Abstract
Molecular dynamics (MD) simulations have become ubiquitous in all areas of life sciences. The size and model complexity of MD simulations are rapidly growing along with increasing computing power and improved algorithms. This growth has led to the production of a large amount of simulation data that need to be filtered for relevant information to address specific biomedical and biochemical questions. One of the most relevant molecular properties that can be investigated by all-atom MD simulations is the time-dependent evolution of the complex noncovalent interaction networks governing such fundamental aspects as molecular recognition, binding strength, and mechanical and structural stability. Extracting, evaluating, and visualizing noncovalent interactions is a key task in the daily work of structural biologists. We have developed PyContact, an easy-to-use, highly flexible, and intuitive graphical user interface-based application, designed to provide a toolkit to investigate biomolecular interactions in MD trajectories. PyContact is designed to facilitate this task by enabling identification of relevant noncovalent interactions in a comprehensible manner. The implementation of PyContact as a standalone application enables rapid analysis and data visualization without any additional programming requirements, and also preserves full in-program customization and extension capabilities for advanced users. The statistical analysis representation is interactively combined with full mapping of the results on the molecular system through the synergistic connection between PyContact and VMD. We showcase the capabilities and scientific significance of PyContact by analyzing and visualizing in great detail the noncovalent interactions underlying the ion permeation pathway of the human P2X3 receptor. As a second application, we examine the protein-protein interaction network of the mechanically ultrastable cohesin-dockering complex.
Introduction
Since the dawn of molecular dynamics (MD) and its application to biomolecular systems over 40 years ago, the field has been rapidly growing, and the methodological toolkit has been vastly expanding (1, 2, 3, 4). This enormous growth has been largely facilitated by the ever faster computational resources, and the complexity of the molecular systems studied has been motivated by development of combined computational and experimental structural biology techniques (5). Simulations of systems with hundreds of thousands of atoms have become routine, and systems on the order of 108 atoms have been successfully simulated (3, 6, 7, 8). In addition, the simulation timescales are gradually increasing (9, 10). As a result, more complex biological systems and processes can now be studied by MD simulations, leading to much larger and denser data sets that need to be analyzed. In the context of molecular binding and recognition, noncovalent interactions play a significant role. Biomolecular recognition heavily relies on a convoluted network of nonbonded interactions such as hydrogen bonding, and ionic or hydrophobic interactions. This holds true for all biomolecular interactions, including protein-protein complexes, protein-DNA complexes, peripheral and integral membrane proteins, and many others (11, 12, 13, 14, 15, 16, 17, 18, 19). Discerning the highly complex networks of interactions is of great interest when performing all-atom MD simulations. The level of detail achievable is unparalleled by other analytic methodologies (2). However, this analysis task is often difficult, tedious, and nonintuitive. Especially protein-protein interfaces are gaining increased significance as potential drug targets, even though they are deemed highly difficult to target because a detailed understanding of noncovalent interaction networks is necessary (20, 21). These types of interactions and their analyses are so abundantly and routinely used that it is astonishing that, to the best of our knowledge, no standalone software package has been developed to address these important analytical tasks in a rapid, intuitive, and accessible manner for a broad user base.
Common biomolecular visualization packages, such as VMD (22) and PyMol (23), aim to provide general and versatile toolkits for MD trajectory analysis. However, graphical user interface (GUI) plugins for customizable and straightforward noncovalent interaction analyses are sparse to date. In addition, several packages for trajectory analysis have been developed, such as MDAnalysis (24, 25), MDTraj (26), g_contacts (27) and the GROMACS tools (28, 29), MAXIMOBY (30), PTRAJ/CPPTRAJ (31), MMTK (32), LOOS (33), and Pteros (34). However, all these tools are text-based and require programming to tailor the analysis to the specific systems under study, which is usually quite time-consuming and somewhat prohibitive for novice users. Further, none of the tools allow for real-time interactive contact analysis with a molecular graphics program.
Here, we present a standalone, GUI-based software package, namely PyContact, for noncovalent interaction analysis of MD simulation trajectories. PyContact screens MD trajectories for noncovalent interactions, also referred to as “contacts,” based on interatomic distances, geometries, and the type of involved molecules. It is intended for a broad user base ranging from novices to experts that seek to streamline the day-to-day analytic task of identifying and visualizing biomolecular contacts in a comprehensible manner. Even though the program is designed to work fully in its standalone mode, we have also fully interfaced it to the powerful molecular visualization program VMD, thus providing the users with a seamless interface between the data and the graphical representation of the molecular system.
