Abstract
Cholangiocarcinoma (CCA) is a highly lethal and aggressive disease. Recently, IDH1/2 mutations have been identified in approximately 20% of CCAs which suggests an involvement of 2-oxoglutarate (2-OG) -dependent dioxygenases in oncogenesis. We investigated if the 2-OG dependent dioxygenase, aspartate beta-hydroxylase (ASPH) was important in tumor development and growth. Immunoassays were used to clarify how ASPH modulates CCA progression by promoting phosphorylation of the retinoblastoma protein (RB1). A xenograft model was employed to determine the role of ASPH on CCA growth. Knockdown of ASPH expression inhibited CCA development and growth by reducing RB1 phosphorylation. Expression of ASPH promoted direct protein interaction between RB1, cyclin-dependent kinases, and cyclins. Treatment with 2-OG-dependent dioxygenase and ASPH inhibitors suppressed the interaction between RB1 and CDK4 as well as RB1 phosphorylation. Knockdown of ASPH expression inhibited CCA progression and RB1 phosphorylation in vivo and they were found to be highly expressed in human CCAs. Knockdown of ASPH expression altered CCA development by modulating RB1 phosphorylation, as one of the major factors regulating the growth of these tumors.
Keywords: alpha-ketoglutarate, cancer metabolism, IDH1 mutations, 2-hydroxyglutarate, bile duct tumors
1. Introduction
Cholangiocarcinoma (CCA) is a devastating disease with a poor prognosis [1]. The 5-year survival rate is only 2% with disease spread outside the liver. Thus, there is a need to develop novel therapeutic targets for these tumors. Recent studies using whole genome sequencing have identified IDH1/2 mutations in about 20 % of CCAs [2, 3] and suggest that targeting such mutations with small molecule inhibitors may be a potential treatment for this disease [4]. The IDH1/2 mutations generate an oncometabolite, (R)2-hydroxyglutarate ((R)2-HG) in the Krebs cycle rather than the intermediate molecule, 2-oxoglutarate (2-OG) [5]. The (R)2-HG has been shown to promote tumorigenesis by modulating epigenetic modification and altering functions of 2-OG-dependent dioxygenases [6–8]. This phenomenon raises the possibility of targeting tumors harboring IDH1/2 mutations by inhibiting (R)2-HG production. Specific inhibitors for IDH1/2 mutations have been developed and exhibit promising therapeutic effects in preclinical models involving gliomas and leukemias [9, 10]. However, a recent study concluded that the inhibitors for IDH1/2 mutations had no suppressive effects on growth of CCA cell lines [11]. Nevertheless, there has been no new therapeutic approach proposed for CCAs with IDH1/2 wildtype genes. It is of interest that clinical studies have demonstrated that CCAs with IDH1/2 mutations have a better prognosis than those with wildtype sequences [12, 13], indicating that 2-OG-dependent dioxygenases could be important for CCA development and growth.
Aspartate beta-hydroxylase (ASPH) is a 2-OG-dependent dioxygenase that functions by hydroxylating aspartyl and asparaginyl residues in EGF-like domains of certain proteins, including Notch1 which has been shown to participate in CCA development and progression [14]. It has been shown to be highly expressed in human CCAs by IHC [15] and knockdown of ASPH substantially inhibits CCA progression by modulating Notch1-mediated cyclin D1 expression [16]. These findings suggest an important role for ASPH in CCA growth. Considering the idea that other 2-OG-dependent dioxygenases also may be required for tumor development and ASPH, as an example, appears to be involved in CCA progression, we sought to clarify how ASPH modulates CCA tumor growth with IDH1/2 wildtype sequences that may yield new targets for therapy. It was hypothesized that the 2-OG-dependent dioxygenase, ASPH may be required for the development and growth of CCAs.
2. Material and Methods
2.1 Animal studies
Six-week-old nude mice were purchased from Charles River Laboratories (Wilmington, MA). The human H1 CCA cells (2×106) were transduced with shRNA-luciferase or shRNA-ASPH and subcutaneously implanted under the right flank of nude mice. The animals were monitored for 2 weeks and the tumor volumes calculated by the formula (width2 × Length/2). The mice were euthanized with CO2 following cervical dislocation. Tumors were removed and dissected for further analysis. All protocols were approved by the Institutional Animal Care and Use Committee of Rhode Island Hospital.
2.2 Cell lines and reagents
Human H1, RBE, SSP25, NEC, and ETK1 CCA cell lines were kindly provided by Dr. Munenori Enjoji at Kyushu University in Japan [17]. TFK1 was purchased from RIKEN cell bank (Japan). The RB1 mutated plasmids including 416 pSG5L HA RB1 del22 (Plasmid #10721), 413 pSG5L HA RB1 (Plasmid #10720), 608 pSG5L HA RB1 del ex4 (Plasmid #10730), and 409 pSG5L HA RB1 (379–928) (Plasmid # 10734) were obtained from Addgene [18]. IDH1 and IDH1R132H associated plasmids were kind gifts from Dr. Yue Xiong [19]. Inducible pSLIK-IDH1-FLAG and pSLIK-IDH1R132H-FLAG were gifts from Christian Metallo (addgene)[20]. IDH1 antibody and ASPH variants were purchased from OriGene Technologies (Rockville, MD) and ASPH variants were reconstructed in pCDH-puro lentivrial vector in our lab. ASPH polyclonal antibody was made as previously described [21]. pRB1s780, pRB1s608, pRB1s807, myc-tag, and flag-tag antibodies used for immunoblotting experiments were from Cell Signaling Technology (Danvers, MA). pRB1s780 and pRB1s608 antibodies used for immunohistochemistry staining as well as trimethylated histone 3 lysine 9 (H3K9Me2), dimethylated-histone 3 lysine 4 (H3K4Me3), and H3 antibodies were from Abcam (Cambridge, MA). Tubulin antibody and iron chelator deferoxamine mesylate salt (DFO) were from Sigma-Aldrich (St. Louis, MO). CDK2, CDK4, CDK6, cyclin D1, cyclin E, RB1, GAPDH antibodies, normal IgG, and protein agarose A/G were purchased from Santa Cruz Biotechnology (Dallas, Texas). The 2-OG antagonist, Dimethyloxallyl Glycine (DMOG) was purchased from Cayman Chemical (Ann Arbor, Michigan).
