Abstract
Intra-axonal protein synthesis has been shown to play critical roles in both development and repair of axons. Axons provide long-range connectivity in the nervous system, and disruption of their function and/or structure is seen in several neurological diseases and disorders. Axonally synthesized proteins or losses in axonally synthesized proteins contribute to neurodegenerative diseases, neuropathic pain, viral transport, and survival of axons. Increasing sensitivity of RNA detection and quantitation coupled with methods to isolate axons to purity has shown that a surprisingly complex transcriptome exists in axons. This extends across different species, neuronal populations, and physiological conditions. These studies have helped to define the repertoire of neuronal mRNAs that can localize into axons and imply previously unrecognized functions for local translation in neurons. Here, we review the current state of transcriptomics studies of isolated axons, contrast axonal mRNA profiles between different neuronal types and growth states, and discuss how mRNA transport into and translation within axons contribute to neurological disorders.
INTRODUCTION
Local translation of axonal mRNAs has been shown to regulate neuronal functions including axon guidance, axon survival, synaptogenesis and injury responses (Figure 1). The possibility of intra-axonal protein synthesis was raised in the late 1960’s (Koenig 1965a; Koenig 1965b; Koenig 1967a; Koenig 1967b), but the methods for detection of mRNAs and newly synthesized proteins at that point lacked necessary sensitivity and specificity. Studies in invertebrate systems pushed the field forward, particularly work on the squid giant axon where large quantities of pure axoplasm could be easily obtained relative to the smaller axons of other organisms (Edstrom and others 1962; Koenig 1967c). Despite evidence for RNA and protein transfer to the squid axon from surrounding glia (Giuditta and others 1980; Giuditta and others 1968; Giuditta and others 1977; Pepe and others 1975), a few groups persisted ultimately showing existence of ribosome-bound mRNAs in these invertebrate axons (Bleher and Martin 2001; Eyman and others 2007; Giuditta and others 1991; Ingoglia and others 1983; Sotelo and others 1999). However, electron micrographs showed no ribosomes in axons of adult rodent hippocampus, while the corresponding dendrites showed prominent ribosomes arrayed in polysome configurations indicative of active protein synthesis (Steward 1983). This led to the conclusion that central nervous system (CNS) axons are not capable of synthesizing proteins in vertebrate organisms. Nonetheless, there were hints that axons of vertebrate neurons, including mammals, contain mRNAs and translate proteins under some conditions (Frankel and Koenig 1977; Funch and others 1981; Koenig 1979; Koenig and Adams 1982). Definitive proof of intra-axonal protein synthesis did not come until methods for in situ hybridization to detect small RNA quantities were refined and methods for amplifying mRNAs by polymerase chain reaction (PCR) advanced.
Figure 1. Mechanisms of axonally mRNA transport.
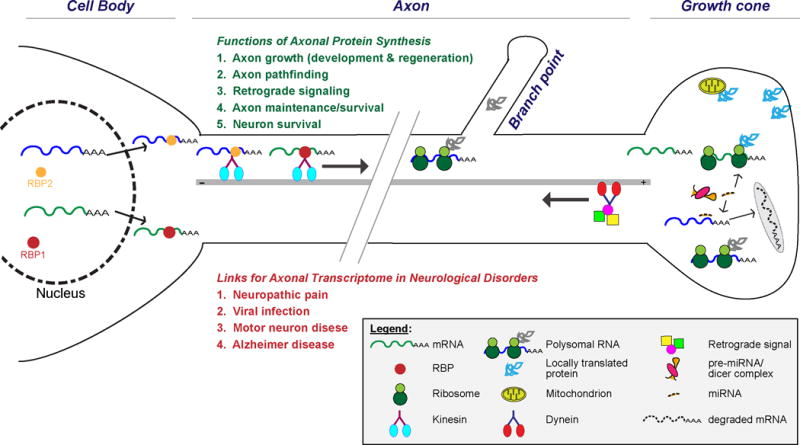
Schematic of neuron showing transport of multiple mRNAs into axons with translation at branch points and within growth cones. Note that translation also likely occurs along axon shaft based on studies from the Holt lab (Zivraj and others 2010). Some of the known functions for axonally translated proteins are noted in green text and examples of altered RNA transport and/or translation in specific neurological disorders is noted in red text.
Ribosome profiles had been detected in axons of the adult mammalian peripheral nervous system (PNS) by electron microscopy (Tennyson 1970; Zelena 1970; Zelena 1972), and work in sympathetic and cortical neurons showed axonal localization of mRNAs and ribosomes (Knowles and others 1996; Olink-Coux and Hollenbeck 1996). Bassell et al. (1998) went on to show that cultures of chick cortical neurons have axonally localized β-actin mRNA, while γ-actin mRNA is retained in the soma of these neurons (Bassell and others 1998). This is identical to the differential localization of actin isoform mRNAs seen in migrating fibroblasts (Kislauskis and others 1993), and pointed to a role for localized β-actin synthesis in cell motility. Since that time, abilities to profile mRNA populations with low input levels and clever methods to isolate axons from cultured neurons were developed to enable unbiased profiling of the axonal transcriptome. Here, we will outline what has been learned from these unbiased transcriptomics approaches and how our views of the functional impact of axonal protein synthesis have changed. These unbiased techniques have included cDNA microarrays, serial analysis of gene expression (SAGE), and next-generation sequencing (RNA-seq), and they continue to advance (Table 1). Together, these studies reveal a surprisingly complex axonal transcriptome that can change as neurons mature and when they respond to different stimuli. While obtaining an unbiased and comprehensive picture of the axon’s transcriptome was the rationale for these studies, the resulting data bring new opportunities to understand how different neurons make use of axonal mRNA transport and translation in growth, function, and disease from the viewpoint of mRNA/protein networks.
Table 1. Axonal Transcriptome Profiles.
| Reference | Axon type (species) | Approach | Estimated axonal mRNA number * | Enriched GO terms for Axonal mRNAs |
|---|---|---|---|---|
| Willis and others 2007 | Injury-conditioned adult dorsal root ganglion (DRG; rat) | Microarray | > 200 | Transmembrane proteins, Translation. |
| Taylor and others 2009 | Embryonic cortical neurons (rat) | Microarray |
Naïve ~ 300 Regenerating ~ 900 |
Naïve – Translation, Mitochondrion, Intracellular Transport, Cytoskeleton. Regenerating – Cell-cell Signaling, Cell Differentiation, Secretion. |
| Zivraj and others 2010 | Embryonic retinal ganglion cell (RGC; Xenopus and mouse) | Microarray |
Xenopus ~ 450 Mouse ~ 1800 |
Xenopus – Translation, Cytoskeletal and Motor, Metabolic and Glycolytic Proteins. Mouse – Translation, Mitochondrion, Axon Guidance. |
| Andreassi and others 2010 | Embryonic superior cervical ganglion (rat) | SAGE | ~350 | Mitochondrial, Ribosomal, RNA Metabolism and Protein Synthesis, Signal Transduction. |
| Gumy and others 2011 | Embryonic adult DRGs (rat) | Microarray |
Embryonic ~ 2600 Adult ~ 2900 (~ 1400 common) |
Embryonic – Protein Synthesis, Mitochondrial, Neurological Disease, Cytoskeletal-related, Transport of Vesicles/trafficking-Related, Axonal elongation and Morphogenesis Adult – Protein Synthesis, Mitochondrial, Neurological Disease, Inflammatory and Immune Response |
| Minis and others 2013 | Embryonic DRG explants (mouse) | RNA-Seq | ~ 6100 | Translation, Mitochondria, Cytoskeleton and Intra-cellular Trafficking (depleted in Transmembrane Protein Encoding) |
| Saal and others 2014 | Embryonic spinal motor neurons (mouse) ± smn siRNA knockdown | Microarray | 1354 axonal probe sets changed with smn depletion (1189 increased and 165 decreased) |
Wild type – Translation, RNA binding, Mitochondrion. SMN-depleted, decreased mRNAs – Synaptic Localization and Function, RNA processing, Axon growth. SMN-depleted, increased mRNAs – Translation, MHC-I components. |
| Baleriola and others 2014 | Embryonic hippocampal neurons (mouse) ± Aβ1-42 treatment. | RNA-Seq | ~ 775 mRNAs changed with Aβ1-42 treatment | Altered levels with Aβ1-42 – Cell death, Transcription, Intracellular Signaling cascade. |
| Briese and others 2015 | Embryonic spinal motor neurons (mouse) | RNA-Seq | ~ 11000 (468 ‘axonally enriched’) axonal | Cell cycle, Nucleosome Organization, Translation. |
| Taliaferro and others 2016 | Embryonic cortical neurons (mouse) and N2A and CAD cell lines | RNA-seq | 778 | Mitochondrion, Ribosome, Translation, RNA processing. |
| Bigler and others 2017 | ESC-derived glutamatergic neurons (human) | Microarray | ~ 4000 axonal mRNAs | Mitochondrion, Neurofilament, Axon Elongation, Translation, Neurotransmission. |
| Rotem and others 2017 | Embryonic spinal motor neurons (mouse) with expression of SOD1G93A, TDP-43A315T vs. control. | RNA-seq |
TDP-43A315T – 176 increased 271 decreased SOD1G93A – 95 increased 80 decreased |
Altered with TDP43A315T and SOD1G93A expression – Calcium binding, extracellular matrix binding, motor proteins, intracellular transport, monosaccharide binding |
Estimated axonal mRNA numbers based on methods used by authors for calling presence.
Breadth of the axonal transcriptome
Early observations in invertebrate systems using cDNA libraries and mRNA differential display estimated that axons contain 200-400 different mRNAs. Gene ontology (GO) analyses showed axonal mRNAs encoding cytoskeletal proteins, translational machinery, nuclear-encoded mitochondrial proteins, and signaling proteins (Gioio and others 1994; Gioio and others 2001; Gioio and others 2004). Subsequent microarray and SAGE studies using rodent sensory and sympathetic neurons revealed hundreds of mRNAs in the vertebrate axons (Andreassi and others 2010; Gumy and others 2011; Willis and others 2007). Methods to physically separate axonal contents from cell bodies in cultured PNS neurons included compartmentalized cultures (‘Campenot chambers’), modified Boyden chambers, and compartmentalized chips (Eng and others 1999; Hsu and others 2015; Zheng and others 2001). It was not feasible to isolate axons from CNS neurons with these culture methods since the dendritic processes admix with axons, but development of microfluidic devices by Taylor and colleagues allowed isolation of axons from cultured CNS neurons (Taylor and others 2009; Taylor and others 2005). This group went on to show similarly complex populations of mRNAs in axons of neocortical neurons by microarrays (Taylor and others 2009; Taylor and others 2005). Xenopus RGC axons and growth cones isolated by laser microdissection indicate comparable axonal transcriptome complexity in vivo (Zivraj and others 2010). These and other microarray studies advanced the numbers of axonal mRNAs from a handful to hundreds (see Table 1).