Methods
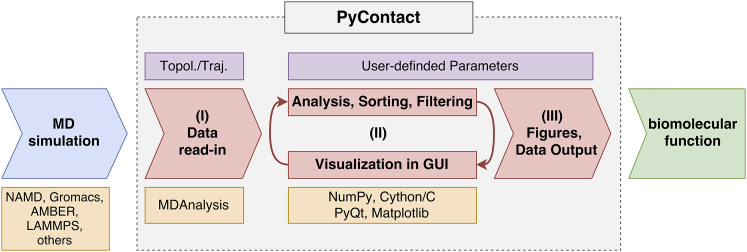
The general workflow for PyContact (Fig. 1) starts with reading in the trajectory and topology of an MD simulation produced by any common MD package, followed by data analysis and visualization of contacts through its GUI. PyContact is highly flexible and can be easily adapted to specific needs of a user through a multitude of powerful selection commands and customization of contact analysis parameters or scoring and filtering features to identify individual contributions of contacts at different levels of detail. Contacts are represented through the contact timeline in the main window (Fig. 2). Color coding is used to distinguish different types of contacts. Detailed statistical information to characterize the evolution and relevance of contacts, e.g., lifetime, contact score, number of hydrogen bonds, or solvent accessible surface areas (SASAs), are provided through histograms or contact maps. Time-dependent representation of the statistical contact analysis connected with the molecular graphics view in VMD (22) allows for interactive inspection of the contacts on the fly. The resulting contact information can be easily shared by exporting data as graphs, images, plain text files, or the current state of the complete session.
Figure 1.
PyContact workflow. In step (I), the read-in of a given MD trajectory is managed. Any file format from common simulation packages that is readable by MDAnalysis (24, 25) can be used. The MDAnalysis library also provides powerful atom selection commands. In step (II), analysis and visualization are performed dynamically, directly in the GUI. The data representation and analysis are fully customizable. In step (III), the analysis results can be directly saved as histograms, graphs, contact maps, or text files. All the steps in the PyContact main box are accessible through the GUI as well as text input via Python. User input is colored purple, used software and libraries are colored orange, and steps executed by PyContact are shown in red and output in green. To see this figure in color, go online.
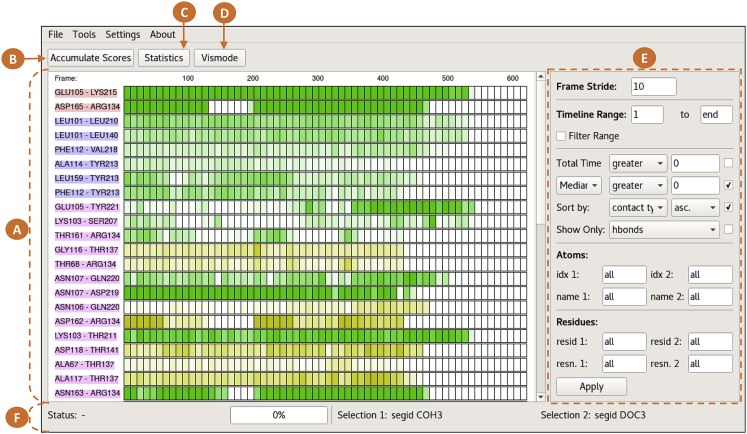
Figure 2.
PyContact main window. (A) Timeline: The first column shows the labels for contacts according to user-defined selections. The color code as determined by amino acid type and geometry is as follows: red, salt bridge; blue, hydrophobic; pink, hydrogen bonds. Additional information is displayed when clicking individual labels. Drawn boxes correspond to equally sized trajectory segments. The interaction scores are coded by color intensity and color-coded based on the nature of the interaction: green, side chain-side chain; yellow, side chain-backbone; blue, backbone-backbone (data not shown in the figure). (B) Accumulate scores: Contact scores can be accumulated to user-defined structural entities of the biomolecule. Thus, the user can switch dynamically between different levels of detail. (C) Statistics button: A panel with general information on the accumulated contacts can be displayed (total scores over time, total mean score, total hydrogen bonds, etc.). (D) Vismode button: Vismode enables clicking on a contact’s timeline and subsequently displaying this contact in molecular graphics in VMD. Clicking and dragging over boxes makes VMD show the respective trajectory frame (Fig. 3). (E) Filter and sorting panel: In the first two segments of the panel, the frame stride, displayed range, and filtered range can be manipulated. The third segment provides filtering and sorting options for the contacts based on their score and type. In the last two panels, contacts can be filtered by residue names, residue ID ranges, atom names, and atom index ranges for both selections. (F) Status and progress panel: This panel shows the application status during data processing and the progress of ongoing calculations. In addition, it displays the current atom selection texts. The depicted data was taken from the steered MD simulation described in the Case Studies section. To see this figure in color, go online.
