Abstract
Despite prognostic grading and staging systems, it is a challenge to predict outcomes for patients with pancreatic neuroendocrine tumors (PanNETs). Sequencing studies of PanNETs have identified alterations in death domain-associated protein (DAXX) and ATRX chromatin remodeler (ATRX). In tumors, mutations in DAXX or ATRX and corresponding loss of protein expression correlate with shorter times of disease-free survival and disease-specific survival of patients. However, DAXX or ATRX proteins were lost in only 50% of distant metastases analyzed. We performed whole-exome sequencing analyses of 20 distant metastases from 20 patients with a single non-syndrome, non-functional PanNET. We found distant metastases contained alterations in MEN1 (n=8), ATRX (n=5), DAXX (n=5), TSC2 (n=3), and DEPDC5 (n=3). We found copy number loss of CDKN2A in 15 metastases (75%) and alterations in genes that regulate chromatin remodeling including SETD2 (n=4), ARID1A (n=2), CHD8 (n=2), and DNMT1 (n=2). In a separate analysis of 347 primary PanNETs, we found loss or deletion of DAXX and ATRX, disruption of SETD2 function (based on loss of H3K36me3), loss of ARID1A expression or deletions in CDKN2A in 81% of primary PanNETs with distant metastases. Among patients with loss or deletion of at least 1 of these proteins or genes, 39% survived disease free for 5 years and 44% had disease-specific survival times of 10 years. Among patients without any of these alterations, 98% survived disease free for 5 years and 95% had disease-specific survival times of 10 years. Therefore, primary PanNETs with loss of DAXX, ATRX, H3K36me3, ARID1A, and/or CDKN2A associate with shorter survival times of patients. Our findings indicate that alterations in chromatin remodeling genes and CDKN2A contribute to metastasis of PanNETs.
Keywords: pancreas, prognosis, prognostic factor, risk
Pancreatic neuroendocrine tumors (PanNETs) are a heterogeneous group of neoplasms with increasing incidence and ill-defined pathobiology. While most PanNETs are indolent and remain stable for years, a subset may behave aggressively and metastasize widely. Thus, the frequent detection of PanNETs presents a treatment dilemma. Current prognostic parameters and systems, such as tumor size and World Health Organization (WHO) grade, are susceptible to interpretation errors, sampling issues and, in a subset of PanNETs, do not accurately reflect the clinical behavior of these neoplasms.1–4 Hence, additional markers are needed to improve the prognostic classification of PanNETs.
Recently, whole-exome and whole-genome sequencing studies have focused on identifying recurrent genetic alterations in primary PanNETs.5, 6 Among these alterations, the most commonly mutated genes are MEN1, DAXX and ATRX. Death domain-associated protein (DAXX) and alpha-thalassemia/mental retardation X-linked (ATRX) genes encode for proteins that participate in chromatin remodeling at telomeres. Mutations in these genes are associated with loss of nuclear expression of their respective proteins by immunohistochemistry and correlate with alternative lengthening of telomeres (ALT), a telomerase-independent telomere maintenance mechanism.7 In addition, loss of DAXX and/or ATRX is associated with shorter times of disease-free survival (DFS) and disease-specific survival (DSS).8–10 Consequently, DAXX and/or ATRX loss is considered to be a driver of metastasis. However, only 50% of distant metastases demonstrate loss of DAXX and/or ATRX.
In contrast to primary PanNETs, the genetic landscape of metastatic PanNETs remains relatively unknown and, therefore, we hypothesized that additional genetic alterations other than those involving DAXX and ATRX account for the metastatic progression of PanNETs and may serve as useful prognostic markers. Thus, we performed whole-exome sequencing of 20 distant metastases from 20 patients with a solitary, non-syndrome and non-functional PanNET (Supplementary Materials and Methods). Similar to sequencing studies of primary PanNETs, whole-exome sequencing of metastatic PanNETs revealed frequent genomic alterations in MEN1 (n=8), ATRX (n=5), DAXX (n=5), TSC2 (n=3), and DEPDC5 (n=3) (Supplementary Table 1 and Supplementary Figure 1).5, 6 Inactivating mutations in DAXX and ATRX were mutually exclusive and correlated with loss of corresponding protein expression and the presence of ALT by telomere FISH. In addition, as described by Heaphy et al7, 2 DAXX-negative and 2 ATRX-negative metastatic PanNETs lacked mutations in DAXX and ATRX, respectively. In contrast to primary PanNETs, MEN1 was not the most commonly altered gene in metastatic PanNETs. CDKN2A copy number loss was found in 15 (75%) cases. Furthermore, genomic alterations in chromatin remodeling genes, such as SETD2 (n=4), ARID1A (n=2), CHD8 (n=2) and DNMT1 (n=2), were seen in 10 (50%) metastatic PanNETs.