The microarray approach is quite good for comparing differences between RNA populations rather than showing absolute presence of an mRNA species. For example, studies from the Sendtner lab showed up and down regulation of mRNA populations in motor axons transfected with siRNAs to spinal motor neuron (Smn) mRNA vs. control siRNAs (Saal and others 2014). Nonetheless, with application of ‘presence calling’ algorithms, Gumy et al. (2010) estimated up to ~2900 mRNAs localize into rat DRG axons using genome wide microarrays. Though it can be difficult to compare between methodologies, detection platforms, and analytical methods used between different studies, most lines of evidence indicate that other neuronal types likely contain similarly complex axonal mRNA populations. Comparing axonal transcriptomes of different neuron types reveals many shared mRNAs and GO terms for the axonal RNA populations. This suggests that these neurons use axonally translated proteins for functions shared between axons, pointing to a common biology for growing axons. Many of the shared mRNAs encode proteins involved in protein synthesis, intracellular transport, calcium regulation, mitochondrial, and cytoskeletal functions (Gioio and others 1994; Gioio and others 2001; Gioio and others 2004; Gumy and others 2011; Saal and others 2014; Taylor and others 2009; Willis and others 2007; Zivraj and others 2010). However, there are clearly discreet differences between neuron types. For example, inositol monophosphatase 1 (IMPA1) mRNA is found in axons of sympathetic neurons but not in cortical axons, and CREB1 mRNA localizes to embryonic sensory axons but not sympathetic axons (Andreassi and others 2010; Taylor and others 2009). Increasing the axon RNA profile depth and breadth will undoubtedly bring more differences to light.
Next-generation sequencing approaches have been applied to subcellular compartments, including the axonal compartment. Not being ‘probe based’, RNA-seq is near completely unbiased and brings the potential to detect different mRNA variants (e.g., splice and untranslated region [UTR] variants). Minis et al. (2014) used cultures of embryonic sensory neurons to generate the first comprehensive view of the mouse axonal transcriptome (Minis and others 2014). This analysis suggested the presence of more than 6000 mRNAs in axons, more than twice what had been estimated from microarrays (Table 1). However, this required 5 μg input axonal RNA, a truly massive quantity relative to the ng yields typical of axon isolations. Briese et al. (2016) showed that axons of cultured spinal motor neuron similarly contain a few thousand axonal mRNAs, but they advanced to a ‘double-random priming’ protocol for cDNA amplification and library preparation that could drop input to pg quantities (Briese and others 2016). 80-90% of the axonal mRNAs identified from previous microarray approaches were detected by these RNA-seq studies, but the increased sensitivity of RNA-Seq brought many new mRNAs to consider and raised more differences between sensory and motor axons. Though there is transcriptome overlap between the motor and sensory, the levels of enrichment compared to cell body/somatodendritic mRNAs in these two studies show relatively low correlation (see Figure 2) (Briese and others 2016; Minis and others 2014). Together, these RNA-seq data sets, as well as others now emerging (Rotem and others 2017), will bring new perspectives on how mRNA transport varies between different neuronal types, different physiological conditions, and disease states (Figure 1). For example, Moradi et al. (2017) recently showed that axonally translated γ- and α-actin regulate different growth morphologies than β-actin in motor axons, while γ-actin mRNA is excluded from sensory and cortical axons (Moradi and others 2017).
Figure 2. Comparison of analytical methods for axonal RNA-Seq studies.
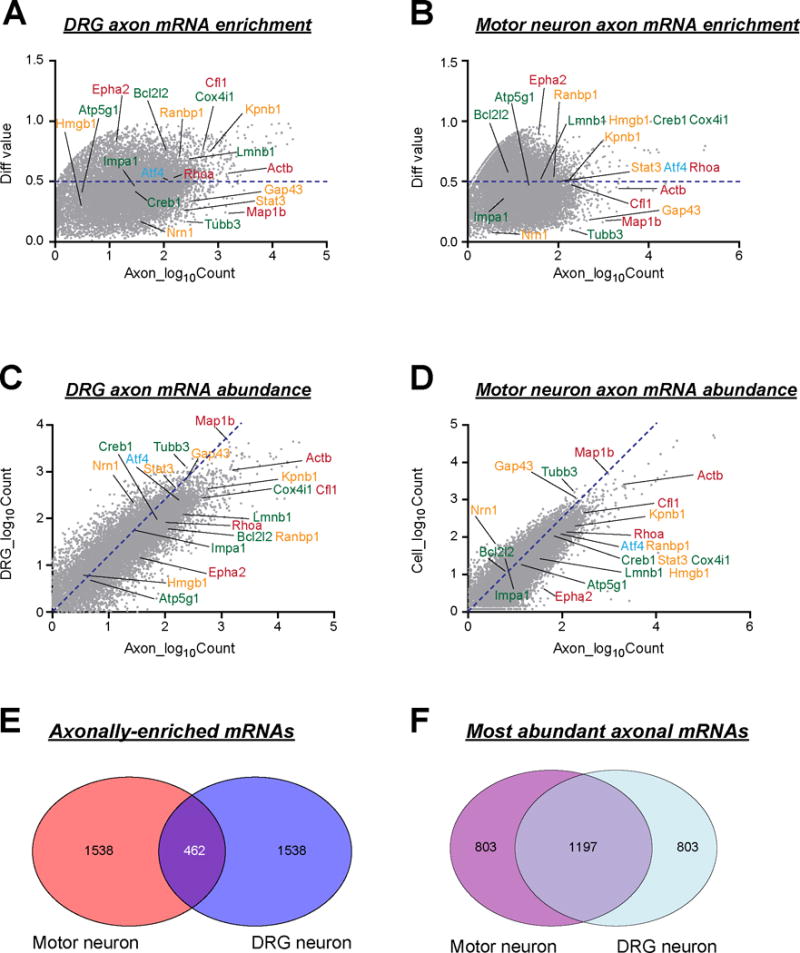
A-B, Axonal mRNAs of embryonic DRG neurons (A) and motor neurons (B) are displayed by relative enrichment in axons over cell bodies (y-axis) and its counts in axons (x-axis) (Briese and others 2016; Minis and others 2014). Raw datasets from the Minis et al. (2014) and Briese et al. (2016) data sets were mapped to Ensembl mouse genome (mm10) using STAR application (Dobin and Gingeras 2015) and counts for each gene was calculated with HTSeq software (Anders and others 2015). FKPM counts were normalized by EdgeR (Robinson and others 2010). These normalized counts were used to calculate the relative enrichment in axons vs. cell bodies, and values for the top 2000 most enriched axonal mRNAs were graphed using R software. Axonal mRNAs with biological roles previously in axon pathfinding (red), injury signaling and regeneration (orange), axon maintenance (green), and neurodegeneration (blue) are shown. The dashed blue line shows cutoff for axonal enrichment.
C-D, In contrast to A-B, the top 2000 most abundant axonal mRNAs from normalized FKPM of the raw data from Minis et al. (2014) and Briese et al. (2016) studies are graphed relative to their cell body normalized FKPM values. Embryonic DRG neurons (C) and motor neurons (D) are shown with dashed blue line representing enrichment cutoff. Axonal mRNAs with previously proven biological functions are annotated as above.
E, Venn diagram of overlap for top 2000 axonally enriched mRNAs from embryonic DRG vs. motor neurons from A-B is shown. Less than one quarter of the mRNAs are shared by this analytical method.
F, In contrast, Venn diagram of overlap between sensory and motor axons for 2000 most abundant axonal mRNAs from C-D, shows much greater overlap between the Minis et al. (2014) and Briese et al. (2016) datasets.
Comparing transcriptomes of different neuronal types and subcellular responses
Each of the mRNA profiling approaches generates large data sets and validation of all identified mRNAs is not feasible. Moreover, pulling these large data sets together into the constraints needed for publication obviously requires categorization and prioritization of the data. Both the DRG and motor axon studies assessed enrichment of mRNAs in the axonal compartment by abundance relative to cell body mRNA populations, rather than absolute abundance in the axon (Briese and others 2016; Minis and others 2014). This approach excluded some axonal mRNAs that have been defined as functional in knockout and overexpression studies (e.g., β-actin and GAP43; Figure 2A). Moreover, there seems to be low overlap (~23%) between the motor and sensory axon mRNAs by this analysis (Figure 2E). However, if we consider only the 2000 most abundant axonal mRNAs based on fragments per kilobase pair per million reads mapped (fpkm) values from the Minis et al. (2014) and Briese et al. (2106) data sets, there is much greater overlap (~60%) between the motor and sensory axon mRNA populations (Figure 2F). This is because the axons contain relatively low levels of some mRNAs species compared to the cell body, but their locally synthesized proteins still serve important functions for the neuron (several of these mRNAs are highlighted in Figure 2A-D). This raises the possibility that analytical methods for these large RNA-seq data sets can exclude some biologically relevant gene products, an important point to bear in mind when deriving conclusions from axonal RNA profiles.
Despite limitations, enrichment of mRNAs in different regions of the neuron and under different physiological conditions can point to previously unrecognized functions for axonally synthesized proteins. For example, mRNAs from microdissected RGC growth cones and axon shafts indicate that different mRNAs are enriched in these two axonal domains (Zivraj and others 2010) (Table 1). This raises the question of how these mRNAs are targeted to different regions of the axon and how the locally synthesized proteins may differentially affect function of the axon. Tubb3 mRNA shifts from axon enriched in embryonic sensory neurons to cell body enriched in adult sensory neurons, suggesting that adult neurons rely on transport of Tubulin-β3 protein from cell bodies (Gumy and others 2011). Populations of growth cone enriched in Xenopus RGC neurons also change as the growth cone arrives at its targets of innervation (Zivraj and others 2010). Thus, the axonal transcriptome responds to the growth status of the neuron, raising the possibility that RNA transport and/or intra-axonal translation brings a new strategy to facilitate neural repair. This bears importance for regeneration of CNS axons, as severed rat spinal cord axons show evidence for protein synthesis when they are provided with a growth promoting extracellular environment (Kalinski and others 2015; Sachdeva and others 2016). Recent work in the Lamprey spinal cord, where some axons can spontaneously regenerate after injury, also points to active protein synthesis in those regenerating CNS axons (Jin and others 2016).
Functional studies for translation of individual proteins in axons points to numerous physiological effects that could impact neural health and regenerative mechanisms. Considering if axonal mRNA transport and translation could contribute to human health precipitates two critical questions: 1) do mRNAs localize into axons of human neurons and 2) is intra-axonal protein synthesis limited to periods of axon growth or do axons translate proteins in their mature, non-growing state? Two very recent works address these questions. First, Bigler et al. (2017) generated CNS glutamatergic neurons from human embryonic stem cells (hESC-neurons). They show over 3900 axonally abundant mRNAs in the hESC-neurons that overlap with rat cortical axon mRNAs in GO terms for translation, ribosome and respiratory chain (Bigler and others 2017) (Table 1). However, the axons of hESC-neurons show differential enrichment of mRNAs encoding cytoskeletal components (microtubule GO term for rat vs. neurofilament GO term for human). This may reflect differences in the growth or maturity status of the neuron cultures, or true species differences in how these neurons target mRNAs into the axons. Second, Shigeoka et al. (2016) used mice with epitope-tagged ribosomal protein (‘RiboTag’ mice) expressed in RGCs to isolate translationally-active mRNAs from optic nerve axons. Comparing nerves from developing to adult animals, there was a decrease in ribosome-bound mRNAs in the adult axons, but many axonal mRNAs were still identified in the mature axons (Shigeoka and others 2016). Thus, both rodents and human neurons have capacity for intra-axonal protein synthesis and, at least in mice, they make use of this mechanism as mature, functional neurons with intact synaptic connections.
Functional consequences of intra-axonal protein synthesis
Stimuli like neurotropic cues and axonal injury have long been known to induce both local effects within axons as well as long-distance effects through retrograde signaling to the neuron’s soma. Such stimuli can trigger translation of ‘axon resident’ mRNAs, which are needed for both local and long-distance effects, as well as alterations in transport of RNA into axons, that change the axon’s transcriptome and translatome (Jung and others 2012; Perry and Fainzilber 2014). Based on GO terms for axonal mRNA cohorts, the transcriptomics studies outlined above imply that axonally translated proteins contribute to neuronal development and physiology. However, in parallel to the axonal RNA profiling studies, analyses of individual axonal mRNAs have clearly shown that axonally synthesized proteins are needed for neuronal development, functional maintenance of axons, and repair of injured axons. We highlight a few of these intra-axonally synthesized proteins below with regards to these functions.