Capabilities and implementation
PyContact is a standalone software package, allowing for out-of-the-box usage of all capabilities without any programming knowledge. It is implemented in Python (https://www.python.org, Version 2.7) and uses PyQt5, the Python bindings to the open-source user interface and application toolkit libraries Qt, for the GUI. The installation is easily managed by the Python package manager pip. PyContact is parallelized using the Python multiprocessing module, ensuring rapid analysis and quick processing of large, complex data sets (Table S1). Furthermore, NumPy is employed for fast numerical calculations (35).
PyContact reads all file formats generated by common MD simulation packages and provides a multitude of atom selection commands, enabled by the MDAnalysis library (24, 25). When loading trajectories, the user defines two atom selections, which enables precise differentiation of individual contributions at different levels of detail to a certain subsystem of interest, such as protein-protein, protein-lipid, protein-nucleic acid, or even protein-small molecule interactions (Figs. 2 and S1).
Further steps are executed on the specified trajectory. For every interatomic distance below a specified cutoff between the two selections, an interatomic contact score is calculated and recorded (Fig. S2) (12, 36, 37, 38). To tweak the analytics to individual scientific questions at hand, customization of parameters (e.g., cutoff values) can be done via the graphical interface. To identify hydrogen bonds correctly, additional geometric criteria (39) are used, i.e., interatomic distances and the angle between a putative hydrogen bond donor, the hydrogen atom, and the acceptor (Fig. S3).
A major feature is the time-dependent representation of the identified contacts within the main PyContact window (Fig. 2 A). Depending on the desired level of detail to be displayed, the user can choose atom-atom interactions or add up the interatomic contact scores to the corresponding atom names, residue names, residue IDs, or segment names (Figs. 2 B and S4). These so-called “accumulated scores” are quickly computed on the fly and can be recalculated during the analysis session (Fig. 1, II). A short demonstration of an analysis session in PyContact is provided as Movie S1.
The contacts are displayed according to their accumulated score, color-coded by intensity, highlighting their evolution (Fig. 2 A). Accordingly, a closer contact will receive a higher score and will be shown with higher color intensity. In practice, this is particularly useful for users to get a quick initial glance at their system and its evolution, and to identify potential key interaction regions. The types of interactions are color-coded to reflect the properties of the contributing amino acids or atomic geometries (Figs. 2 A and S5). Furthermore, the color coding distinguishes between side chain-side chain, backbone-backbone, and side chain-backbone contacts. Detailed information about a contact, including time averages of the contact score, lifetime, and others, can be accessed by simply clicking on its label in the timeline (Figs. 2 A and S6).
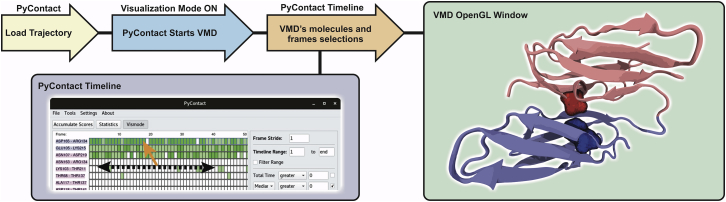
Moreover, the intuitive GUI environment provides a multitude of contact filtering and sorting capabilities, which narrows the information to the most important details (Fig. 2 E). Inspired by the VMD Timeline plugin (40), the capability to visualize selected contacts from PyContact in VMD was included for three-dimensional visual inspection of the contacts. This visualization is achieved by remote control of VMD through the PyContact user interface (Figs. 2 D and 3). The aforementioned add-on interfaces are readily accessible from the top application toolbar. A quick tour of contact analysis using the interactive PyContact and VMD connection can be found in the Movie S1. SASA, which provides a quantitative metric for the overall contact area at an interface, is commonly used to quantify the extent of noncovalent interactions of macromolecular complexes. In PyContact, the SASA is calculated with the Shrake-Rupley algorithm (41) with user-customizable parameters available (Fig. S7). To ensure high performance through fast neighbor-list generation, we adapted an algorithm for Cartesian grid search from VMD (22). The C/C++ code was wrapped by Cython (42) to make it accessible for PyContact.
Figure 3.