Considering genomic alterations in SETD2, ARID1A and CDKN2A were previously described in primary PanNETs, but at a significantly lower prevalence than in metastatic PanNETs, the status of SETD2, ARID1A and CDKN2A was reevaluated using orthogonal methods. The SETD2 gene encodes for a histone methyltransferase that is specific for H3 lysine 36 trimethylation (H3K36me3) and loss-of-function mutations result in the absence of H3K36me3 expression by immunohistochemistry.11 ARID1A inactivating mutations are associated with loss of the corresponding protein.12 Genomic deletions in CDKN2A can be assayed using dual-color FISH.13 An analysis of the sequenced metastatic PanNETs revealed loss of H3K36me3 and ARID1A by immunohistochemistry and deletions for CDKN2A by dual-color FISH had a 100% concordance with alterations in their respective genes (Supplementary Figure 2).
In order to determine the prognostic significance of SETD2, ARID1A and CDKN2A alterations in relationship to DAXX/ATRX loss, the status of DAXX, ATRX, H3K36me3, ARID1A and CDKN2A was evaluated in 347 solitary, non-syndromic, primary PanNETs (Supplementary Tables 2 and 3). Loss or deletion of DAXX/ATRX, H3K36me3, ARID1A, CDKN2A was identified in 80 (23%), 28 (8%), 10 (3%) and 25 (7%) of primary PanNETs, respectively, and associated with larger mean tumor size, higher WHO grade, lymphovascular invasion, higher pathologic tumor stage, synchronous distant metastases and metachronous distant metastases (p < 0.05). Of note, DAXX/ATRX loss correlated with deletion in CDKN2A, but not the absence of H3K36me3 or ARID1A.
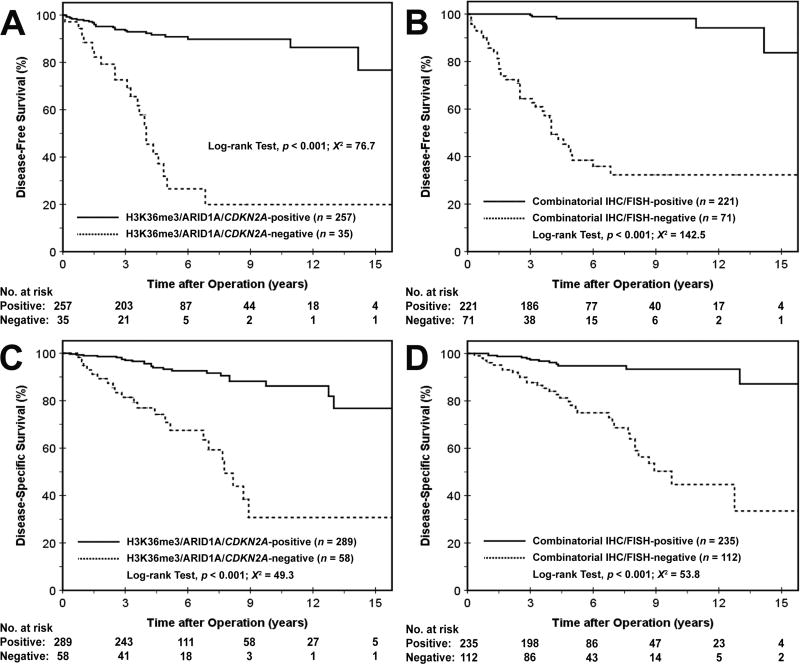
Overall, DAXX, ATRX, H3K36me3, ARID1A and/or CDKN2A loss or deletion was identified in 112 (32%) PanNETs. Further, loss or deletion of at least 1 marker was detected in 81 (81%) primary PanNETs from patients, who either had synchronous metastases (41 of 55, 75%) or developed metachronous metastases (without synchronous metastases) on follow-up (40 of 45, 89%). The DFS and DSS for patients with PanNETs demonstrating either loss or deletion in DAXX, ATRX, H3K36me3, ARID1A and/or CDKN2A were 63% at 3-years and 39% at 5-years (p < 0.001, X2 = 142.5), and 75% at 5-years and 44% at 10-years (p < 0.001, X2 = 53.9), respectively (Figure 1 and Supplementary Figure 3). Conversely, patients with PanNETs that lacked alterations in DAXX, ATRX, H3K36me3, ARID1A and CDKN2A had a DFS of 99% at 3-years and 98% at 5-years, and DSS of 96% at 5-years and 95% at 10-years. Univariate and multivariate Cox regression analysis were used to determine the prognostic significance of these 5 markers. Loss or deletion in any 1 marker was an independent prognostic factor for DFS with a hazard ratio (HR) of 14.44 (p < 0.001) and DSS with a HR of 3.12 (p = 0.006) (Table 1).
