Abstract
We used time-resolved Förster resonance energy transfer, circular dichroism, and molecular dynamics simulation to investigate the structural dependence of synaptotagmin 1’s intrinsically disordered region (IDR) on phosphorylation and dielectric constant. We found that a peptide corresponding to the full-length IDR sequence, a ∼60-residue strong polyampholyte, can sample structurally collapsed states in aqueous solution, consistent with its κ-predicted behavior, where κ is a sequence-dependent parameter that is used to predict IDR compaction. In implicit solvent simulations of this same sequence, lowering the dielectric constant to more closely mimic the environment near a lipid bilayer surface promoted further sampling of collapsed structures. We then examined the structural tendencies of central region residues of the IDR in isolation. We found that the exocytosis-modulating phosphorylation of Thr112 disrupts a local disorder-to-order transition induced by trifluoroethanol/water mixtures that decrease the solution dielectric constant and stabilize helical structure. Implicit solvent simulations on these same central region residues testing the impact of dielectric constant alone converge on a similar result, showing that helical structure is formed with higher probability at a reduced dielectric. In these helical conformers, lysine-aspartic acid salt bridges contribute to stabilization of transient secondary structure. In contrast, phosphorylation results in formation of salt bridges unsuitable for helix formation. Collectively, these results suggest a model in which phosphorylation and compaction of the IDR sequence regulate structural transitions that in turn modulate neuronal exocytosis.
Introduction
Synaptotagmin 1 (Syt 1) is the calcium ion (Ca2+) sensor for neurotransmitter release, chelating Ca2+ upon action potential-induced entry and consequently triggering fast synchronous release (1). The underlying mechanisms by which Syt 1 mediates this biological event remain incompletely understood. Our goal has been to investigate the allosteric network of Syt 1 to elucidate regulatory mechanisms that underlie controlled neurotransmitter release (2, 3, 4). Our group found that an intrinsically disordered region (IDR) within Syt 1 (residues ∼80–141) exerts allosteric influence over the adjacent C2 domain referred to as “C2A” (Fig. 1 A) (4). Until recently, this IDR had received little attention and its structural biology still represents a significant gap in our understanding of Syt 1 function. Because the Syt 1 IDR exerts allosteric influence over C2A, structural or disordered ensemble transitions that occur there may be important for modulating C2 domain functions that in turn influence exocytosis (5). Additionally, the IDR was recently shown to bind to the PH domain of dynamin 1, implicating its sequence in an important protein-protein interaction of endocytosis (6).
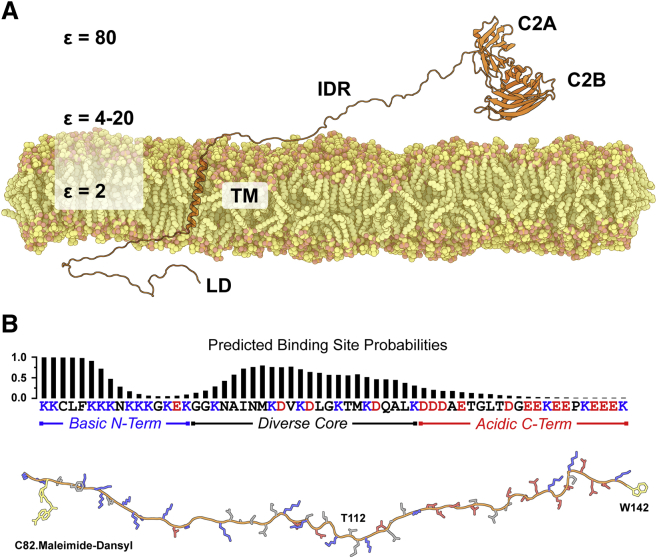
Figure 1.
Model of Syt 1 and amino acid sequence of its IDR. (A) Syt 1 consists of a short lumenal domain (LD), a single transmembrane helix (TM), a ∼60-residue IDR, and two calcium ion- and phospholipid-binding C2 domains in tandem, C2A and C2B. Approximate dielectric constants for the bilayer core (ε = 2), interfacial region (ε = 4–20), and bulk solution (ε = 80) are shown to indicate environments the polyampholytic IDR may experience. (B) The IDR sequence (residues 80–141) is shown with the basic residues in blue and the acidic residues in red. The distribution of charged residues partially delineates the sequence into three segments as indicated. Above the sequence is the ANCHOR-predicted probability for each residue potentially contributing to a binding site as described in (13, 14). The modeled peptide below the sequence shows the location for covalent attachment of acceptor dyes as well as location of the added tryptophan (yellow) used in FRET experiments. Thr112, the phosphorylation site examined in this study, is also indicated. To see this figure in color, go online.
When examining the amino acid sequence of this IDR to make initial structural predictions, it appears to be a strong polyampholyte with ∼50% of its residues consisting of an almost equal number of oppositely charged amino acids (Fig. 1 B) (7, 8). However, the distribution of charged residues divides the IDR into three main segments: a highly positive N-terminal region (∼80–96), a more sequence-diverse central region with dispersed lysine-aspartic acid repeat motifs (∼97–120), and a highly negative C-terminal region (∼121–141) (Fig. 1 B). Intrinsically disordered proteins (IDPs) that have a high degree of charge separation sample more compact structural states, and the degree of compactness can be predicted with the calculated term κ. This κ term varies from 0 (less compact) to 1 (most compact) and, in model 50-mer peptides consisting of an equal number of lysine and glutamic acid residues, correlates to radii of gyration (Rg) that range from 3.0 to 1.7 nm (9). When we performed this calculation for Syt 1’s IDR sequence, we determined a κ-value of 0.22, suggesting that the IDR can sample more compact structural states whose Rg approaches 2.3 nm (Tables S1 and S2; Supporting Materials and Methods). Further application of the sequence-dependent analysis software, CIDER (http://pappulab.wustl.edu/CIDER/), places the Syt 1 IDR in a chimeric region of the IDP phase diagram, predicting exploration of both coil and hairpin structures (Fig. S1) (10). Recently it was proposed that this IDR functions in Syt 1 as an “electrostatic zipper” that opens and closes to mediate docking of synaptic vesicles and fusion pore opening, respectively, although no distance measurement was made (11). Chimeric behavior of the IDR would be appropriate for such opening and closing and can be experimentally measured using Förster resonance energy transfer (FRET), an approach we have explored here (Fig. 1 B).
Whereas the calculated κ term for the Syt 1 IDR probably stems from the N- and C-terminal regions with high charge density, the central region has its charged residues more evenly distributed, suggestive of more local coil disorder. Within this central region there is a series of KD-repeats with the exception of a KT sequence at residue positions 111 and 112. Many of these charged residues are in i,i+4 positions, suggesting they could stabilize helical secondary structure through salt-bridge formation. Intriguingly, Thr112 is a site of phosphorylation that was recently shown to promote PKC-mediated potentiation of synaptic transmission (Fig. 1 B) (12). Addition of a phosphate at this site would complete a positive-negative repeat pattern in this central region of the IDR, impact the local electrostatics of the region, and probably alter its structural propensity. Furthermore, this central region contains a predicted binding site that is adjacent to Thr112 (Fig. 1 B), suggesting that phosphorylation of Thr112 could initiate a structural transition that modulates binding (13, 14). Such a mechanism could help explain how phosphorylation in vivo promotes synaptic plasticity. Collectively, this differential distribution of charged residues suggests that electrostatics serve more than one structural role in IDR-modulation of neuronal exocytosis.
The polyampholyte nature of this IDR is of further significance because it is part of an integral membrane protein. The lipid bilayer core and interfacial regions have low dielectric constants (ε = 2–20) compared to bulk aqueous solution (ε = 80), so the potential structural impact of charged residues in the IDR sequence is likely to be more pronounced in Syt 1’s native local environment (Fig. 1 A). In this study, we aimed to answer the above questions in an attempt to establish basic principles of this IDR’s behavior.