During development of the nervous system, axons sense attractant and repulsant cues in their environment while navigating to their targets for innervation. Studies using Xenopus and rodent neurons have shown that axon turning in response to ‘pathfinding cues’ requires new protein synthesis in axons. Axonally synthesized proteins that impact the axon’s cytoskeleton play a central role in this process, including β-actin, RhoA, and ADF/cofilin (Piper and others 2006; Wu and others 2005; Zhang and others 2001; Zhang and others 1999). β-actin mRNA is enriched in growth cones where it is locally translated in response to the attractant cues, Netrin-1 and neurotrophins (Campbell and Holt 2001; Zhang and others 1999). When these cues are asymmetrically presented to axons, β-actin concentrates and is translated on the side of the growth cone with the highest stimulus concentration (Leung and others 2006; Yao and others 2006). On the other hand, repellent cues like semaphorin-3A and Slit2B appear to block translation of β-actin and activate translation of RhoA and/or ADF/cofilin mRNAs in growth cones to promote actin filament depolymerization (Piper and others 2006; Piper and others 2005; Wu and others 2005). Intra-axonal translation of MAP1B mRNA in distal axons is also up-regulated in response to Sema-3A (Li and others 2009). Local translation of the MKK7 mRNA, which encodes a protein kinase that impacts MAP1b protein function, also supports neurite outgrowth (Feltrin and others 2012). Additionally, navigation of spinal cord commissural axons to appropriately navigate to their targets requires intra-axonal translation of the Eph receptor A2 (EphA2) (Brittis and others 2002). These and other reports point to the importance of axonally synthesized proteins for axon pathfinding during development of the nervous system.
Axonally synthesized proteins contribute to other modes of axon growth, both during development and following injury. Collateral branching of axons in response to nerve growth factor (NGF) requires actin filament polymerization and intra-axonal protein synthesis, and NGF triggers increased intra-axonal translation of CTTN, WASF1, and Arp2 mRNAs, whose protein products can impact actin polymerization (Spillane and others 2011). Overexpression of axonally-targeted β-actin mRNA increases axon branching (Donnelly and others 2013a), and β-actin mRNA is translated adjacent to branch points where filopodia can mature into collateral branches (Spillane and others 2013). On the other hand, overexpression of axonally-targeted GAP43 mRNA decreases axon branching and increases axon length (Donnelly and others 2013a). These observations suggest that the neuron can balance the translation of mRNAs in distal axons to alter its growth pattern. Indeed, localized translation of mRNAs encoding translational machinery, eIF2B2 and eIF4G2, was shown to influences axon growth and elongation (Kar and others 2013). Transport of mRNAs can also be linked to their transcriptional activity. For example in adult sensory neurons, GAP43 and β-actin compete for binding to limited quantities of ZPB1 protein (also called IGF2BP1 and IMP1) that is needed for their localization into axons (Donnelly and others 2011), and elevations in one relative to the other mRNA can impact the axon’s transcriptome and growth (Yoo and others 2013).
Translation of specific proteins in axons has also been shown to contribute to further maturation as well as maintenance/survival of the axon. Axonal β-catenin mRNA concentrates in the presynaptic terminals of CNS neurons in culture, where its translation product regulates synaptic vesicle release during synaptogenesis (Taylor and others 2013). Tyrosine hydroxylase mRNA, which encodes the rate-limiting enzyme in catecholamine biosynthesis, localizes to sympathetic axons to presumably regulate neurotransmitter synthesis (Gervasi and others 2016a). Axonal translation of mRNAs for the nuclear-encoded mitochondrial proteins, COXIV and ATP5G1, key components of the oxidative phosphorylation chain, regulate axonal mitochondrial ATP production to support sympathetic axon survival (Aschrafi and others 2010; Aschrafi and others 2008; Gioio and others 2001; Gioio and others 2004; Natera-Naranjo and others 2012; Wen and others 2002). Axonal translation of Lamin B2 (lb2) mRNA in RGC axons also promotes axonal survival by maintaining mitochondrial function and axon transport (Jung and others 2012). NGF stimulation of embryonic DRG axons triggers transcription and subsequent axonal transport of Bclw mRNA, with axonally synthesized Bclw protein promoting axonal survival and blocking activation of caspase locally in the axons (Cosker and others 2013). Axonally synthesized proteins have also been shown to support neuronal survival through intra-nuclear effects. NGF-dependent synthesis of IMPA1 in the axons of cultured sympathetic neurons helps maintain axon viability through activation of CREB1 in the nucleus (Andreassi and others 2010). On the other hand, NGF-dependent survival of embryonic DRG neurons was shown to be regulated by axonal translation of CREB1 mRNA with retrograde transport of the protein to the nucleus (Cox and others 2008).
In cultured neurons, axonal protein synthesis is needed for growth cone formation following axotomy (Verma and others 2005). Though which proteins are needed remains unknown, in vivo axotomy triggers translation of several mRNAs in PNS axons. Importin β1 (Impβ1) mRNA was shown to be rapidly translated in sciatic nerve axons following crush injury through a Ca2+-dependent mechanism (Hanz and others 2003). Impβ1 protein forms an obligate heterodimer with Importin α3 protein, and the Importin α/β complex is used for nuclear import of cargo proteins. The axonal Importin heterodimer is retrogradely transported by dynein to deliver NLS-containing proteins to the nucleus from distal axons (Hanz and others 2003). Consistent with this mechanism, selective knockout of axonal Impβ1 attenuates activation of regeneration-associated genes following PNS axotomy (Perry and others 2012). Intra-axonal translation of RanBP1 following PNS axotomy facilitates Importin α3 protein’s interaction with newly synthesized Impβ1 (Yudin and others 2008). In addition to CREB1 noted above, a number of different transcription factors seem to be retrogradely transported in injured PNS axons (Ben-Yaakov and others 2012). Among these, Stat3α and PPARγ mRNAs are locally translated in injured PNS axons and retrogradely transported to DRGs; axonally-synthesized Stat3α protein supports post-injury survival of sensory neurons and PPARγ protein is needed for axon growth (Ben-Yaakov and others 2012; Lezana and others 2016). Axonal translation of vimentin mRNA, which encodes an intermediate filament protein, similarly supports regeneration but does so by serving as a scaffold to link activated extracellular-regulated kinases (Erk) 1/2 to Importins for retrograde transport (Perlson and others 2005). Interestingly, axonal vimentin is proteolized shortly after its translation, and a proteolytic fragment of vimentin serves this scaffold, providing a mechanistic link between intra-axonal protein synthesis and the well established activation of proteases in injured axons. Taken together, these and other studies indicate a coordinated response to axonal injury that converges on intra-axonal mRNAs.
Intra-axonal translation in neurological disorders
As functions for axonally-synthesized proteins are increasingly recognized and methodologies for detection of mRNAs advance, axonal mRNAs and their protein products have now been linked to a several neurological disorders. These include motor neuron diseases, neurodegenerative diseases, neuropathic pain, and infection by neurotropic viruses (Table 2). These disease manifestations may represent a failure of the growth or axonal maintenance functions of intra-axonal protein synthesis. On the other hand, an increase in synthesis of some proteins locally may contribute to or even precipitate or propagate a neurological disease. For example, the prominent role of intra-axonal protein synthesis in response to axotomy in the PNS outlined above likely reflects an adaptive response to injury that may be shared with some neurodegenerative diseases. In this section, we summarize studies linking axonal protein synthesis to neurological disorders, and speculate roles for axonally-synthesized proteins in disease pathophysiology. The reader should bear in mind that this is a rapidly evolving area and additional diseases and/or disease mechanisms linked to axonal protein synthesis will undoubtedly be uncovered in the near future.
Table 2. Axonally translated proteins in neurological disorders.
| Disease/Disorder | mRNAs Altered | Mechanism | References |
|---|---|---|---|
| Alzheimer disease | ATF4 | Aβ1-42 induced transcription and anterograde transport | (Baleriola and others 2014) |
| Amyotrophic lateral sclerosis (ALS) | Nefl | Decreased transport with TDP-43 mutation | (Alami and others 2014) |
| 271 decreased + 176 increased | Altered axonal localization with TDP-43A315T expression | (Rotem and others 2017) | |
| 80 decreased + 95 increased | Altered axonal localization with SOD1G93A expression | ||
| APC mRNA ligands | Altered axonal localization with FUS/TLS expression | (Yasuda and others 2017) | |
|
| |||
| Spinal Muscular Atrophy (SMA) | β-actin, GAP43, Cav2.2 | Decreased axonal levels with SMN depletion | (Fallini and others 2016; Jablonka and others 2007) |
| Increased and decreased mRNAs | SMN depletion from motor neurons substantially alters the axonal transcriptome | (Saal and others 2014) | |
|
| |||
| Neuropathic pain | CREB1 | Capsaicin and IL-6 injection increase intra-axonal translation through mTOR pathways | (Melemedjian and others 2010; Melemedjian and others 2013; Melemedjian and others 2014) |
|
| |||
| Neurotropic viral infection | ? | Retrograde transport of pseudorabies virus requires axonal translation | (Koyuncu and others 2013) |
Infection of sensory neurons by herpes family viruses requires retrograde transport of virus from distal axons to the soma where a chronic, latent infection is established. Several groups have studied this retrograde transport of viral particles (for review see (Enquist 2012)). The Enquist group took advantage of compartmentalized culture methods to show that efficient retrograde transport of pseudorabies virus requires axonally-synthesized proteins (Koyuncu and others 2013). Although it is not clear which proteins are needed, it is tempting to speculate that the products of axonal Impβ1 and RanBP1 may be involved. Similar candidates are the dynein-modulating proteins Lis1 and p150glued that the Hengst lab recently showed are synthesized in axons (Villarin and others 2016). Interestingly, Song et al. (2016) showed that infection-diminishing effects of interferon-β (IFN-β) are linked to axon-intrinsic mechanisms, while protective effects from IFN-γ require transcription in the soma (Song and others 2016). The authors showed that axonal translation is increased by treatment with IFN-β compared to IFN-γ. However, knowing whether the IFN-β induced axonal protein synthesis is needed to prevent retrograde transport of virus will require targeting specific proteins. For example, can pseudorabies virus be retrogradely transported in the Impβ1 3’UTR knockout mice that the Fainzilber lab developed (Perry and others 2012)?
Neurological disorders have also been linked to altered translation in distal axons. Jimenez-Diaz et al. (2008) showed that myelinated axons in the rodent foot pad contain components of the translational machinery and inhibition of protein synthesis with rapamycin prevents capsaicin-induced hyperalgesia, an experimental model of neuropathic pain (Jimenez-Diaz and others 2008). This local effect of rapamycin suggests involvement of the mTOR pathway for induction of neuropathic pain. Consistent with this, hyperalgesia seen after injection of interleukin 6 (IL-6) into the paw pad requires mTOR complex 1 (mTORC1), ERK 1/2, and eukaryotic initiation factor 4E (eIF4E) activities (Melemedjian and others 2010; Melemedjian and others 2011; Melemedjian and others 2013). Interestingly, CREB1 is one of the locally translated proteins induced by IL-6, and the axonally-synthesized CREB1 is retrogradely transported to the DRG where it regulates gene expression (Melemedjian and others 2014). Though no axon transcriptome analyses have been published for neuropathic pain states, transcriptomes for growing adult sensory axons have detected a number of mRNAs that show altered levels in sensory ganglia with neuropathic pain states (e.g., ATF3, NPY) (Gumy and others 2011; LaCroix-Fralish and others 2011; Willis and others 2007). Thus, it is quite feasible that the axonal transcriptome is altered with neuropathic pain, which could subsequently alter signaling or plasticity at sensory nerve terminals.