VMD remote control. PyContact users can load an MD trajectory into VMD through the graphical interface. Through the interactive connection between PyContact and VMD, users can click (orange mouse pointer on timeline) on a contact’s timeline to visualize the respective contact in the molecular graphics session with VMD. Swiping through the timeline (dashed arrow) makes VMD advance to the respective trajectory frame. Thus, the connection supplies users with simultaneous statistical and visual analysis in a time-dependent manner. In the displayed VMD window, a trajectory of human Filamin B domains 16 (blue) and 17 (red) is shown. As shown in (16), F1747 (blue) and V1832 (red) establish a highly correlated contact at the protein-protein interface. To see this figure in color, go online.
To represent and share the results of the contact analysis, figure generation and data export are enabled through an additional interface, built on the Matplotlib environment (Fig. 1, III; Fig. S8) (43). This interface supports saving images from the contact timeline, histograms, and contact maps for specified parameters (e.g., hydrogen bond percentages). When desired, the acquired contacts can be saved as plain text files to use the data in any other plotting software. It is also possible to share an entire analysis session by saving and reloading the current state, allowing one to easily resume an analysis session. In addition to simplification of routine noncovalent interaction analysis for beginners, advanced users are given the possiblity to automate more complex analyses through Python scripting and to add their own analysis procedures (Fig. S9). For example, high-level statistics, such as principal component analysis and clustering, could be directly performed with the scikit-learn package (44).
Results and Discussion
Case studies
PyContact has been thoroughly tested on several diverse examples, including protein-protein, protein-membrane, and protein-small molecule systems (45). To illustrate the versatile applicability and capabilities of PyContact, we performed case studies on two different systems previously investigated with MD simulations (18, 46, 47). In the first case study, we analyze the protein-lipid, protein-ion, and lipid-ion interactions involved in ion permeation through the human P2X3 receptor. MD simulations of the P2X3 ion permeation have already been published (18), however, the data have not been analyzed in detail for the aforementioned non-covalent interactions with respect to the precise permeation pathway and mechanism. In the second case, we analyze a steered MD simulation (48) of a type III X-module:Dockerin:Cohesin (XDoc-Coh) complex (46, 47).
Lipid-guided ion permeation through P2X3
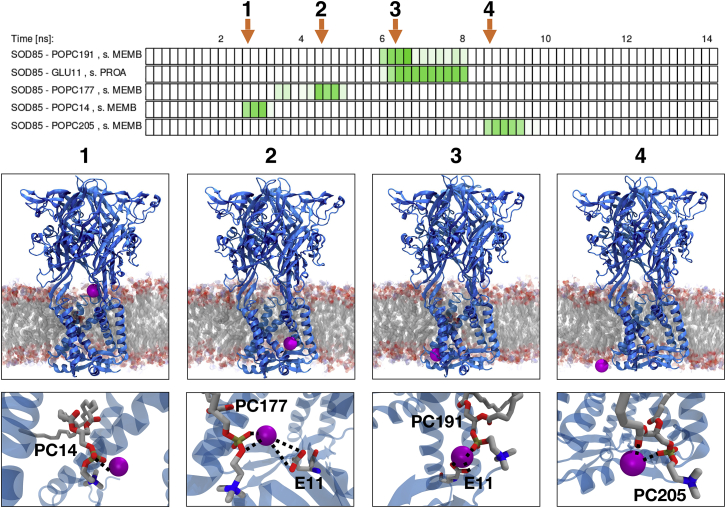
The P2X purinergic receptors are nonselective cation channels activated by extracellular nucleotides, such as ATP (49). These receptors are ubiquitously found in eukaryotes, modulating diverse processes, such as synaptic transmission, hearing, taste, and inflammation (50, 51). Recent x-ray structures of human P2X3 did not explain the complete pathway of ion permeation, but enabled MD simulations which then provided insights into lipid- and protein-driven ion permeation (18). By using PyContact, interactively connected to VMD, we further analyzed the simulation data, characterizing the permeation pathway in great detail. Intriguingly, the pathway of sodium ions inside the protein is governed mainly by precisely positioned lipids, interacting with hydrophobic patches of the protein itself. For the ion permeation simulation, P2X3 was embedded in a lipid bilayer of dioleoylphosphatidylcholine (DOPC). The individual steps of permeation and corresponding biomolecular interactions are depicted in Fig. 4. In all steps, Na+ is coordinated by a DOPC phosphate group. Lipid-ion interactions are much more abundant than protein-ion interactions; however, the protein plays a pivotal role in positioning the DOPC molecules inside the grooves: hydrophobic patches (Table S2), previously defined as key residues in gate opening (18), directly interact with the hydrophobic lipid tails. The headgroups are coordinated by polar residues. Residue T330 has been suggested to bind Na+ upon channel entering in step 1 (18). Our contact analysis, however, shows that T330 rather interacts with DOPC through hydrogen bonding. This interaction fixates DOPC positioning inside the protein such that Na+ can directly bind to the DOPC headgroup. In step 2, breakage of a salt link between choline of DOPC and E11 makes the lipid headgroup rotate such that Na+ is incorporated between phosphate and E11. The positively charged trimethylamino group points away from Na+. In the third step of permeation, Na+ exhibits a similar contact pattern to step 2. Ion exit (step 4) is guided by interaction of Na+ with a DOPC proximal to the egress pathway.