2.3 Human samples (supplemental information)
2.4 Immunoblotting (supplemental information)
2.5 Immunofluorescence (supplemental information)
2.6 Immunohistochemistry (supplemental information)
2.7 Cell cycle assays (supplemental information)
2.8 Cell growth assay (supplemental information)
2.9 Senescence assay (supplemental information)
2.10 Statistical analysis (supplemental information)
3. Results
3.1 Targeting 2-OG-dependent dioxygenases by an antagonist, an iron chelator, and (R)2-HG reduced CCA cell growth with wildtype IDH1/2
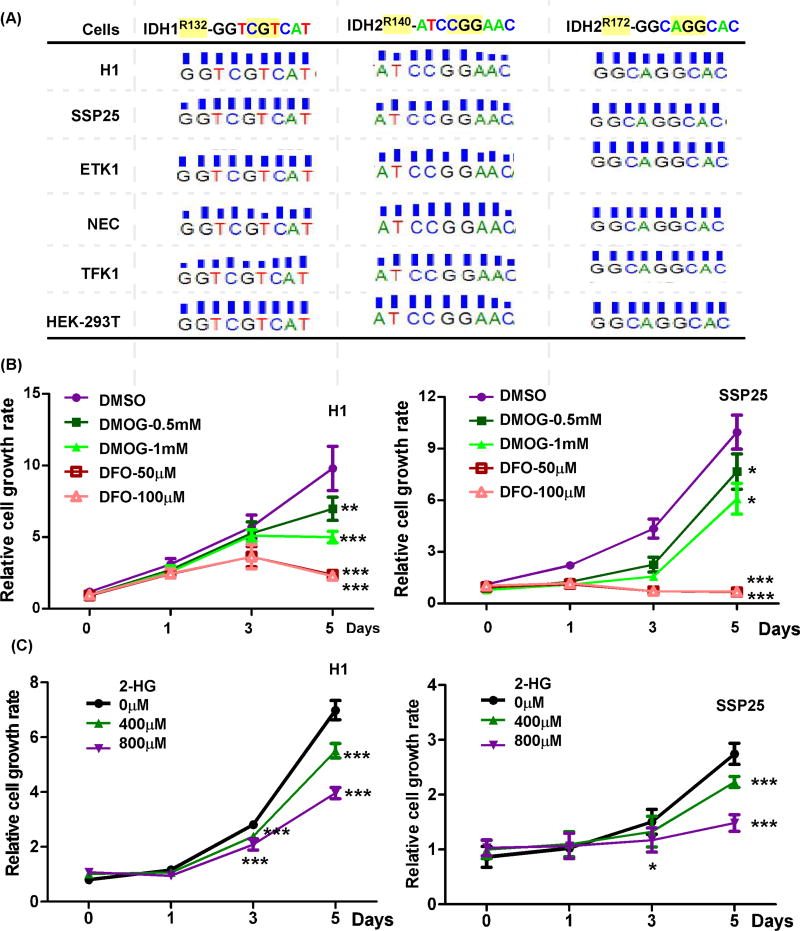
To investigate if targeting of 2-OG-dependent dioxygenases may inhibit CCA IDH1/2 wildtype tumor growth, we used three different strategies that include a 2-OG antagonist, an iron chelator, and (R)2-HG to suppress the endogenous 2-OG-dependent dioxygenases activity. The IDH1 and IDH2 genotypes were determined in several CCA cell lines by genomic sequencing and the available databases (The Cancer Cell Line Encyclopedia, https://portals.broadinstitute.org/ccle/home). It was found that several CCA cell lines have the IDH1/2 wildtype sequence (Fig. 1A). The IDH1/2 mutated (IDH1/2mut) plasmids and the genomic DNA of HEK293T cells were used as positive and negative controls, respectively. When H1 and SSP25 CCA cells were challenged with the 2-OG antagonist (DMOG) or the iron chelator (DFO), their cell growth rates were substantially reduced (Fig. 1B and sFig. 1). To evaluate if inhibiting 2-OG-dependent dioxygenases by the oncometabolite generated by the IDH1 mutation would reduce CCA cell growth, we challenged different CCA cell lines with (R)2-HG and found treatment significantly decreased cell growth as well [22] (Fig. 1C). These results imply that targeting 2-OG-dependent dioxygenases inhibits cell growth of CCA cells with wild type IDH1/2 sequences.
Figure 1. The 2-OG-dependent dioxygenases are involved in cell growth of CCAs with wildtype IDH1/2.
(A) Genomic sequences of HEK-293T cells and CCA cell lines, including H1, SSP25, ETK1, NEC, and TFK1. (B) Treatments with an iron chelator (DFO) and a 2-OG antagonist (DMOG) inhibited cell growth of CCA cells with wildtype IDH1/2, including H1 and SSP25. (C) Challenge with the IDH1mut oncometabolite, (R)2-HG reduced cell growth of CCAs. *, p<0.05; **, p<0.01; ***, p<0.001.
3.2 The 2-OG-dependent dioxygenase ASPH is involved in CCA growth, progression and induction of cell senescence
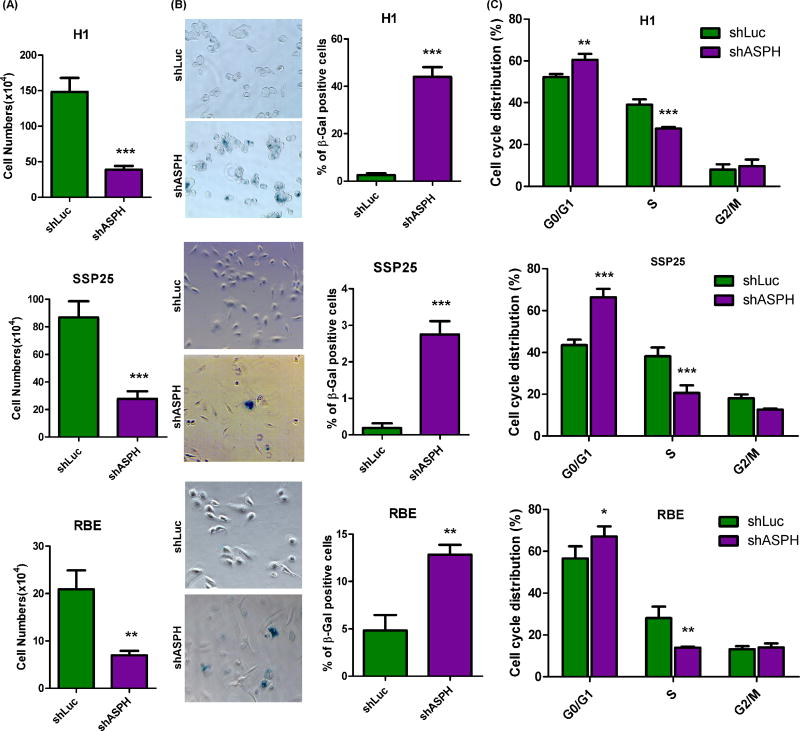
The IDH1mut produced (R)2-HG and this phenomenon has been linked to malignant transformation by regulating several 2-OG-dependent dioxygenases, including the Ten Eleven Translocation family proteins, as well as HIF, prolyl 4-hydroxylases and JumonjiC (JmjC)-domain–containing histone demethylases [6–8, 13, 23]. However, there has been no previous investigation clarifying how inhibiting 2-OG-dependent dioxygenases by IDH1 mutant protein or (R)2-HG suppresses tumor progression. In this context, ASPH is one of the 2-OG-dependent dioxygenase where overexpression has been shown to participate in malignant transformation of CCA cells [24]. To determine the role of ASPH in this regard, we evaluated the cellular effects by knockdown experiments in the H1, SSP25, and RBE with IDH1mut phenotype. Knockdown of ASPH substantially suppressed cell proliferation of CCAs independent of the genotype of IDH1 (Fig. 2A). As the IDH1mut has been suggested to induce senescence of glioma cells [25], we evaluated the effect of knocking down ASPH levels on induction of cell senescence. It was observed that knockdown of ASPH promoted development of senescence in H1, SSP25, and to a lesser extent in RBE CCA cells (Fig. 2B). Since cell proliferation and senescence are both regulated by the cell cycle, we investigated if knockdown of ASPH influences these events. Knockdown of ASPH significantly altered cell cycle progression by accumulating CCA cells in the G0/G1 phase to retard their entering into S phase (Fig. 2C).
