Abstract
Genomic regions of autozygosity (ROAs) represent segments of individual genomes that are homozygous for haplotypes inherited identical-by-descent (IBD) from a common ancestor. ROAs are nonuniformly distributed across the genome, and increased ROA levels are a reported risk factor for numerous complex diseases. Previously, we hypothesized that long ROAs are enriched for deleterious homozygotes as a result of young haplotypes with recent deleterious mutations—relatively untouched by purifying selection—being paired IBD as a consequence of recent parental relatedness, a pattern supported by ROA and whole-exome sequence data on 27 individuals. Here, we significantly bolster support for our hypothesis and expand upon our original analyses using ROA and whole-genome sequence data on 2,436 individuals from The 1000 Genomes Project. Considering CADD deleteriousness scores, we reaffirm our previous observation that long ROAs are enriched for damaging homozygotes worldwide. We show that strongly damaging homozygotes experience greater enrichment than weaker damaging homozygotes, while overall enrichment varies appreciably among populations. Mendelian disease genes and those encoding FDA-approved drug targets have significantly increased rates of gain in damaging homozygotes with increasing ROA coverage relative to all other genes. In genes implicated in eight complex phenotypes for which ROA levels have been identified as a risk factor, rates of gain in damaging homozygotes vary across phenotypes and populations but frequently differ significantly from non-disease genes. These findings highlight the potential confounding effects of population background in the assessment of associations between ROA levels and complex disease risk, which might underlie reported inconsistencies in ROA-phenotype associations.
Keywords: autozygosity, consanguinity, homozygosity, identity by descent, inbreeding, human populations, deleterious variation, genetic association, disease risk, pharmacogenomics
Introduction
Genomic regions of autozygosity (ROAs) reflect homozygosity for haplotypes inherited identical-by-descent (IBD) from a recent ancestor. Long ROAs most likely derive from a more recent ancestor; shorter ones from a more distant ancestor. Their patterns in individual genomes therefore reflect the long-term effects of both population history, such as founder effects and isolation, and cultural practices, such as endogamy and prescribed inbreeding. Investigations in worldwide human populations have found ROAs to be ubiquitous and frequent even in ostensibly outbred populations,1, 2, 3, 4, 5 where ROAs of different lengths have different continental and population patterns, both with regards to their total lengths in individual genomes1, 2, 3, 4, 5 and in their nonuniform genomic distribution that forms hotspots and coldspots.2, 5 Moreover, long ROAs, which arise most frequently via recent inbreeding, are enriched for deleterious variation carried in homozygous form,6, 7 providing a potential mechanistic basis for the general reduction in fitness and health of individuals that are the outcome of prolonged inbreeding, a phenomenon commonly referred to as inbreeding depression.8
Over the past decade, it has been increasingly apparent that the ROA load in individual genomes is an important genetic risk factor for numerous complex diseases and contributing factor to variation in multiple complex traits. Early studies using Wright’s inbreeding coefficient9 identified associations between inbreeding and variation in standing height, weight, blood pressure, and body mass index,10, 11, 12 as well as increased risks for coronary artery disease, stroke, and cancer.13, 14 The increasing availability of dense genotype data on large disease cohorts enabled the use of more accurate genomic inbreeding estimates based upon ROAs, confirming previous associations with standing height15, 16, 17, 18 and coronary artery disease (CAD),19 as well as identifying new associations with thyroid20 and colorectal21 cancer and multiple neurodegenerative disorders: Alzheimer disease,22, 23 Parkinson disease,24 and amyotrophic lateral sclerosis25 (ALS). These associations are consistent with the view that variants with individually small effect sizes associated with complex diseases and traits are more likely to be rare than to be common,26, 27 distributed abundantly rather than sparsely in the genome,28, 29 and recessive rather than dominant.29, 30 As the fraction of the genome in ROAs increases as a consequence of prescribed and unintentional inbreeding,31 the number of deleterious alleles carried in homozygous form would also be expected to increase,6, 7 thereby elevating the long-term probability of negative health outcomes. However, for some complex diseases the association between ROA load and genetic risk remains unclear, with both positive and negative suggestions of association reported for schizophrenia32, 33 and cognitive ability.17, 34, 35 These discrepancies highlight the genomic complexities underlying observed associations between ROA load and genetic disease risk, where the population and cultural backgrounds of study subjects would be expected to be important determining factors.