Alterations in axonal transport have been implicated in the neuronal degeneration in the CNS and PNS, both for causality and affecting disease progression (Millecamps and Julien 2013; Pareyson and others 2015). Exposing cultured neurons to β amyloid peptides (Aβ), whose accumulation in amyloid plaques results in Alzheimer disease (AD), has been shown to trigger neurite degeneration (LaFerla and others 1995; Pike and others 1993). Imaging studies in an AD mouse model showed evidence for alterations in dendritic mRNA translation adjacent to amyloid plaques (Meyer-Luehmann and others 2009). By selectively applying peptides to the axonal compartment of cortical neurons in microfluidic chambers, Baleroia et al. (2014) showed that levels of some axonal mRNAs are uniquely increased after Aβ1-42 exposure. Proteins encoded by these Aβ1-42 axon enriched mRNAs included ones that alter both amyloid production and tau modifications that are associated with AD (e.g., APP, Clu, and Fermt2). The transcription factor ATF4 (also called CREB2) was also included in the Aβ1-42-induced axonal mRNA cohort, and the mRNA was also detected in post-mortem brains from AD patients. In culture, the axonally-synthesized ATF4 protein is retrogradely transported to the soma where it triggers cell death by increasing transcription of the UPR-associated CCAAT-enhancer-binding protein homologous protein (CHOP) (Baleriola and Hengst 2015). This result links axon-to-soma and soma-to-axon signaling in AD through axonal mRNA transport/translation.
Mutations in RNA binding proteins are causal for the motor neuron diseases, spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS). Consequently, alterations in localization of mRNAs into motor axons have been linked to both diseases. SMA is caused by loss of the SMN1 gene, which encodes an obligate component of the protein complex used to assemble small nuclear RNP (snRNP) complex core that is needed for RNA splicing in the nucleus (Rage and others 2013). SMN protein also localizes to axons where it interacts with other RBPs and mRNAs (Akten and others 2011; Fallini and others 2011; Rossoll and others 2003; Zhang and others 2006; Zhang and others 2003). Axonal functions of SMN are distinct from its role in snRNP biogenesis (Carrel and others 2006). Loss of SMN protein causes defects in clustering of axonal voltage-gated Ca2+ channels, which decreases axon growth (Jablonka and others 2007). Decreased motor axon growth has also been detected after knockdown of zebrafish SMN (McWhorter and others 2003). Using microfluidic devices to isolate axons from motor neurons, Saal et al. (2014) showed that the axon’s transcriptome changes with depletion of SMN protein. Consistent with the axon growth deficits, SMN depletion significantly decreased levels of axonal mRNAs encoding proteins implicated in axon growth and synaptic activity (Saal and others 2014). The perturbations in the levels of these mRNAs may ultimately contribute to the altered neuronal transmission and defective pre-synaptic excitability observed in Smn-deficient motor neurons (Jablonka and others 2007; Kong and others 2009; Ruiz and others 2010).
Mutations in Tar DNA binding protein-43 (TDP-43) and fused in sarcoma/translocated in liposarcoma (FUS/TLS) protein can cause familial ALS (Kwiatkowski and others 2009; Lagier-Tourenne and Cleveland 2009; Sreedharan and others 2008; Vance and others 2009). Mutation in these genes can also be causative in frontotemporal dementia (FTD), so the cellular mechanisms leading to neuronal degeneration may be shared between motor and cortical neurons. Disease-causing mutations in TDP-43 decrease mobility of the RBP in axons, and decrease anterograde transport of low molecular weight neurofilament (Nefl) mRNA (Alami and others 2014). Rotem et al. (2017) most recently published axonal RNA-Seq analyses of motor neurons expressing mutant TDP-43 (TDP-43A315T) and SOD1 (SOD1G93A). SOD1G93A mutation is similarly causal for familial ALS, but SOD1 has no RNA interacting motifs like TDP-43 and FUS/TLS. Overall, more mRNAs were dysregulated in the TDP-43A351T mutants than SODG93A, which likely points to a more direct role for TDP-43 in RNA metabolism. However, both TDP-43A315T and SOD1G93A altered the axonal transcriptome, with the mRNAs in the ALS motor axons depleted for synapse assembly, axon extension regulation, and transcription factor mRNAs (Rotem and others 2017). Depletion of synapse- and axon extension-associated mRNAs are shared between SMA and these ALS-causing mutations, suggesting that altered axonal translation may similarly affect axon growth and function in these two diseases.
Similar unbiased profiles for the axonal transcriptome of neurons expressing mutant FUS/TLS have not been reported, but functional studies point to alterations in axonal mRNAs with FUS/TLS mutation. Both FUS/TLS and TDP-43 are low-complexity domain proteins, whose aggregation is thought to impart a gain-of-function to the proteins that eventually leads to cell death (Johnson and others 2009; Scekic-Zahirovic and others 2016; Shiihashi and others 2016). Wild-type FUS/TLS was shown to regulate subcellular translation of adenomatous polyposis complex (APC) protein-associated mRNAs in fibroblasts (Yasuda and others 2013). APC interacts with several mRNAs in fibroblast pseudopodia (Mili and others 2008), and it binds to mRNAs with roles in neural development and microtubule organization that localize to axonal growth cones (Preitner and others 2014). Interestingly, neuronal inclusions formed by mutant FUS/TLS protein specifically decrease axonal mRNA levels through sequestering APC-RNPs and decreasing kinesin function (Yasuda and others 2017). This advances the appealing hypothesis that these motor neuron disease-associated genes have both gain-of-function and loss-of-function mechanisms that can impact the axonal transcriptome. However, it should be noted that FUS/TLS, TDP-43, and SMN affect RNA splicing and other post-transcriptional mechanisms that could indirectly affect the axonal transcriptome. Further, FUS/TLS interacts with SMN, and mutations in FUS/TLS could obviously affect mRNA splicing through effects on SMN (Sun and others 2015). Finally, a hexanucleotide repeat expansion in the C9orf72 gene was shown in a relatively higher proportion of familial ALS than TDP-43, FUS/TLS, and SOD1 mutations (Majounie and others 2012). Disruption of nuclear pore function is seen in neurons carrying the C9orf72 expansion, and this could impact axon-to-soma signaling that regulates injury-induced gene expression through Importin α3/β1 complex (Donnelly and others 2013b; Haeusler and others 2014; Zhang and others 2015). Interestingly, work from Fainzilber lab suggests that continual soma-to-axon transport of Impβ1 mRNA and axon-to-soma transport of Impβ1 protein is used for cell size-sensing in neurons (Perry and others 2016). So disruptions of nuclear pore function due to C9orf72 mutation could alter gene expression and impact soma-to-axon signaling derived from mRNA transport/translation.
Regulation of axonal transport of mRNAs
The link between TDP-43, FUS/TLS and SMN proteins and axonal RNA transport emphasizes a central role for RBPs in determining the axonal transcriptome. Distribution of mRNAs to different subcellular regions is driven by RBP-mRNA interactions that are regulated a number of different levels. For example, newly transcribed β-actin mRNA binds to ZBP2 (also called MARTA2 and FUBP3) in the nucleus, which subsequently recruits ZBP1 to β-actin mRNA (Chao and others 2010; Farina and others 2003). The ZBP1-β-actin complex then translocates to the cytoplasm and is directed to subcellular domains like axons. The RNA elements needed for mRNA localization have mostly been found in 3’UTR, but occasionally 5’UTR and coding sequences are needed (Meer and others 2012; Rihan and others 2017; Wang and others 2009). These localization motifs can be defined by their primary, secondary, or tertiary structures (Aschrafi and others 2010; Bolognani and others 2010; Di Liegro and others 2014; Khvorova and Wolfson 2012; Li and others 2010). Although a number of axonal localization motifs have now been uncovered, these have not shown any consensus nucleotide sequence for axonal localization (Aronov and others 1999; Ben-Yaakov and others 2012; Cosker and others 2016; Cosker and others 2013; Gervasi and others 2016b; Kaplan and others 2009; Merianda and others 2015; Merianda and others 2013a; Vuppalanchi and others 2010; Wu and others 2005; Yoo and others 2013; Yoon and others 2012; Yudin and others 2008; Zhang and others 2001). It should be noted that most of the approaches looking for commonality between RNA motifs have been limited to asking if the motif is shared with other axonal mRNAs. However, recent RNA-seq studies have uncovered motifs shared between axonal mRNAs (Rotem and others 2017), so the increased depth of next generation sequencing coupled with bioinformatics resources will likely shed more light on this topic.
Only a handful of axonal RBPs needed for transport of axonal mRNAs are now known. In addition to SMN, TDP-43, FUS/TLS and ZBP1 mentioned above, HuD (also known as Elav4), hnRNP R, FMRP, Nucleolin, and Splicing factor poly-glutamine rich (SFPQ) have been implicated in transport of mRNAs into axons (Akten and others 2011; Antar and others 2006; Cosker and others 2016; Dombert and others 2014; Fallini and others 2012; Glinka and others 2010; Perry and others 2016; Yoo and others 2013). HuD binds to AU-rich elements (ARE) in 3’UTRs and is well known to stabilize mRNAs (Beckel-Mitchener and others 2002). However, HuD interaction is also needed for axonal localization of GAP43 (Yoo and others 2013). hnRNP R was identified as SMN protein interactor (Rossoll and others 2002). hnRNP R along with ZBP1 appears to be needed for transport of β-actin into motor axons (Glinka and others 2010), and both SMN and hnRNP R are detected in pre-synaptic terminals of developing and mature motor neurons (Dombert and others 2014). Though better characterized as a dendritic RBP, FMRP also localizes to axons where it binds to MAP1b (Antar and others 2006). Growth cones of cortical neurons from FMRP knockout mice show increased filopodia similar to the increased dendritic spine density seen with loss of FMRP (Antar and others 2006). Nucleolin was recently shown to bind to Impβ1, and this nucleolin-Impβ1 complex binds directly to kinesin motor protein (Perry and others 2016). SFPQ protein is needed for assembly of Bclw and lb2 mRNA granules for axonal localization (Cosker and others 2016). SFPQ is typically considered for its roles in transcription and splicing, but the cytosolic version of SFPQ is needed for motor development in zebrafish where the protein localizes to motor axons (Thomas-Jinu and others 2017). Nucleolin is similarly well characterized for nuclear functions (Tajrishi and others 2011), and its role in transport of Impβ1 emphasize that the neuron uses nuclear RBPs for multiple functions that include post-transcriptional regulation in the distal reaches of the cytoplasm. It will be of high interest to determine how the neuron coordinates RNP assembly and subsequent modifications through interactions of nuclear RBPs and mRNAs. This has not been well defined for neurons, but initiation of RBP formation in the nucleus for localizing mRNAs with subsequent modifications as the RNP moves into different subcellular domains was elegantly demonstrated for Xenopus oocytes (Kress and others 2004).