Materials and Methods
Reagents
Potassium chloride (KCl), sodium phosphate monobasic (NaH2PO4), and sodium phosphate dibasic (Na2HPO4) were obtained from Thermo Fisher Scientific (Waltham, MA). 2,2,2-trifluoroethanol (TFE) ≥ 99.0% was obtained from Sigma-Aldrich (St. Louis, MO). N-[2-(dansylamino)ethyl]maleimide and N-(iodoacetaminoethyl)-1-naphthylamine-5-sulfonic acid (AEDANS) acceptor labels were obtained from Sigma-Aldrich. Three peptides corresponding to the full-length Syt 1 IDR (residues 80–141) with a C-terminal tryptophan as well as the core region of the IDR with some of the acidic C-terminus (residues 97–130) in both unphosphorylated and phosphorylated form (phosphorylated at threonine 112) were obtained from Selleck Chemicals (Houston, TX) at a purity of 95% based both on mass spectrometry and HPLC. Numbering of residues assumes start codon methionine is removed. The amino acid sequences for each of the three peptides correspond to the following: (full-length IDR) KKCLFKKKNKKKGKEKGGKNAINMKDVKDLGKTMKDQALKDDDAETGLTDGEEKEEPKEEEKW; (unphosphorylated core of IDR) GKNAINMKDVKDLGKTMKDQALKDDDAETGLTDG; (phosphorylated core of IDR) GKNAINMKDVKDLGK(pT)MKDQALKDDDAETGLTDG. The core region peptides included a few residues from the acidic terminus of the IDR to help ensure adequate solubility. Despite this inclusion of C-terminal residues, these peptides are referred to as the “core region” throughout the article. In an attempt to avoid potential perturbations to helicity (15), the peptides did not include N- or C-terminal caps.
In the full-length IDR construct, the tryptophan was added both to enable UV measurement for concentration determination and to act as a donor probe in time-resolved FRET experiments. Use of tryptophan and dansyl was also chosen as a way of minimizing structural perturbation to the system with fluorescent probes, only requiring addition of one residue and exploiting the single native cysteine.
Tryptophan lifetime FRET
For the time-resolved FRET, we first reconstituted full-length IDR peptide in 10 mM NaH2PO4/Na2HPO4 buffer with the pH adjusted to 7.4. Measuring the A280 and using a calculated extinction coefficient of 5600 cm−1 M−1, the concentration of peptide was determined. A fourfold excess of maleimide-dansyl (in the case of the full-length synthetic peptide) or AEDANS (in the case of the recombinant full-length IDR) acceptor probe that had been reconstituted in dimethylformamide was then added to the peptide solution and allowed to incubate with the full-length IDR for 3 h at room temperature (∼23°C) in the presence of 1 mM TCEP reducing agent to prevent cysteine cross linking. During the reconstitution of full-length IDR, the peptide forms a dimer that is in equilibrium with its monomeric form. To isolate the acceptor-labeled monomer for FRET and simultaneously separate labeled peptide from free dye, we subjected the peptide solution to size exclusion chromatography using a prepacked S100 Sephadex column from GE Healthcare Life Sciences (Marlborough, MA). Because of the large molecular weight difference between the peptide dimer (∼14 kDa), peptide monomer (∼7 kDa), and free maleimide-dansyl probe (∼370 Da), all three species can be resolved into separate fractions. The elution fractions containing just acceptor-labeled full-length IDR peptide were identified by UV absorption, pooled, and concentrated using an Amicon Ultra-4 centrifugal filter unit from Merck Millipore (Burlington, MA) whose molecular mass cutoff was 3 kDa. The final concentration of full-length peptide and the extent of labeling with maleimide-dansyl were determined with UV absorption using the approach described by Gustiananda et al. (16). In this approach, the peptide concentration is determined by measuring the donor-acceptor peptide and subtracting the contribution from dansyl with a correction factor previously described (17):
| (1) |
| (2) |
The ratio of [IDR peptide] and [maleimide-dansyl] enables an assessment of labeling efficiency and for the full-length IDR peptide, this ratio was near unity, indicating complete labeling. This seemed reasonable considering the IDR is unfolded, allowing for the cysteine to be well exposed to free acceptor dye in bulk solution.
Tryptophan lifetimes were measured on a custom-made spectrometer from Fluorescence Innovations (Bozeman, MT) (2, 18), which uses an Nd:Yag laser for excitation of pyrromethene 597 with a 532-nm wavelength. The emission from pyrromethene passes through a doubling crystal to create a 295-nm wavelength specific for tryptophan excitation. Syt 1 IDR with a C-terminal tryptophan was synthesized (or expressed recombinantly) and subjected to lifetime measurements at 23°C in the absence and presence of dansyl (or AEDANS in the case of the recombinant IDR) acceptor probe irreversibly linked to the N-terminal cysteine residue present in the natural sequence (Fig. 1 B). Tryptophan emission from samples at concentrations of 1–3 μM was monitored at 340 nm.
Tryptophan waveforms with and without covalently attached acceptor dye were fit to multiexponential decays using a custom-made fitting program, FargoFit (19). The general description of the waveform is a sum of exponentials:
| (3) |
where ai represents the amplitude of each ith component, n represents the number of components, and τi is the fluorescence lifetime. The tryptophan waveforms were sufficiently described by two exponentials (Fig. 2 A). To determine the distance R between tryptophan and dansyl probes, amplitude-weighted average lifetimes 〈τ〉 were calculated and subsequently used to determine the efficiency of energy transfer (E), which was then used in a rearranged Förster equation with 2.1 nm for the tryptophan-dansyl R0:
| (4) |
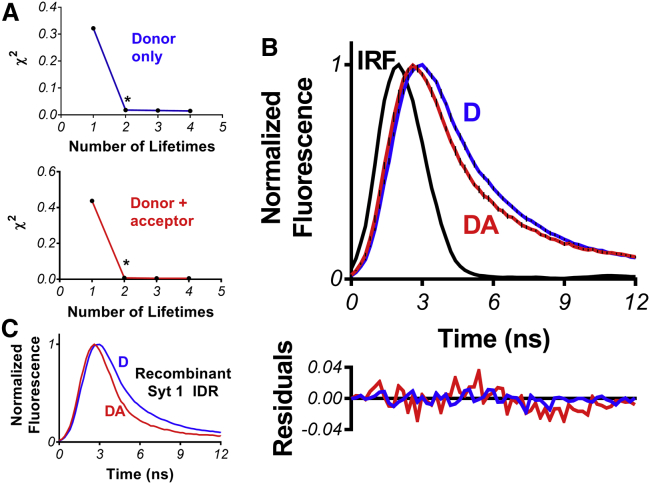
Figure 2.
Sampling of a structurally collapsed state by the Syt 1 IDR in aqueous solution. (A) Determination of the number of lifetimes needed to fit tryptophan decays without (blue) and with (red) acceptor dyes is shown. In both cases, two lifetimes were considered necessary and sufficient (asterisk). (B) Time-resolved FRET measurement of Syt 1 IDR synthesized peptide shows Trp-dansyl undergoing energy transfer in 10 mM sodium phosphate buffered to a pH of 7.4. (Blue) Donor-only. (Red) Donor-acceptor. (Error bars) SD of four replicates. (Black) Instrument response function (IRF). The distance derived from the change in average lifetime (〈τD〉 = 1.53 ± 0.11, 〈τDA〉 = 1.06 ± 0.10) is 2.40 ± 0.05 nm (Eq. 4). Shown below the time-resolved data are residuals of fitting to biexponential functions. (C) A similar FRET result was obtained from a recombinantly expressed Syt 1 IDR construct that was labeled with AEDANS acceptor. The slight increase in FRET in the recombinant IDR is thought to be due to inclusion of a his-tag, which tend to promote compaction in IDPs (20). To see this figure in color, go online.
Circular dichroism
Peptides were reconstituted in buffer containing 10 mM sodium phosphate (pH 7.4), and stock solution concentrations were determined using BCA microplates and UV absorption (for full-length peptides). TFE was added at incremental percentages v/v over a range of 0–60%. Each raw data set of absorption had a corresponding buffer scan subtracted from it to remove any absorption contributions from buffer or TFE cosolvent. Resulting data sets were plotted as mean residue ellipticity (MRE) according to the following equation:
| (5) |
where θ is raw ellipticity, MW is the molecular weight of the peptide, N is the number of residues, 1 is the cuvette path length, and c is the peptide concentration in mg/mL. Concentrations of core region peptide samples ranged from 18 to 20 μM for all experiments, whereas full-length IDR peptides were at a concentration of 10 μM. To extract the free energy of folding for Syt 1 IDR peptides, a simultaneous nonlinear least squares fit of 198- and 222-nm wavelengths was performed (mathematical derivation in Supporting Materials and Methods). Four replicate measurements of TFE gradients for each Syt 1 IDR peptide were individually fit and then averaged to determine standard deviations. Estimates of helicity were obtained by fitting data sets to linear combinations of α-helix, β-sheet, and random coil secondary structures as described previously (21, 22).
