.02820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Nagle J.F. Diffuse scattering provides material parameters and electron density profiles of biomembranes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;69:040901. doi: 10.1103/PhysRevE.69.040901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyatskaya Y., Liu Y., Nagle J.F. Method for obtaining structure and interactions from oriented lipid bilayers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2001;63:011907. doi: 10.1103/PhysRevE.63.011907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, Y. 2003. New method to obtain structure of biomembranes using diffuse x-ray scattering: application to fluid phase DOPC lipid bilayers. Ph.D. thesis. Carnegie Mellon University, Pittsburgh, PA.
- 35.Mills T.T., Toombes G.E.S., Nagle J.F. Order parameters and areas in fluid-phase oriented lipid membranes using wide angle x-ray scattering. Biophys. J. 2008;95:669–681. doi: 10.1529/biophysj.107.127845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boscia A.L., Akabori K., Tristram-Nagle S. Membrane structure correlates to function of LLP2 on the cytoplasmic tail of HIV-1 gp41 protein. Biophys. J. 2013;105:657–666. doi: 10.1016/j.bpj.2013.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adler A.J., Greenfield N.J., Fasman G.D. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 1973;27:675–735. doi: 10.1016/s0076-6879(73)27030-1. [DOI] [PubMed] [Google Scholar]
- 38.Greenfield N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan J., Mills T.T., Nagle J.F. Cholesterol perturbs lipid bilayers nonuniversally. Phys. Rev. Lett. 2008;100:198103. doi: 10.1103/PhysRevLett.100.198103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brahms S., Brahms J. Determination of protein secondary structure in solution by vacuum ultraviolet circular dichroism. J. Mol. Biol. 1980;138:149–178. doi: 10.1016/0022-2836(80)90282-x. [DOI] [PubMed] [Google Scholar]
- 41.Rothemund S., Beyermann M., Sönnichsen F.D. Structure effects of double D-amino acid replacements: a nuclear magnetic resonance and circular dichroism study using amphipathic model helices. Biochemistry. 1995;34:12954–12962. doi: 10.1021/bi00040a005. [DOI] [PubMed] [Google Scholar]
- 42.Krause E., Beyermann M., Bienert M. Conformation of a water-soluble β-sheet model peptide. A circular dichroism and Fourier-transform infrared spectroscopic study of double D-amino acid replacements. Int. J. Pept. Protein Res. 1996;48:559–568. doi: 10.1111/j.1399-3011.1996.tb00875.x. [DOI] [PubMed] [Google Scholar]
- 43.Lee D.L., Powers J.P., Hodges R.S. Effects of single D-amino acid substitutions on disruption of β-sheet structure and hydrophobicity in cyclic 14-residue antimicrobial peptide analogs related to gramicidin S. J. Pept. Res. 2004;63:69–84. doi: 10.1046/j.1399-3011.2003.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scolnik Y., Portnaya I., Shinitzky M. Subtle differences in structural transitions between poly-L- and poly-D-amino acids of equal length in water. Phys. Chem. Chem. Phys. 2006;8:333–339. doi: 10.1039/b513974k. [DOI] [PubMed] [Google Scholar]
- 45.Wade D., Boman A., Merrifield R.B. All-D amino acid-containing channel-forming antibiotic peptides. Proc. Natl. Acad. Sci. USA. 1990;87:4761–4765. doi: 10.1073/pnas.87.12.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pristovsek P., Kidric J. Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: an NMR and molecular modeling study. J. Med. Chem. 1999;42:4604–4613. doi: 10.1021/jm991031b. [DOI] [PubMed] [Google Scholar]
- 47.Bhattacharjya S., David S.A., Balaram P. Polymyxin B nonapeptide: conformations in water and in the lipopolysaccharide-bound state determined by two-dimensional NMR and molecular dynamics. Biopolymers. 1997;41:251–265. [Google Scholar]
- 48.Tristram-Nagle S., Nagle J.F. HIV-1 fusion peptide decreases bending energy and promotes curved fusion intermediates. Biophys. J. 2007;93:2048–2055. doi: 10.1529/biophysj.107.109181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan J., Tieleman D.P., Tristram-Nagle S. Alamethicin in lipid bilayers: combined use of x-ray scattering and MD simulations. Biochim. Biophys. Acta. 2009;1788:1387–1397. doi: 10.1016/j.bbamem.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O’Leary W.M., Wilkinson S.G. Microbial Lipids. In: Ratledge C., Wilkinson S.G., editors. Vol. 1. Academic Press; San Diego, CA: 1989. [Google Scholar]
- 51.O’Leary W.M., Wilkinson S.G. Microbial Lipids. In: Ratledge C., Wilkinson S.G., editors. Vol. 2. Academic Press; San Diego, CA: 1989. [Google Scholar]
- 52.Neidhardt F.H., editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Vol. 1. ASM Press; Washington, DC: 1996. [Google Scholar]
- 53.Mileykovskaya E., Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Epand R.M., Rotem S., Epand R.F. Bacterial membranes as predictors of antimicrobial potency. J. Am. Chem. Soc. 2008;130:14346–14352. doi: 10.1021/ja8062327. [DOI] [PubMed] [Google Scholar]
- 55.Seddon J.M. Structure of the inverted hexagonal (HII) phase, and non-lamellar phase transitions of lipids. Biochim. Biophys. Acta. 1990;1031:1–69. doi: 10.1016/0304-4157(90)90002-t. [DOI] [PubMed] [Google Scholar]
