Abstract
Objective
Obesity is strongly linked to genes regulating neuronal signaling and function, implicating the central nervous system in the maintenance of body weight and energy metabolism. Genome-wide association studies identified significant associations between body mass index (BMI) and multiple loci near Cell adhesion molecule2 (CADM2), which encodes a mediator of synaptic signaling enriched in the brain. Here we sought to further understand the role of Cadm2 in the pathogenesis of hyperglycemia and weight gain.
Methods
We first analyzed Cadm2 expression in the brain of both human subjects and mouse models and subsequently characterized a loss-of-function mouse model of Cadm2 for alterations in glucose and energy homeostasis.
Results
We show that the risk variant rs13078960 associates with increased CADM2 expression in the hypothalamus of human subjects. Increased Cadm2 expression in several brain regions of Lepob/ob mice was ameliorated after leptin treatment. Deletion of Cadm2 in obese mice (Cadm2/ob) resulted in reduced adiposity, systemic glucose levels, and improved insulin sensitivity. Cadm2-deficient mice exhibited increased locomotor activity, energy expenditure rate, and core body temperature identifying Cadm2 as a potent regulator of systemic energy homeostasis.
Conclusions
Together these data illustrate that reducing Cadm2 expression can reverse several traits associated with the metabolic syndrome including obesity, insulin resistance, and impaired glucose homeostasis.
Keywords: Cadm2/SynCAM2, Energy homeostasis, Insulin sensitivity, Genome-wide association studies, Leptin signaling
Highlights
-
•
Risk variant rs13078960 associates with increased CADM2 expression.
-
•
Leptin treatment reduces hypothalamic Cadm2 expression in obese mice.
-
•
Deletion of Cadm2 protects from genetically and diet-induced obesity.
1. Introduction
The central nervous system is widely known to regulate energy expenditure and hormone sensitivity; however, the full extent to which our metabolism is managed by the brain and the key genes involved remain to a large degree unknown [1], [2]. Recent studies have begun to identify neurons in regions of the brain beyond the hypothalamus which engage circuitry of the arcuate nucleus and mediate effects on energy balance [3]. While neuron-specific disruption of the insulin and leptin receptors both significantly influence body mass and glucose homeostasis, these key signaling mediators are widely expressed throughout the brain including the hippocampus, cerebral cortex, and cerebellum, indicating that the role of many independent subpopulations in energy homeostasis remains to be described [4], [5], [6]. Moreover, it has long been established that both insulin and leptin are released systemically in proportion to body fat mass; however it is of great interest to understand the extent to which these pathways coordinately regulate the cellular networks that influence metabolism by identifying molecular determinants which contribute to both signaling cascades [7].
Cadm2 (also known as SynCAM2, Igsf4d, and Nectin-like molecule 3) has been shown to be enriched throughout the central nervous system, and forms oligomers via its extracellular domain [8]. As immunoglobulin domain-containing adhesion proteins, Cadm2 and its related family members, including Cadm1, have been established to mediate the assembly of pre-synaptic specializations in neurons in the brain to direct homo- and heterophilic interactions across the nascent and mature synaptic cleft [9]. Genome-wide association studies meta-analysis for body mass index (BMI) recently identified several susceptibility loci which associate with body mass index and map near genes known to function in the central nervous system including CADM1 and CADM2 [10], [11], [12]. We recently showed neuronal Cadm1 regulates body weight and energy homeostasis via its expression within the hippocampus and hypothalamus [13]. Here we illustrate loss of Cadm2 protects mice from obesity and hyperglycemia by regulating locomotor activity and thermogenesis further underlining the functional role of this gene family in systemic energy homeostasis via the brain.
2. Materials and methods
2.1. Animals
Mice were housed in groups of 3–5 animals and maintained on a 12-hour light/dark cycle with ad libitum access to regular chow food or high fat diet (containing 60% kcal fat, cat. no. E15741-347, ssniff Spezialdiäten GmbH), in accordance with the Landesamt für Gesundheit und Soziales (LAGeSo). All experimental procedures were approved under protocols G 0216/16, G 0357/10, G 0204/14, O 0405/09, and T 0436/08. Cadm2KO mice were generated by the trans-NIH Knock-Out Mouse Project (KOMP) and obtained from the KOMP Repository (www.komp.org). Cadm2KO mice were characterized after backcrossing for four generations to C57BL/6 and then crossed to Lepob/ob mice (Jackson Labs). Results were consistent in both genders; however, data from female mice is not shown.
2.2. Analytic procedures
2.2.1. Quantification of metabolic parameters
Blood glucose levels were obtained with the One Touch glucometer (Bayer) and plasma insulin measurements were measured by ELISA as described (Crystal Chem) [14].
2.2.2. Antibodies for western blot analysis
The following primary antibodies were used for western blotting at 1:1000 dilution: Cadm1 (MBL CM004-3), Cadm2 (Synaptic systems, 243203), UCP-1 (Cell Signaling, #14670), STAT3 (Cell Signaling, #9139), p-STAT3 (Cell Signaling, #4113), TH (Cell Signaling, #2792), β-Actin (Sigma, A1978), GAPDH (Abcam, ab8245), α-Tubulin (Sigma, T6557). Variance in the banding patterns of Cadm proteins in different panels may result from variation in acrylamide percentage or levels of protein glycosylation. Image densitometry of 16-bit TIF images for all western blots was performed using ImageJ.