RBP-mRNA interactions are regulated at several levels. RNP complexes have been shown to contain single copies of an RNA (Amrute-Nayak and Bullock 2012; Mikl and others 2011). Changes in how much of an mRNA is targeted into axons can be determined by level or activity of its RBP, with different cellular stimuli increasing or decreasing axonal transport of target mRNAs (Merianda and others 2013a; Merianda and others 2013b; Willis and others 2007). Some mRNAs also appear to compete for binding to limited quantities of an RBP, such that changes in transcription of mRNAs that share RBPs can alter transport of other mRNAs. This is seen for β-actin and GAP43 that compete for binding to ZBP1 for their localization into sensory axons (Donnelly and others 2011; Kim and others 2015; Yoo and others 2013). Interestingly, HuD and KH-type splicing regulatory factor (KHSRP) proteins compete for binding to GAP43, where HuD interaction stabilizes and localizes GAP43 and KHSRP interaction targets GAP43 for degradation (Bird and others 2013). So different RBPs can also compete for binding to the same mRNA species. These competitions likely extend to other mRNAs as ZBP1 and HuD are known to bind to many mRNAs (Bolognani and others 2010; Jonson and others 2007). Other axonal RBPs have been shown to similarly bind to many different mRNAs, but due to technical limitations on materials obtained from isolated axons, these studies have overwhelmingly been limited to whole cell or tissue extracts rather than testing interactions within the axonal compartment. Better methodologies to test for interactions directly within axons will be needed to fully understand the dynamics of axonal RBP-mRNA interactions. Additionally, the affinity of different mRNAs for binding to an RBP and different components within an RNP will need to be considered.
A final consideration for the RBP-mRNA interactions is that some mRNAs are transported into axons and then stored until needed. κ-opioid receptor (KOR) mRNA is translationally-silenced in axons until its protein product is needed (Tsai and others 2009; Tsai and Wei 2010). Intra-axonal translation of Importin β1, Stat3α, RanBP1, and Vimentin rapidly increases in PNS axons following axotomy, suggesting that these mRNAs are similarly be stored until needed (Ben-Yaakov and others 2012; Hanz and others 2003; Perlson and others 2005; Yudin and others 2008). Likewise, HMGB1 is constitutively transported into sensory axons and its translation upregulated during axon regeneration (Merianda and others 2015). It is not clear how such mRNA storage or sequestration is accomplished in axons. Since the transport RNP provides a level of translational suppression, the mRNA could remain with its transport RNP for storage. This would require that the RNP dock with some structure in the axons for safe-keeping (Figure 3A). The RNP contents could also be modified to effectively generate an axonal stress granule-like structure, as suggested for KOR (Tsai and others 2009; Tsai and Wei 2010).
Figure 3. Unresolved mechanisms to regulate axonal mRNA dynamics.
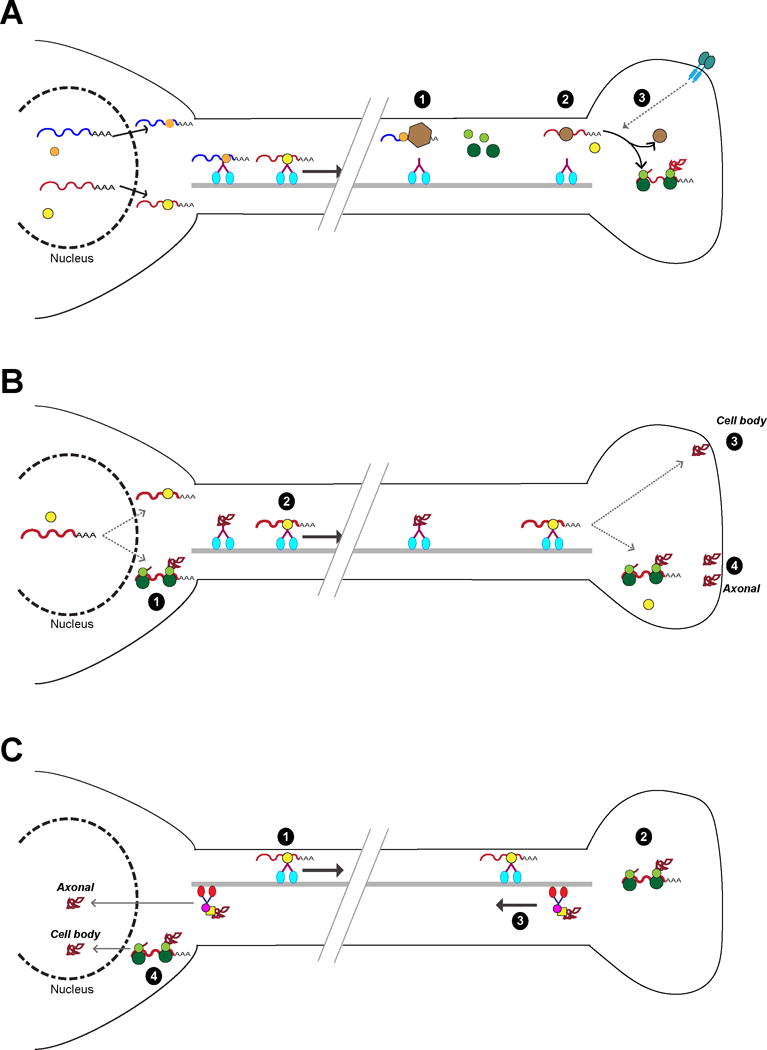
A, Where are mRNAs stored in axons? There is clear evidence for storage of mRNAs in axons, where they are somehow sequestered away from the translational machinery and kept dormant until needed. The mechanisms underlying this are not known. How does an RNP disengage from the motor proteins used for transport into axons and avoid translation of its mRNA? Binding of additional proteins to the RNP (1) or replacement of RNP proteins with other RBPs (2) are possible mechanisms. Additionally, there must be stimulus-dependent mechanisms to recruit axonal mRNAs from their storage site (3).
B, Does local translation impart unique function(s) to an axonal protein? For some proteins the axon has a dual source – cell body-synthesized protein that is transported down the axon (1) and the protein derived from the axonally translated mRNA. It is not clear how or if these two protein sources (3,4) are functionally distinct.
C, Does axonal translation impart unique function(s) to retrogradely transported proteins? Several transcription factors are known to be translated in axons (1,2) and retrogradely transported to the neuronal cell body (3). For all identified to date, there is already a cell body (or nuclear) pool of the transcription factor resident in the neuron that is derived from protein synthesis in the cell body (4). It is not clear how or if the neuron distinguishes these two sources of protein. Do they have distinct functional roles, perhaps regulating different gene cohorts? For legend refer to Figure 1.
CONCLUSIONS
As outlined above, the field of axonal protein synthesis has rapidly advanced. The initial candidate mRNA analyses and functional studies for intra-axonal protein synthesis provided rationales for generating in-depth, unbiased profiles of axonal transcriptomes. Developments in RNA detection, including approaches for genome wide analyses, and culture methods to isolate axons made these profiles feasible. Profiles of axons from cultured neurons, further provided rationale to tackle the in vivo axonal transcriptome as the Holt lab recently published (Shigeoka and others 2016). The knowledge coming from these approaches has raised the exciting possibility of novel functions for axonally-synthesized proteins beyond the growth, injury responses, maintenance/survival, and disease links outlined above. Indeed, there is a high potential for additional links between axonally-synthesized proteins and neurological diseases, both for disease causality and progression. For example, altered anterograde and retrograde transport has been implicated in pathophysiology of peripheral axonopathies, and the cargoes showing altered soma-to-axon and axon-to-soma transport would certainly include axonal mRNAs as well as their locally generated protein products. Moreover, additional disease genes directly affecting RNA transport and/or translation may come to light as we gain more mechanistic insight into RNA-protein interactions required for axonal mRNA transport and translation. Better recognition of localization motifs as well as other functional RNA motifs may also uncover sequence variants in human genome that affect axonal mRNA localization and translation. Likewise, RNA-seq approaches bring the potential to recognize RNA splice and UTR variants that impart distinct subcellular localization and translational control for transcript variants. Analyses of single gene products has shown examples of UTR variants imparting differential localization for both axons and dendrites (An and others 2008; Perry and others 2012), and RNA-seq data for differential localization of splice and UTR variants from mRNA profiles of neurites was recently published (Taliaferro and others 2016).
Despite obvious advances in the field, many questions remain unanswered for regulation of axonal mRNA transport and translation. We have tried to point out some of these unresolved issues in the sections above. However, we have only briefly touched on functions of the axonally-synthesized proteins, and this is an area for much growth. A critical question for several axonal mRNAs is whether the locally generated protein has distinct functions from that which is synthesized in the cell body. For example, only a small fraction of axonal β-actin protein is synthesized locally in axons – most is transported from the cell body, with less than 10% derived from the axonal mRNA (Eng and others 1999; Zheng and others 2001). Does the axonally synthesized β-actin differ from the cell body synthesized β-actin that is transported down the axon as protein (Figure 3B)? If not, why would the neuron bother with two sources for this and other abundant proteins? The axonally-synthesized proteins that are retrogradely transported raise a similar question. For Impβ1, the cargo(es) that Importin α3/β1 heterodimer delivers from axons would likely be the distinguishing feature. But for others like transcription factors, there will be protein delivered from the axon and a resident soma/nuclear protein (e.g., Stat3α, CREB, PPARγ; Figure 3C). How does the neuron distinguish between these protein populations and is there any contextual information conveyed from axonally synthesized version that imparts a unique transcriptional profile?
For space limitations, we have not discussed axonal localization of non-coding RNAs. Non-coding RNAs (ncRNA) have also been detected in sympathetic, motor, and cortical axons as well as peripheral nerve in vivo (Kye and others 2007; Natera-Naranjo and others 2010; Sasaki and others 2014). In part these were uncovered by RNA-seq approaches (Phay and others 2015; Phay and others 2016), but targeted profiles for micro-RNAs (miRNA) and other, small ncRNAs have been used (Rotem and others 2017). ncRNAs are an emerging front for transcriptomics, and these bring potential for multiple levels of post-transcriptional regulation. For example, miRNAs can regulate mRNA survival and translation. So these can impact both the axonal transcriptome as well as which proteins are generated from those axonal mRNAs. Some miRNAs, pi-RNAs, and other non-coding RNAs are enriched in axons (Kye and others 2007; Natera-Naranjo and others 2010; Phay and others 2015; Phay and others 2016; Sasaki and others 2014). miRNAs biogenesis begins in the nucleus with processing of primary miRNA (pri-miRNA) into a precursor miRNA (pre-miRNA). Pre-miRNA to miRNA processing can occur within axons, which could provide a localized mechanism for regulating miRNA activity (Aschrafi and others 2008; Kim and others 2015; Phay and others 2015). However, how and if this processing is regulated is yet to be determined. Nonetheless, this emerging area of the non-coding transcriptome in axons is an exciting development that will bring a better understanding of post-transcriptional control mechanisms that the neuron uses.
Acknowledgments
The authors were supported by grants from the National Institutes of Health (R01-NS041596, R01-NS086993 and P01-NS055976), Department of Defense/US Army Congressionally Mandated Research Program (W81XWH-2013-1-308), and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. JL Twiss is the SC SmartState Endowed Chair in Childhood Neurotherapeutics at the University of South Carolina.