Figure 4.
Cation pathway through P2X3. The top timeline shows PyContact output of protein-ion and membrane-ion interactions. The image sequences below show the respective events by molecular graphics in VMD (22), where an overview of the position of Na+ in the protein is given in the upper image sequence, and the lipid-ion interactions guiding permation are shown in the lower sequence. In step 1, Na+ enters the protein lumen, coordinated by a DOPC (PC) phosphate group. The Na+ ion loses contact with DOPC in step 2 and migrates toward the lower part of the channel. Na+ is subsequently coordinated by E11 and the DOPC phosphate group. In step 3, Na+ moves toward a fenestration on the opposite channel side, where it establishes similar contacts as in step 2. Exit of the Na+ ion in step 4 is guided by a proximal lipid residue transporting Na+ out of the protein via interaction with the carbonyl oxygen atom. The protein chains, represented by cartoons, are colored light blue. The Na+ ion is drawn as a purple sphere. To see this figure in color, go online.
Investigation of the P2X3 trajectory with PyContact in combination with VMD revealed key protein-lipid and ion-lipid interactions, which are crucial for proper lipid positioning and underly molecular events of Na+ permeation. These interactions were immediately visible from the PyContact timeline, and the synergistic molecular graphics investigation in VMD of the respective rearrangements complemented the analysis procedure.
Ultrastable cellulosome-adhesion complex
Dockerins and cohesins are the main building blocks of cellulosomes being responsible for the cellulosomal assembly in a multimodular complexation. It was revealed with atomic force microscopy-based single-molecule force spectroscopy experiments that the type III XDoc-Coh complex can withstand forces in the remarkable range of 600–750 pN (46, 47).
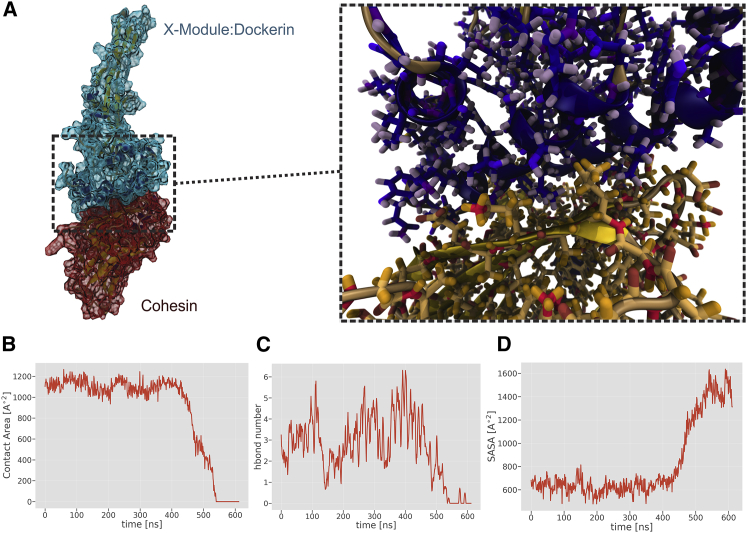
Steered MD simulations, where XDoc and Coh were pulled apart, mimicked the atomic force microscopy experiment and revealed the key amino acids stabilizing the interface of the XDoc-Coh complex (46). Remarkably, it was previously observed that contact area slightly increases during pulling simulations when compared with equilibrium simulations (46, 47). This phenomenon happens due to amino acid side-chain reorganization under mechanical load, with many of these residues establishing much larger contact areas with their counterpart in the other subunit. The XDoc-Coh complex therefore provides an excellent example whereby noncovalent interactions play a key role in the biomechanical properties. With PyContact, it was possible to analyze the trajectory and generate publication-grade figures in a short time without any scripting (Fig. 5). The time-dependent evolution of the contact surface area during the pulling simulation was analyzed with the SASA module (Fig. 5, B and D), whereas the rearrangement of the hydrogen bonding array is already directly observable in the main window timeline (Fig. 2). Using the various filtering options of PyContact, we could easily differentiate between permanent hydrogen bonds, e.g., N107-D219 and D165-R134, and hydrogen bonds that are only formed upon mechanical stress, i.e., T68-T137, K156-D219, and K103-T211, established between XDoc and Coh, respectively.