2.2.3. Primary cell cultures
Primary neurons were derived from hippocampi of mice at age post-natal day 2 and dissected in cold HBSS (Invitrogen), followed by digestion with papain. After centrifugation, tissue pellet was resuspended in Neurobasal (Invitrogen) supplemented with B27 (Invitrogen) and Glutamax (Invitrogen) for plating. Hippocampal cells were cultured in 24-well dishes with 12 mm coverslips (5000 cells per coverslip) coated with poly-d-lysine and laminin and immunohistochemistry was performed as described previously [15]. After post-fixation in 4% PFA, sections were blocked with 10% BSA/PBS, then incubated with primary antibody overnight at 4 °C, followed by the fluorochrome-conjugated secondary antibodies for 1 h. Fluorescence was imaged under a confocal microscope (Zeiss LSM700). Digital images were analyzed with Fiji/ImageJ.
2.3. Mouse phenotyping
All phenotyping analysis was performed in a ‘blinded’ manner; genotypes were unknown to the investigator during the experimentation and age of animals is stated in figure legends. All genotypes were present during all experiments and randomization was implemented to the extent that all animals were identified by number prior to analysis.
2.3.1. Body temperature, composition, and energy expenditure analysis
Body temperature was measured by rectal probe thermometry at ambient room temperature. Body composition analysis was measured using Minispec Model LF90 II (6.5 mHz) (Bruker Instruments). VO2, VCO2, food intake, and locomotor activity were measured using the PhenoMaster System (TSE, Germany), and energy expenditure was analyzed as previously described using ANCOVA [16]. Animals were placed into individual cages with weight sensors quantifying ad libitum access to food. VO2 and VCO2 level were measured for 1 min in a 9 min interval for 4 consecutive days and locomotor activity was measured continuously by breaks of light beams. The first 24 h of measuring time was excluded from the analysis to allow for acclimation to the new cage environment. Measurement of energy expenditure was normalized to lean body mass as previously described [13].
2.3.2. Tolerance tests
Glucose and pyruvate tolerance tests were performed following an overnight fast (16 h) and injected intraperitoneally with either glucose (2 g/kg body weight) or pyruvate (2 g/kg body weight in saline) as described [14]. Insulin tolerance tests were performed after same day fast (6 h) by injecting insulin (Sigma) intraperitoneally (0.75 U/kg body weight). Murine leptin (Peprotech) (0.75 μg/g body weight) was injected intraperitoneally twice daily (09:00 and 19:00) for 3 days. Body weight and food intake were measured daily at 08:30.
2.4. eQTL analysis
We have downloaded currently unpublished eQTL data from the GTEx consortium analysis version 6, including results from ten distinct brain regions [17]. We specifically queried the data for association tests that were performed for the gene CADM2 against SNP rs13078960. We adjusted for multiple hypothesis testing using the method of Benjamini and Hochberg required false discovery rate (FDR) below 15% to call associations significant [18]. Boxplots were obtained through the GTEx portal.
2.5. Statistical analysis
All results are expressed as mean ± standard error (SEM), and statistical analysis is summarized in Supplementary Table 1. Comparisons between data sets with two groups were evaluated using an unpaired Student's t-test. One-way and two-way repeated-measures ANOVA analysis has been performed using GraphPad Prism Software Version 7 for comparisons of three or more groups. Post hoc statistics were performed using Sidak's multiple comparison test. A P-value of less than or equal to 0.05 was considered statistically significant. The presented data met the assumptions of the statistical tests used. Normality and equal variances were tested using GraphPad Prism software. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications [13].