Figure 2. Targeting the 2-OG-dependent dioxygenase, ASPH, suppressed growth, cell cycle progression and inhibited senescence of CCA cell lines.
(A) Cell numbers were counted at day 4 in 3 CCA cell lines transduced with shRNA-luciferase (shLuc) or shRNA-ASPH (shASPH), including H1 and SSP25 as well as RBE with the IDH1mut sequences. (B) Senescence associated β-gal expression and staining was used to evaluate senescence in CCA cell lines treated as indicated. (C) Cell cycle progression was analyzed in CCA cell lines 48 hours post sub-culture by using flow cytometry. *, p<0.05; **, p<0.01; ***, p<0.001.
3.3 ASPH expression modulates phosphorylation of RB1
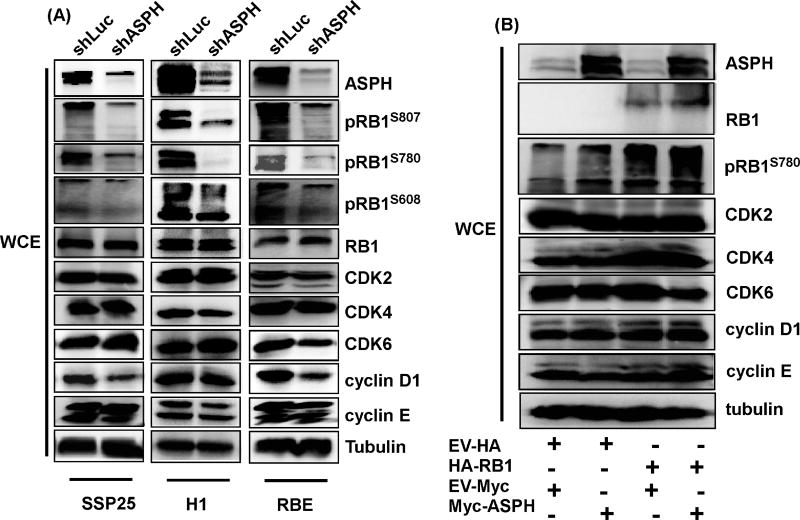
Cell proliferation, senescence, and cell cycle progression are mediated by multiple molecular signaling pathways [26–28]. Among the most important, the RB1 cascade has been shown to be one of the main driving factors for cell cycle progression from G0/G1 to S phase [27]. Additionally, RB1 has been demonstrated to be critically involved in the generation of cell senescence [26]. Therefore, RB1 expression was assessed in three CCA cell lines, including H1, SSP25, and RBE transduced with control (shLuc) or shASPH. Knockdown of ASPH did not affect total RB1 protein expression in these cells (Fig. 3A). We also ectopically expressed ASPH in HEK-293T cells to determine if expression of ASPH increases the RB1 level. Expression of ASPH did not increase total RB1 protein levels but enhanced the expression of phosphorylated RB1 (Fig. 3B). Since phosphorylation of RB1 is one of the most extensively investigated post-translational modifications which inactivate the tumor suppressor functions of RB1, it is a formal possibility that ASPH may suppress RB1 phosphorylation as a mechanism to inhibit CCA growth and progression. Thus, we further analyzed different RB1 phosphorylation sites, including pRB1s780, pRB1s608, and pRB1s807 in human CCA cell lines following ASPH knockdown. The results revealed that knockdown of ASPH suppressed phosphorylation of RB1 at all these sites (Fig. 3A). A similar result illustrating that ASPH enhanced pRB1s780 was obtained in HEK-293T cells following ectopic expression of ASPH (Fig. 3B). As RB1 modulates cell cycle progression mainly by controlling E2F1 transcriptional activity, the E2F1 target gene, cdc2, expression level was determined in to the context of ASPH expression. It was determined that cdc2 was down-regulated with knockdown of ASPH in CCA cells (sFig.2).
Figure 3. Expression of ASPH is correlated with protein phosphorylation of RB1.
(A) Immunoblotting blot (IB) results of ASPH, pRB1s807, pRB1s608, pRB1s780, RB1, CDK2, CDK4, CDK6, cyclin D1, cyclin E, and tubulin were determined in H1, SSP25, and RBE CCA cells transduced with shLuc and shASPH. Knockdown of ASPH decreased RB1 phosphorylation. (B) Ectopic expression of ASPH for 48 hours promoted RB1 phosphorylation.
3.4 ASPH affects RB1 phosphorylation by interaction with cyclin-dependent kinases
The protein complex composed of cyclin D1, CDK4, and CDK6 as well as CDK2/cyclin E protein complexes have been demonstrated to be important factors affecting phosphorylation of RB1 [27]. To determine if ASPH modulates RB1 phosphorylation by altering the expression of these proteins, we evaluated levels in three CCA cell lines with ASPH knockdown as well as in HEK-293T cells with ectopic over-expression of ASPH. As shown in Fig. 3, altering ASPH expression levels did not influence the concentrations of these proteins, suggesting that other mechanisms may be involved in the regulation of RB1 phosphorylation by ASPH.