Figure 5.
Analysis of the Cohesin-Dockerin interface with PyContact. In the steered MD simulation, the XDoc-Coh complex was mechanically pulled apart. (A) Structure of the XDoc-Coh complex (left) and the interfacial residues (right). The image was created with VMD (22). (B) Time-evolution of the contact area. The contact area of Cohesin to Dockerin was calculated for every frame from within the PyContact SASA module. (C) The hydrogen bond number per frame (hbond number) was plotted from the PyContact Statistics widget. (D) The SASA of the Cohesin interface residues was also calculated directly from the SASA module, showing how the residues become solvent-accessible when the XDoc-Coh complex is broken. To see this figure in color, go online.
Conclusions
Recognition and conformational changes of biomolecules mediated by noncovalent interactions are key for cellular function and integrity. MD simulations provide a tool to extensively study such interactions at an atomic resolution. PyContact provides a tool to extract, evaluate, and visualize noncovalent interaction networks from MD simulation trajectories. Through a plethora of analysis features, PyContact enables customizability to the individual needs of a scientific question, and the graphical interface ensures access to all features and functions in an easy-to-use manner without the necessity of programming. The implementation in Python and its free availability and distribution allows scientists to adjust parameters or even expand the existing code to their needs. Particularly, job automation is easily accessible through PyContact. The time-dependent representation of the contact analysis connected with the molecular graphics view in VMD allows for interactive inspection and visualization of contacts on the fly. The obtained contact information can be easily shared by exporting data as graphs, images, pure text files, or the current state of the complete session. We showcased the capabilities of analyzing protein-protein, protein-lipid, protein-ion, and lipid-ion interactions with case studies of ion permeation through the human P2X3 receptor and the XDoc-Coh complex. In the P2X3 case, the time-resolved statistical analysis of protein-ion, protein-lipid, and lipid-ion contacts during ion permeation enables easy identification of key steps and provides a comprehensible representation of the pathway. For cohesin and dockerin, the load-dependent evolution of the interaction network within the protein-protein complex was analyzed and represented in a comprehensible way with just a few mouse clicks. In the future, we plan to implement customizable scoring functions and smooth scoring functions for hydrogen bonds. Further, we will extend the analysis features to work even more readily for protein-lipid and protein-small molecule interactions. Identifying protein-small molecule interactions will gain more importance through advances in computer-aided drug design. Therefore, the need for tools to identify these interactions will even increase in the future, and PyContact provides a basis for more features and broader applicability yet to come.
Software availability
PyContact is freely available under the GPLv3 license. The codes for Cartesian grid search and SASA calculations were derived from VMD, which is available under a GPL-compatible license (http://www.ks.uiuc.edu/Research/vmd/current/LICENSE.html). The most recent information and a tutorial are available on our project websites (http://www.ks.uiuc.edu/Research/pycontact and https://pycontact.github.io) and the development is hosted on https://github.com/maxscheurer/pycontact. The latest version is 1.0.2 and has been archived at 10.5281/zenodo.1041419.
Author Contributions
M. Scheurer and T.R. conceived the program. M. Scheurer and P.R. programmed the software and designed the graphical user interface. M. Scheurer, M. Siggel, R.C.B., E.T., and T.R. wrote and edited the manuscript. M. Scheurer performed the case studies. M. Scheurer, P.R., M. Siggel, R.C.B., K.S., E.T., and T.R. discussed PyContact features. K.S., E.T., and T.R. supervised the project.
Acknowledgments
The authors thank U. Höweler, J. Vermaas, Z. Wu, and R. Beck for helpful discussions and M. Shekhar for providing the P2X MD trajectory. Simulations also made use of the Argonne Leadership Computing Facility (ALCF)/Mira supercomputer as part of the Department of Energy (DoE) Advanced Scientific Computing Research Leadership Computing Challenge program.
The authors also acknowledge support by the state of Baden-Württemberg through bwHPC (bwForCluster MLS&WISO) and the German Research Foundation through grant INST 35/1134-1 FUGG, which enabled testing trajectories for several MD simulation systems. This work was partially supported by a grant from the National Institutes of Health (P41-GM104601). This research used resources of the Argonne Leadership Computing Facility, which is a DoE Office of Science User Facility, supported under contract DE-AC02-06CH11357. R.C.B. is partially supported by the National Science Foundation grant MCB-1616590. T.R. acknowledges support as a Feodor Lynen von Humboldt Postdoctoral Fellow.
Editor: Monika Fuxreiter.
Footnotes
Klaus Schulten is deceased.