3. Results
3.1. SNP associates with increased CADM2 in human subjects
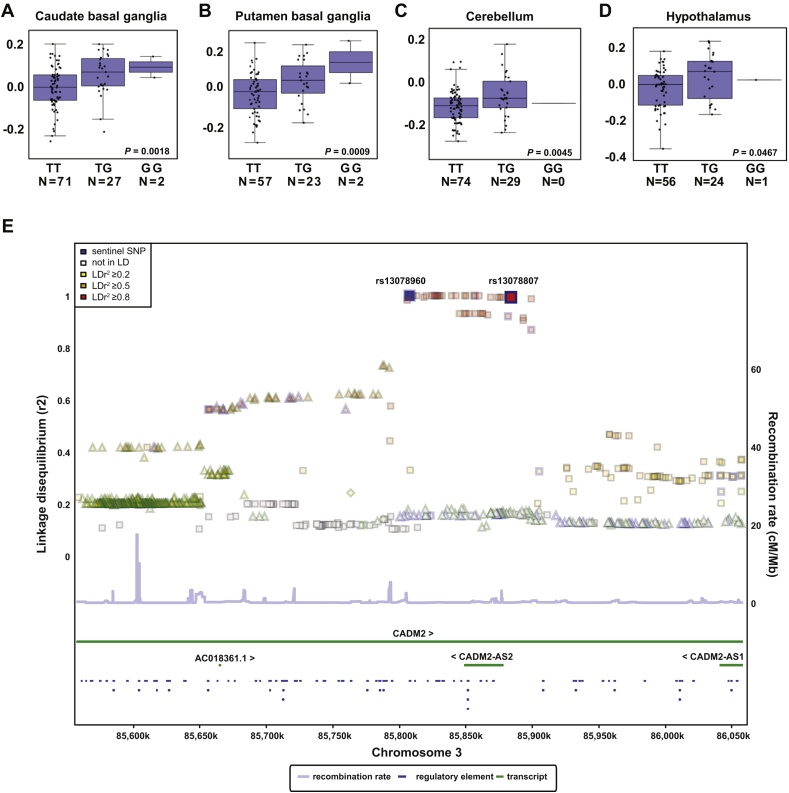
Recent genome-wide association studies (GWAS) identified a single nucleotide polymorphism (SNP) (rs13078960) in proximity to CADM2, which associates with increased body mass index (BMI) in human subjects [12]. We analyzed expression quantitative trait locus (eQTL) data from 10 distinct regions of the brain available from the GTEx consortium and observed that this risk allele (the allele associated with increased BMI) is associated with increased expression of its proximal gene in the multiple brain regions of human subjects: CADM2 (caudate basal ganglia (P = 0.0018), putamen basal ganglia (P = 0.0009), cerebellum (P = 0.0045), and hypothalamus (P = 0.0467)) (Figure 1A–D) [19]. We note that the lead SNP rs13078960 is in strong linkage disequilibrium (R2 = 0.99 in 1000 genomes) with rs13078807, which has been identified in an independent cohort previously [12] and likely tags a shared haplotype containing the antisense transcript CADM2-AS2 (Figure 1E).
Figure 1.
BMI risk SNPs associate with increased CADM2 expression in several brain regions of human subjects. Boxplots show the 25% and 75% quantiles of normalized gene expression levels (y-axis), solid horizontal lines indicate the median, and whiskers indicate the 10% and 90% quantiles. A-D, Elevated expression of CADM2 associates with risk allele (G) of rs13078960 in caudate basal ganglia, putamen basal ganglia, cerebellum, and hypothalamus, respectively. The risk allele (G) is associated with higher expression levels in human brain. E, Plot showing the linkage disequilibrium (R2) in the 1000 genomes population between the lead SNP rs13078960 (blue box) [12] on the (left) y-axis for all SNPs in a region around the lead SNP plotted against the position of the SNP (x-axis). Genes locations are shown as green horizontal lines. The strand of each gene is indicated by the arrows ‘>’ and ‘<’ next to the gene symbols. Regulatory elements annotated by the SNIPa webserver [25] are indicated as blue lines. The right y-axis indicates the recombination rate, which is shown as light blue line. Color-coding of each SNP reflects the strength of linkage disequilibrium. The SNP highlighted in blue is the lead SNP identified in [12].
3.2. Cadm2 mediates leptin sensitivity
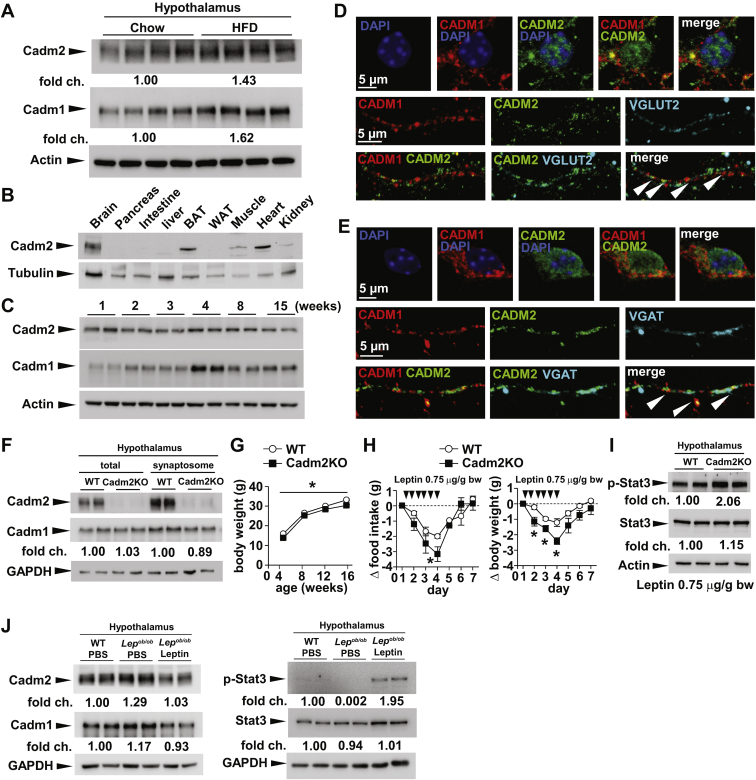
Consistent with our eQTL analysis, we observed increased Cadm2 protein levels in the hypothalamus of mice on high-fat diet for 20 weeks compared to lean chow-fed, littermate controls (Figure 2A). These results coincide with our earlier study showing increased Cadm2 expression in several brain regions of obese and insulin-resistant ob/ob mice (Lepob/ob) [13]. Together these observations further support that increased expression of Cadm2 in the human and mouse brain associates with elevated body weight and indicate that this gene plays a significant role in maintaining energy balance. Analysis by western blotting indicates that the brain is the highest site of expression, and Cadm2 levels are stable from birth through post-natal development (Figure 2B and C). Interestingly, administration of the high fat diet also incurred an increase in BAT and skeletal muscle; however it is not known in which cell type Cadm2 is present within these tissues (Supplementary Figure 1A). Within primary hippocampal neurons, we observed that Cadm2 co-localizes with its related family member Cadm1 in both glutamatergic and GABAergic neurons in vitro (Figure 2D and E).