Atomistic molecular dynamics simulation of Syt 1 IDR
Peptides corresponding to both the full-length Syt 1 IDR and the core region were generated in Discovery Studio 2.0 (Accelrys, San Diego, CA). Molecular dynamics (MD) simulations were performed with the software NAMD 2.9 (http://www.ks.uiuc.edu/Research/namd/) (23) using the CHARMM36 force field (http://mackerell.umaryland.edu/charmm_ff.shtml) (24), the generalized Born implicit solvent model (25), and 2-fs time steps. Peptide termini were left charged to mimic the state of the peptides used in circular dichroism (CD) experiments. The peptide systems were maintained at constant temperature with a Langevin thermostat (298 K). Peptides corresponding to residues 80–141 in the unphosphorylated state or residues 97–129 of the Syt 1 IDR with and without phosphorylation at Thr112 were subjected to energy minimization and gradually warmed up over a period of 200 ps before equilibration. All systems subsequently achieved root mean square deviation convergence within 50 ns (Fig. S2). We performed a total of eight different simulations. For the full-length Syt 1 IDR sequence, the peptide was subjected to two different salt concentrations (10 mM and 100 mM) at two generalized Born implicit solvent dielectric constants (ε = 80 and ε = 20). For the core region residues alone, we simulated the peptide with and without phosphorylation of Thr112 at the two dielectric constant values described above, at a salt concentration of 100 mM. An ε-value of 80 was used to mimic an aqueous solution environment, whereas a value of 20 was used to mimic the upper limit of the membrane interfacial region (26, 27). The full-length IDR peptide was simulated at ε = 80 for 970 ns with either a 10 mM salt concentration or a 100 mM salt concentration. At ε = 20, the full-length peptide was simulated for 480 and 760 ns at salt concentrations of 10 and 100 mM, respectively. The unphosphorylated core region peptide was simulated with ε = 20 for 1 μs and with ε = 80 for 1.08 μs. The phosphorylated core region peptide was simulated with ε = 20 for 1.0 μs and with ε = 80 for 1.1 μs. Trajectories were analyzed using VMD 1.9.1 (28). The program STRIDE, included within VMD (http://www.ks.uiuc.edu/Research/vmd/), was used to monitor secondary structure throughout each trajectory (29). When calculating the probability of existing in different types of secondary structure (coil, β-sheet, turn, and helix), we combine 310-helicies with α-helices under the assumption that 310-helical structure is an on-path transition point between coil and α-helix (30).
Protein purification
For FRET studies on the recombinant IDR construct, a nucleotide sequence encoding residues 80–141 with a C-terminal tryptophan followed by a His-tag was cloned into a pET28a vector and subsequently transformed into BL21 Escherichia coli using a heat-shock protocol from New England BioLabs (Ipswich, MA). Transformed cells were plated onto a kanamycin-containing agar plate with an antibiotic concentration of 50 μg/mL and subsequent colonies were used to generate inoculate for large-scale bacterial growth and protein expression, induced with 1 mM IPTG. After 12 h at 25°C, cells were harvested by centrifugation. Cell pellets were resuspended in 20 mM MOPS, 200 mM KCl, 4 M urea, and 1 mM PMSF at pH 7.4 (4°C), then lysed by sonication. The resulting lysate was treated with DNase and 1 mM MgCl2 and allowed to stir at 4°C for 1 h, followed by centrifugation to remove cellular debris. The supernatant was passed through a Nalgene prefilter plus GFP sterile filter (Thermo Fisher Scientific) with a pore size of 0.2 μm to remove further particulate matter. The filtered solution was then slurried with TALON metal affinity resin (Clontech, Mountain View, CA) buffered in 20 mM MOPS, 200 mM KCl, and 4 M urea, pH 7.4. The slurry was gently stirred at 4°C for 2 h, then transferred into a column to remove unbound protein and nucleic acid. Settled resin was subsequently rinsed with four column volumes of buffer containing 20 mM MOPS, 200 mM KCl, 4 M urea, and 10 mM imidazole (to remove nonspecifically bound proteins) at pH 7.4. Resin was then rinsed with a high salt buffer (20 mM MOPS, 1 M KCl, 4 M urea, and 10 mM imidazole at pH 7.4) to separate nucleic acid bound to linker region peptide (specifically, the region containing a high number of lysine residues). The resin was then rinsed with four column volumes of buffer containing 20 mM MOPS, 200 mM KCl, 4 M urea, and 10 mM imidazole at pH 7.4. The linker region protein was then eluted with buffer containing 20 mM MOPS, 100 mM KCl, 4 M urea, and 300 mM imidazole at pH 7.4. Fractions of eluted protein were run on SDS-PAGE and pure fractions whose 260:280 ratio was <0.7 (which indicates >95% purity with respect to nucleic acid contamination) were pooled and concentrated. Syt 1 IDR was then dialyzed against 4 L of buffer containing 10 mM sodium phosphate at pH 7.4 four times, each with an equilibration time of 6 h to ensure complete exchange. The Syt IDR was then subjected to size-exclusion chromatography as described above for the Syt IDR synthesized peptide to isolate monomers.
Results
Full-length IDR peptide can sample compact structural states in aqueous solution
To first test the hypothesis that the Syt 1 IDR can sample compact structural states consistent with an electrostatic zipper, we used tryptophan (Trp)-dansyl FRET, placing the aromatic amino acid at the C-terminus of an IDR peptide and dansyl at the N-terminus using the sequence’s single native cysteine for irreversible attachment (Fig. 1 B). The Trp-dansyl probe pair has an R0 of 2.1 nm, a value suitable for detecting IDR compaction based on the κ-prediction if it occurs. We measured the Trp lifetime decay with and without acceptor dansyl and found the lifetime to be shortened by proximity of the acceptor probe. When fit to biexponential functions (Fig. 2 A), it was found that the Trp and dansyl probes were a mean distance of 2.40 ± 0.05 nm apart (Fig. 2 B), which represents the average interprobe distance but only for compact conformers detected by this dye pair.
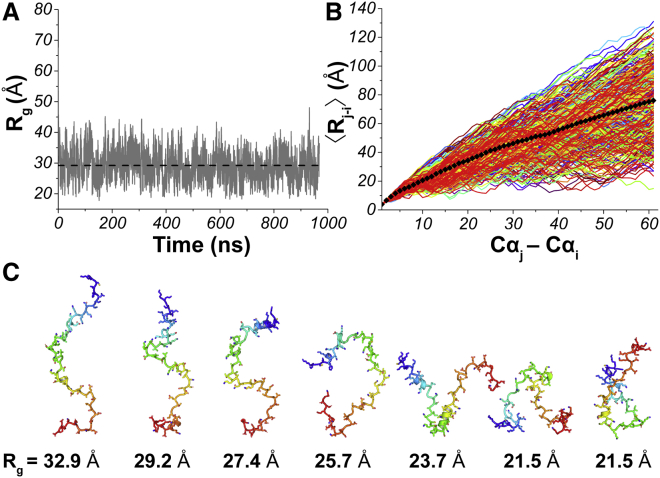
Given the ∼60-residue separation between the two probes, such a measurement indicates that the IDR peptide can sample compact structural states in aqueous solution, although this measurement cannot differentiate between hairpin and globular structures. Additionally, by using our minimally perturbing probe pair, we are not able to measure occupation of conformers adopted beyond a ∼3.4-nm interprobe distance, where FRET efficiency between Trp-Dansyl is <5%. Thus, to complement our FRET measurements characterizing the structural behavior of the IDR in an aqueous environment with low salt, we next performed MD simulations on a peptide corresponding to the full-length Syt 1 IDR sequence (residues 80–141). We used an implicit solvent model with a dielectric constant of ε = 80 and a 10 mM salt concentration. The simulated IDR structure sampled a wide range of conformers under these conditions as indicated by its fluctuating Rg during the trajectory (Fig. 3 A). Intriguingly, although FRET indicates that sampling of compact conformers does occur (Fig. 2), MD simulations suggest that compact conformers are not the dominant structural species in the overall aqueous environment ensemble (Fig. 3 B). On average, the IDR sequence samples extended coils more frequently (Fig. 3 C). Collectively, FRET and MD results correlate with the IDR’s predicted behavior based on amino acid sequence alone and is consistent with the previously proposed electrostatic zipper model in which the IDR samples both open (extended) and closed (compact) conformers (10, 11).
Figure 3.
MD simulation of the full-length Syt 1 IDR (residues 80–141) with an implicit solvent (ε = 80) and salt concentration of 10 mM. (A) Radius of gyration (Rg) as a function of simulation time indicates that the IDR sequence samples both extended and compact states. Dotted line indicates average Rg. (B) Interresidue distance (〈Rj-i〉) between first (i) and each subsequent (j) residue (Cαj-Cαi) in the Syt 1 IDR is plotted as a function of simulation time. Plots suggest weak long-range interactions promote sampling of more compact conformations. However, compact conformers are not the dominant structural species in an aqueous environment, as indicated by the location of the average interresidue distances (black diamonds). (C) Representative conformers are given of different Rg. Structures, N- to C-terminus, are color-coded blue to red. To see this figure in color, go online.