Figure 1.
Kaplan-Meier curves comparing the cumulative probability of disease-free survival (A and B, n=282) and disease-specific survival (C and D, n=347) after surgical resection among PanNET patients with regards to H3K36me3/ARID1A/CDKN2A and DAXX/ATRX/H3K36me3/ARID1A/CDKN2A status. Disease-free survival was calculated in patients that did not present with distant metastases at the time of surgical resection.
Table 1.
Univariate and multivariate Cox regression analysis for disease-free survival and disease-specific survival.
| Patient or Tumor Characteristics | Univariate Cox Regression Analysis | Multivariate Cox Regression Analysis | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Disease-Free Survival HR (95% CI) |
p | Disease-Specific Survival HR (95% CI) |
p | Disease-Free Survival HR (95% CI) |
p | Disease-Specific Survival HR (95% CI) |
p | |
| Gender, male vs. female | 1.10 (0.61 – 1.99) | 0.752 | 0.94 (0.52 – 1.68) | 0.824 | ||||
| Age, years | 1.03 (1.01 – 1.06) | 0.008 | 1.03 (1.00 – 1.05) | 0.038 | ||||
| Tumor size, > 2.0 cm vs. ≤ 2.0 cm | 37.79 (5.20 – 274.50) | < 0.001 | 27.57 (3.80 – 200.16) | 0.001 | CRIP10.30 (1.39 – 76.64) | 0.023 | 5.21 (0.69 – 39.62) | 0.111 |
| Functional vs. nonfunctional | 0.18 (0.02 – 1.28) | 0.086 | 0.58 (0.18 – 1.89) | 0.584 | ||||
| Location, head and uncinate vs. body and tail | 1.25 (0.69 – 2.27) | 0.459 | 1.67 (0.93 – 2.99) | 0.088 | ||||
| WHO grade, G2 or G3 vs. G1 | 4.74 (2.50 – 8.98) | < 0.001 | 5.79 (2.91 – 11.53) | < 0.001 | 1.54 (0.77 – 3.07) | 0.215 | 2.05 (0.99 – 4.27) | 0.054 |
| Lymphovascular invasion, presence vs. absence | 6.60 (3.38 – 12.90) | < 0.001 | 11.73 (4.91 – 28.00) | < 0.001 | ||||
| Perineural invasion, presence vs. absence | 5.43 (2.95 – 9.98) | < 0.001 | 4.05 (2.23 – 7.35) | < 0.001 | ||||
| Tumor stage (pT), pT3 vs. pT1 and pT2 | 5.51 (3.01 – 10.08) | < 0.001 | 8.60 (4.23 – 17.47) | < 0.001 | ||||
| Lymph node metastasis, presence vs. absence | 5.31 (2.84 – 9.93) | < 0.001 | 5.21 (2.69 – 10.10) | < 0.001 | 1.56 (0.80 – 3.04) | 0.190 | 2.12 (1.07 – 4.20) | 0.031 |
| Distant metastases at presentation, presence vs. absence | 10.02 (5.43 – 18.49) | < 0.001 | 3.64 (1.91 – 6.91) | < 0.001 | ||||
| DAXX/ATRX/H3K36me3/ARID1A/CDKN2A, loss/deletion vs. preserved/wild type | 35.45 (13.91 – 90.34) | < 0.001 | 9.55 (4.58 – 19.91) | < 0.001 | 14.43 (5.37 – 38.75) | < 0.001 | 3.12 (1.39 – 7.00) | 0.030 |
Notwithstanding, our study has a number of limitations. It is retrospective by design and not all patients received the same form of treatment. Among patients with primary PanNETs, 8% underwent enucleation or central pancreatectomy, and, as a result, regional lymphadenectomy may be suboptimal. Removing these patients from our analyses would have little impact on the statistical correlations associated with DAXX, ATRX, H3K36me3, ARID1A and CDKN2A status. Of note, enucleation and central pancreatectomy are typically done in the setting of small PanNETs (≤2 cm) because they often have an indolent clinical course.14 However, studies suggest a subset of small PanNETs can behave aggressively.2, 3 Within our cohort, 2% of small PanNETs developed distant metastases and in each case harbored loss or deletion of at least 1 marker. Although additional studies are necessary, loss or deletion in DAXX, ATRX, H3K36me3, ARID1A and/or CDKN2A in preoperative biopsies may indicate an increased risk of developing metastatic disease and, in turn, prompt a change in surgical management to ensure complete regional lymph node dissection.