Figure 2.
Loss of Cadm2 expression results in decreased body weight and improved leptin sensitivity. A, Western blot analysis of Cadm2 and Cadm1 in total lysates from hypothalamus of wild-type mice on normal chow diet and littermate controls on high fat diet (HFD) feeding. B, Western blot analysis of Cadm2 in brain, pancreas, intestine, liver, BAT, WAT, muscle, heart in the wild-type mice. C, Western blot analysis of Cadm2 expression in wild-type brain from 1 week to 15 weeks. D, Representative confocal images of Cadm1 and Cadm2 expression in VGLUT2-positive primary hippocampal neurons. Immunostaining for Cadm1 (red), Cadm2 (green), and VGLUT2 (cyan) identify points of co-localization in dendritic branch (white arrows). E, Representative confocal images of Cadm1 and Cadm2 expression in VGAT-positive primary hippocampal neurons. Immunostaining for Cadm1 (red), Cadm2 (green), and VGAT (cyan) identify points of co-localization in dendritic branch (white arrows). F, Western blot analysis of Cadm1 and Cadm2 in total and synaptosome-enriched lysates from hypothalamus of 12-week-old Cadm2KO mice and littermate controls. G, Body weight curves of Cadm2KO mice (n = 9) and littermate controls (n = 19). H, Quantification of food intake and body weight change in 11-week old Cadm2KO (n = 4) and littermate controls (n = 4) during leptin challenge. Daily food intake and body weight was measured for 5 days prior to leptin administration for base line. I, Western blot analysis of STAT3 phosphorylation in the hypothalamus of Cadm2KO and wild-type (WT) littermates after leptin injection (0.75 μg/g body weight). p-STAT3, phosphorylated STAT3. J, Western blot analysis of Cadm1, Cadm2, and p-STAT3 in the hypothalamus of 12-week-old wild-type mice, Lepob/ob mice after 5 days PBS or leptin injection. Results are presented as mean ± SEM. *P < 0.05.
We previously showed that global loss-of-function of Cadm2 in mice (Cadm2KO mice) resulted in decreased body weight and improved insulin sensitivity and energy expenditure rate (Figure 2F and G) [13]. Similar to global loss-of-function of Cadm1 in mice, Cadm2KO animals exhibited lower body weight and food intake in comparison to littermate control mice after intraperitoneal administration of leptin (Figure 2H and Supplementary Figure 1B). Leptin treatment also resulted in increased hypothalamic expression of phosphorylated-STAT3 in Cadm2KO mice, further indicating that total loss of Cadm2 expression resulted in increased leptin sensitivity (Figure 2I). Lastly, intraperitoneal administration of leptin to obese and insulin-resistant ob/ob mice (Lepob/ob) reversed the increased hypothalamic expression of Cadm2, suggesting expression of this protein is regulated by this hormone and impacts the downstream effects of this signaling pathway (Figure 2J and Supplementary Figure 1C). These results further underline the relevance of Cadm2 to body weight regulation, and we next evaluated the contribution of this gene to weight gain in a mouse model of obesity and hyperglycemia.