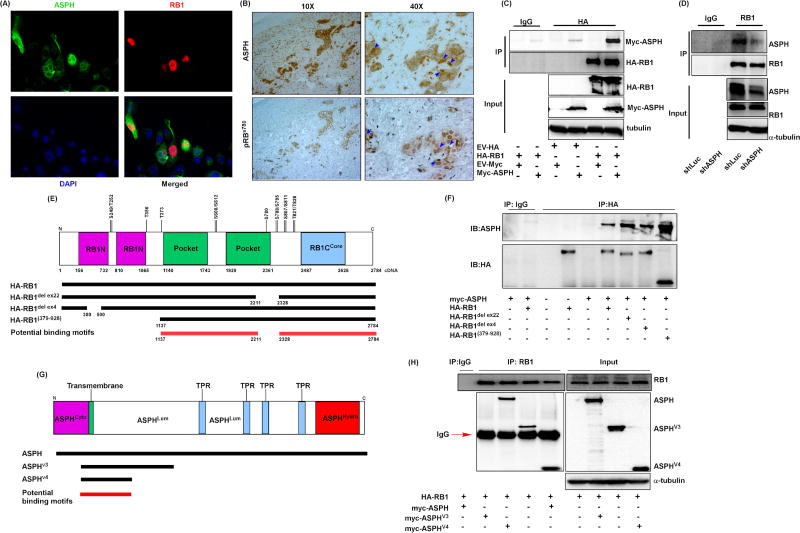
The ASPH protein has been previously shown to modulate phosphorylation of GSK3β through interactions between GSK3β and AKT [29] and suggest that ASPH may directly bind to RB1 as well. To determine if ASPH and RB1 interact, we transiently transfected RB1 and ASPH expression plasmids into HEK-293T cells and monitored cellular localization by immunofluorescence experiments. Indeed, it was found that ASPH and RB1 co-localize to the nucleus (Fig. 4A). This observation was also observed in human CCA tissue samples. In this context, ASPH and pRB1s780 co-localize in tumors as indicated by the blue arrow heads (Fig. 4B). This phenotype was validated by examining ASPH and RB1 in the cytoplasm and nuclear fractions.
Figure 4. ASPH co-localizes and directly interacts with RB1 protein.
(A) Immunofluorescence staining of RB1 and ASPH in HEK-293T cells transfected with myc-tagged ASPH and HA-tagged RB1 for 48 hours. Green fluorescence indicates the ASPH signal, red fluorescence shows RB1 localization, and DAPI is used for nuclear staining. (B) Immunohistochemical staining of ASPH and pRBs780 was performed in human CCA tissue. The blue arrow heads indicate co-localization of ASPH and RB1. (C) IB results of myc-tagged ASPH and HA-tagged RB1 were determined in the IP products of HEK-293T cells 48 hours post transfection as indicated. HA-RB1, myc-ASPH, and tubulin were determined in the whole cell lysate (WCL) for ensuring even protein loading of inputs. (D) Knockdown of ASPH suppresses the interaction between RB1 and ASPH. ASPH, RB1, and tubulin were evaluated in the WCL of RBE cells treated with shLuc or shASPH. (E) Illustration of RB1 protein domains. (F) IB results of myc-tagged ASPH and HA-tagged RB1 in the IP products of HEK-293T cells transfected with different RB1 mutant plasmids. (G) Illustration of ASPH protein domains. (H) IB results of myc-ASPH were determined in the IP products of HEK-293T cells transfected as indicated. RB1, myc-ASPH, myc-ASPH variant 3 (ASPHV3), myc-ASPH variant 4 (ASPHV4), and tubulin were measured in the WCL.
The RB1 was mainly localized in the nucleoplasm, but ASPH was found both in the cytoplasm and nucleoplasm fractions (sFig. 3A). We transiently transfecting HA-tagged RB1 and Myc-tagged ASPH in HEK-293T cells, and used HA-tagged antibody to pull down the RB1 protein complex; ASPH was detected in the RB1 protein complex by a Myc-tagged antibody. The results of input protein concentration demonstrate successful transfection and immunoprecipitation (IP) following pull-down of the RB1 protein complex and revealed ASPH in the RB1 protein complex (Fig. 4C). Consistently, the IP results performed on the nucleoplasm fraction further verified the interaction of ASPH and RB1 in the nucleus of CCA cells (sFig. 3B). These observations suggest a direct protein-protein interaction between ASPH and RB1. This event was able to be inhibited in the human RBE CCA cell line which has high endogenous levels of ASPH and RB1 (Fig. 4D).
Experiments were performed to determine which protein regions were responsible for the interaction between RB1 and ASPH. The RB1 protein has N-terminal (RB1N), pocket, and C-terminal Core domains (RB1Ccore) (Fig. 4E). We transfected ASPH in combination with different mutated HA-tagged RB1 plasmids expressed into HEK-293T cells and performed IP experiments with HA-tagged antibodies to pull down different RB1 protein complexes. It was noteworthy that ASPH protein was detectable in all different RB1 protein complexes (Fig. 4F), suggesting that ASPH has a direct interaction with all mutated RB1 proteins studied thus far. By peptide mapping, we suggest that the potential protein motif of RB1 for ASPH binding may be located within the region between the pocket and RB1Ccore domains (Fig. 4E, red line). To determine the protein domain of ASPH responsible for this protein interaction, Myc-tagged ASPH variant forms and HA-tagged RB1 were transfected in HEK-293T cells and the RB1 containing protein complex was pulled down by a HA-tag antibody. Similarly, all ASPH variant forms could be detected in the RB1 protein complex (Fig. 4H), suggesting that RB1 physically interacted with ASPH. The protein motif of ASPH likely to be responsible for RB1 binding appeared to be located within the ASPH variant form 4 (Fig. 4G, red line).
3.5 The protein interaction between RB1 and the CDK complexes is modulated by ASPH expression
The protein interactions between RB1 and the CDK complexes were investigated by transiently transfecting HA-tagged RB1 and Myc-tagged ASPH in HEK-293T cells. As determined by co-IP experiments, expression of ASPH resulted in increased protein-protein interaction between RB1, CDK2, and CDK4 as well as RB1, cyclin D1, and cyclin E (Fig. 5A and B), which in turn led to enhanced RB1 phosphorylation. In contrast, the expression of ASPH variant form 3 without the enzymatic domain of ASPH did not promote this interaction between RB1 and CDK4 (Fig. 5C). As the enzymatic activity of ASPH is critical for its transforming function, we determined if using a mutant ASPH with 20% of original enzymatic activity [30] would inhibit the protein interactions between RB1 and the CDK complexes. Interestingly, expression of ASPH consistently increased the protein interaction between RB1 and CDK4 in HEK-293T cells, but expression of the mutant forms of ASPH with low enzymatic activity lost this property (Fig. 5D), suggesting that the enzymatic activity of ASPH was required for modulating RB1 phosphorylation. Since ASPH is a 2-OG-dependent dioxygenase, general inhibitors for 2-OG-dependent dioxygenases were useful to confirm this observation. As expected, DMOG and DFO treatments suppressed the effect of ASPH on this protein-protein interaction (Fig. 5E). We also evaluated if another specific and direct ASPH inhibitor (MO-I-1151), kindly provided by Dr. Mark Olsen [16], may alter the ASPH-mediated protein-protein interaction with CDK4. This ASPH inhibitor, which also reduces its enzymatic activity by 90% [16], suppressed the protein interaction between RB1 and CDK4 as well (Fig. 5E).
Figure 5. The enzymatic activity of ASPH is involved in the protein-protein interaction between RB1 and CDK complexes.