References
- Akten B, Kye MJ, Hao le T, Wertz MH, Singh S, Nie D, et al. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc Natl Acad Sci U S A. 2011;108(25):10337–42. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, Han SS, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81(3):536–43. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrute-Nayak M, Bullock SL. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol. 2012;14(4):416–23. doi: 10.1038/ncb2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134(1):175–87. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Saiardi A, et al. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13(3):291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol Cell Neurosci. 2006;32(1-2):37–48. doi: 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Aronov S, Marx R, Ginzburg I. Identification of 3′UTR region implicated in tau mRNA stabilization in neuronal cells. J Mol Neurosci. 1999;12(2):131–45. doi: 10.1007/BF02736927. [DOI] [PubMed] [Google Scholar]
- Aschrafi A, Natera-Naranjo O, Gioio AE, Kaplan BB. Regulation of axonal trafficking of cytochrome c oxidase IV mRNA. Mol Cell Neurosci. 2010;43(4):422–30. doi: 10.1016/j.mcn.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28(47):12581–90. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baleriola J, Hengst U. Targeting axonal protein synthesis in neuroregeneration and degeneration. Neurotherapeutics. 2015;12(1):57–65. doi: 10.1007/s13311-014-0308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baleriola J, Walker CA, Jean YY, Crary JF, Troy CM, Nagy PL, et al. Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. Cell. 2014;158(5):1159–72. doi: 10.1016/j.cell.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, et al. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18(1):251–65. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckel-Mitchener AC, Miera A, Keller R, Perrone-Bizzozero NI. Poly(A) tail length-dependent stabilization of GAP-43 mRNA by the RNA-binding protein HuD. J Biol Chem. 2002;277(31):27996–8002. doi: 10.1074/jbc.M201982200. [DOI] [PubMed] [Google Scholar]
- Ben-Yaakov K, Dagan SY, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, et al. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 2012;31(6):1350–63. doi: 10.1038/emboj.2011.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigler RL, Kamande JW, Dumitru R, Niedringhaus M, Taylor AM. Messenger RNAs localized to distal projections of human stem cell derived neurons. Sci Rep. 2017;7(1):611. doi: 10.1038/s41598-017-00676-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird CW, Gardiner AS, Bolognani F, Tanner DC, Chen CY, Lin WJ, et al. KSRP modulation of GAP-43 mRNA stability restricts axonal outgrowth in embryonic hippocampal neurons. PLoS One. 2013;8(11):e79255. doi: 10.1371/journal.pone.0079255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleher R, Martin R. Ribosomes in the squid giant axon. Neuroscience. 2001;107(3):527–34. doi: 10.1016/s0306-4522(01)00366-9. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Contente-Cuomo T, Perrone-Bizzozero NI. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res. 2010;38(1):117–30. doi: 10.1093/nar/gkp863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese M, Saal L, Appenzeller S, Moradi M, Baluapuri A, Sendtner M. Whole transcriptome profiling reveals the RNA content of motor axons. Nucleic Acids Res. 2016;44(4):e33. doi: 10.1093/nar/gkv1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittis PA, Lu Q, Flanagan JG. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell. 2002;110(2):223–35. doi: 10.1016/s0092-8674(02)00813-9. [DOI] [PubMed] [Google Scholar]
- Campbell D, Holt C. Chemotropic responses of reginal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1016. doi: 10.1016/s0896-6273(01)00551-7. [DOI] [PubMed] [Google Scholar]
- Carrel TL, McWhorter ML, Workman E, Zhang H, Wolstencroft EC, Lorson C, et al. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J Neurosci. 2006;26(43):11014–22. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24(2):148–58. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker KE, Fenstermacher SJ, Pazyra-Murphy MF, Elliott HL, Segal RA. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat Neurosci. 2016;19(5):690–6. doi: 10.1038/nn.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker KE, Pazyra-Murphy MF, Fenstermacher SJ, Segal RA. Target-derived neurotrophins coordinate transcription and transport of bclw to prevent axonal degeneration. J Neurosci. 2013;33(12):5195–207. doi: 10.1523/JNEUROSCI.3862-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10(2):149–59. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Liegro CM, Schiera G, Di Liegro I. Regulation of mRNA transport, localization and translation in the nervous system of mammals (Review) Int J Mol Med. 2014;33(4):747–62. doi: 10.3892/ijmm.2014.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Gingeras TR. Mapping RNA-seq Reads with STAR. Curr Protoc Bioinformatics. 2015;51:11–14. 1–19. doi: 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombert B, Sivadasan R, Simon CM, Jablonka S, Sendtner M. Presynaptic localization of Smn and hnRNP R in axon terminals of embryonic and postnatal mouse motoneurons. PLoS One. 2014;9(10):e110846. doi: 10.1371/journal.pone.0110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Park M, Spillane M, Yoo S, Pacheco A, Gomes C, et al. Axonally Synthesized beta-Actin and GAP-43 Proteins Support Distinct Modes of Axonal Growth. J Neurosci. 2013a;33(8):3311–22. doi: 10.1523/JNEUROSCI.1722-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, et al. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30(22):4665–77. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013b;80(2):415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edstrom JE, Eichner D, Edstrom A. The ribonucleic acid of axons and myelin sheaths from Mauthner neurons. Biochim Biophys Acta. 1962;61:178–84. doi: 10.1016/0926-6550(62)90080-4. [DOI] [PubMed] [Google Scholar]
- Eng H, Lund K, Campenot RB. Synthesis of beta-tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J Neurosci. 1999;19:1–9. doi: 10.1523/JNEUROSCI.19-01-00001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist LW. Five questions about viral trafficking in neurons. PLoS Pathog. 2012;8(2):e1002472. doi: 10.1371/journal.ppat.1002472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyman M, Cefaliello C, Ferrara E, De Stefano R, Lavina ZS, Crispino M, et al. Local synthesis of axonal and presynaptic RNA in squid model systems. Eur J Neurosci. 2007;25(2):341–50. doi: 10.1111/j.1460-9568.2007.05304.x. [DOI] [PubMed] [Google Scholar]
- Fallini C, Bassell GJ, Rossoll W. The ALS disease protein TDP-43 is actively transported in motor neuron axons and regulates axon outgrowth. Hum Mol Genet. 2012;21(16):3703–18. doi: 10.1093/hmg/dds205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Donlin-Asp PG, Rouanet JP, Bassell GJ, Rossoll W. Deficiency of the Survival of Motor Neuron Protein Impairs mRNA Localization and Local Translation in the Growth Cone of Motor Neurons. J Neurosci. 2016;36(13):3811–20. doi: 10.1523/JNEUROSCI.2396-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallini C, Zhang H, Su Y, Silani V, Singer RH, Rossoll W, et al. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J Neurosci. 2011;31(10):3914–25. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH. Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol. 2003;160(1):77–87. doi: 10.1083/jcb.200206003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltrin D, Fusco L, Witte H, Moretti F, Martin K, Letzelter M, et al. Growth cone MKK7 mRNA targeting regulates MAP1b-dependent microtubule bundling to control neurite elongation. PLoS Biol. 2012;10(12):e1001439. doi: 10.1371/journal.pbio.1001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel R, Koenig E. Identification of major indigenous protein components in mammalian axons and locally synthesized axonal protein in hypoglossal nerve. Exp Neurol. 1977;57:282–295. doi: 10.1016/0014-4886(77)90064-4. [DOI] [PubMed] [Google Scholar]
- Funch PG, Kinsman SL, Faber DS, Koenig E, Zottoli SJ. Mauthner axon diameter and impulse conduction velocity decrease with growth of goldfish. Neurosci Lett. 1981;27(2):159–64. doi: 10.1016/0304-3940(81)90261-5. [DOI] [PubMed] [Google Scholar]
- Gervasi NM, Scott SS, Aschrafi A, Gale J, Vohra SN, MacGibeny MA, et al. The local expression and trafficking of tyrosine hydroxylase mRNA in the axons of sympathetic neurons. RNA. 2016a doi: 10.1261/rna.053272.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervasi NM, Scott SS, Aschrafi A, Gale J, Vohra SN, MacGibeny MA, et al. The local expression and trafficking of tyrosine hydroxylase mRNA in the axons of sympathetic neurons. RNA. 2016b;22(6):883–95. doi: 10.1261/rna.053272.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioio A, Chun J-T, Crispino M, Capano C, Giuditta A, Kaplan B. Kinesin mRNA is present in the squid giant axon. J Neurochem. 1994;63:13–18. doi: 10.1046/j.1471-4159.1994.63010013.x. [DOI] [PubMed] [Google Scholar]
- Gioio AE, Eyman M, Zhang H, Lavina ZS, Giuditta A, Kaplan BB. Local synthesis of nuclear-encoded mitochondrial proteins in the presynaptic nerve terminal. J Neurosci Res. 2001;64(5):447–53. doi: 10.1002/jnr.1096. [DOI] [PubMed] [Google Scholar]
- Gioio AE, Lavina ZS, Jurkovicova D, Zhang H, Eyman M, Giuditta A, et al. Nerve terminals of squid photoreceptor neurons contain a heterogeneous population of mRNAs and translate a transfected reporter mRNA. Eur J Neurosci. 2004;20(4):865–72. doi: 10.1111/j.1460-9568.2004.03538.x. [DOI] [PubMed] [Google Scholar]
- Giuditta A, Cupello A, Lazzarini G. Ribosomal RNA in the axoplasm of the squid giant axon. J Neurochem. 1980;34(6):1757–60. doi: 10.1111/j.1471-4159.1980.tb11271.x. [DOI] [PubMed] [Google Scholar]
- Giuditta A, Dettbarn WD, Brzin M. Protein synthesis in the isolated giant axon of the squid. Proc Natl Acad Sci U S A. 1968;59(4):1284–7. doi: 10.1073/pnas.59.4.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuditta A, Menichini E, Perrone Capano C, Langella M, Martin R, Castigli E, et al. Active polysomes in the axoplasm of the squid giant axon. J Neurosci Res. 1991;28(1):18–28. doi: 10.1002/jnr.490280103. [DOI] [PubMed] [Google Scholar]
- Giuditta A, Metafora S, Felsani A, Del Rio A. Factors for protein synthesis in the axoplasm of squid giant axons. J Neurochem. 1977;28(6):1393–5. doi: 10.1111/j.1471-4159.1977.tb12339.x. [DOI] [PubMed] [Google Scholar]
- Glinka M, Herrmann T, Funk N, Havlicek S, Rossoll W, Winkler C, et al. The heterogeneous nuclear ribonucleoprotein-R is necessary for axonal beta-actin mRNA translocation in spinal motor neurons. Hum Mol Genet. 2010;19(10):1951–66. doi: 10.1093/hmg/ddq073. [DOI] [PubMed] [Google Scholar]
- Gumy LF, Yeo GS, Tung YC, Zivraj KH, Willis D, Coppola G, et al. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17(1):85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507(7491):195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, et al. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40(6):1095–104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Hsu W, HW C, Wu C, Wu H, Lee Y, Chen E, et al. Glutamate Stimulates Local Protein Synthesis in the Axons of Rat Cortical Neurons by Activating α-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors and Metabotropic Glutamate Receptors. J Biol Chem. 2015;290(34):20748–60. doi: 10.1074/jbc.M115.638023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingoglia NA, Giuditta A, Zanakis MF, Babigian A, Tasaki I, Chakraborty G, et al. Incorporation of 3H-amino acids into proteins in a partially purified fraction of axoplasm: evidence for transfer RNA-mediated, post-translational protein modification in squid giant axons. J Neurosci. 1983;3(12):2463–73. doi: 10.1523/JNEUROSCI.03-12-02463.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka S, Beck M, Lechner BD, Mayer C, Sendtner M. Defective Ca2+ channel clustering in axon terminals disturbs excitability in motoneurons in spinal muscular atrophy. J Cell Biol. 2007;179(1):139–49. doi: 10.1083/jcb.200703187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Diaz L, Geranton SM, Passmore GM, Leith JL, Fisher AS, Berliocchi L, et al. Local translation in primary afferent fibers regulates nociception. PLoS ONE. 2008;3(4):e1961. doi: 10.1371/journal.pone.0001961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin LQ, Pennise CR, Rodemer W, Jahn KS, Selzer ME. Protein synthetic machinery and mRNA in regenerating tips of spinal cord axons in lamprey. J Comp Neurol. 2016;524(17):3614–3640. doi: 10.1002/cne.24020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284(30):20329–39. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonson L, Vikesaa J, Krogh A, Nielsen LK, Hansen T, Borup R, et al. Molecular composition of IMP1 ribonucleoprotein granules. Mol Cell Proteomics. 2007;6(5):798–811. doi: 10.1074/mcp.M600346-MCP200. [DOI] [PubMed] [Google Scholar]
- Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13(5):308–24. doi: 10.1038/nrn3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinski AL, Sachdeva R, Gomes C, Lee SJ, Shah Z, Houle JD, et al. mRNAs and Protein Synthetic Machinery Localize into Regenerating Spinal Cord Axons When They Are Provided a Substrate That Supports Growth. J Neurosci. 2015;35(28):10357–70. doi: 10.1523/JNEUROSCI.1249-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan BB, Gioio AE, Hillefors M, Aschrafi A. Axonal protein synthesis and the regulation of local mitochondrial function. Results Probl Cell Differ. 2009;48:225–42. doi: 10.1007/400_2009_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar AN, Macgibeny MA, Gervasi NM, Gioio AE, Kaplan BB. Intra-axonal Synthesis of Eukaryotic Translation Initiation Factors Regulates Local Protein Synthesis and Axon Growth in Rat Sympathetic Neurons. J Neurosci. 2013;33(17):7165–74. doi: 10.1523/JNEUROSCI.2040-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Wolfson A. New competition in RNA regulation. Nat Biotechnol. 2012;30(1):58–9. doi: 10.1038/nbt.2092. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kim P, Phay M, Yoo S. Identification of precursor microRNAs within distal axons of sensory neuron. J Neurochem. 2015;134(2):193–9. doi: 10.1111/jnc.13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Li Z, Singer RH, Taneja KL. Isoform-specific 3′-untranslated sequences sort alpha-cardiac and beta-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J Cell Biol. 1993;123(1):165–72. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, et al. Translocation of RNA granules in living neurons. J Neurosci. 1996;16(24):7812–20. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig E. Synthetic mechanisms in the axon - I: local axonal synthesis of acetylcholinesterase. J Neurochem. 1965a;12:343–355. doi: 10.1111/j.1471-4159.1965.tb04235.x. [DOI] [PubMed] [Google Scholar]
- Koenig E. Synthetic mechanisms in the axon - II: RNA in myelin-free axons of the cat. J Neurochem. 1965b;12:357–361. doi: 10.1111/j.1471-4159.1965.tb04236.x. [DOI] [PubMed] [Google Scholar]
- Koenig E. Synthetic mechanisms in the axon - III: Stimulation of acetylcholinesterase synthesis by actinomycin-D in the hypoglossal nerve. J Neurochem. 1967a;14(4):429–35. doi: 10.1111/j.1471-4159.1967.tb09541.x. [DOI] [PubMed] [Google Scholar]
- Koenig E. Synthetic mechanisms in the axon - IV: In vitro incorporation of [3H]precursors into axonal protein and RNA. J Neurochem. 1967b;14(4):437–46. doi: 10.1111/j.1471-4159.1967.tb09542.x. [DOI] [PubMed] [Google Scholar]
- Koenig E. Synthetic mechanisms in the axon. IV. In vitro incorporation of [3H]precursors into axonal protein and RNA. J Neurochem. 1967c;14(4):437–46. doi: 10.1111/j.1471-4159.1967.tb09542.x. [DOI] [PubMed] [Google Scholar]
- Koenig E. Ribosomal RNA in Mauthner axon: Implications for a protein synthesizing machinery in the myelinated axon. Brain Research. 1979;174:95–107. doi: 10.1016/0006-8993(79)90806-0. [DOI] [PubMed] [Google Scholar]
- Koenig E, Adams P. Local protein synthesizing activity in axonal fields regenerating in vitro. J Neurochem. 1982;39(2):386–400. doi: 10.1111/j.1471-4159.1982.tb03960.x. [DOI] [PubMed] [Google Scholar]
- Kong L, Wang X, Choe DW, Polley M, Burnett BG, Bosch-Marce M, et al. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci. 2009;29(3):842–51. doi: 10.1523/JNEUROSCI.4434-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyuncu OO, Perlman DH, Enquist LW. Efficient retrograde transport of pseudorabies virus within neurons requires local protein synthesis in axons. Cell Host Microbe. 2013;13(1):54–66. doi: 10.1016/j.chom.2012.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TL, Yoon YJ, Mowry KL. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J Cell Biol. 2004;165(2):203–11. doi: 10.1083/jcb.200309145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323(5918):1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Kye MJ, Liu T, Levy SF, Xu NL, Groves BB, Bonneau R, et al. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13(8):1224–34. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCroix-Fralish ML, Austin JS, Zheng FY, Levitin DJ, Mogil JS. Patterns of pain: meta-analysis of microarray studies of pain. Pain. 2011;152(8):1888–98. doi: 10.1016/j.pain.2011.04.014. [DOI] [PubMed] [Google Scholar]
- LaFerla FM, Tinkle BT, Bieberich CJ, Haudenschild CC, Jay G. The Alzheimer’s A beta peptide induces neurodegeneration and apoptotic cell death in transgenic mice. Nat Genet. 1995;9(1):21–30. doi: 10.1038/ng0195-21. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136(6):1001–4. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9(10):1247–56. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezana JP, Dagan SY, Robinson A, Goldstein RS, Fainzilber M, Bronfman FC, et al. Axonal PPARgamma promotes neuronal regeneration after injury. Dev Neurobiol. 2016;76(6):688–701. doi: 10.1002/dneu.22353. [DOI] [PubMed] [Google Scholar]
- Li C, Bassell GJ, Sasaki Y. Fragile X Mental Retardation Protein is Involved in Protein Synthesis-Dependent Collapse of Growth Cones Induced by Semaphorin-3A. Front Neural Circuits. 2009;3:11. doi: 10.3389/neuro.04.011.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Quon G, Lipshitz HD, Morris Q. Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. RNA. 2010;16(6):1096–107. doi: 10.1261/rna.2017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11(4):323–30. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162(5):919–31. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meer EJ, Wang DO, Kim S, Barr I, Guo F, Martin KC. Identification of a cis-acting element that localizes mRNA to synapses. Proc Natl Acad Sci U S A. 2012;109(12):4639–44. doi: 10.1073/pnas.1116269109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melemedjian OK, Asiedu MN, Tillu DV, Peebles KA, Yan J, Ertz N, et al. IL-6- and NGF-Induced Rapid Control of Protein Synthesis and Nociceptive Plasticity via Convergent Signaling to the eIF4F Complex. J Neurosci. 2010;30(45):15113–23. doi: 10.1523/JNEUROSCI.3947-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melemedjian OK, Asiedu MN, Tillu DV, Sanoja R, Yan J, Lark A, et al. Targeting adenosine monophosphate-activated protein kinase (AMPK) in preclinical models reveals a potential mechanism for the treatment of neuropathic pain. Mol Pain. 2011;7:70. doi: 10.1186/1744-8069-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melemedjian OK, Khoutorsky A, Sorge RE, Yan J, Asiedu MN, Valdez A, et al. mTORC1 inhibition induces pain via IRS-1-dependent feedback activation of ERK. Pain. 2013;154(7):1080–91. doi: 10.1016/j.pain.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melemedjian OK, Tillu DV, Moy JK, Asiedu MN, Mandell EK, Ghosh S, et al. Local translation and retrograde axonal transport of CREB regulates IL-6-induced nociceptive plasticity. Mol Pain. 2014;10:45. doi: 10.1186/1744-8069-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda TT, Coleman J, Kim HH, Kumar Sahoo P, Gomes C, Brito-Vargas P, et al. Axonal amphoterin mRNA is regulated by translational control and enhances axon outgrowth. J Neurosci. 2015;35(14):5693–706. doi: 10.1523/JNEUROSCI.3397-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda TT, Gomes C, Yoo S, Vuppalanchi D, Twiss JL. Axonal localization of neuritin/CPG15 mRNA in neuronal populations through distinct 5′ and 3′ UTR elements. J Neurosci. 2013a;33(34):13735–42. doi: 10.1523/JNEUROSCI.0962-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merianda TT, Vuppalanchi D, Yoo S, Blesch A, Twiss JL. Axonal transport of neural membrane protein 35 mRNA increases axon growth. J Cell Sci. 2013b;126(Pt 1):90–102. doi: 10.1242/jcs.107268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Mielke M, Spires-Jones TL, Stoothoff W, Jones P, Bacskai BJ, et al. A reporter of local dendritic translocation shows plaque- related loss of neural system function in APP-transgenic mice. J Neurosci. 2009;29(40):12636–40. doi: 10.1523/JNEUROSCI.1948-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikl M, Vendra G, Kiebler MA. Independent localization of MAP2, CaMKIIalpha and beta-actin RNAs in low copy numbers. EMBO Rep. 2011;12(10):1077–84. doi: 10.1038/embor.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453(7191):115–9. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S, Julien JP. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci. 2013;14(3):161–76. doi: 10.1038/nrn3380. [DOI] [PubMed] [Google Scholar]
- Minis A, Dahary D, Manor O, Leshkowitz D, Pilpel Y, Yaron A. Subcellular transcriptomics-dissection of the mRNA composition in the axonal compartment of sensory neurons. Dev Neurobiol. 2014;74(3):365–81. doi: 10.1002/dneu.22140. [DOI] [PubMed] [Google Scholar]
- Moradi M, Sivadasan R, Saal L, Luningschror P, Dombert B, Rathod RJ, et al. Differential roles of alpha-, beta-, and gamma-actin in axon growth and collateral branch formation in motoneurons. J Cell Biol. 2017;216(3):793–814. doi: 10.1083/jcb.201604117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Aschrafi A, Gioio AE, Kaplan BB. Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA. 2010;16(8):1516–29. doi: 10.1261/rna.1833310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natera-Naranjo O, Kar AN, Aschrafi A, Gervasi NM, Macgibeny MA, Gioio AE, et al. Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol Cell Neurosci. 2012;49(3):263–70. doi: 10.1016/j.mcn.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Olink-Coux M, Hollenbeck PJ. Localization and active transport of mRNA in axons of sympathetic neurons. J Neurosci. 1996;16:1346–1358. doi: 10.1523/JNEUROSCI.16-04-01346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareyson D, Saveri P, Sagnelli A, Piscosquito G. Mitochondrial dynamics and inherited peripheral nerve diseases. Neurosci Lett. 2015;596:66–77. doi: 10.1016/j.neulet.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Pepe IM, Giuditta A, Cimarra P. Inhibition of neuronal protein synthesis in the giant fibre system of the squid by a high potassium concentration. J Neurochem. 1975;24(6):1271–3. doi: 10.1111/j.1471-4159.1975.tb03911.x. [DOI] [PubMed] [Google Scholar]
- Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45(5):715–26. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Perry RB, Fainzilber M. Local translation in neuronal processes–in vivo tests of a “heretical hypothesis”. Dev Neurobiol. 2014;74(3):210–7. doi: 10.1002/dneu.22115. [DOI] [PubMed] [Google Scholar]