3.3. Loss of Cadm2 protects from obesity and diabetes
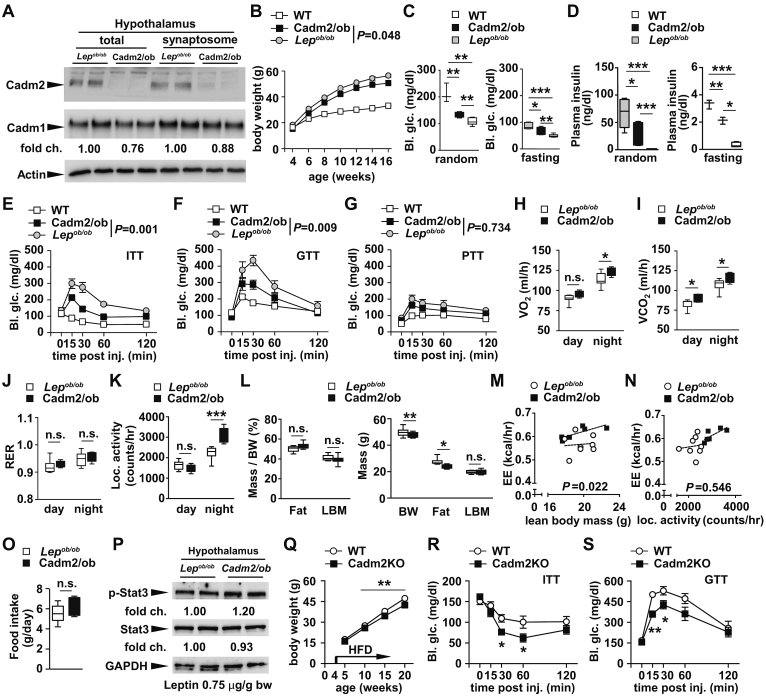
In light of the increased expression of Cadm2 in several brain regions of mouse models of both genetically and diet-induced obesity, we crossed Cadm2KO mice onto the Lepob/ob background (Cadm2/ob) and observed lower body mass, circulating random glucose, and plasma insulin levels compared to Lepob/ob littermates (Figure 3A–D). We next performed insulin, glucose, and pyruvate tolerance tests (ITT, GTT, and PTT, respectively) and observed lower glucose levels in Cadm2/ob mice in comparison to littermate controls indicating systemic insulin sensitivity was improved (Figure 3E–G). To address whether the energy expenditure rates were altered in Cadm2/ob animals, we quantified O2 consumption, CO2 production, energy expenditure, and locomotor activity, and all were higher in Cadm2-deficent mice, while nutrient partitioning (based on respiratory exchange rate (RER)) was normal (Figure 3H–K). Cadm2/ob exhibited a reduction in fat mass and evaluation of energy expenditure using analysis of covariance (ANCOVA), as plotted in relation to lean body mass, confirmed the increased expenditure rates in Cadm2-deficient animals compared to littermate Lepob/ob mice (Figure 3L and M) [16]. When energy expenditure was plotted in relation to locomotor activity, this comparison showed that these parameters are not significantly correlated suggesting several factors may contribute to the role of Cadm2 in energy homeostasis (Figure 3N). Additionally, loss of Cadm2 did not affect food intake of Lepob/ob mice compared to littermate controls (Figure 3O). We observed increased hypothalamic expression of phosphorylated-STAT3 in Cadm2/ob mice, indicating loss of Cadm2 can impact this signaling cascade even in the absence of leptin (Figure 3P). Lastly, after administration of high fat diet, Cadm2-deficient mice exhibited reduced body weight and improved insulin sensitivity and glucose tolerance in comparison to control littermates (Figure 3Q–S). Together these results indicate loss of Cadm2 protected mice from both genetically and diet-induced obesity and insulin resistance by improving energy homeostasis and insulin sensitivity independent of any change in food intake.
Figure 3.
Cadm2-deficient mice are protected from genetic obesity. A, Western blot analysis of Cadm1 and Cadm2 in total and synaptosome-enriched lysates from hypothalamus of 12-week-old Lepob/ob mice and Cadm2/ob. B, Body weight in male Cadm2/ob mice (n = 8), Lepob/ob mice (n = 13) and littermate controls (WT, n = 19) from 4 to 16 weeks of age. C, Random and fasting glucose measurements in 16-week-old Cadm2/ob mice, Lepob/ob mice and littermate controls (n = 3–7). D, Plasma insulin measurements in 16-week-old Cadm2/ob mice, Lepob/ob mice and littermate controls (n = 3–6). E, Glucose measurements during an insulin tolerance test on 12-week old Cadm2/ob mice, Lepob/ob mice and littermate controls (n = 4–6). F, Glucose measurements during a glucose tolerance test on 11-week old Cadm2/ob mice, Lepob/ob mice and littermate controls (n = 4–6). G, Glucose measurements during pyruvate tolerance test on 12-week old Cadm2/ob mice, Lepob/ob mice and littermate controls (n = 4–6). H-K, Quantification of O2 consumption, CO2 production, RER, and locomotor activity in 12-week old Cadm2/ob mice and Lepob/ob mice (n = 6–8). L, Body composition analysis of male Cadm2/ob mice and Lepob/ob mice (n = 6–9). M, Energy expenditure of individual animals plotted against lean body mass from 12-week old Cadm2/ob mice (n = 6) and Lepob/ob mice (n = 8). N, Energy expenditure of individual animals plotted against locomotor activity from 12-week old Cadm2/ob mice (n = 6) and Lepob/ob mice (n = 8). O, Quantification of daily food intake of Cadm2/ob mice (n = 6) and Lepob/ob mice (n = 8). P, Western blot analysis of STAT3 phosphorylation in the hypothalamus of Cadm2/ob mice and Lepob/ob mice after leptin injection (0.75 μg/g body weight). p-STAT3, phosphorylated STAT3. Q, Body weight in male Cadm2 mice and littermate controls (WT) from 5 to 20 weeks of age (n = 6–9). High fat diet feeding was initiated on 4 weeks old. R, Glucose measurements during an insulin tolerance test on 21-week old Cadm2 mice and littermate controls (n = 6–9). High fat diet feeding was initiated on 4 weeks-old. S, Glucose measurements during a glucose tolerance test on 22-week old Cadm2 mice and littermate controls (n = 6–9). High fat diet feeding was initiated on 4 weeks-old. Results are presented as mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001.