TFE-water mixtures promote partial helicity in core IDR residues
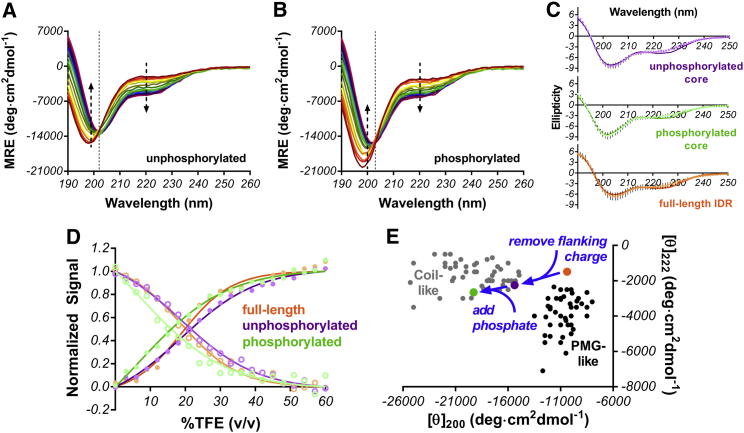
Next, to assess the structural potential of the more sequence-diverse central region, we used CD and the helix-promoting cosolvent trifluoroethanol (TFE) (Fig. 4; Fig. S3). Several of the residues in the central region, because they have hydrophobic side chains, would likely have low free energy barriers to desolvation, which often occurs upon protein-protein binding. This, coupled with the core region’s predicted binding site and phosphorylation (Fig. 1 B), make it a potential site of ordering. Moreover, TFE/water mixtures allow for creation of low dielectric environments that more closely mimic that found near a lipid bilayer surface. For these reasons, TFE is a useful perturbing agent to study structural transitions of a membrane-localized polyampholyte IDR (31). In the unphosphorylated state, increasing TFE concentration results in partial folding into helical structure (Fig. 4 A). This transition exhibits two-state behavior exemplified by the isodichroic point (vertical dotted line). When we fit the transition to a two-state model (Fig. 4 D, purple, and see Supporting Materials and Methods) we measured a folding free energy (ΔG0) of 1.0 ± 0.2 kcal/mole (Table 1).
Figure 4.
Impact of Thr112 phosphorylation on Syt 1 IDR core region (GKNAINMKDVKDLGKTMKDQALKDDDAETGLTDG). (A) MRE of unphosphorylated Syt 1 IDR core and (B) MRE of phosphorylated Syt 1 IDR core is shown as a function of increasing TFE (0–60% v/v going from red to violet) in 10 mM sodium phosphate at a pH of 7.4. (C) Fitting of absorption profiles to linear combinations of α-helix, β-sheet, and random coil in each IDR construct is shown at a maximal TFE concentration of 60% v/v. (D) Simultaneous fitting is shown of the 198-nm coil minimum (solid circles, dark line) and 222-nm (open circles, lighter line) signals during folding transition for unphosphorylated (purple), phosphorylated (green), and full-length (orange) peptides. (E) Comparison of 222:200-nm ratio from full-length (orange), unphosphorylated core (purple), and phosphorylated core (green) is shown. Removal of flanking charge shifts IDR from its near premolten globule-like state to a more coil-like state and phosphorylation accentuates this effect. This plot was created using data reported in Ref. (34). To see this figure in color, go online.
Table 1.
Fit Parameters from TFE-Induced Folding of Syt 1 IDR Constructs
| m-Value (cal/mol·TFE) | ΔG0 (cal/mol) | [θ]U (198 nm) | [θ]F (198 nm) | [θ]U (222 nm) | [θ]F (222 nm) | |
|---|---|---|---|---|---|---|
| Unphosphorylated | −55 ± 2 | 1000 ± 200 | −12 ± 1 | −4.9 ± 0.2 | −0.8 ± 0.2 | −4.3 ± 0.2 |
| Phosphorylated | −48 ± 4 | 360 ± 90 | −14.9 ± 0.9 | −7.3 ± 0.9 | −1.1 ± 0.5 | −4.2 ± 0.7 |
| Full-length IDR | −82 ± 20 | 1500 ± 300 | −9.3 ± 0.5 | −3.6 ± 0.3 | −2.1 ± 0.4 | −4.8 ± 0.2 |
To assess the impact of phosphorylation, we repeated the TFE titration with a phosphorylated peptide (phosphothreonine, pThr112) of identical sequence (Fig. 4 B). This approach of using TFE-induced folding has been successfully applied in other disordered systems to discern differences in folding propensity upon phosphorylation (32, 33). Upon inclusion of the posttranslational modification, TFE-induced folding was somewhat restricted, as evident in the absorption profile (Fig. 4 B; Fig. S3). When the structural transition was fit to a two-state model as before, a ΔG0 of 0.36 ± 0.09 kcal/mole was found (Fig. 4 D, green), indicating that phosphorylation increases the ease with which helical structure forms with increasing TFE concentration (the ΔΔG0 is equal to −0.6 ± 0.2 kcal/mole). However, when we quantified the helical content of unphosphorylated and phosphorylated peptides in the presence of maximal TFE (Fig. 4 C), the phosphorylated peptide did not reach the same level of helicity as that of the unphosphorylated peptide (Table 2), giving rise to an apparent lowering of the folding free energy. Collectively, these findings indicate that phosphorylation interferes with TFE-induced helix formation in the core region residues.
Table 2.
Estimated Helical Content of Unphosphorylated and Phosphorylated Syt 1 IDR Core Regions
| Fraction Helix (Experiment) | Fraction Total Helix (MD Simulation) | Fraction α-Helix (MD Simulation) | Fraction 310-Helix (MD Simulation) | |
|---|---|---|---|---|
| Unphosphorylated | 0.42 ± 0.01 | 0.16 | 0.12 | 0.04 |
| Phosphorylated | 0.37 ± 0.03 | 0.09 | 0.04 | 0.05 |
Given for maximum (60% v/v) TFE concentration (experiment) or lowest (ε = 20) dielectric constant (MD simulation).
When examining the CD absorption profile of IDPs, another useful parameter in assessing structure is the 222:200-nm ratio, which can help discern whether or not a particular IDP is in a more compact premolten globule state or in a disordered coil state (34, 35). When we compare the full-length Syt 1 IDR with the unphosphorylated and phosphorylated core region peptides, a trend becomes apparent (Fig. 4 E).
In the full-length IDR, the 222:200-nm ratio is consistent with its predicted chimeric behavior existing in a region of space between purely coil and purely premolten globule, which is consistent with the FRET measurement of compactness (Fig. 2) and MD simulation sampling of extended coils (Fig. 3). However, when the flanking positive and negative regions are largely removed to generate a peptide that includes core region residues, the 222:200-nm ratio is shifted more definitively in the direction of coil disorder, and phosphorylation shifts the ratio further still (Fig. 4 E). This indicates that the central region, where positive and negative charges are more evenly distributed, imparts more coil behavior and phosphorylation enhances that behavior. However, with the flanking densely positive and densely negative regions, structural states intermediate between premolten globule and coil are preferred.
This suggests that, if the central region transitions to structural ensembles containing more ordered elements, the flanking charged regions that promote sampling of compact structural states may exert regulatory control over accessibility of those core region conformers. TFE-induced folding of full-length Syt 1 IDR has a larger ΔG0 of 1.5 ± 0.3 kcal/mole compared to the unphosphorylated core peptide, which supports the hypothesis that flanking basic and acidic termini can limit core region folding (Fig. 4 D, orange; Table 1). Also consistent with this hypothesis is the observation that estimated helical content of the full-length peptide is statistically indistinguishable from that of the unphosphorylated core peptide (fraction helical in 60% v/v TFE, full-length 0.45 ± 0.02 vs. core 0.42 ± 0.01). However, because we did not measure TFE-induced folding of the flanking basic and acidic regions in isolation, we cannot conclusively say the core region residues are the only residues that become helical in the full-length IDR peptide. Regardless, mechanisms where structural compaction in IDPs limit accessibility of internal regions of sequence have been proposed in other systems (36, 37) and may still bear relevance to the Syt 1 IDR structure within the context of the whole protein embedded in a synaptic vesicle.