We previously hypothesized that long ROAs—being comprised of younger haplotypes carrying more low-frequency and rare alleles than older haplotypes—would contain a larger fraction of all genome-wide damaging homozygotes compared to nondamaging homozygotes.6 While our results supported this hypothesis, their generalizability was severely limited by small sample size and the approaches available at that time to interrogate and interpret alleles with apparent functional importance. Here, we expand upon our original analyses examining the relationship between genomic patterns in ROAs and deleterious variation using the 2,436 unrelated individuals from 26 populations included in Phase 3 of The 1000 Genomes Project36 on which previously reported whole-genome sequence (WGS)-based genetic and ROA datasets are available.5 These data provide us with ∼100-fold more individuals (2,436 versus 27) and genetic data that are not biased by the limitations of the exome capture platform used in our original 2013 study.37 We extend our original analyses that were based upon the PolyPhen238 and SIFT39 deleteriousness categorization methods to consider a recently reported composite measure of potential functional consequence, Combined Annotation Dependent Depletion40 (CADD), that provides a deleteriousness score rather than categorization for each nonreference allele, enabling a more fine-grained investigation of the relationship between ROAs and deleterious variation patterns. We further analyze the relationship between ROAs and deleterious variation in specific sets of genes reported to cause Mendelian disease, to contribute to complex diseases and traits, or to encode Federal Drug Administration (FDA)-approved drug targets, to provide a more focused assessment of the involvement of ROAs in the determination of genetic risk for disease through modification of the patterns and properties of deleterious allele loads present in individual genomes.
Subjects and Methods
Genetic and ROA Data
We examined autosomal single-nucleotide variants (SNV) in publicly available phased genotypes for the 2,436 unrelated individuals from 26 populations included in Phase 3 of The 1000 Genomes Project that were obtained through a combination of low-coverage whole-genome sequencing (WGS) and high-coverage whole-exome sequencing (WES) approaches.36 We used the WGS dataset described in Blant et al.5 that had undergone quality-control checks for relatedness and SNV quality, but restricted our analyses to only the 40,637,503 SNVs located in the transcribed regions of genes as defined in build hg19 of the University of California, Santa Cruz (UCSC) human genome database and that have a PHRED-scaled deleteriousness “C-score” (“PSC score” henceforth) in the CADD database.40 We used the WGS-based ROA dataset described in Blant et al.5 that was inferred using a weighted likelihood approach and classified into five length-based classes with a Gaussian mixture model applied to their genetic map lengths with the GARLIC software tool.41
Damaging Homozygote Comparisons
Comparisons of the rates of gain of damaging and nondamaging homozygotes inside ROAs in individual genomes were performed using Equation 10 of Szpiech et al.6 while comparisons of rates of gain of damaging homozygotes among ROA classes and gene sets were performed using Equation 13. Unless stated otherwise, we considered a PSC score threshold of 15 to distinguish damaging SNV (≥15) from nondamaging SNV (<15).42
Disease Gene Sets
Lists of autosomal genes associated with autosomal-dominant and autosomal-recessive Mendelian diseases in the Online Mendelian Inheritance in Man43 (OMIM) database, associated with clinically significant diseases in the ClinVar44 database, encoding FDA-approved drug targets,45 and located nearest to reported genome-wide association study (GWAS) hits46 were obtained from Daniel MacArthur’s group at Massachusetts General Hospital (Boston, MA). The MacArthur OMIM lists represent the union of two previously reported lists of genes associated with autosomal-dominant (669) and autosomal-recessive (1,130) diseases in the OMIM database,47, 48 which were used to create a list containing autosomal genes not associated with dominant or recessive diseases (24,260; “non-OMIM” henceforth); genes associated with both dominant and recessive disease were ignored. Similarly, we used the lists of autosomal genes in the ClinVar database (3,078), encoding FDA-approved drug targets (270), and located nearest to reported GWAS hits (3,205) to create lists of non-ClinVar (22,970), non-FDA-approved drug target (25,778), and non-GWAS (22,843) autosomal genes.
The GWAS catalog49 was used to create lists of autosomal genes associated with eight complex diseases and traits for which ROA levels have been identified as a risk factor: standing height (568 genes), CAD (327), ALS (262), Alzheimer disease (375), Parkinson disease (186), schizophrenia (1,104), colorectal cancer (181), and thyroid cancer (25). For each disease and trait, we created a comparative gene set that contained all autosomal genes not associated with that disease or trait in the GWAS catalog.
For each individual and for each ROA class, we calculated the fraction of the total length of the transcribed regions of each gene set that overlapped ROAs and tabulated how many damaging and nondamaging homozygotes are present inside and outside ROAs in each gene set, based upon the genomic position of each gene in build hg19 of the UCSC human genome database.