(A) IB results of CDK2, CDK4, and RB1 were determined in the IP products of HEK-293T treated as indicated. CDK2, CDK4, and ASPH were measured in the WCL. (B) IB results of cyclin D1, cyclin E, and RB1 were determined in HEK-293T cells treated as indicated. GAPDH, cyclin D1, cyclin E, and RB1 were measured in the WCL. (C) HA-RB1 and CDK4 were determined in the IP products of HEK-293T cells transfected with HA-RB1 in combination with EV, myc-ASPH, or myc-ASPH variant 3 (Myc-ASPHv3) which does not contain the enzymatic domain of ASPH. Myc tag, CDK4, and tubulin were measured in the WCL. The WCL was collected from HEK-293T cells 48 hours post sub-culture. (D) ASPH, HA-RB1, and CDK4 were analyzed in the IP products of HEK-293T cells transfected with HA-RB1 in combination with myc-EV, myc-ASPH, or myc-ASPHH675R which only has 20 % of enzymatic activity. The WCL was collected 48 hours post transfection. (E) The effects of DFO, DMOG, and specific ASPH inhibitor, MO-I-1151 on the protein interaction between CDK4 and RB1. HA-RB1 and CDK4 were measured in the IP products of HEK-293T transduced with myc-EV or myc-ASPH. HA-RB1 was transiently transfected in the HEK-293T-EV or HEK-293T-ASPH for 48 hours, and the treated HEK-293T cells were sub-cultured. 48 hours later, the WCL was collected from the treated HEK-293T cells which were challenged with different concentrations of DMOG, DFO, and MO-I-1151 24 hours before harvesting.
3.6 Targeting ASPH suppresses RB1 phosphorylation without significant impact on histone methylation
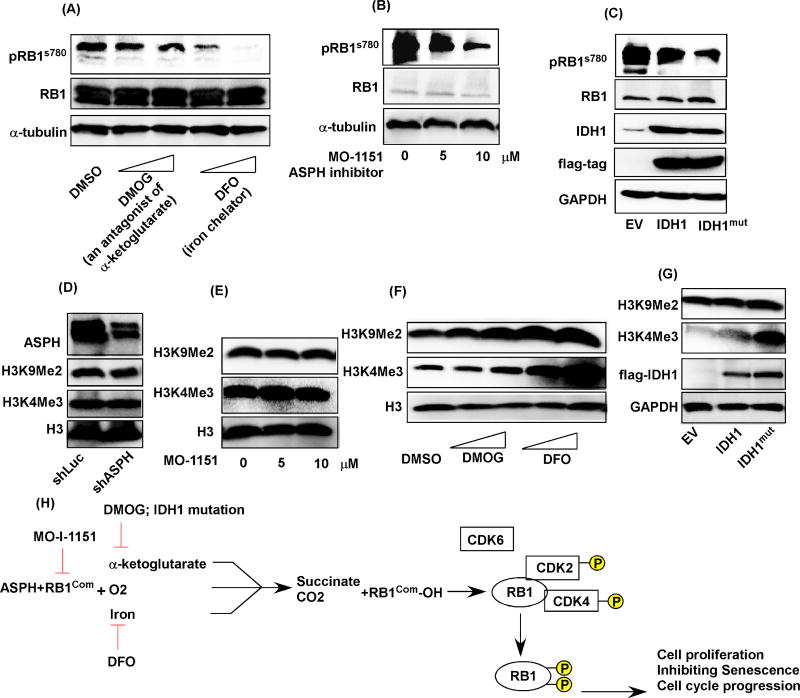
As knockdown of ASPH substantially suppressed development and progression of CCAs as well as RB1 phosphorylation, it would be important to determine if targeting ASPH by other inhibitors may exert a similar effect. Targeting ASPH by DFO and DMOG significantly inhibited CCA cell proliferation (Fig. 1B). Targeting ASPH with MO-I-1151 also suppressed CCA growth (sFig. 4). To further investigate whether targeting ASPH by these compounds may modulate CCA progression by affecting RB1 phosphorylation, we determined the effects of IDH1 mutation, DFO, DMOG, and MO-I-1151 on RB1 phosphorylation. The results suggested that DMOG, DFO, and MO-I-1151 significantly inhibited RB1 phosphorylation in the H1 CCA cell line (Fig. 6A and B). Similarly, transfection with IDH1mut expression plasmid in HEK-293T cells suppressed RB1 phosphorylation (Fig. 6C). Considering the possibility that IDH1 mutation may promote cellular transformation by affecting histone methylation [8, 23], we investigated whether inhibiting ASPH enzymatic activity by the IDH1 mutation, DFO, DMOG, and MO-I-1151 influences histone methylation.
Figure 6. The ASPH specific inhibitor, MO-I-1151 suppressed RB1 phosphorylation without affecting histone methylation in contrast to other inhibitors of 2-OG-dependent dioxygenases.
The pRB1s780, RB1, and α-tubulin proteins were determined in serum starved H1 cells which were harvested 4 hours post challenges with (A) 0.5mM, 1mM DMOG, 50µM, 100µM DFO, (B) 5µM, or 10µM MO-I-1151. (C) pRB1s780, RB1, IDH1, flag-tag, and GAPDH were measured in HEK-293T cells transfected with EV, flag-IDH1, flag-IDH1 mutated (IDH1mut). (D) ASPH, H3K9Me2, H3K4Me3, and H3 were determined in H1 cells transduced with shLuc or shASPH. The expression levels of H3K9Me2, H3K4Me3, and H3 were evaluated in H1 cells treated with (E) 5, 10 µM MO-I-1151, (F) 0.5, 1mM DMOG, 50, or 100µM DFO for 24 hours. (G) The results of H3K9Me2, H3K4Me3, flag-IDH1, and GAPDH in HEK-293T cells transfected with EV, IDH1, or IDH1mut for 24 hours were shown. (H) A cartoon illustrates the actions of targeting ASPH by different strategies in the protein interaction between RB1 and CDK complexes (RB1com) as well as RB1 phosphorylation.
Importantly, it was found that targeting ASPH by shRNA knockdown and MO-I-1151, an inhibitor of enzymatic activity [16], did not affect histone methylation of H3K9Me3 and H3K4Me2 (Fig. 6D and E). However, treatment with DMOG, DFO, and IDH1 mutation all led to increased histone methylation of H3K9Me2 and H3K4Me3 (Fig. 6F), consistent with previous observations [21, 23]. These results suggest reduced ASPH enzymatic activity results in decreased protein-protein interaction between RB1 and the CDK complexes, which was associated with reduced RB1 phosphorylation and subsequently leads to diminished CCA proliferation and cell cycle progression, as well as induction of cellular senescence; the postulated signaling pathways that may be involved are described in Fig. 6H.