- 56.Bergstrom C.L., Beales P.A., Groves J.T. Cytochrome c causes pore formation in cardiolipin-containing membranes. Proc. Natl. Acad. Sci. USA. 2013;110:6269–6274. doi: 10.1073/pnas.1303819110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schröder G., Brandenburg K., Seydel U. Polymyxin B induces transient permeability fluctuations in asymmetric planar lipopolysaccharide/phospholipid bilayers. Biochemistry. 1992;31:631–638. doi: 10.1021/bi00118a001. [DOI] [PubMed] [Google Scholar]
- 58.Lopez Cascales J.J., Garro A., Enriz R.D. The dynamic action mechanism of small cationic antimicrobial peptides. Phys. Chem. Chem. Phys. 2014;16:21694–21705. doi: 10.1039/c4cp02537g. [DOI] [PubMed] [Google Scholar]
- 59.Neidhardt F.H., editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Vol. 2. ASM Press; Washington, DC: 1996. [Google Scholar]
- 60.Labischinski H., Barnickel G., Giesbrecht P. High state of order of isolated bacterial lipopolysaccharide and its possible contribution to the permeation barrier property of the outer membrane. J. Bacteriol. 1985;162:9–20. doi: 10.1128/jb.162.1.9-20.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu E.L., Engström O., Im W. Molecular dynamics and NMR spectroscopy studies of E. coli lipopolysaccharide structure and dynamics. Biophys. J. 2013;105:1444–1455. doi: 10.1016/j.bpj.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brandenburg K., Moriyon I., Seydel U. Biophysical investigations into the interaction of lipopolysaccharide with polymyxins. Thermochim. Acta. 2002;382:189–198. [Google Scholar]
- 63.Domingues M.M., Inácio R.G., Santos N.C. Biophysical characterization of polymyxin B interaction with LPS aggregates and membrane model systems. Biopolymers. 2012;98:338–344. doi: 10.1002/bip.22095. [DOI] [PubMed] [Google Scholar]
- 64.Schindler M., Osborn M.J. Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry. 1979;18:4425–4430. doi: 10.1021/bi00587a024. [DOI] [PubMed] [Google Scholar]
- 65.Berglund N.A., Piggot T.J., Khalid S. Interaction of the antimicrobial peptide polymyxin B1 with both membranes of E. coli: a molecular dynamics study. PLoS Comput. Biol. 2015;11:e1004180. doi: 10.1371/journal.pcbi.1004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brahms S., Brahms J., Brack A. Identification of β, β-turns and unordered conformations in polypeptide chains by vacuum ultraviolet circular dichroism. Proc. Natl. Acad. Sci. USA. 1977;74:3208–3212. doi: 10.1073/pnas.74.8.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reed J., Reed T.A. A set of constructed type spectra for the practical estimation of peptide secondary structure from circular dichroism. Anal. Biochem. 1997;254:36–40. doi: 10.1006/abio.1997.2355. [DOI] [PubMed] [Google Scholar]
- 68.Tristram-Nagle S., Nagle J.F. Lipid bilayers: thermodynamics, structure, fluctuations, and interactions. Chem. Phys. Lipids. 2004;127:3–14. doi: 10.1016/j.chemphyslip.2003.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kucerka N., Nagle J.F., Katsaras J. Lipid bilayer structure determined by the simultaneous analysis of neutron and x-ray scattering data. Biophys. J. 2008;95:2356–2367. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tristram-Nagle S., Liu Y., Nagle J.F. Structure of gel phase DMPC determined by x-ray diffraction. Biophys. J. 2002;83:3324–3335. doi: 10.1016/S0006-3495(02)75333-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagle J.F., Tristram-Nagle S. Structure of lipid bilayers. Biochim. Biophys. Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boscia A.L., Treece B.W., Tristram-Nagle S. X-ray structure, thermodynamics, elastic properties and MD simulations of cardiolipin/dimyristoylphosphatidylcholine mixed membranes. Chem. Phys. Lipids. 2014;178:1–10. doi: 10.1016/j.chemphyslip.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murzyn K., Pasenkiewicz-Gierula M. Structural properties of the water/membrane interface of a bilayer built of the E. coli lipid A. J. Phys. Chem. B. 2015;119:5846–5856. doi: 10.1021/jp5119629. [DOI] [PubMed] [Google Scholar]
- 74.Molecular Modeling Pro. 2017. Norgwyn Montgomery Software, Inc. (NGMSI), North Wales, PA. http://www.norgwyn.com/mmpplus.html.
- 75.Mills T.T., Tristram-Nagle S., Feigenson G.W. Liquid-liquid domains in bilayers detected by wide angle x-ray scattering. Biophys. J. 2008;95:682–690. doi: 10.1529/biophysj.107.127910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McGillivray D.J., Valincius G., Lösche M. Molecular-scale structural and functional characterization of sparsely tethered bilayer lipid membranes. Biointerphases. 2007;2:21–33. doi: 10.1116/1.2709308. [DOI] [PubMed] [Google Scholar]
- 77.Heinrich F., Lösche M. Zooming in on disordered systems: neutron reflection studies of proteins associated with fluid membranes. Biochim. Biophys. Acta. 2014;1838:2341–2349. doi: 10.1016/j.bbamem.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Snyder S., Kim D., McIntosh T.J. Lipopolysaccharide bilayer structure: effect of chemotype, core mutations, divalent cations, and temperature. Biochemistry. 1999;38:10758–10767. doi: 10.1021/bi990867d. [DOI] [PubMed] [Google Scholar]
- 79.Soares T.A., Straatsma T.P. Assessment of the convergence of molecular dynamics simulations of lipopolysaccharide membranes. Mol. Simul. 2008;34:295–307. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.