Results
Numbers of Damaging Homozygotes inside ROAs
At the worldwide scale, as the fraction of the genome covered by ROAs increases, the number of damaging homozygotes falling within ROAs also increases (Pearson’s r = 0.959, p < 10−16; Figure 1A), while the number of damaging homozygotes falling outside of ROAs instead decreases (r = −0.952, p < 10−16). However, while numbers of damaging homozygotes present inside and outside ROAs are generally predicted well by the fraction of an individual’s genome in ROAs, a small number of individuals at the upper end of observed ROA fractions deviate noticeably from this relationship (Figure 1A). Most of these individuals are from the Dai (CDX) population, with the rest coming from assorted other populations (Figure S1). This observation potentially reflects the combined effects of the Dai’s small population size (∼1.2 million in Yunnan province,50 China, where The 1000 Genomes Project samples were collected), complex history,51, 52 and practices of endogamy (see entry for Dai in Web Resources) and consanguinity.53 These have led to elevated ROA levels compared with the other populations included in The 1000 Genomes Project,5 creating an environment where increases in the damaging homozygote load inside ROAs are not well tolerated. In one possibility, by damaging alleles found in homozygous form inside ROAs being more deleterious than damaging alleles found outside ROAs, and therefore having a greater cumulative detrimental effect on fitness. Consequently, we observe fewer damaging homozygotes inside ROAs—both those of newly arisen strongly damaging alleles and of weaker damaging alleles segregating on the haplotypes upon which they arise—since individuals with elevated numbers tend not to exist in the extant population. In this view, higher numbers of damaging homozygotes found outside ROAs could reflect an overall enrichment of the Dai for older and weaker damaging alleles.
Figure 1.
Number of Damaging Homozygotes versus the Fraction of the Genome in ROAs
(A) Scatterplot depicting how the number of damaging nonreference homozygotes outside (gray) and inside (black) ROA changes with the fraction of the genome in ROAs. Dashed lines depict the regression line for damaging homozygotes outside (gray; r = −0.952, p < 10−16) and inside (black; r = 0.959, p < 10−16) ROAs with outlier individuals excluded.
(B) The baseline number of damaging homozygotes present in the genomes of noninbred individuals in each population.
(C) The net change in damaging homozygotes in each population with a 10% increase in the fraction of the genome covered by ROAs.
In (B) and (C), each bar is colored by the geographic region of the population it represents.
When we compare the linear relationship between the fraction of the genome covered by ROAs and the rate of change in numbers of damaging homozygotes inside and outside ROAs, consistent with our original study,6 the decreasing slope for non-autozygous regions is shallower than the increasing slope for autozygous regions (Table 1). Thus, the rise in damaging homozygotes inside ROAs outpaces the decline of damaging homozygotes outside ROAs. The fitted lines predict that an average noninbred individual carries ∼7,252 damaging homozygotes and that increasing genomic ROA coverage by 10% results in a net increase of ∼246. These values are ∼40 times the baseline number of damaging homozygotes and ∼6 times the net increase reported in our original study (181 and 44, respectively), likely reflecting significant improvements in genomic coverage and in availability of deleteriousness predictions in this study. The WGS-based dataset used here has ∼100-fold more individuals (2,436 versus 27) and provides a more comprehensive gene set than the exome-capture-based dataset used in our original study. Moreover, the CADD deleteriousness prediction method40 used here provides a prediction for all observed SNVs, in contrast to the PolyPhen238 and SIFT39 methods used in our original study that only provide predictions for SNVs causing amino acid substitutions, greatly increasing the number of SNVs available for the calculations. Nevertheless, these patterns highlight how cultural and population processes that increase ROA levels in the genome can elevate numbers of damaging homozygotes carried by the general population, with potential negative consequences for general health.
Table 1.