Implicit solvent simulations show increased helicity in unphosphorylated core residues upon reducing dielectric constant
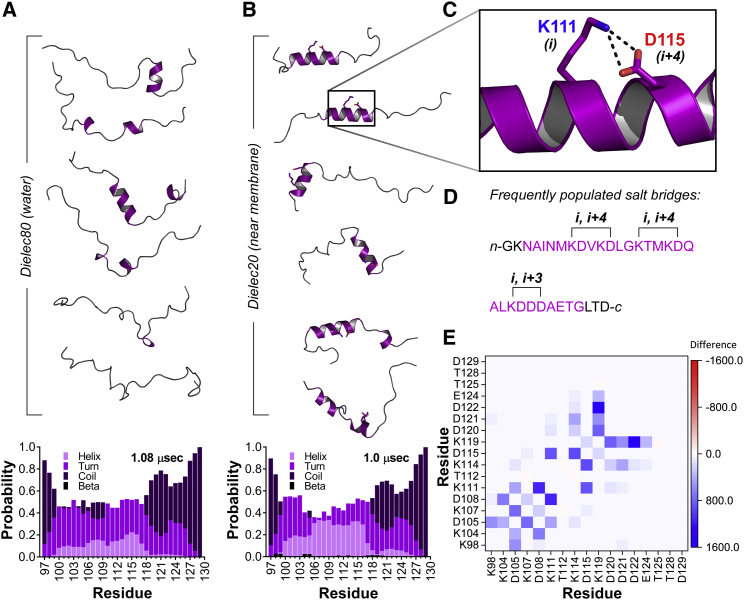
Whereas CD provides global information on Syt 1 IDR core region structure, we also sought atomistic details from MD simulation similar to our FRET experiments (23, 24). In the in vitro studies, both unphosphorylated and phosphorylated peptides formed helical structures in the lower dielectric environments of TFE-water mixtures (Fig. 4). Although TFE is known to stabilize helical structure (38, 39), helical conformers would be a reasonable form of secondary structure for this IDR to adopt. Several of its residues have high to intermediate helical propensity (40, 41). Moreover, several oppositely charged residues in the core region are positioned in i, i+4 locations, suggesting a role for salt bridges in the stabilization of helical structure. In the low dielectric environment of the interfacial region of the bilayer, there should be a higher propensity to form such salt bridges. We tested this hypothesis using an implicit solvent, first simulating the IDR core region at ε = 80 (similar to that of water) and then with ε = 20 (closer to that of the interfacial region). In both simulations, salt concentration was 100 mM to more closely mimic in vivo conditions. At ε = 80, the unphosphorylated peptide largely occupied coil or turn secondary structure (Fig. 5 A; Table S3), consistent with CD measurements of the synthetic peptide (Fig. 4). When the unphosphorylated peptide was simulated again with ε = 20, there was a large increase in the probability of helical conformers (Fig. 5 B; Table S3). When examining the 1.0 μs trajectory, the helical conformers formed were often stabilized by lysine-aspartic acid salt bridges (Fig. 5 C). In particular, K111 and D115 as well as K104 and D108 were among the longer-lived interactions within continuous helices (Fig. 5 D); however, several other electrostatic interactions were also observed (Fig. 5 E), not all of which were helix-stabilizing.
Figure 5.
Simulated structural ensemble of unphosphorylated Syt 1 IDR core region. (A) Representative conformers of the peptide at a dielectric constant of 80 with secondary structure probability per residue shown below (histogram). (B) Representative conformers of the peptide at a dielectric constant of 20 and secondary structure probability per residue shown below (histogram). Note increase in helical probability. (C) Closeup is given of example salt bridge that forms to stabilize helix in low dielectric environment. (D) IDR core region sequence shows sample salt-bridge interactions found in helices at a dielectric constant of ε = 20. Residues highlighted in purple occupy helical secondary structure. (E) Heat map shows change in all salt-bridge interactions in going from dielectric 80 to dielectric 20. An increased frequency of interaction between two charged residues is indicated by gradations of blue, whereas decreased interactions are indicated by gradations of red. Note increased frequency of i, i+3 and i, i+4 contacts. To see this figure in color, go online.
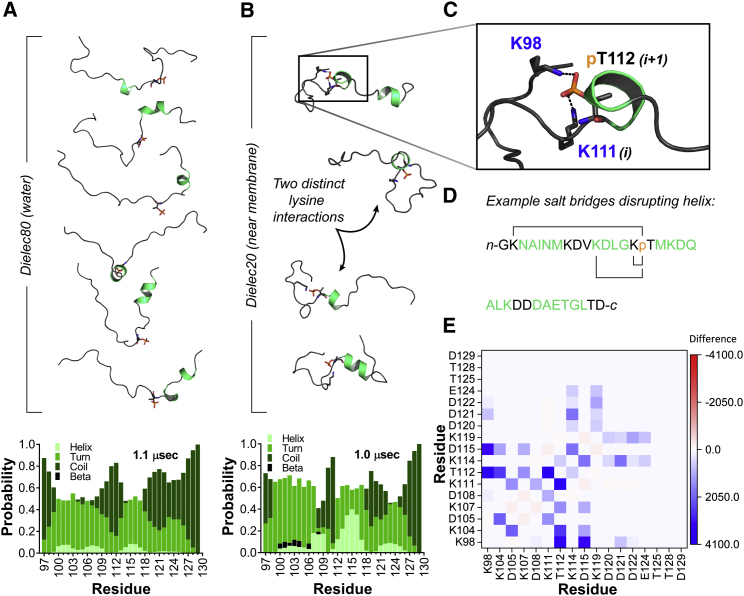
Implicit solvent simulations indicate Thr112 phosphorylation disrupts formation of helical conformers in IDR core residues
To assess the impact of phosphorylation on the structural propensity of the IDR core residues, we repeated the implicit solvent simulations on a phosphorylated peptide of identical sequence. At ε = 80 and 100 mM salt, the peptide was largely disordered and residual helical content was minimal (Fig. 6 A; Table S3). When the phosphorylated peptide was simulated at ε = 20, helical structure formed but to a lesser degree compared to the unphosphorylated peptide (Fig. 6 B; Table S3), which is consistent with CD data (Fig. 4). This difference is more pronounced when the total helical content is separated back into α- and 310-helices. The unphosphorylated peptide’s total helical content is largely dominated by α-helix (Table 2). In the phosphorylated peptide, the ratio of α- to 310-helices is closer to 50:50, with a slight preference toward the 310-helix (Table 2).
Figure 6.
Simulated structural ensemble of phosphorylated Syt 1 IDR core. (A) Representative conformers are shown of the peptide at a dielectric constant of 80. Secondary structure probability per residue is shown below (histogram). Note the reduced helicity compared to unphosphorylated peptide. (B) Representative conformers of the peptide at a dielectric constant of 20 are shown. Secondary structure probability per residue is shown below (histogram). Note that residues 107–118, which formed a continuous helix in the unphosphorylated peptide, are disrupted by coil secondary structure immediately adjacent to phosphorylation site. (C) Closeup shown of example salt bridge that distorts peptide backbone and limits helix formation in low dielectric environment. (D) IDR core region sequence shows salt-bridge interactions inhibitory toward helices at a dielectric constant of ε = 20. Green residues are those that occupy helical conformers during the trajectory. (E) Heat map shows change in salt-bridge interactions in going from dielectric 80 to dielectric 20. An increased frequency of interaction between two charged residues is indicated by gradations of blue, whereas decreased interactions are indicated by gradations of red. To see this figure in color, go online.
The region of highest helicity in the unphosphorylated peptide was significantly disrupted by phosphorylation, as indicated by a sharp rise in turn/coil secondary structures in the residues immediately adjacent to and including Thr112 (Fig. 6, A and B). Similar to the unphosphorylated peptide under ε = 20 simulation conditions, several salt bridges were prevalent during the course of the 1.0 μs trajectory. In this case, however, the salt bridges involved the phosphate and several different lysine residues, none of which were in optimal positions for stabilizing helical structure (Fig. 6, C and D). For example, K111 (i, i+1), K107 (i, i+5), K104 (i, i+8), and K98 (i, i+14) all form salt bridges with the phosphate (Fig. 6 E). In addition to these unfavorable intramolecular interactions, phosphothreonine itself is thought to have a lower helical propensity that may further contribute to reduced helix formation (42). Moreover, in other model helical peptides, phosphorylation events that are centrally localized in the helix are more destabilizing to structure (43).
