Main Text
Cells constantly remodel the extracellular matrix (ECM) of connective tissues during development, aging, and injury. This is achieved by mechanically engaging scaffolding components of the ECM, including fibronectin fibers and collagen fibrils, and by regulating diverse protein expression and activity. Proteolysis is a crucial aspect of ECM remodeling, though it is typically tested by solution-based assays that do not reproduce the native ECM environment. Many bacterial and mammalian enzymes readily cleave collagen triple helices in solution. However, collagen molecules within the ECM are densely packed within fibrils, cross-linked to each other, and exposed to mechanical stresses. Even at the molecular level, stress and the specific collagenase can qualitatively affect collagen proteolysis (1). To make reliable progress, high-throughput force-loaded single-molecule approaches are needed to obtain high-quality data for a range of proteolytic enzymes that act on collagen.
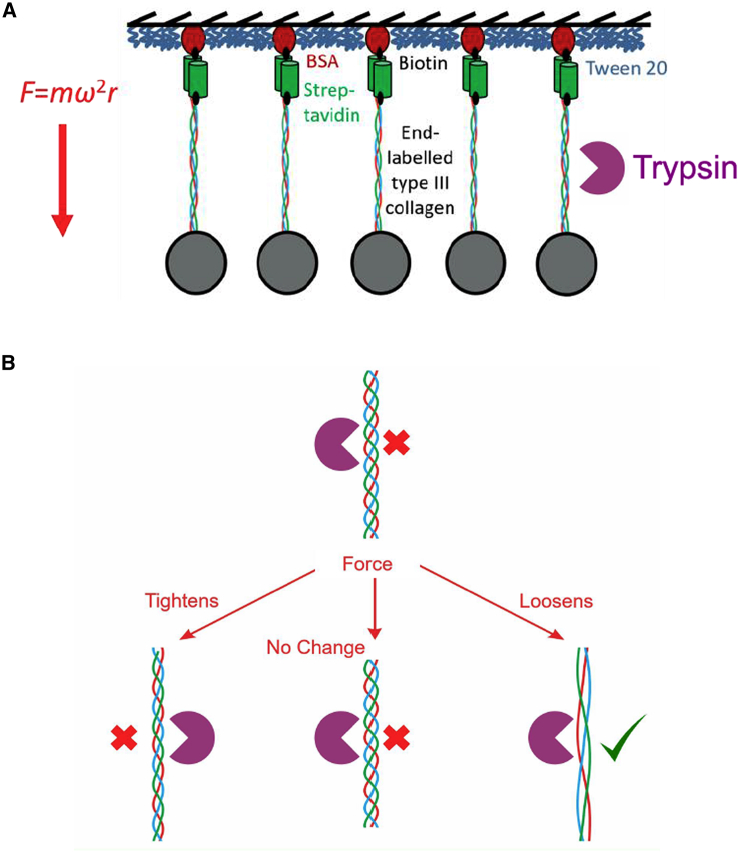
Kirkness and Forde (2) adapted centrifuge force microscopy (CFM), a technique introduced to manipulate DNA (3), to pull thousands of recombinant collagen-III triple helices in parallel (Fig. 1 A). Using this approach, they have demonstrated, for the first time, force-induced collagen cleavage by trypsin (Fig. 1 B), which is not a specialized collagenase. The applied force, 9 pN, was not enough to denature (unfold) the collagen triple helix (4), so the enhanced cleavage activity that is observed cannot be explained by the belief that trypsin cleaves only denatured collagen molecules.
Figure 1.
Centrifuge force microscopy coupled with careful surface chemistry to achieve single-molecule collagen-tethering (A) opens the road for proteolytic assays under constant force. In the case of collagen-III (B), this assay demonstrates force-induced proteolytic activity of trypsin (2). To see this figure in color, go online.
In overloaded tendons that are then relaxed the collagen fibril structure remains significantly disturbed (4, 5). Since trypsin digests the surface of these disturbed fibrils, this had been interpreted as evidence of surface denaturation (4, 6). Assuming that Kirkness and Forde’s result also applies to collagen-I, which is the main component of tendon fibrils, then trypsin may instead be cleaving collagen molecules that are held under tension by the disturbed fibril structure. If confirmed, this would fundamentally change the way the community interprets trypsin digestion data on damaged tissues.
Extension of Kirkness and Forde’s low-cost CFM-based approach to a variety of collagenases is natural and desirable. Earlier experimental studies have shown decreased or increased collagenase activity with forces in the 1–15 pN range for bacterial collagenases or matrix-metalloproteinases. For matrix-metalloproteinase 1 (7), and now for trypsin, such moderate forces on the collagen molecule seem to increase access and activity. For collagenase from the bacterium Clostridium hystolyticum a similar range of forces either inhibits or has no measurable effect on activity (7, 8). Clearly, systematic studies with multiple force levels on each of multiple enzymes are necessary and now practical.
Even further, observing the length change due to the applied force in real time would provide additional information on the state of the collagen molecule during cleavage. For this purpose, Kirkness and Forde’s compact battery-powered CFM embodiment might be improved by incorporating holographic lens-free imaging (9). This approach could track bead position rather than just bead release due to collagen proteolysis, and so could assess collagen molecular length. The combined effects of applied force and molecular length on the cleavage rate of a given collagenase should then be the complete data set needed to constrain and refine molecular dynamics simulations (4) and so finally unlock the detailed mechanism(s) of collagen cleavage under force.
Editor: Alexander Dunn.
References
- 1.Chang S.W., Buehler M.J. Molecular biomechanics of collagen molecules. Mater. Today. 2014;17:70–76. [Google Scholar]
- 2.Kirkness M.W.H., Forde N.R. Single-molecule assay for proteolytic susceptibility: force-induced collagen destabilization. Biophys. J. 2018;114:570–576. doi: 10.1016/j.bpj.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halvorsen K., Wong W.P. Massively parallel single-molecule manipulation using centrifugal force. Biophys. J. 2010;98:L53–L55. doi: 10.1016/j.bpj.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zitnay J.L., Li Y., Weiss J.A. Molecular level detection and localization of mechanical damage in collagen enabled by collagen hybridizing peptides. Nat. Commun. 2017;8:14913. doi: 10.1038/ncomms14913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veres S.P., Lee J.M. Designed to fail: a novel mode of collagen fibril disruption and its relevance to tissue toughness. Biophys. J. 2012;102:2876–2884. doi: 10.1016/j.bpj.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willett T.L., Labow R.S., Lee J.M. Increased proteolysis of collagen in an in vitro tensile overload tendon model. Ann. Biomed. Eng. 2007;35:1961–1972. doi: 10.1007/s10439-007-9375-x. [DOI] [PubMed] [Google Scholar]
- 7.Adhikari A.S., Glassey E., Dunn A.R. Conformational dynamics accompanying the proteolytic degradation of trimeric collagen I by collagenases. J. Am. Chem. Soc. 2012;134:13259–13265. doi: 10.1021/ja212170b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camp R.J., Liles M., Ruberti J.W. Molecular mechanochemistry: low force switch slows enzymatic cleavage of human type I collagen monomer. J. Am. Chem. Soc. 2011;133:4073–4078. doi: 10.1021/ja110098b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLeod E., Ozcan A. Microscopy without lenses. Phys. Today. 2017;70:51–56. [Google Scholar]