Net Change in Damaging Homozygotes with 10% Increase in Genomic ROA Coverage
| C-score Category |
Outside ROA |
Inside ROA |
10% Increase in ROA Coverage |
|||||
|---|---|---|---|---|---|---|---|---|
| Slope | Intercept | Slope | Intercept | Outside | Inside | Net Change | % Change | |
| C ≥ 15 | −8,833.07 | 6,826.64 | 11,288.57 | 425.62 | −883.31 | 1,128.86 | 245.55 | 3.39 |
| 15 ≤ C < 20 | −7779.32 | 5943.77 | 9663.67 | 355.21 | −777.93 | 966.37 | 188.44 | 2.99 |
| 20 ≤ C < 25 | −1,001.53 | 822.78 | 1454.80 | 60.10 | −100.15 | 145.48 | 45.33 | 5.13 |
| C ≥ 25 | −52.22 | 60.09 | 170.10 | 10.30 | −5.22 | 17.01 | 11.80 | 16.57 |
The larger sample sizes of The 1000 Genomes Project populations—ranging between 55 and 109 individuals5—allow us to explore for the first time how the net change in damaging homozygotes varies across populations. Performing the linear regression analyses separately in each population, we find appreciable variability in baseline numbers of damaging homozygotes in noninbred individuals (Figure 1B) and the net change with a 10% increase in genomic ROA coverage (Figure 1C). Across populations, baseline numbers are greatest in East Asian populations and generally lowest in admixed Amerindian-European populations, while net changes are highest in Amerindian-European populations and lowest in European and East Asian populations. The highest net change is observed in Peruvians (568; PEL) and lowest in the Luhya (−1; LWK), with 6 of the 26 populations having a negative change: LWK, British (GBR), Gujarati (GIH), Bengali (BEB), Southern Han (CHS), and Afro-Caribbeans from Barbados (ACB). In general, net change is inversely proportional to the baseline number of damaging homozygotes in noninbred individuals (Figure S2; r = −0.651, p = 1.58 × 10−4), indicating that inbreeding will have the greatest effect on the number of damaging homozygotes present in individual genomes in populations where individuals generally possess fewer damaging homozygotes. In one possibility, this might reflect a ceiling effect, where any large increase in the damaging homozygote burden of inbred individuals from populations where individual burdens of damaging homozygotes are generally high will have a significant impact on their fitness and are thus not well tolerated. Nevertheless, the observed variability across populations in the damaging homozygote burden of noninbred individuals and in the net change with increasing genomic ROA coverage emphasizes how associations between ROA load and disease risk may be extremely sensitive to population background and highlights how increased ROA levels cannot always be used as an indicator for increased numbers of damaging alleles carried in homozygous form.
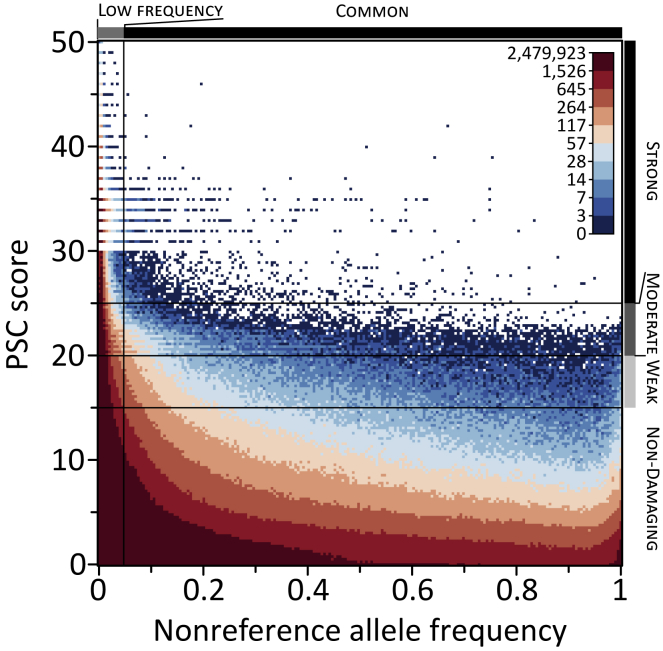
Unlike the PolyPhen238 and SIFT39 deleteriousness categorization methods used in our original study,6 the CADD deleteriousness prediction method used here provides a numerical score, where larger scores indicate a more deleterious mutation.40 This enables us to ask the following question: does the net increase in damaging homozygotes inside ROAs influence numbers of strongly deleterious homozygotes to a greater extent than numbers of weakly damaging homozygotes? We consider three deleteriousness categories—weak (PSC scores between 15 and 20), moderate (PSC scores between 20 and 25), and strong (PSC scores greater than 25)—and explore how numbers of damaging homozygotes in each category inside and outside ROAs change as the fraction of the genome covered by ROAs increases. Numbers of damaging homozygotes decrease outside ROAs and increase inside ROAs with increasing genomic ROA coverage for each deleteriousness category (Figure 2). Across populations, the net change observed for each category with a 10% increase in genomic ROA coverage (Figure S3A) generally mirror those observed when all categories are combined (Figure 1C). However, at a worldwide scale, individuals with 10% of their genome covered by ROAs carry 16.57% more strongly damaging homozygotes, 5.13% more moderately damaging homozygotes, and 2.99% more weakly damaging homozygotes than noninbred individuals (Table 1); this pattern of a proportionally larger increase in the number of strongly damaging homozygotes relative to numbers of moderately and weakly damaging homozygotes also persists at the population level (Figure S3B). These findings indicate that as the genome is progressively covered by ROAs, the rate of gain of homozygotes for strongly damaging alleles outpaces those for less damaging alleles, providing a potential mechanism by which increased ROA levels can elevate risk for disease.
Figure 2.