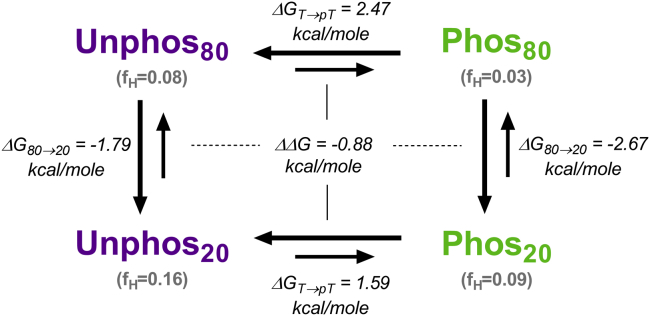
After examining unphosphorylated and phosphorylated peptides individually, we next considered them simultaneously as part of a thermodynamic cycle for the core region structural states (Fig. 7). Using the simulation-derived secondary structure probabilities, we calculated energetics for structural transitions that could then be compared to those derived from CD. Overall, the energetic trends were similar to those extracted from TFE folding experiments. Transitioning from high to low dielectric constant was energetically favorable for both unphosphorylated and phosphorylated states. However, like the CD experiments, the phosphorylated peptide had a more energetically favorable transition (ΔΔGU→F = −0.88 kcal/mole) despite having less overall helicity in each dielectric environment. In both dielectric environments, phosphorylation was energetically unfavorable for helix formation.
Figure 7.
Simulation-derived thermodynamic cycle of structural states adopted by Syt 1 IDR core region (residues 97–129). Folding free energies of each structural state (Unphos80, Phos80, Unphos20, Phos20) were determined by applying the equation ΔG = −RTln(fH/fNH), where fH is the fraction of helical conformers and fNH is the fraction of nonhelical conformers. The free energy changes associated with transitioning between each state were then calculated and are shown in the figure above. Note that for both unphosphorylated and phosphorylated peptides, lowering the dielectric constant is favorable toward helix formation. The fraction of helical content is shown parenthetically below each structural state. At a given dielectric constant, phosphorylation is generally unfavorable toward helix formation. Note that the apparent ΔΔG computed from MD structural data is of similar magnitude to that derived from CD. To see this figure in color, go online.
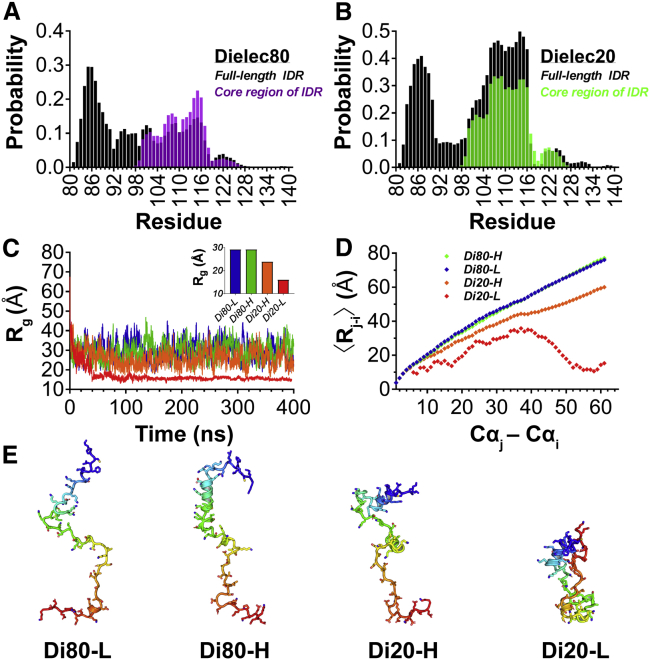
Sampling compact structural states in full-length IDR simulations can limit core region folding
After obtaining data on full-length IDR compaction and helix formation in the core region residues separately, we next considered how sampling of compact structural states may influence core region helix propensity. We simulated the full-length IDR sequence at ε = 80 and ε = 20 at high (100 mM) and low (10 mM) salt concentrations and compared their structural behaviors. As suggested by the CD TFE-folding data, inclusion of the flanking N- and C-terminal regions seem to reduce probability of core region helix formation (Fig. 8 A). This conclusion stems in part from comparison of the helix probability seen for the core region alone (Fig. 5 A) with the same core region residues within the full-length sequence. Comparison of core region and full-length peptides at ε = 20 was less supportive in this regard (Fig. 8 B). However, when full-length IDR peptides at ε = 80 and ε = 20 are examined at high and low salt concentrations, the correlation becomes more apparent. At ε = 80 and 100 mM salt, there is a decrease in the sampling of compact structural states (Fig. S4) that coincides with a modest increase in helix probability in the core region residues (Fig. S5). The decrease in sampling of compact structural states is still subtle at ε = 80 (Fig. 8, C–E), which may in part explain why we did not measure a significantly different change in FRET efficiency under increasing salt concentration (Fig. S6 A). This same salt-induced change was, however, more pronounced in simulations performed at ε = 20 (Figs. S4 and S5). In general, the IDR more readily samples compact/hairpin structural states at ε = 20, as evidenced by Rg and polymer-chain behaviors (Fig. 8, C–E).
Figure 8.
Structural impact of dielectric constant and salt on full-length Syt 1 IDR. Comparison of core region helix propensity is given in full-length (black histograms) and core region (purple or green histograms) peptides at (A) ε = 80 and (B) ε = 20 when salt concentration is 100 mM. The full-length IDR shows (C) variable Rg depending on simulation conditions where L and H represent 10 and 100 mM salt, respectively. (Inset) Shown here is the average Rg for the entire trajectory. The average inter-residue distance for all conditions is shown in (D) along with representative conformers in (E). Structures in (E), N- to C-terminus, are color-coded blue to red. To see this figure in color, go online.
Although salt influences compaction at ε = 20, another likely competitor for intramolecular interactions within the IDR is intermolecular interactions with the lipid bilayer. Previous EPR spin label accessibility measurements indicate that the basic N-terminal region can partially penetrate into a POPC/POPS lipid bilayer when reconstituted (44). This suggests that the basic N-terminus is the IDR region most likely responsible for direct lipid binding. To assess this potential contribution, we measured membrane binding of IDR peptides representing either the full-length IDR sequence or core region (residues 97–130) to large unilamellar vesicles whose lipid composition mimics that of a synaptic vesicle. Consistent with our hypothesis, only the full-length IDR peptide that includes the basic N-terminus was found to bind membrane (Fig. S6, B and C, Supporting Materials and Methods). Although the core region undergoes structural changes when the full-length IDR is associated with a synaptic vesicle mimic (4), these cosedimentation results suggest that the basic N-terminus is largely responsible for lipid binding. Because of this binding and the high local concentration of acidic phospholipids normally experienced by Syt 1 in the bilayer, lipid binding may be able to compete with intramolecular association of N- and C-terminal regions. Collectively, the simulations of the full-length IDR and binding results point to the possibility of a structural interplay between IDR compaction and occupation of transient helical conformers in the core region residues.
Discussion
In this study, we probed the structural propensity of Syt 1’s IDR, examining the dependence on dielectric constant and phosphorylation. These are important goals because, in the former case, the type of intrinsic disorder exploited by Syt 1 (strong polyampholyte) is naturally subjected to a low dielectric environment by virtue of being embedded in a lipid bilayer. This makes the impact of electrostatics more pronounced, enhancing any effect charged residues exert on local protein structure. In the latter case, phosphorylation at Thr112 has been shown to potentiate synaptic transmission in hippocampal neurons (12), but a molecular rationale for this cellular change is lacking.
With regard to dielectric constant, we found both through experiment (Fig. 4) and simulation (Figs. 5 and 6) that reduced dielectric constant promotes helix formation in core region residues. These results are of biological interest because we previously reported that the IDR remains largely disordered when associated with a lipid bilayer (4). Although we could not rule out the possibility of the IDR peptide undergoing oligomerization on the membrane surface, which could alter structural propensity, another possibility for lack of structure is lipid-induced unfolding. The bilayer provides a constant low dielectric environment that should promote folding in the core region of the IDR, but other chemical factors undoubtedly still influence secondary structure. The vesicles we used previously were composed of nine lipid species in specific mole fractions designed to mimic the outer leaflet of the native organelle. IDR interaction with this cocktail of lipid headgroups may favor a more disordered state as part of a regulatory mechanism. Many IDPs and IDRs that interact with membranes are sensitive to lipid composition and have their secondary structure altered by different lipid mixtures (45). In the single-pass integral membrane protein phospholamban, for example, this type of lipid modulation of structure has been directly linked to altered functionality. In response to cationic phospholipids, phospholamban’s cytoplasmic domain becomes more disordered and this structural shift consequently decreases the protein’s inhibitory potency toward the sarcoplasmic reticulum Ca2+-ATPase that it normally regulates (46).