- Perry RB, Rishal I, Doron-Mandel E, Kalinski AL, Medzihradszky KF, Terenzio M, et al. Nucleolin-Mediated RNA Localization Regulates Neuron Growth and Cycling Cell Size. Cell Rep. 2016;16(6):1664–1676. doi: 10.1016/j.celrep.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RB-T, Doron E, Iavnilovitch E, Rishal I, Dagan S, Tsoory M, et al. Subcellular Knockout of Importin β1 Perturbs Axonal Retrograde Signaling. Neuron. 2012;75:294–305. doi: 10.1016/j.neuron.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phay M, Kim HH, Yoo S. Dynamic Change and Target Prediction of Axon-Specific MicroRNAs in Regenerating Sciatic Nerve. PLoS One. 2015;10(9):e0137461. doi: 10.1371/journal.pone.0137461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phay M, Kim HH, Yoo S. Analysis of piRNA-Like Small Non-coding RNAs Present in Axons of Adult Sensory Neurons. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-0340-2. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike CJ, Burdick D, Walencewicz AJ, Glabe CG, Cotman CW. Neurodegeneration induced by beta-amyloid peptides in vitro: the role of peptide assembly state. J Neurosci. 1993;13(4):1676–87. doi: 10.1523/JNEUROSCI.13-04-01676.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Anderson R, Dwivedy A, Weinl C, van Horck F, Leung KM, et al. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron. 2006;49(2):215–28. doi: 10.1016/j.neuron.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Salih S, Weinl C, Holt CE, Harris WA. Endocytosis dependent desensitization and protein synthesis-dependent resensitization in retional growth cone adaptation. Nat Neurosci. 2005;8:179–186. doi: 10.1038/nn1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Quan J, Nowakowski DW, Hancock ML, Shi J, Tcherkezian J, et al. APC is an RNA-binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell. 2014;158(2):368–82. doi: 10.1016/j.cell.2014.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rage F, Boulisfane N, Rihan K, Neel H, Gostan T, Bertrand E, et al. Genome-wide identification of mRNAs associated with the protein SMN whose depletion decreases their axonal localization. RNA. 2013;19(12):1755–66. doi: 10.1261/rna.040204.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihan K, Antoine E, Maurin T, Bardoni B, Bordonne R, Soret J, et al. A new cis-acting motif is required for the axonal SMN-dependent Anxa2 mRNA localization. RNA. 2017 doi: 10.1261/rna.056788.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll W, Jablonka S, Andreassi C, Kroning AK, Karle K, Monani UR, et al. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003;163(4):801–12. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll W, Kroning AK, Ohndorf UM, Steegborn C, Jablonka S, Sendtner M. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum Mol Genet. 2002;11(1):93–105. doi: 10.1093/hmg/11.1.93. [DOI] [PubMed] [Google Scholar]
- Rotem N, Magen I, Ionescu A, Gershoni-Emek N, Altman T, Costa CJ, et al. ALS Along the Axons - Expression of Coding and Noncoding RNA Differs in Axons of ALS models. Sci Rep. 2017;7:44500. doi: 10.1038/srep44500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz R, Casanas JJ, Torres-Benito L, Cano R, Tabares L. Altered intracellular Ca2+ homeostasis in nerve terminals of severe spinal muscular atrophy mice. J Neurosci. 2010;30(3):849–57. doi: 10.1523/JNEUROSCI.4496-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saal L, Briese M, Kneitz S, Glinka M, Sendtner M. Subcellular transcriptome alterations in a cell culture model of spinal muscular atrophy point to widespread defects in axonal growth and presynaptic differentiation. RNA. 2014;20(11):1789–802. doi: 10.1261/rna.047373.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva R, Farrell K, McMullen M-K, Twiss JL, Houle JD. Dynamic Changes in Local Protein Synthetic Machinery in Regenerating Central Nervous System Axons after Spinal Cord Injury. Neural Plasticity. 2016;2016:4087254. doi: 10.1155/2016/4087254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Gross C, Xing L, Goshima Y, Bassell GJ. Identification of axon-enriched microRNAs localized to growth cones of cortical neurons. Dev Neurobiol. 2014;74(3):397–406. doi: 10.1002/dneu.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scekic-Zahirovic J, Sendscheid O, El Oussini H, Jambeau M, Sun Y, Mersmann S, et al. Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. EMBO J. 2016;35(10):1077–97. doi: 10.15252/embj.201592559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, et al. Dynamic Axonal Translation in Developing and Mature Visual Circuits. Cell. 2016;166(1):181–92. doi: 10.1016/j.cell.2016.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiihashi G, Ito D, Yagi T, Nihei Y, Ebine T, Suzuki N. Mislocated FUS is sufficient for gain-of-toxic-function amyotrophic lateral sclerosis phenotypes in mice. Brain. 2016;139(Pt 9):2380–94. doi: 10.1093/brain/aww161. [DOI] [PubMed] [Google Scholar]
- Song R, Koyuncu OO, Greco TM, Diner BA, Cristea IM, Enquist LW. Two Modes of the Axonal Interferon Response Limit Alphaherpesvirus Neuroinvasion. MBio. 2016;7(1):e02145–15. doi: 10.1128/mBio.02145-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo JR, Kun A, Benech JC, Giuditta A, Morillas J, Benech CR. Ribosomes and polyribosomes are present in the squid giant axon: An immunocytochemical study. Neuroscience. 1999;90(2):705–715. doi: 10.1016/s0306-4522(98)00587-9. [DOI] [PubMed] [Google Scholar]
- Spillane M, Ketschek A, Jones SL, Korobova F, Marsick B, Lanier L, et al. The actin nucleating Arp2/3 complex contributes to the formation of axonal filopodia and branches through the regulation of actin patch precursors to filopodia. Develop Neurobiol. 2011;71(9):747–58. doi: 10.1002/dneu.20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillane M, Ketschek A, Merianda TT, Twiss JL, Gallo G. Mitochondria coordinate sites of axon branching through localized intra-axonal protein synthesis. Cell Rep. 2013;5(6):1564–75. doi: 10.1016/j.celrep.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–72. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O. Polyribosomes at the base of dendritic spines of central nervous system neurons–their possible role in synapse construction and modification. Cold Spring Harb Symp Quant Biol. 1983;48(Pt 2):745–59. doi: 10.1101/sqb.1983.048.01.077. [DOI] [PubMed] [Google Scholar]
- Sun S, Ling SC, Qiu J, Albuquerque CP, Zhou Y, Tokunaga S, et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat Commun. 2015;6:6171. doi: 10.1038/ncomms7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajrishi MM, Tuteja R, Tuteja N. Nucleolin: The most abundant multifunctional phosphoprotein of nucleolus. Commun Integr Biol. 2011;4(3):267–75. doi: 10.4161/cib.4.3.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro JM, Vidaki M, Oliveira R, Olson S, Zhan L, Saxena T, et al. Distal Alternative Last Exons Localize mRNAs to Neural Projections. Mol Cell. 2016;61(6):821–33. doi: 10.1016/j.molcel.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Berchtold NC, Perreau VM, Tu CH, Li Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29(15):4697–707. doi: 10.1523/JNEUROSCI.6130-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2(8):599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Wu J, Tai HC, Schuman EM. Axonal translation of beta-catenin regulates synaptic vesicle dynamics. J Neurosci. 2013;33(13):5584–9. doi: 10.1523/JNEUROSCI.2944-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson VM. The fine structure of the axon and growth cone of the dorsal root neuroblast of the rabbit embryo. J Cell Biol. 1970;44(1):62–79. doi: 10.1083/jcb.44.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Jinu S, Gordon PM, Fielding T, Taylor R, Smith BN, Snowden V, et al. Non-nuclear Pool of Splicing Factor SFPQ Regulates Axonal Transcripts Required for Normal Motor Development. Neuron. 2017;94(2):322–336. doi: 10.1016/j.neuron.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai NP, Tsui YC, Wei LN. Dynein motor contributes to stress granule dynamics in primary neurons. Neuroscience. 2009;159:647–656. doi: 10.1016/j.neuroscience.2008.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai NP, Wei LN. RhoA/ROCK1 signaling regulates stress granule formation and apoptosis. Cell Signal. 2010;22(4):668–75. doi: 10.1016/j.cellsig.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Chierzi S, Codd AM, Campbell DS, Meyer RL, Holt CE, et al. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J Neurosci. 2005;25(2):331–42. doi: 10.1523/JNEUROSCI.3073-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarin JM, McCurdy EP, Martinez JC, Hengst U. Local synthesis of dynein cofactors matches retrograde transport to acutely changing demands. Nat Commun. 2016;7:13865. doi: 10.1038/ncomms13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuppalanchi D, Coleman J, Yoo S, Merianda TT, Yadhati AG, Hossain J, et al. Conserved 3′-untranslated region sequences direct subcellular localization of chaperone protein mRNAs in neurons. J Biol Chem. 2010;285(23):18025–38. doi: 10.1074/jbc.M109.061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, et al. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324(5934):1536–40. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Jurkovicova D, Pickel VM, Gioio AE, Kaplan BB. Identification of a novel membrane-associated protein expressed in neurons of the squid and rodent nervous systems. Neuroscience. 2002;114(4):995–1004. doi: 10.1016/s0306-4522(02)00362-7. [DOI] [PubMed] [Google Scholar]
- Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, et al. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178(6):965–80. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, et al. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436(7053):1020–4. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca(2+)-dependent growth cone guidance. Nat Neurosci. 2006;9(10):1265–73. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Clatterbuck-Soper SF, Jackrel ME, Shorter J, Mili S. FUS inclusions disrupt RNA localization by sequestering kinesin-1 and inhibiting microtubule detyrosination. J Cell Biol. 2017;216(4):1015–1034. doi: 10.1083/jcb.201608022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Zhang H, Loiselle D, Haystead T, Macara IG, Mili S. The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. J Cell Biol. 2013;203(5):737–46. doi: 10.1083/jcb.201306058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S, Kim HH, Kim P, Donnelly CJ, Kalinski AL, Vuppalanchi D, et al. A HuD-ZBP1 ribonucleoprotein complex localizes GAP-43 mRNA into axons through its 3′ untranslated region AU-rich regulatory element. J Neurochem. 2013;126(6):792–804. doi: 10.1111/jnc.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BC, Jung H, Dwivedy A, O’Hare CM, Zivraj KH, Holt CE. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148(4):752–64. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, et al. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59(2):241–52. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelena J. Ribosome-like particles in myelinated axons of the rat. Brain Res. 1970;24:359–363. doi: 10.1016/0006-8993(70)90120-4. [DOI] [PubMed] [Google Scholar]
- Zelena J. Ribosomes in myelinated axons of dorsal root ganglia. Zeitschrift fur Zellforschung und Mikroskopische Anatomie. 1972;124(2):217–29. doi: 10.1007/BF00335680. [DOI] [PubMed] [Google Scholar]
- Zhang H, Xing L, Rossoll W, Wichterle H, Singer RH, Bassell GJ. Multiprotein Complexes of the Survival of Motor Neuron Protein SMN with Gemins Traffic to Neuronal Processes and Growth Cones of Motor Neurons. J Neurosci. 2006;26(33):8622–8632. doi: 10.1523/JNEUROSCI.3967-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, et al. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31(2):261–75. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Pan F, Hong D, Shenoy SM, Singer RH, Bassell GJ. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J Neurosci. 2003;23(16):6627–37. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of beta-actin mRNA and protein localization within growth cones. J Cell Biol. 1999;147(1):59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525(7567):56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, et al. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J Neurosci. 2001;21(23):9291–303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivraj KH, Tung YC, Piper M, Gumy L, Fawcett JW, Yeo GS, et al. Subcellular Profiling Reveals Distinct and Developmentally Regulated Repertoire of Growth Cone mRNAs. J Neurosci. 2010;30(46):15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]