Number of Damaging Homozygotes versus the Fraction of the Genome in ROAs
Scatterplots depicting how numbers of damaging nonreference homozygotes outside (gray) and inside (black) ROAs changes with the fraction of the genome in ROAs for different deleteriousness categories.
(A) Variants with PSC scores between 15 and 20 (r = −0.956 and r = 0.974, respectively).
(B) Variants with PSC scores between 20 and 25 (r = −0.926 and r = 0.952, respectively).
(C) Variants with PSC scores greater than 25 (r = −0.556 and r = 0.838, respectively).
All Pearson’s correlations have p < 10−16. Figure format follows Figure 1A. Net changes in damaging homozygotes in each deleteriousness category are provided in Table 1.
Relationship between Damaging Homozygotes and ROA Age
Our original study6 considered ROAs classified into three classes based upon their physical map length. Here, we instead consider five ROA classes that are defined based upon their genetic rather than physical lengths,5 providing a more fine-scaled relationship between ROA classification and age.54, 55 Our choice of five classes was motivated by the observation that the Bayesian Information Criterion for the Gaussian Mixture Model used for classification plateaued at five for all populations present in our WGS dataset.5 In this context, we consider long class 5 ROAs to likely arise from recent inbreeding, while intermediate-length class 4 ROAs likely arise from population processes that influence effective population size, and shorter class 1 to 3 ROAs likely arise from linkage disequilibrium (LD) patterns on different evolutionary timescales. That is, class 3 ROAs are formed from haplotypes that were created through more recent events influencing genome-wide LD patterns than shorter class 1 ROAs. These interpretations are consistent with worldwide patterns in the total lengths and numbers of each ROA class as well as correlations observed between their genomic distributions and spatially variable genomic properties such as recombination rate and signals of natural selection.2, 5
First considering a single damaging category (PSC score ≥ 15), at the worldwide scale we find the rate of gain of damaging homozygotes inside ROAs with increasing genomic ROA coverage to significantly outpace the rate of gain of nondamaging homozygotes (PSC score < 15) in class 3 to 5 ROAs (Figure 3, Table 2). Conversely, the rate of gain of damaging homozygotes inside class 1 ROAs is significantly lower than nondamaging homozygotes (Figure 3A), while rates of gain of damaging and nondamaging homozygotes are similar for class 2 ROAs (Figure 3B). When the rate of gain in damaging homozygotes inside each class of ROA are compared (Figure 3F, Table S1), the rate is highest for class 5 ROAs and lowest for class 1 ROAs, generally decreasing inversely with ROA class (i.e., expected haplotype age). This pattern is consistent with the expectation that, relative to nondamaging alleles, damaging alleles will be recent in origin since purifying selection has not yet removed them from the gene pool, and thus will appear in homozygous form most often within longer ROAs that arise through recent inbreeding. When considering homozygotes in general, we find those made of low-frequency alleles (<5% in the population) to decrease in number inside class 1 to 4 ROAs with increasing genomic ROA coverage (Figures S4B–S4E), while numbers inside class 5 ROAs instead increase (Figure S4F). The patterns with homozygotes made of common alleles (frequencies ≥ 5%; Figure S5) mirror those observed when we do not stratify alleles by frequency (Figure 3).
Figure 3.
Fraction of Nonreference Homozygotes Falling in ROAs versus the Fraction of the Genome in ROAs
Scatterplots depicting how the fraction of nondamaging (gray) and damaging (in color) nonreference homozygotes in ROAs changes with the fraction of the genome in ROAs.
(A) Class 1 ROAs (r = 0.887 and r = 0.818, respectively).
(B) Class 2 ROAs (r = 0.942 and r = 0.910, respectively).
(C) Class 3 ROAs (r = 0.973 and r = 0.963, respectively).
(D) Class 4 ROAs (r = 0.980 and r = 0.975, respectively).
(E) Class 5 ROAs (r = 0.974 and r = 0.959, respectively).
(A–E) All Pearson’s correlations have p < 10−16. A comparison of slopes and intercepts of the regressions for nondamaging and damaging homozygotes are provided in Table 2.
(F) Regression lines for the fraction of damaging homozygotes in each ROA class taken from (A)–(E). A comparison between ROA classes of the slopes and intercepts of the regressions for damaging homozygotes is provided in Table S1.
(A–F) The black dotted line depicts the identity line.
Table 2.