3.7 Overexpression of CDK2 and CDK4 does not reverse the effect of ASPH knockdown on RB1 phosphorylation and CCA cell growth
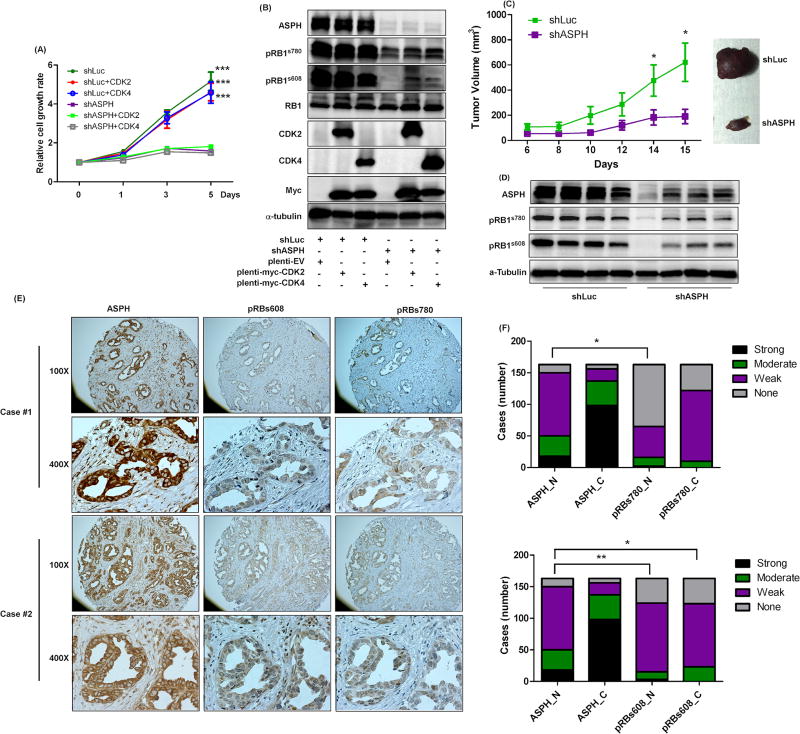
Although we have identified that knockdown of ASPH modulates CCA progression by affecting RB1 phosphorylation, the consequent signal transduction pathways were not completely defined. As the CDKs are major kinases for RB1 phosphorylation and expression of ASPH promotes the interaction between RB1 and CDK4, overexpression of CDKs may reverse the effects of ASPH knockdown on CCA progression. To determine the rescue effects of CDKs on CCA development and growth, Myc-tagged CDK2 and CDK4 were transduced into H1 CCA cells treated with shLuc (control) or shASPH via lentiviral transfection system. Overexpression of CDK2 and CDK4 were verified by CDK2, CDK4 and Myc-tag antibodies (Fig. 7B). Overexpression of CDK2 and CDK4 did not reverse the inhibition effect of ASPH knockdown on CCA cell growth (Fig. 7A and sFig. 5A). Furthermore, overexpression of CDK2 and CDK4 did not rescue RB1 phosphorylation at pRB1s780 and pRB1s608 sites (Fig. 7B and sFig. 5B). This finding supports the hypothesis (Fig. 6H) of the importance of ASPH in CCA development and growth by the mechanisms proposed.
Figure 7. The correlations between ASPH, phosphorylated RB1, and CDKs in CCAs.
(A) Relative cell growth rates were determined in H1-shLuc and H1-shASPH transduced with plenti-myc-EV, plenti-myc-CDK2, or plenti-myc-CDK4 at days 0, 1, 3, and 5. (B) ASPH, pRB1s780, pRB1s608, RB1, CDK2, CDK4, myc, and α-tubulin were determined in H1 cells treated as indicated. (C) Tumor growth of H1-shLuc and H1-shASPH was evaluated in vivo by calculating tumor volumes of subcutaneous tumors implanted into nude mice. (D) The representative IB results of ASPH, pRBs780, pRBs608, and α-tubulin were obtained in the WCL harvested from the tumors of experimental nude mice. (E) The representative images (100× and 400×) of ASPH, pRBs608, and pRBs780 are shown in 2 cases of human patients with CCA tumors. (F) The intensities of nuclear ASPH, cytoplasmic ASPH, nuclear pRB1s780, cytoplasmic pRB1s780, nuclear pRB1s608, and cytoplasmic pRB1s608 were determined in human CCA samples, n=163. *, p<0.05; **, p<0.01; ***, p<0.001. The student t test was used to analyze statistical difference for (A) and (B). The Kendall tau-b rank test was used to evaluate if ASPH, pRB1s608, and pRB1s780 have significant correlation in human CCA tumors tested for (F).
3.8 Expression of ASPH is correlated with RB1 phosphorylation in vivo
To determine whether targeting ASPH inhibits CCA progression, the shLuc-treated and shASPH-treated H1 cells were subcutaneously implanted into nude mice; the ASPH knockdown substantially inhibited CCA growth and progression in vivo (Fig. 7C). As ASPH appears to modulate CCA progression by affecting RB1 phosphorylation, the tumor tissue samples were used to examine the effect of ASPH knockdown on RB1 phosphorylation in vivo. ASPH knockdown was validated using immunoblotting (Fig. 7D); reduced expression consistently suppressed RB1 phosphorylation at pRB1s780 and pRB1s608 in vivo. To further evaluate if ASPH expression was correlated with generation of pRB1s780 and pRB1s608 in human CCA, 163 human tumor samples were examined using IHC. ASPH was detectable in most CCA tumors (95.7%) (Fig. 7E and F). In addition, these CCA tumors also stained positive for pRB1s780 and pRB1s608 as shown by the representative examples displayed in Fig. 7E. Consistently, the expression of ASPH was highly correlated with either pRB1s780 (p<0.01) or pRB1s608 (p<0.05), suggesting that ASPH participates in CCA progression by regulating RB1 phosphorylation in human disease.
4. Discussion
Several investigations have identified IDH1/2 mutations in about 20% of CCA tumors [2, 3, 12]. As IDH1/2 mutations have been previously identified in gliomas (13); inhibitors have been developed and currently are under clinical trials (9, 10). A recent study indicated that an IDH1 inhibitor did not suppress the progression of CCAs having IDH1 mutation, however, these investigations also screened several compounds and identified that CCA having IDH1mut may be highly sensitive to treatment with a multi-kinase inhibitor, dasatinib (11). Nevertheless, there are still approximately 80% of CCAs with wildtype IDH1/2 where there is no effective therapy.