3.4. Cadm2 regulates thermogenesis and energy metabolism
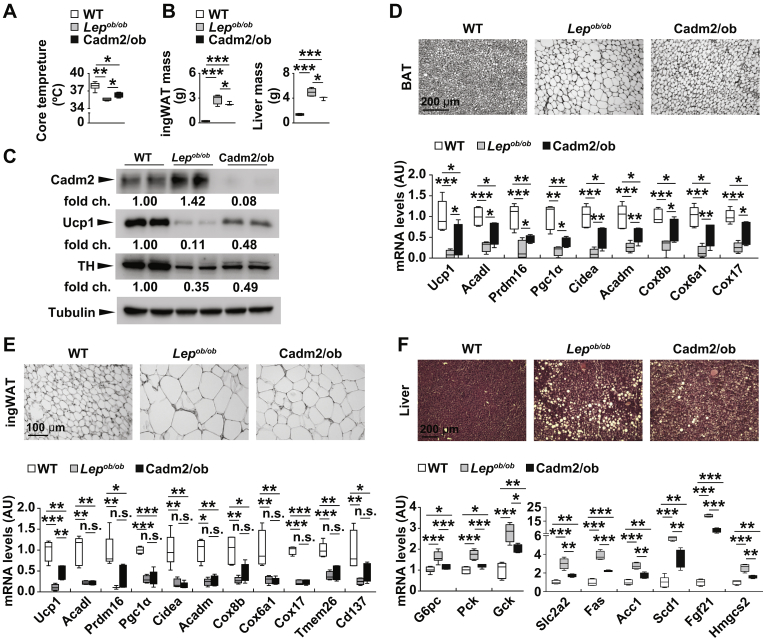
To further understand the underlying causes for the decrease in body weight, we next measured the core body temperature of Cadm2/ob mice and observed a higher mean temperature compared to littermate Lepob/ob animals (Figure 4A). Additionally, inguinal white adipose tissue (ingWAT) and liver mass were both lower in Cadm2/ob mice, indicating Cadm2 contributes body mass by regulating systemic energy metabolism (Figure 4B). Consistent with increased core body temperature, Ucp1 expression in BAT was higher in Cadm2/ob animals compared to littermate Lepob/ob mice (Figure 4C). Meanwhile, expression of tyrosine hydroxylase (TH), a marker for noradrenergic parenchymal nerve fibers, was also higher in Cadm2/ob mice compared to littermate Lepob/ob mice and correlates with both thermogenesis and an increase in activity of the sympathetic nerves of this fat depot (Figure 4C).
Figure 4.
Increased thermogenesis in Cadm2-deficient Lepob/obmice. A, Core body temperature in 12-week-old mice (n = 4–16). B, Inguinal WAT (ingWAT) and liver mass in 12-week-old mice (n = 3–5). C, Western blot analysis of interscapular BAT extracts from 12-week-old wild-type (WT), Lepob/ob mice and Cadm2/ob mice using antibodies against Cadm2, Ucp1, tyrosine hydroxylase (TH) and α-tubulin. D–F, Haematoxilin and eosin staining of interscapular BAT, ingWAT, and liver and qRT-PCR gene expression analysis from 12-week-old wild-type (WT), Lepob/ob mice and Cadm2/ob mice (n = 3–6). Results are presented as mean ± SEM. *P < 0.05, **P < 0.01 and ***P < 0.001.
Histological analysis of interscapular brown adipose tissue (BAT) from Cadm2/ob mice revealed a reduction in lipid accumulation compared to Lepob/ob littermates in line with increased energy expenditure in this tissue (Figure 4D). Consistent with these observations, gene expression analysis showed several genes enriched in BAT (including Ucp1, Acadl, Prdm16, Pgc1a, Cidea, Acadm, Cox8b, Cox6a1, and Cox17) are decreased in Lepob/ob mice compared to littermate controls. Meanwhile, the expression of these genes in the BAT of Cadm2/ob is significantly increased in comparison to Lepob/ob littermates, indicating Cadm2 contributes to the expression of these BAT-selective genes (Figure 4D). The restoration of expression of these BAT-selective genes in Cadm2/ob mice indicates loss of Cadm2 can reverse weight gain by promoting thermogenesis and energy expenditure in BAT [9], [20]. Histological analysis of WAT tissue suggests a similar reduction in lipid storage; however, expression analysis in WAT of many of these BAT-enriched genes was unaltered in Cadm2/ob in comparison to Lepob/ob littermate mice, indicating loss of Cadm2 does not impact browning in this fat depot (Figure 4E). Lastly, image analysis of liver also shows loss of Cadm2 reverses lipid accumulation in liver (Figure 4F). Gene expression analysis in liver for markers of gluconeogenesis (G6pc and Pck), glucose metabolism (Gck and Slc2a2), lipogenesis (Fas, Acc1, and Scd1), and ketogenesis (Hmgcs2 and Fgf21) showed increased expression in Lepob/ob mice compared to wild-type littermates was reversed in the livers of Cadm2/ob mice.