Another possible explanation for the full-length IDR being more disordered when associated with a membrane could relate to an increased tendency to sample compact structural states (Fig. 8). Increased population of compact states due to low dielectric constant was inversely correlated with core region helix formation. This suggests that, in addition to possible lipid-regulation of folding, there may also be suppression of structure from intramolecular interactions between basic and acidic termini.
Another finding of biological interest in this study is the site of apparent helix formation (Figs. 5 and 6). The position where helical structure forms is intriguing because it correlates with a disorder-based predicted binding site (Fig. 1). In a study by de Jong et al. (12), residues 109–116 were identified as being inhibitory toward synaptic release when unphosphorylated, and permissive when phosphorylated. Our results suggest a potential structural model for this altered function. In the absence of phosphorylation, these residues (which form part of a predicted binding site) have a tendency to form an α-helix that may be necessary for association with an as-yet unidentified partner. When phosphorylated, however, there is a local disruption of helix formation that may then limit binding. This mode of regulation for binding interactions is common. There are examples in other IDP systems where single phosphorylation events similarly break helices (47) or, in extreme cases, sequester a partially helical binding site into a different form of secondary structure (48). In some instances, phosphorylation does not have a significant impact on secondary structure, but rather shifts local and global disordered ensembles to regulate interactions (36). And in other cytoplasmic regions of integral membrane proteins, phosphorylation can rearrange salt-bridge interaction networks to switch local structure (33). Alternatively, a core region helix may form part of an interaction surface that contributes to Syt oligomerization (44), in which case helix disruption could alter the oligomeric state. These possibilities are also worth considering in the context of other synaptotagmin isoforms. In synaptotagmin 7, for instance, phosphorylation of Ser103 (a residue in its putative IDR) also potentiates exocytosis (in this case, of insulin (49)). Such a similarity suggests that findings on Syt 1’s IDR may help inform future study of other synaptotagmin IDR isoforms.
The structural differences between unphosphorylated and phosphorylated core region residues in Syt 1 also provide potential insight into a recent report of the IDR interacting with the PH domain of dynamin 1, a GTPase known for facilitating fission of vesicles that are being endocytosed (6). That study showed that a T112E missense mutation in the IDR abolishes the binding interaction, suggesting that phosphorylation is a regulatory mechanism for the interaction. However, that study also showed that deletion of residues 117–119 (ALK) or 111–119 (KTMKDQALK) did not disrupt binding of the IDR to the PH domain. Given that the KTMKDQALK residues are among those that adopt helical conformers in the absence of phosphorylation, the binding interaction with dynamin’s PH domain may not involve a specific structured helix in the Syt 1 IDR, but instead rely on a mechanism involving local electrostatic interactions.
On a more global level, the full-length IDR seems to sample collapsed structures (Figs. 2 and 8), consistent with both computational predictions (9, 10) and effects of site-directed mutagenesis on the basic and acidic regions of the IDR in vesicle-vesicle docking/fusion assays (11). The other two sites of phosphorylation within the IDR (Thr125 and Thr128) are located in the acidic C-terminal region, suggesting their placement could enhance compaction (50, 51). This may be particularly relevant in a low dielectric environment like that near the membrane surface, which already enhances occupation of compact conformations (Fig. 8).
When comparing the CD and MD estimates of helical structure in lowest dielectric environments, there are differences in the absolute fractions determined by each method. Although CD and MD do not quantitatively converge, they both indicate that phosphorylation is inhibitory toward helix formation. The lack of quantitative agreement is not surprising; the MD simulations consider only the change in dielectric constant, ignoring other helix-stabilizing properties of TFE that involve alterations of peptide-water interactions, which would likely require explicit treatment of both TFE and water (52, 53, 54).
Conclusions
We examined the structural impact of dielectric constant and phosphorylation on Syt 1’s IDR. The general sequestration of positive and negative residues at the N- and C-termini promotes sampling of collapsed structural states in aqueous solution (Fig. 2), although compact states are more minor compared to extended coil states (Fig. 3). The more sequence-diverse core region of the IDR, where charge is more uniformly distributed, can form short helices when subjected to TFE (Fig. 4) or a simulated low dielectric environment (Fig. 5). Phosphorylation at Thr112, however, disrupts this helical tendency both in TFE folding experiments and in implicit solvent simulations (Figs. 4 and 6). In addition to phosphorylation alteration of folding (Fig. 7), the helical propensity of these same core region residues may also be limited in the full-length sequence as compact structural states are sampled (Fig. 8). Collectively, these results argue that the Syt 1 IDR is influenced by the low dielectric environment near the membrane surface, and that altered charge distribution through posttranslational modification can exert local alterations to its structural propensity.
Author Contributions
M.E.F. performed FRET and CD experiments. M.E.F. and B.P.B. performed MD simulations. M.E.F. and D.D.T. analyzed FRET data. M.E.F., B.P.B., and D.D.T. analyzed MD simulations. M.E.F., V.N.U., and A.H. analyzed CD data. M.E.F., A.H., and D.D.T. wrote the manuscript. All authors approved the final version of the manuscript.
Acknowledgments
The authors acknowledge the Minnesota Supercomputing Institute (MSI) at the University of Minnesota for providing resources that contributed to the research results reported within this article. The authors acknowledge Octavian Cornea for his technical assistance during manuscript preparations.
This material is based upon work supported by the National Science Foundation (NSF) (MCB-1616854) to D.D.T. and A.H. as well as Chancellor’s Small Grants (University of Minnesota – Duluth) to A.H. M.E.F. was supported by the National Institutes of Health (NIH) (T32 AR007612) to D.D.T.
Editor: David Eliezer.
Footnotes
Supporting Materials and Methods, six figures, and three tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)35093-2.
Supporting Citations
Supporting Material
References
- 1.Südhof T.C. A molecular machine for neurotransmitter release: synaptotagmin and beyond. Nat. Med. 2013;19:1227–1231. doi: 10.1038/nm.3338. [DOI] [PubMed] [Google Scholar]
- 2.Fealey M.E., Gauer J.W., Hinderliter A. Negative coupling as a mechanism for signal propagation between C2 domains of synaptotagmin I. PLoS One. 2012;7:e46748. doi: 10.1371/journal.pone.0046748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fealey M.E., Hinderliter A. Allostery and instability in the functional plasticity of synaptotagmin I. Commun. Integr. Biol. 2013;6:e22830. doi: 10.4161/cib.22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fealey M.E., Mahling R., Hinderliter A. Synaptotagmin I’s intrinsically disordered region interacts with synaptic vesicle lipids and exerts allosteric control over C2A. Biochemistry. 2016;55:2914–2926. doi: 10.1021/acs.biochem.6b00085. [DOI] [PubMed] [Google Scholar]
- 5.Hilser V.J., Thompson E.B. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc. Natl. Acad. Sci. USA. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McAdam R.L., Varga K.T., Sossin W.S. The juxtamembrane region of synaptotagmin 1 interacts with dynamin 1 and regulates vesicle fission during compensatory endocytosis in endocrine cells. J. Cell Sci. 2015;128:2229–2235. doi: 10.1242/jcs.161505. [DOI] [PubMed] [Google Scholar]
- 7.Dobrynin A.V., Colby R.H., Rubinstein M. Polyampholytes. J. Polym. Sci. Pol. Phys. 2004;42:3513–3538. [Google Scholar]
- 8.Higgs P.G., Joanny J.F. Theory of polyampholyte solutions. J. Chem. Phys. 1991;94:1543–1554. [Google Scholar]