Differences in Regression Slopes and Intercepts for Damaging and Nondamaging Homozygotes inside ROAs
| ROA Class |
Difference in Intercept |
Difference in Slope |
||
|---|---|---|---|---|
| β2 | p | β3 | p | |
| 1 | 0.0017 | 0.0123 | −0.0931 | 1.03 × 10−9 |
| 2 | 0.0072 | 7.24 × 10−11 | 0.0061 | 0.6538 |
| 3 | 0.0079 | <10−16 | 0.0375 | 7.31 × 10−5 |
| 4 | 0.0032 | 6.56 × 10−10 | 0.0492 | 1.36 × 10−9 |
| 5 | 1.83 × 10−4 | 0.5130 | 0.1490 | <10−16 |
Reported β and p values were calculated with Equation 10 of Szpiech et al.6
The decreasing trends observed with low-frequency allele homozygotes in class 1 to 4 ROAs that arise via background relatedness that is due to evolutionary history within a population accords with the expected effects of genetic drift on such alleles in the presence of inbreeding, while the increasing trend with long class 5 ROAs instead highlights how numbers of homozygotes for low-frequency alleles can be amplified considerably in the presence of recent inbreeding. Indeed, if we consider the proportion of genome-wide homozygotes made of low-frequency alleles that fall inside ROAs, we see almost no change per unit increase in genomic coverage for class 1 to 3 ROAs made of older haplotypes (Figures S6A and S6B), a moderate increase for class 4 ROAs made of haplotypes of intermediate age (Figures S6C and S6D), and the sharpest increase for class 5 ROAs made of young haplotypes (Figure S6E). This is consistent with the expectation that low-frequency alleles are unlikely to be found in homozygous form genome-wide, but when they do form they tend to be concentrated in autozygous regions that arise through recent parental relatedness.
Alleles in our strongly damaging category are mostly present at low frequencies (Figure 4), with PSC scores generally decreasing as the frequency of the nonreference allele increases. Focusing on alleles with PSC scores ≥15, 93.14% have a mean frequency of <5% across the populations in which they are present, and 72.81% have a mean frequency of <1%. Within each deleteriousness category, 98.61%, 94.26%, and 92.25% of strongly, moderately, and weakly damaging alleles have a mean frequency <5%, respectively, while 86.40%, 75.22%, and 70.76% have a mean frequency <1%. When we consider the rate of gain of damaging homozygotes inside ROAs with increasing genomic ROA coverage separately for each of the three deleteriousness categories (Figure S7), several interesting patterns emerge that generally accord with the observed relationships of nonreference allele frequency with deleteriousness (Figure 4) and numbers of homozygotes in each ROA class (Figures S4 and S5). First, the rate of gain for strongly damaging homozygotes is significantly greater than for nondamaging homozygotes with long class 5 ROAs, while it is significantly lower with class 2 to 4 ROAs (Table 3). Second, the rate of gain for moderately damaging homozygotes is significantly greater than that for nondamaging homozygotes with long class 5, while it is significantly lower with class 1 ROAs (Table 3). Third, the rate of gain for weakly damaging homozygotes is significantly greater than that for nondamaging homozygotes with class 3 to 5 ROAs, while it is significantly lower with class 1 ROAs (Table 3). Fourth, the rate of gain for strongly damaging homozygotes significantly outpaces those for moderately and weakly damaging homozygotes with class 2 to 5 ROAs (Table S2), which themselves differ significantly only with class 2 to 4 ROAs.
Figure 4.
Relationship between Deleteriousness and Nonreference Allele Frequency
A heatmap comparing PSC scores assigned to each nonreference allele by the CADD method40 and the average frequency of the nonreference allele in those populations in which it was observed. Cells are colored by decile.
Table 3.