Supporting Materials and Methods, nine figures, two tables, and one movie are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)35051-8.
Contributor Information
Emad Tajkhorshid, Email: emad@life.illinois.edu.
Till Rudack, Email: till.rudack@rub.de.
Supporting Material
The movie demonstrates the main contact analysis features of PyContact and the common workflow.
References
- 1.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 2.Dror R.O., Dirks R.M., Shaw D.E. Biomolecular simulation: a computational microscope for molecular biology. Annu. Rev. Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- 3.Perilla J.R., Goh B.C., Schulten K. Molecular dynamics simulations of large macromolecular complexes. Curr. Opin. Struct. Biol. 2015;31:64–74. doi: 10.1016/j.sbi.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ribeiro J.V., Bernardi R.C., Schulten K. QwikMD-integrative molecular dynamics toolkit for novices and experts. Sci. Rep. 2016;6:26536. doi: 10.1038/srep26536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goh B.C., Hadden J.A., Schulten K. Computational methodologies for real-space structural refinement of large macromolecular complexes. Annu. Rev. Biophys. 2016;45:253–278. doi: 10.1146/annurev-biophys-062215-011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao G., Perilla J.R., Zhang P. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature. 2013;497:643–646. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandler D.E., Strümpfer J., Schulten K. Light harvesting by lamellar chromatophores in Rhodospirillum photometricum. Biophys. J. 2014;106:2503–2510. doi: 10.1016/j.bpj.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu I., Mori T., Feig M. Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. eLife. 2016;5:e19274. doi: 10.7554/eLife.19274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindorff-Larsen K., Piana S., Shaw D.E. How fast-folding proteins fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 10.Jensen M.Ø., Jogini V., Shaw D.E. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y., Vukovi L., Schulten K. Recognition of Poly-Ubiquitins by the proteasome through protein refolding guided by electrostatic and hydrophobic interactions. J. Phys. Chem. B. 2016;33:8137–8146. doi: 10.1021/acs.jpcb.6b01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vermaas J.V., Tajkhorshid E. Conformational heterogeneity of α-synuclein in membrane. Biochim. Biophys. Acta. 2014;1838:3107–3117. doi: 10.1016/j.bbamem.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vermaas J.V., Tajkhorshid E. Differential membrane binding mechanics of synaptotagmin isoforms observed in atomic detail. Biochemistry. 2017;56:281–293. doi: 10.1021/acs.biochem.6b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Contreras F.-X., Ernst A.M., Brügger B. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature. 2012;481:525–529. doi: 10.1038/nature10742. [DOI] [PubMed] [Google Scholar]
- 15.Baylon J.L., Vermaas J.V., Tajkhorshid E. Atomic-level description of protein-lipid interactions using an accelerated membrane model. Biochim. Biophys. Acta. 2016;1858:1573–1583. doi: 10.1016/j.bbamem.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan H., Hu J.P., Chang S. Molecular dynamics simulations of DNA-free and DNA-bound TAL effectors. PLoS One. 2013;8:e76045. doi: 10.1371/journal.pone.0076045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etheve L., Martin J., Lavery R. Dynamics and recognition within a protein-DNA complex: a molecular dynamics study of the SKN-1/DNA interaction. Nucleic Acids Res. 2016;44:1440–1448. doi: 10.1093/nar/gkv1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mansoor S.E., Lü W., Gouaux E. X-ray structures define human P2X(3) receptor gating cycle and antagonist action. Nature. 2016;538:66–71. doi: 10.1038/nature19367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seppälä J., Bernardi R.C., Pentikäinen U. Skeletal dysplasia mutations effect on human filamins’ structure and mechanosensing. Sci. Rep. 2017;7:4218. doi: 10.1038/s41598-017-04441-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wells J.A., McClendon C.L. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 21.Jain V., Maingi V., Baker J. Molecular dynamics simulations of PPI dendrimer–drug complexes. Soft Matter. 2013;9:6482. [Google Scholar]
- 22.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 23.Schrödinger LLC. 2015. The PyMOL Molecular Graphics System, Version 1.8.
- 24.Michaud-Agrawal N., Denning E.J., Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gowers, R. J., M. Linke, …, O. Beckstein. 2016. MDAnalysis: a python package for the rapid analysis of molecular dynamics simulations. Proc. 15th Python Sci. Conf. 98–105.
- 26.McGibbon R.T., Beauchamp K.A., Pande V.S. MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blau C., Grubmüller H. g_contacts: fast contact search in bio-molecular ensemble data. Comput. Phys. Commun. 2013;184:2856–2859. [Google Scholar]
- 28.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abraham M.J., Murtola T., Lindah E. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 30.Höweler, U. 2007. MAXIMOBY (CHEOPS).