Several follow-up clinical studies have observed that patients with tumors harboring IDH1/2mut have better prognosis than those having tumors with wildtype IDH1/2 [31, 32], including CCA [12]. It has been proposed that IDH1/2 mutations promote tumor growth and progression mainly by targeting 2-OG-dependent dioxygenases through generation of (R)2-HG. Thus, (R)2-HG may inhibit other 2-OG-dependent enzymes one of which is highly expressed in CCAs such as ASPH, and important in pathogenesis, since overexpression promotes cell proliferation, migration, invasion and metastases. Indeed, a previous study has found that ASPH was highly expressed in CCAs [15] and this finding has been confirmed in the current study (95.7%) so that targeting ASPH may suppress development, growth and progression of CCAs [16]. Since ASPH is one of the 2-OG-dependent enzymes, it is likely that (R)2-HG may suppress CCA oncogenesis by targeting ASPH. Indeed, our findings demonstrated that targeting ASPH with (R)2-HG, an iron chelator-DFO, and a 2-OG antagonist DMOG all of which lead to reduced enzymatic activity and/or expression will inhibit cell growth in CCAs with both IDH1mut or IDH1/2 wildtype phenotypes.
Additionally, knockdown of ASPH substantially suppressed cell proliferation, cell cycle progression, and induced the development of senescence in CCAs with wildtype IDH1/2 but to a lesser extent in CCA with IDH1mut sequence. These results further support the hypothesis that ASPH is highly associated with the progression of CCAs having IDH1/2 wildtype genes. The CCA cell line with IDH1mut shows less responsiveness to ASPH knockdown, since it is grown under conditions of low ASPH enzymatic activity related to high (R)2-HG production. However, knockdown of ASPH still significantly inhibits its progression, suggesting that ASPH may be required for CCA tumorigenesis no matter what are the genotypes of IDH1/2.
These tumors represent transformed bile duct epithelial cells and have subsequently overcome the process of cell senescence [26, 33]. Given the observation that knockdown of ASPH substantially suppresses cell growth, cell cycle progression, and elicits senescence, this 2-OG enzyme may influence tumorigenesis by modulating senescence through phosphorylation of RB1. Although a RB1 mutation has been rarely described in CCAs (4.5%) [34], altered RB1 function by post-translational phosphorylation has been suspected to be involved in CCA progression [35]. Interestingly, our observations also indicate the importance of RB1 function in CCA oncogenesis and reveal a novel molecular mechanism by which ASPH modulates CCAs through influencing RB1 phosphorylation via affecting the protein interaction between RB1 and CDK complexes. More important, this idea was strengthened by suppressing ASPH enzymatic activity by using IDH1mut expressed protein, DFO, DMOG, and a specific inhibitor (MO-I-1151) further suggesting the possible link between ASPH and oncometabolite ((R)2-HG) of cancer metabolism.
Additionally, our study may partially explain why CCAs having IDH1mut phenotype may have better prognosis than patients having solid tumors with IDH1 wildtype [12, 31, 32], because IDH1mut, DFO, and DMOG treatments all result in reduced RB1 phosphorylation by suppressing the protein interaction between RB1 and CDK complexes, mediated by the 2-OG-dependent ASPH enzyme; the net results may be CCA cell growth inhibition and a better clinical prognosis.
Although the current study provides a hypothesis to partially explain how ASPH may be involved in a better prognosis of CCA patients having IDH1/2mut by inhibiting function due to over production of (R)-2HG, further experiments will be needed in order to clarify which other, if any, 2-OG-dependent dioxygenase may participate in pathogenesis. Nevertheless, the current study proposes a novel molecular mechanism by which ASPH promotes CCA development, growth, and progression by modulating RB1 phosphorylation via affecting the protein interaction between RB1 and CDK complexes. Indeed, clinical IHC studies on tumor specimens collected from three cohorts in different areas of the world, including Asia and two North America sites, further support the correlation between ASPH expression and RB1 phosphorylation in vivo and suggest that ASPH may be a potential therapeutic target in CCA patients having the IDH1/2 wildtype genotype.
Supplementary Material
Highlights.
ASPH, a 2-OG dependent dioxygenease, is highly expressed in human cholangiocarcinomas.
ASPH promotes phosphorylation of the RB1 protein in vitro and in vivo.
Overexpression of ASPH facilitates RB1 and cyclin dependent kinase interactions.
The enzymatic activity of ASPH is necessary for RB1 phosphorylation.
Phosphorylation of RB1 enhances the growth and progression of cholangiocarcinoma.
Acknowledgments
Financial Support: This work was supported by National Institutes of Health (grant numbers R01CA-123544 and P20GM103430-12) American Association for the Study of Liver Diseases (Pinnacle Research Award in Liver Disease), and Rhode Island Foundation (grant number 134279).
Abbreviations (in order of appearance)
- CCA
Cholangiocarcinoma
- ASPH
aspartate beta-hydroxylase
- RB1
retinoblastoma protein
- (R)2-HG
(R)2-hydroxyglutarate
- DFO
deferoxamine mesylate salt
- DMOG
Dimethyloxallyl Glycine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: There is no conflict of interest among the authors.
References
- 1.Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383:2168–2179. doi: 10.1016/S0140-6736(13)61903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, Offerhaus GJ, Roa JC, Roberts LR, Gores GJ, Popescu I, Alexandrescu ST, Dima S, Fassan M, Simbolo M, Mafficini A, Capelli P, Lawlor RT, Ruzzenente A, Guglielmi A, Tortora G, de Braud F, Scarpa A, Jarnagin W, Klimstra D, Karchin R, Velculescu VE, Hruban RH, Vogelstein B, Kinzler KW, Papadopoulos N, Wood LD. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nature genetics. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan-On W, Nairismagi ML, Ong CK, Lim WK, Dima S, Pairojkul C, Lim KH, McPherson JR, Cutcutache I, Heng HL, Ooi L, Chung A, Chow P, Cheow PC, Lee SY, Choo SP, Tan IB, Duda D, Nastase A, Myint SS, Wong BH, Gan A, Rajasegaran V, Ng CC, Nagarajan S, Jusakul A, Zhang S, Vohra P, Yu W, Huang D, Sithithaworn P, Yongvanit P, Wongkham S, Khuntikeo N, Bhudhisawasdi V, Popescu I, Rozen SG, Tan P, Teh BT. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nature genetics. 2013;45:1474–1478. doi: 10.1038/ng.2806. [DOI] [PubMed] [Google Scholar]