Differences in Regression Slopes and Intercepts for Damaging and Nondamaging Homozygotes in Each Deleteriousness Category
| ROA Class | Deleteriousness Category |
Difference in Intercept |
Difference in Slope |
||
|---|---|---|---|---|---|
| β2 | p | β3 | p | ||
| 1 | weak | 0.0012 | 0.0864 | −0.0975 | 1.43 × 10−10 |
| moderate | 0.0039 | 4.87 × 10−6 | −0.0873 | 5.50 × 10−6 | |
| strong | 0.0227 | <10−16 | −0.0205 | 0.6681 | |
| 2 | weak | 0.0063 | 1.08 × 10−8 | 0.0078 | 0.5683 |
| moderate | 0.0111 | <10−16 | −0.0050 | 0.7578 | |
| strong | 0.0374 | <10−16 | −0.1095 | 0.0015 | |
| 3 | weak | 0.0066 | 2.37 × 10−15 | 0.0450 | 2.09 × 10−6 |
| moderate | 0.0132 | <10−16 | 0.0030 | 0.7943 | |
| strong | 0.0486 | <10−16 | −0.1897 | 1.30 × 10−14 | |
| 4 | weak | 0.0018 | 2.80 × 10−7 | 0.0663 | <10−16 |
| moderate | 0.0070 | <10−16 | −0.0016 | 0.8698 | |
| strong | 0.0323 | <10−16 | −0.0881 | 0.0018 | |
| 5 | weak | −2.60 × 10−4 | 0.2677 | 0.1505 | <10−16 |
| moderate | 7.21 × 10−4 | 0.0252 | 0.1927 | <10−16 | |
| strong | 0.0060 | 3.24 × 10−11 | 0.8164 | <10−16 | |
Reported β and p values were calculated with Equation 10 of Szpiech et al.6
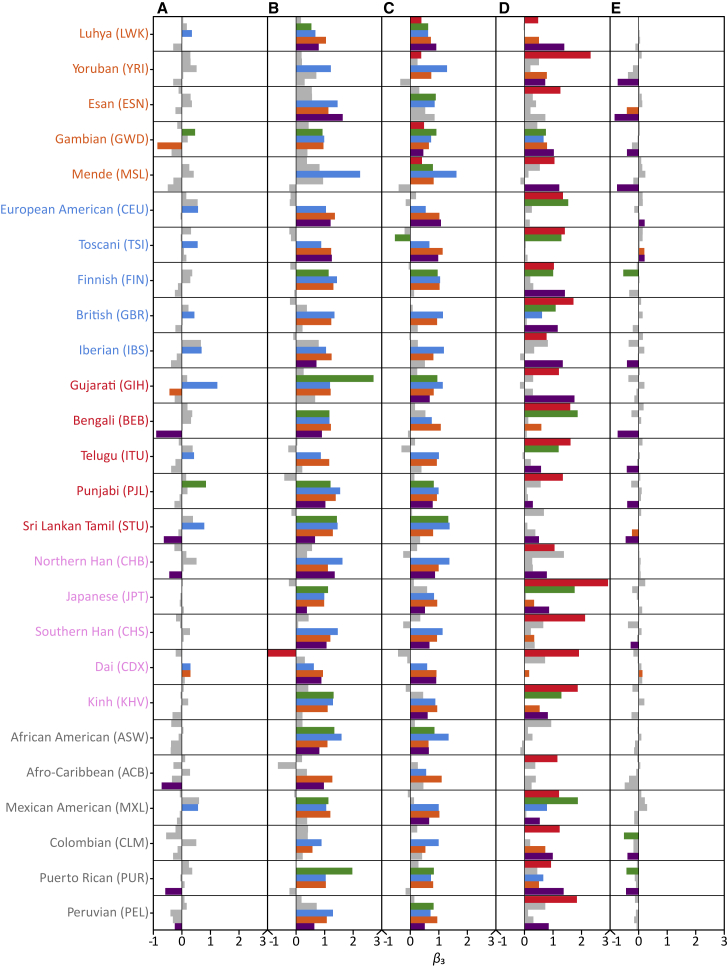
At the population level, there was again great variability in the differential rates of gain of damaging and nondamaging homozygotes inside ROAs with increasing genomic ROA coverage (Figure 5, Table S3). Rates of gain for damaging homozygotes significantly outpace those for nondamaging homozygotes in a majority of populations for longer ROAs that arise due to recent parental relatedness, whereas in shorter ROAs that arise due to more distant background parental relatedness only a few populations show the same pattern of increased rates of gain for damaging homozygotes. Indeed, significant increases in rates of gain for damaging homozygotes were seen in ∼69% (18 of 26) populations with long class 5 ROAs and ∼92% (24 of 26) of populations with intermediate-length class 4 ROAs. In contrast, among shorter class 1 to 3 ROAs, significant increases in rates of gain for damaging homozygotes were seen in ∼46% (12 of 26) populations with class 3 ROAs, ∼12% (3 of 26) with class 2 ROAs, and ∼23% (6 of 26) with class 1 ROAs. Notably, for all ROA classes, the rate of gain for damaging homozygotes did not differ significantly from the rate for nondamaging homozygotes in the Yoruba (YRI) population. These observations again underscore the potential confounding effects of population background on potential associations between genomic ROA levels and risk for complex disease due to differences in the relationship between damaging homozygote load and ROA across populations that likely reflect the cumulative effects of their distinct evolutionary and cultural histories.
Figure 5.
Differences in the Rate of Gain of Damaging and Nondamaging Homozygotes in Each Population
A bar plot showing for each population and ROA class the magnitude of β3 from regressions comparing the rates of gain of damaging and non-damaging nonreference homozygotes with increasing genomic ROA coverage. Bars depicting β3 with p < 0.05 are shown in color, while those with p ≥ 0.05 are shown in gray. β3 values and their associated p value can be found in Table S3.