4. Discussion
It is now widely known that the brain is a central regulator of body weight and systemic energy metabolism; however, the precise cellular mechanisms and pathways mediating energy homeostasis remain to be fully elucidated [2]. This effort has been aided by genome-wide association studies, which continue to implicate genes enriched in the central nervous system in the susceptibility to obesity [12]. Here we illustrate the association of the risk variant rs13078960 with increased CADM2 expression in multiple brain regions of human subjects which further underlines the link between CADM genes and weight gain [13]. These observations importantly coincide with our results in mouse models of obesity showing increased hypothalamic expression of both Cadm2 and its related family member Cadm1. Interestingly, we showed that the increased expression of Cadm1 and 2 in this region is reversed by leptin treatment, suggesting that specific subpopulations within the brain may maintain a complex dynamic with their metabolic environment by altering the expression of these cell adhesion proteins. We previously showed that increased expression of Cadm1 and Cadm2 in several brain regions of Lepob/ob mice could be restored by administration of a ketogenic diet, further strengthening the notion that these genes are sensitive to changes in the physiologic state. The administration of leptin and ketogenic diet both result in reduced consumption of glucose and the lowering of body weight in Lepob/ob mice further implicating Cadm proteins in energy homeostasis.
As mediators of synapse formation and morphology, the modulatory nature of Cadm proteins in response to leptin may suggest they function to regulate neuronal activity and that their role in key circuits that impact body weight and adiposity may be transient [9]. The effects of leptin may allude to previous studies by Biederer and colleagues identifying Cadm proteins as regulators of synaptic plasticity and network excitation [21], [22]. Future studies may determine that Cadm proteins ‘fine-tune’ cell signaling according to changes in metabolic state and its expression in several brain regions ‘coordinate’ pathways that contribute to systemic energy homeostasis. In light of the effect on leptin sensitivity, the increase in the expression of Cadm proteins in the brain may also suggest that these synaptic proteins in addition to their interacting partners contribute to leptin resistance in the brain. To date, it is unclear whether the expression of Cadm proteins reflects changes in systemic glucose levels or whether their function is relevant to specific glucose-sensing neurons. It should also be noted that the impact of loss of Cadm2 on body weight was greater on the Lepob/ob background in comparison to the high fat diet challenge, further suggesting the physiologic role of Cadm2 overlaps with leptin-sensitive pathways. As leptin has been long known to impact the development of neuronal circuits within the arcuate nucleus, it is plausible that the deletion of Cadm2 or its related proteins impacts the early network organization regulating energy expenditure and behaviors affecting systemic metabolism [23].
We previously showed loss of Cadm1 impacted inhibitory post-synaptic currents of POMC neurons in addition to the number of excitatory synaptic contacts that project to this population [13]. Given this, future studies should determine whether the functional role of Cadm2 in energy homeostasis is also mediated by POMC neurons. In light of our observations showing increased expression of Cadm1 and 2 in several brain regions of human subjects, it is possible that Cadm proteins within multiple cellular networks spread across the brain act in a coordinated manner in the context of energy expenditure. Future experiments should also address the function of Cadm proteins in regions recently implicated in the regulation of energy metabolism including the cerebellum, hindbrain, brainstem, and amygdala [24]. In the absence of the identification of other neurons in addition to POMC neurons potently regulating energy expenditure, it is reasonable to hypothesize that information from multiple circuits ‘funnel’ into the arcuate nucleus and ultimately to POMC neurons to impact energy homeostasis. Likewise, it is also possible that independent circuits may function ‘in parallel’ and drive separate and specific mechanisms (i.e. thermogenesis, locomotor activity, insulin sensitivity, hepatic glucose production), contributing to energy homeostasis. Lastly, Cadm proteins are present in non-neuronal cell-types of the brain (i.e. oligodendrocytes, astrocytes) and their specific contributions within each of these subpopulations should be determined in the context of energy homeostasis. The functional role of Cadm proteins in systemic physiology has been shown to be complex and given the robust association of several risk variants with the increased expression of CADM genes in the brain of human subjects, understanding their role in diverse circuits will undoubtedly provide new insight into the pathways driving energy homeostasis in the brain.
5. Conclusions
To date, multiple SNPs in proximity to the CADM2 gene have now been shown to associate with both increased BMI and increased expression of CADM2 in several brain regions of human subjects thereby establishing its potential role in human obesity. Our observations using mouse models show reducing Cadm2 expression in Lepob/ob mice will reverse the hyperglycemia, insulin resistance, and adiposity and further underlines the relevance of studying the Cadm gene family in the context of metabolic diseases. Improving our understanding of Cadm proteins in synaptic organization and physiology within the brain may facilitate mapping of distinct cellular networks that regulate energy metabolism. In summary, these observations on Cadm2 identify it as a potent regulator of insulin and leptin sensitivity in addition to energy homeostasis and provide insight into the mechanisms underlying the progression of diabetes and obesity.
Funding
This work was funded by the Helmholtz Gemeinschaft Metabolic Dysfunction Consortium, the European Foundation for the Study of Diabetes (EFSD), and the Federal Ministry of Education and Research (BMBF, Germany) in the project eMed:symAtrial (01ZX1408D to M.H.), the European Research Council (ERC-2013-ADG-335692, to T.W.).