- 4.Saha SK, Parachoniak CA, Ghanta KS, Fitamant J, Ross KN, Najem MS, Gurumurthy S, Akbay EA, Sia D, Cornella H, Miltiadous O, Walesky C, Deshpande V, Zhu AX, Hezel AF, Yen KE, Straley KS, Travins J, Popovici-Muller J, Gliser C, Ferrone CR, Apte U, Llovet JM, Wong KK, Ramaswamy S, Bardeesy N. Mutant IDH inhibits HNF-4alpha to block hepatocyte differentiation and promote biliary cancer. Nature. 2014;513:110–114. doi: 10.1038/nature13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, Gross S, Travins J, Weiss S, Looper R, Ligon KL, Verhaak RG, Yan H, Kaelin WG., Jr Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, Kunii K, Pedraza A, Schalm S, Silverman L, Miller A, Wang F, Yang H, Chen Y, Kernytsky A, Rosenblum MK, Liu W, Biller SA, Su SM, Brennan CW, Chan TA, Graeber TG, Yen KE, Mellinghoff IK. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, Straley K, Kernytsky A, Liu W, Gliser C, Yang H, Gross S, Artin E, Saada V, Mylonas E, Quivoron C, Popovici-Muller J, Saunders JO, Salituro FG, Yan S, Murray S, Wei W, Gao Y, Dang L, Dorsch M, Agresta S, Schenkein DP, Biller SA, Su SM, de Botton S, Yen KE. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 11.Saha SK, Gordan JD, Kleinstiver BP, Vu P, Najem MS, Yeo JC, Shi L, Kato Y, Levin RS, Webber JT, Damon LJ, Egan RK, Greninger P, McDermott U, Garnett MJ, Jenkins RL, Rieger-Christ KM, Sullivan TB, Hezel AF, Liss AS, Mizukami Y, Goyal L, Ferrone CR, Zhu AX, Joung JK, Shokat KM, Benes CH, Bardeesy N. Isocitrate Dehydrogenase Mutations Confer Dasatinib Hypersensitivity and SRC Dependence in Intrahepatic Cholangiocarcinoma. Cancer discovery. 2016;6:727–739. doi: 10.1158/2159-8290.CD-15-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang P, Dong Q, Zhang C, Kuan PF, Liu Y, Jeck WR, Andersen JB, Jiang W, Savich GL, Tan TX, Auman JT, Hoskins JM, Misher AD, Moser CD, Yourstone SM, Kim JW, Cibulskis K, Getz G, Hunt HV, Thorgeirsson SS, Roberts LR, Ye D, Guan KL, Xiong Y, Qin LX, Chiang DY. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32:3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer discovery. 2013;3:730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 14.Zender S, Nickeleit I, Wuestefeld T, Sorensen I, Dauch D, Bozko P, El-Khatib M, Geffers R, Bektas H, Manns MP, Gossler A, Wilkens L, Plentz R, Zender L, Malek NP. A critical role for notch signaling in the formation of cholangiocellular carcinomas. Cancer cell. 2013;23:784–795. doi: 10.1016/j.ccr.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 15.Lavaissiere L, Jia S, Nishiyama M, de la Monte S, Stern AM, Wands JR, Friedman PA. Overexpression of human aspartyl(asparaginyl)beta-hydroxylase in hepatocellular carcinoma and cholangiocarcinoma. The Journal of clinical investigation. 1996;98:1313–1323. doi: 10.1172/JCI118918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang CK, Iwagami Y, Aihara A, Chung W, de la Monte S, Thomas JM, Olsen M, Carlson R, Yu T, Dong X, Wands J. Anti-Tumor Effects of Second Generation beta-Hydroxylase Inhibitors on Cholangiocarcinoma Development and Progression. PloS one. 2016;11:e0150336. doi: 10.1371/journal.pone.0150336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda T, Sepe P, Lahousse S, Tamaki S, Enjoji M, Wands JR, de la Monte SM. Antisense oligodeoxynucleotides directed against aspartyl (asparaginyl) beta-hydroxylase suppress migration of cholangiocarcinoma cells. Journal of hepatology. 2003;38:615–622. doi: 10.1016/s0168-8278(03)00052-7. [DOI] [PubMed] [Google Scholar]
- 18.Sellers WR, Novitch BG, Miyake S, Heith A, Otterson GA, Kaye FJ, Lassar AB, Kaelin WG., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes & development. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, Ding J, Lei Q, Guan KL, Xiong Y. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Molecular cell. 2014;55:253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohr L, Yeung A, Aloman C, Wittrup D, Wands JR. Antibody-directed therapy for human hepatocellular carcinoma. Gastroenterology. 2004;127:S225–231. doi: 10.1053/j.gastro.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 22.Fu X, Chin RM, Vergnes L, Hwang H, Deng G, Xing Y, Pai MY, Li S, Ta L, Fazlollahi F, Chen C, Prins RM, Teitell MA, Nathanson DA, Lai A, Faull KF, Jiang M, Clarke SG, Cloughesy TF, Graeber TG, Braas D, Christofk HR, Jung ME, Reue K, Huang J. 2-Hydroxyglutarate Inhibits ATP Synthase and mTOR Signaling. Cell metabolism. 2015;22:508–515. doi: 10.1016/j.cmet.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, Cowley GS, Root DE, Ebert BL, Kaelin WG., Jr (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ince N, de la Monte SM, Wands JR. Overexpression of human aspartyl (asparaginyl) beta-hydroxylase is associated with malignant transformation. Cancer research. 2000;60:1261–1266. [PubMed] [Google Scholar]
- 25.Stoczynska-Fidelus E, Piaskowski S, Bienkowski M, Banaszczyk M, Hulas-Bigoszewska K, Winiecka-Klimek M, Radomiak-Zaluska A, Och W, Borowiec M, Zieba J, Treda C, Rieske P. The failure in the stabilization of glioblastoma-derived cell lines: spontaneous in vitro senescence as the main culprit. PloS one. 2014;9:e87136. doi: 10.1371/journal.pone.0087136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narita M, Nunez S, Heard E, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 27.Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 28.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nature reviews. Molecular cell biology. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 29.Iwagami Y, Huang CK, Olsen MJ, Thomas JM, Jang G, Kim M, Lin Q, Carlson RI, Wagner CE, Dong X, Wands JR. Aspartate beta-hydroxylase modulates cellular senescence through glycogen synthase kinase 3beta in hepatocellular carcinoma. Hepatology. 2016;63:1213–1226. doi: 10.1002/hep.28411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong X, Lin Q, Aihara A, Li Y, Huang CK, Chung W, Tang Q, Chen X, Carlson R, Nadolny C, Gabriel G, Olsen M, Wands JR. Aspartate beta-Hydroxylase expression promotes a malignant pancreatic cellular phenotype. Oncotarget. 2015;6:1231–1248. doi: 10.18632/oncotarget.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, Zhao X, Li Y, Li Q, Wang H, Hu J, Kong G, Wu M, Ding C, Chen N, Hu H. Mutational landscape of intrahepatic cholangiocarcinoma. Nature communications. 2014;5:5696. doi: 10.1038/ncomms6696. [DOI] [PubMed] [Google Scholar]
- 35.Zheng T, Hong X, Wang J, Pei T, Liang Y, Yin D, Song R, Song X, Lu Z, Qi S, Liu J, Sun B, Xie C, Pan S, Li Y, Luo X, Li S, Fang X, Bhatta N, Jiang H, Liu L. Gankyrin promotes tumor growth and metastasis through activation of IL-6/STAT3 signaling in human cholangiocarcinoma. Hepatology. 2014;59:935–946. doi: 10.1002/hep.26705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.