In summary, metastatic PanNETs not only harbor frequent genetic alterations in MEN1, DAXX and ATRX, but also in SETD2, ARID1A and CDKN2A. Loss or deletion of DAXX/ATRX, H3K36me3 (as a surrogate for SETD2), ARID1A and CDKN2A in primary PanNETs correlated with several adverse clinicopathologic features. In addition, loss or deletion of at least 1 marker was associated with shorter DFS and DSS times, and a negative, prognostic factor for DFS and DSS, independent of tumor size and WHO grade. While further studies are required, the assessment of these 5 markers by immunohistochemistry and FISH offers an objective method of evaluating prognostic risk using pathologic specimens. Moreover, our observations highlight the potential role of chromatin remodeling genes and CDKN2A in the metastatic progression of PanNETs.
Supplementary Material
Acknowledgments
Funding support: This project was supported in part by a grant from the National Pancreas Foundation, Western Pennsylvania Chapter (to A. D. Singhi).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure/conflict of interest: The authors have no conflicts of interest to declare.
Author Contributions
Study Concept and Design: SR, WAF, HZ and ADS
Acquisition of Data: SR, WAF, TCL, DC, AL, CM, MAL, RJO, AHZ, MEH, AT, KKL, NB, REB, KM, MNN, AS, HJZ, and ADS
Analysis and Interpretation of Data: SR, WAF, HZ and ADS
Drafting of the Manuscript: SR, WAF, HZ and ADS
References
- 1.Reid MD, et al. Mod Pathol. 2016;29:93. [Google Scholar]
- 2.Kuo EJ, et al. Ann Surg Oncol. 2013;20:2815–21. doi: 10.1245/s10434-013-3005-7. [DOI] [PubMed] [Google Scholar]
- 3.Haynes AB, et al. Arch Surg. 2011;146:534–8. doi: 10.1001/archsurg.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz Del Arco C, et al. Diagn Cytopathol. 2017;45:29–35. doi: 10.1002/dc.23635. [DOI] [PubMed] [Google Scholar]
- 5.Jiao Y, et al. Science. 2011;331:1199–203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scarpa A, et al. Nature. 2017;543:65–71. doi: 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]
- 7.Heaphy CM, et al. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singhi AD, et al. Clin Cancer Res. 2017;23:600–609. doi: 10.1158/1078-0432.CCR-16-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JY, et al. Clin Cancer Res. 2017;23:1598–1606. doi: 10.1158/1078-0432.CCR-16-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marinoni I, et al. Gastroenterology. 2014;146:453–60. e5. doi: 10.1053/j.gastro.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 11.Ho TH, et al. Oncogene. 2016;35:1565–74. doi: 10.1038/onc.2015.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson WJ, et al. Nat Genet. 2016;48:848–55. doi: 10.1038/ng.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singhi AD, et al. Mod Pathol. 2016;29:14–24. doi: 10.1038/modpathol.2015.121. [DOI] [PubMed] [Google Scholar]
- 14.Kulke MH, et al. J Natl Compr Canc Netw. 2015;13:78–108. doi: 10.6004/jnccn.2015.0011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.