- 31.Roe D.R., Cheatham T.E., 3rd PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 32.Hinsen K. The molecular modeling toolkit: a new approach to molecular simulations. J. Comput. Chem. 2000;21:79–85. [Google Scholar]
- 33.Romo T.D., Grossfield A. LOOS: an extensible platform for the structural analysis of simulations. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2009;2009:2332–2335. doi: 10.1109/IEMBS.2009.5335065. [DOI] [PubMed] [Google Scholar]
- 34.Yesylevskyy S.O. Pteros: fast and easy to use open-source C++ library for molecular analysis. J. Comput. Chem. 2012;33:1632–1636. doi: 10.1002/jcc.22989. [DOI] [PubMed] [Google Scholar]
- 35.Van Der Walt S., Colbert S.C., Varoquaux G. The NumPy array: a structure for efficient numerical computation. Comput. Sci. Eng. 2011;13:22–30. [Google Scholar]
- 36.Best R.B., Hummer G., Eaton W.A. Native contacts determine protein folding mechanisms in atomistic simulations. Proc. Natl. Acad. Sci. USA. 2013;110:17874–17879. doi: 10.1073/pnas.1311599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vermaas J.V., Petridis L., Smith J.C. Mechanism of lignin inhibition of enzymatic biomass deconstruction. Biotechnol. Biofuels. 2015;8:217. doi: 10.1186/s13068-015-0379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheinerman F.B., Brooks C.L., 3rd Calculations on folding of segment B1 of streptococcal protein G. J. Mol. Biol. 1998;278:439–456. doi: 10.1006/jmbi.1998.1688. [DOI] [PubMed] [Google Scholar]
- 39.Torshin I.Y., Weber I.T., Harrison R.W. Geometric criteria of hydrogen bonds in proteins and identification of “bifurcated” hydrogen bonds. Protein Eng. 2002;15:359–363. doi: 10.1093/protein/15.5.359. [DOI] [PubMed] [Google Scholar]
- 40.Isralewitz, B., 2012. Timeline: a VMD plugin for trajectory analysis (NIH Center for Macromolecular Modeling and Bioinformatics, University of Illinois at Urbana-Champaign).
- 41.Shrake A., Rupley J.A. Environment and exposure to solvent of protein atoms. Lysozyme and insulin. J. Mol. Biol. 1973;79:351–371. doi: 10.1016/0022-2836(73)90011-9. [DOI] [PubMed] [Google Scholar]
- 42.Behnel S., Bradshaw R., Smith K. Cython: the best of both worlds. Comput. Sci. Eng. 2011;13:31–39. [Google Scholar]
- 43.Hunter J.D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- 44.Pedregosa F., Varoquaux G., Duchesnay É. Scikit-learn: machine learning in python. J. Mach. Learn. Res. 2012;12:2825–2830. [Google Scholar]
- 45.Scheurer M., Brisker-Klaiman D., Dreuw A. Molecular mechanism of flavin photoprotection by archaeal dodecin: photoinduced electron transfer and Mg2+-promoted proton transfer. J. Phys. Chem. B. 2017;121:10457–10466. doi: 10.1021/acs.jpcb.7b08597. [DOI] [PubMed] [Google Scholar]
- 46.Schoeler C., Malinowska K.H., Nash M.A. Ultrastable cellulosome-adhesion complex tightens under load. Nat. Commun. 2014;5:5635. doi: 10.1038/ncomms6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoeler C., Bernardi R.C., Gaub H.E. Mapping mechanical force propagation through biomolecular complexes. Nano Lett. 2015;15:7370–7376. doi: 10.1021/acs.nanolett.5b02727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Isralewitz B., Gao M., Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 2001;11:224–230. doi: 10.1016/s0959-440x(00)00194-9. [DOI] [PubMed] [Google Scholar]
- 49.Valera S., Hussy N., Buell G. A new class of ligand-gated ion channel defined by P2x receptor for extracellular ATP. Nature. 1994;371:516–519. doi: 10.1038/371516a0. [DOI] [PubMed] [Google Scholar]
- 50.Burnstock G., Krügel U., Illes P. Purinergic signalling: from normal behaviour to pathological brain function. Prog. Neurobiol. 2011;95:229–274. doi: 10.1016/j.pneurobio.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Surprenant A., North R.A. Signaling at purinergic P2X receptors. Annu. Rev. Physiol. 2009;71:333–359. doi: 10.1146/annurev.physiol.70.113006.100630. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The movie demonstrates the main contact analysis features of PyContact and the common workflow.