Availability of data and material
All primary data supporting the findings of this study are available on reasonable request.
Author's contributions
X.Y., Z.W., V.S., A.G., T.W., M.H., and M.P. contributed to the conception and design of the study, and M.P wrote the manuscript. All authors approved the final version of this manuscript.
Acknowledgments
The authors would like to thank N. Zampieri for helpful discussions and assistance in the conduct of this work.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.molmet.2017.11.010.
Conflict of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Mattson M.P. Energy intake and exercise as determinants of brain health and vulnerability to injury and disease. Cell Metabolism. 2012;16:706–722. doi: 10.1016/j.cmet.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waterson M.J., Horvath T.L. Neuronal regulation of energy homeostasis: beyond the hypothalamus and feeding. Cell Metabolism. 2015;22:962–970. doi: 10.1016/j.cmet.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 3.Kong D., Tong Q., Ye C., Koda S., Fuller P.M., Krashes M.J. GABAergic RIP-cre neurons in the arcuate nucleus selectively regulate energy expenditure. Cell. 2012;151:645–657. doi: 10.1016/j.cell.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleinridders A., Ferris H.A., Cai W., Kahn C.R. Insulin action in brain regulates systemic metabolism and brain function. Diabetes. 2014;63:2232–2243. doi: 10.2337/db14-0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Luca C., Kowalski T.J., Zhang Y., Elmquist J.K., Lee C., Kilimann M.W. Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. Journal of Clinical Investigation. 2005;115:3484–3493. doi: 10.1172/JCI24059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derakhshan F., Toth C. Insulin and the brain. Current Diabetes Reviews. 2013;9:102–116. [PubMed] [Google Scholar]
- 7.Niswender K. Early and aggressive initiation of insulin therapy for type 2 diabetes: what is the evidence? Clinical Diabetes. 2009;27:60–68. [Google Scholar]
- 8.Thomas L.A., Akins M.R., Biederer T. Expression and adhesion profiles of SynCAM molecules indicate distinct neuronal functions. Journal of Comparative Neurology. 2008;510:47–67. doi: 10.1002/cne.21773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fogel A.I., Akins M.R., Krupp A.J., Stagi M., Stein V., Biederer T. SynCAMs organize synapses through heterophilic adhesion. Journal of Neuroscience. 2007;27:12516–12530. doi: 10.1523/JNEUROSCI.2739-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nature Genetics. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nature Genetics. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rathjen T., Yan X., Kononenko N., Ku M.C., Song K., Ferrarese L. Regulation of body weight and energy homeostasis by neuronal cell adhesion molecule 1. Nature Neuroscience. 2017;20:1096–1103. doi: 10.1038/nn.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tattikota S.G., Rathjen T., McAnulty S.J., Wessels H.H., Akerman I., van de Bunt M. Argonaute2 mediates compensatory expansion of the pancreatic β cell. Cell Metabolism. 2014;19:122–134. doi: 10.1016/j.cmet.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glynn M.W., McAllister A.K. Immunocytochemistry and quantification of protein colocalization in cultured neurons. Nature Protocols. 2006;1:1287–1296. doi: 10.1038/nprot.2006.220. [DOI] [PubMed] [Google Scholar]
- 16.Tschöp M.H., Speakman J.R., Arch J.R., Auwerx J., Bruening J.C., Chan L. A guide to analysis of mouse energy metabolism. Nat Methods. 2011;9:57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S. The genotype-tissue expression (GTEx) project. Nature Genetics. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B, Statistical Methodology. 1995;57:289–300. [Google Scholar]
- 19.Ardlie K.G., Deluca D.S., Segre A.V., Sullivan T.J., Young T.R., Gelfand E.T. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duteil D., Tosic M., Lausecker F., Nenseth H.Z., Mueller J.M., Urban S. Lsd1 ablation triggers metabolic reprogramming of brown adipose tissue. Cell Reports. 2016;17:1008–1021. doi: 10.1016/j.celrep.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park K.A., Ribic A., Laage Gaupp F.M., Coman D., Huang Y., Dulla C.G. Excitatory synaptic drive and feedforward inhibition in the hippocampal CA3 circuit are regulated by SynCAM 1. Journal of Neuroscience. 2016;36:7464–7475. doi: 10.1523/JNEUROSCI.0189-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robbins E.M., Krupp A.J., Perez de Arce K., Ghosh A.K., Fogel A.I., Boucard A. SynCAM 1 adhesion dynamically regulates synapse number and impacts plasticity and learning. Neuron. 2010;68:894–906. doi: 10.1016/j.neuron.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouret S.G. Trophic action of leptin on hypothalamic neurons that regulate feeding. Science. 2004;304:108–110. doi: 10.1126/science.1095004. [DOI] [PubMed] [Google Scholar]
- 24.Grill H.J., Hayes M.R. Hindbrain neurons as an essential hub in the neuroanatomically distributed control of energy balance. Cell Metabolism. 2012;16:296–309. doi: 10.1016/j.cmet.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnold M., Raffler J., Pfeufer A., Suhre K., Kastenmueller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics (Oxford, England) 2015;31:1334–1336. doi: 10.1093/bioinformatics/btu779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.