- 9.Das R.K., Pappu R.V. Conformations of intrinsically disordered proteins are influenced by linear sequence distributions of oppositely charged residues. Proc. Natl. Acad. Sci. USA. 2013;110:13392–13397. doi: 10.1073/pnas.1304749110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holehouse A.S., Das R.K., Pappu R.V. CIDER: resources to analyze sequence-ensemble relationships of intrinsically disordered proteins. Biophys. J. 2017;112:16–21. doi: 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai Y., Lou X., Shin Y.K. The synaptotagmin 1 linker may function as an electrostatic zipper that opens for docking but closes for fusion pore opening. Biochem. J. 2013;456:25–33. doi: 10.1042/BJ20130949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jong A.P., Meijer M., Verhage M. Phosphorylation of synaptotagmin-1 controls a post-priming step in PKC-dependent presynaptic plasticity. Proc. Natl. Acad. Sci. USA. 2016;113:5095–5100. doi: 10.1073/pnas.1522927113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mészáros B., Simon I., Dosztányi Z. Prediction of protein binding regions in disordered proteins. PLoS Comput. Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dosztányi Z., Mészáros B., Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakrabartty A., Doig A.J., Baldwin R.L. Helix capping propensities in peptides parallel those in proteins. Proc. Natl. Acad. Sci. USA. 1993;90:11332–11336. doi: 10.1073/pnas.90.23.11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustiananda M., Liggins J.R., Gready J.E. Conformation of prion protein repeat peptides probed by FRET measurements and molecular dynamics simulations. Biophys. J. 2004;86:2467–2483. doi: 10.1016/S0006-3495(04)74303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haughland R.P. Molecular Probes; Eugene, OR: 2001. Handbook of Fluorescent Probes and Research Products. [Google Scholar]
- 18.Gauer J.W., Sisk R., Hinderliter A. Mechanism for calcium ion sensing by the C2A domain of synaptotagmin I. Biophys. J. 2012;103:238–246. doi: 10.1016/j.bpj.2012.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kast D., Espinoza-Fonseca L.M., Thomas D.D. Phosphorylation-induced structural changes in smooth muscle myosin regulatory light chain. Proc. Natl. Acad. Sci. USA. 2010;107:8207–8212. doi: 10.1073/pnas.1001941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marsh J.A., Forman-Kay J.D. Sequence determinants of compaction in intrinsically disordered proteins. Biophys. J. 2010;98:2383–2390. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adler A.J., Greenfield N.J., Fasman G.D. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 1973;27:675–735. doi: 10.1016/s0076-6879(73)27030-1. [DOI] [PubMed] [Google Scholar]
- 22.Greenfield N., Fasman G.D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 23.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Best R.B., Zhu X., MacKerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanner D.E., Chan K.Y., Schulten K. Parallel generalized Born implicit solvent C with NAMD. J. Chem. Theory Comput. 2011;7:3635–3642. doi: 10.1021/ct200563j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cherepanov D.A., Feniouk B.A., Mulkidjanian A.Y. Low dielectric permittivity of water at the membrane interface: effect on the energy coupling mechanism in biological membranes. Biophys. J. 2003;85:1307–1316. doi: 10.1016/S0006-3495(03)74565-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raudino A., Mauzerall D. Dielectric properties of the polar head group region of zwitterionic lipid bilayers. Biophys. J. 1986;50:441–449. doi: 10.1016/S0006-3495(86)83480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 29.Frishman D., Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 30.Millhauser G.L. Views of helical peptides: a proposal for the position of 310-helix along the thermodynamic folding pathway. Biochemistry. 1995;34:3873–3877. doi: 10.1021/bi00012a001. [DOI] [PubMed] [Google Scholar]
- 31.Uversky V.N. Protein misfolding in lipid-mimetic environments. Adv. Exp. Med. Biol. 2015;855:33–66. doi: 10.1007/978-3-319-17344-3_2. [DOI] [PubMed] [Google Scholar]
- 32.Zor T., Mayr B.M., Wright P.E. Roles of phosphorylation and helix propensity in the binding of the KIX domain of CREB-binding protein by constitutive (c-Myb) and inducible (CREB) activators. J. Biol. Chem. 2002;277:42241–42248. doi: 10.1074/jbc.M207361200. [DOI] [PubMed] [Google Scholar]
- 33.Pavšič M., Ilc G., Lenarčič B. The cytosolic tail of the tumor marker protein Trop2—a structural switch triggered by phosphorylation. Sci. Rep. 2015;5:10324. doi: 10.1038/srep10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uversky V.N. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J., Motlagh H.N., Hilser V.J. Thermodynamic dissection of the intrinsically disordered N-terminal domain of human glucocorticoid receptor. J. Biol. Chem. 2012;287:26777–26787. doi: 10.1074/jbc.M112.355651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Y., Chen Y., Orban J. Phosphorylation-induced conformational ensemble switching in an intrinsically disordered cancer/testis antigen. J. Biol. Chem. 2015;290:25090–25102. doi: 10.1074/jbc.M115.658583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stott K., Watson M., Thomas J.O. Structural insights into the mechanism of negative regulation of single-box high mobility group proteins by the acidic tail domain. J. Biol. Chem. 2014;289:29817–29826. doi: 10.1074/jbc.M114.591115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers J.K., Pace C.N., Scholtz J.M. Trifluoroethanol effects on helix propensity and electrostatic interactions in the helical peptide from ribonuclease T1. Protein Sci. 1998;7:383–388. doi: 10.1002/pro.5560070219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roccatano D., Colombo G., Mark A.E. Mechanism by which 2,2,2-trifluoroethanol/water mixtures stabilize secondary-structure formation in peptides: a molecular dynamics study. Proc. Natl. Acad. Sci. USA. 2002;99:12179–12184. doi: 10.1073/pnas.182199699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luque I., Mayorga O.L., Freire E. Structure-based thermodynamic scale of α-helix propensities in amino acids. Biochemistry. 1996;35:13681–13688. doi: 10.1021/bi961319s. [DOI] [PubMed] [Google Scholar]
- 41.Pace C.N., Scholtz J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szilák L., Moitra J., Vinson C. Phosphorylation destabilizes α-helices. Nat. Struct. Biol. 1997;4:112–114. doi: 10.1038/nsb0297-112. [DOI] [PubMed] [Google Scholar]
- 43.Andrew C.D., Warwicker J., Doig A.J. Effect of phosphorylation on α-helix stability as a function of position. Biochemistry. 2002;41:1897–1905. doi: 10.1021/bi0113216. [DOI] [PubMed] [Google Scholar]
- 44.Lu B., Kiessling V., Cafiso D.S. The juxtamembrane linker of full-length synaptotagmin 1 controls oligomerization and calcium-dependent membrane binding. J. Biol. Chem. 2014;289:22161–22171. doi: 10.1074/jbc.M114.569327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartels T., Ahlstrom L.S., Beyer K. The N-terminus of the intrinsically disordered protein α-synuclein triggers membrane binding and helix folding. Biophys. J. 2010;99:2116–2124. doi: 10.1016/j.bpj.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., James Z.M., Thomas D.D. Structural and functional dynamics of an integral membrane protein complex modulated by lipid headgroup charge. J. Mol. Biol. 2012;418:379–389. doi: 10.1016/j.jmb.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinmetz M.O., Jahnke W., van Oostrum J. Phosphorylation disrupts the central helix in Op18/stathmin and suppresses binding to tubulin. EMBO Rep. 2001;2:505–510. doi: 10.1093/embo-reports/kve105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bah A., Vernon R.M., Forman-Kay J.D. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519:106–109. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- 49.Wu B., Wei S., Han W. Synaptotagmin-7 phosphorylation mediates GLP-1-dependent potentiation of insulin secretion from β-cells. Proc. Natl. Acad. Sci. USA. 2015;112:9996–10001. doi: 10.1073/pnas.1513004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hilfiker S., Pieribone V.A., Czernik A.J. Regulation of synaptotagmin I phosphorylation by multiple protein kinases. J. Neurochem. 1999;73:921–932. doi: 10.1046/j.1471-4159.1999.0730921.x. [DOI] [PubMed] [Google Scholar]
- 51.Vrljic M., Strop P., Brunger A.T. Post-translational modifications and lipid binding profile of insect cell-expressed full-length mammalian synaptotagmin 1. Biochemistry. 2011;50:9998–10012. doi: 10.1021/bi200998y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fioroni M., Diaz M.D., Berger S. Solvation phenomena of a tetrapeptide in water/trifluoroethanol and water/ethanol mixtures: a diffusion NMR, intermolecular NOE, and molecular dynamics study. J. Am. Chem. Soc. 2002;124:7737–7744. doi: 10.1021/ja0259335. [DOI] [PubMed] [Google Scholar]
- 53.Starzyk A., Barber-Armstrong W., Decatur S.M. Spectroscopic evidence for backbone desolvation of helical peptides by 2,2,2-trifluoroethanol: an isotope-edited FTIR study. Biochemistry. 2005;44:369–376. doi: 10.1021/bi0481444. [DOI] [PubMed] [Google Scholar]
- 54.Kentsis A., Sosnick T.R. Trifluoroethanol promotes helix formation by destabilizing backbone exposure: desolvation rather than native hydrogen bonding defines the kinetic pathway of dimeric coiled coil folding. Biochemistry. 1998;37:14613–14622. doi: 10.1021/bi981641y. [DOI] [PubMed] [Google Scholar]
- 55.Rice A.M., Mahling R., Hinderliter A. Randomly organized lipids and marginally stable proteins: a coupling of weak interactions to optimize membrane signaling. Biochim. Biophys. Acta. 2014;1838:2331–2340. doi: 10.1016/j.bbamem.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 56.Santoro M.M., Bolen D.W. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.