Relationship between Damaging Homozygotes and ROAs in Disease-Associated Gene Sets
The relationship between deleterious variation and ROAs might be most apparent, and might be most important for understanding associations between ROAs and disease, in genomic regions known to cause disease when disrupted. We therefore investigated how ROA levels and their relationship with damaging homozygotes differed between genes included in the OMIM43 and ClinVar44 databases and genes not included in these databases, as well as between sets of genes located nearest to reported GWAS hits46 or encoding FDA-approved drug targets45 compared with genes not in these sets.
ROA Coverage
Strikingly, we find the fraction of each disease-associated gene set in ROAs to be significantly lower than the fraction for their comparative gene set (Figure 6; p < 10−16 for all comparisons, Wilcoxon signed-rank test). The difference is greatest for OMIM recessive (Figure 6A) and ClinVar (Figure 6B) genes, and smallest for genes nearest reported GWAS hits (Figure 6D). Moreover, despite their appreciably lower overall ROA levels, rates of gain for damaging homozygotes inside ROAs for each disease-associated gene set are significantly greater than rates of gain for their comparative gene set (Figure 7, Table 4). The difference is greatest for OMIM recessive and ClinVar genes and smallest for genes nearest to GWAS hits, while those of OMIM dominant and FDA-approved drug target genes are intermediate between these two extremes (Table 4). In general, observed rates of gain in damaging homozygotes appear inversely related to the degree of reduction in ROA levels around genes. Relative to their comparative gene set, OMIM recessive and ClinVar genes experience the greatest gain in damaging homozygotes with increasing ROA coverage while also exhibiting the greatest reduction in overall ROA coverage. Conversely, genes nearest to GWAS hits exhibit the smallest gain in damaging homozygotes and reduction in overall ROA coverage.
Figure 6.
Fraction of Disease Gene Sets in ROAs Relative to Non-Disease Gene Sets
Scatterplots comparing the fraction of the total length of transcribed regions in any size ROA for disease-associated gene sets with the same fraction for genes not in the disease-associated set in individual genomes.
(A) OMIM autosomal dominant (red; r = 0.990, p < 10−16) and recessive (blue; r = 0.994, p < 10−16) genes against non-OMIM genes.
(B) ClinVar against non-ClinVar genes (r = 0.996, p < 10−16).
(C) FDA-approved drug target against non-drug-target genes (r = 0.947, p < 10−16).
(D) Genes nearest to reported GWAS hits against all other genes (r = 0.995, p < 10−16).
The identity line is shown in gray, while trend lines of the linear regression fit for each comparison is shown in black or in color.
Figure 7.
Fraction of Nonreference Homozygotes in ROAs versus the Fraction of Transcribed Regions in ROAs
Scatterplots depicting how the fraction of damaging nonreference homozygotes within any size ROA changes with the fraction of the transcribed regions in ROAs in individual genomes shown separately for each pair of disease-associated (black or color) and non-disease-associated (gray) gene sets.
(A) OMIM autosomal dominant (red; r = 0.901, p < 10−16) and recessive (blue; r = 0.926, p < 10−16) genes against non-OMIM (r = 0.960, p < 10−16) genes.
(B) ClinVar (r = 0.950, p < 10−16) against non-ClinVar (r = 0.959, p < 10−16) genes.
(C) FDA-approved drug target (r = 0.887, p < 10−16) against non-drug-target (r = 0.959, p < 10−16) genes.
(D) Genes nearest to reported GWAS hits (r = 0.948, p < 10−16) against all other genes (r = 0.958, p < 10−16).
Trend lines of the linear regression fit for each gene set is shown in the color of that gene set. The black dotted line depicts the identify line. A comparison of slopes and intercepts of the regressions for the disease-associated gene set and the non-disease-associated gene set are provided in Table 4.
Table 4.
Differences in Regression Slopes and Intercepts for Damaging Homozygotes in Disease Gene Sets and Non-Disease Gene Sets
| Disease Gene Set |
Difference in Intercept |
Difference in Slope |
||
|---|---|---|---|---|
| β2 | p | β3 | p | |
| OMIM dominant | −0.0032 | 0.3587 | 0.1121 | 1.01 × 10−13 |
| OMIM recessive | 0.0130 | 5.42 × 10−5 | 1.0175 | <10−16 |
| ClinVar | −0.0062 | 0.0299 | 0.8715 | <10−16 |
| FDA-approved drug targets | −0.0321 | <10−16 | 0.2532 | <10−16 |
| GWAS (nearest gene) | −0.0137 | 2.45 × 10−6 | 0.0398 | 6.34 × 10−4 |
Reported β and p values were calculated with Equation 13 of Szpiech et al.6
Rates of Gain in Damaging Homozygotes
When we instead consider rates of gain in damaging homozygotes separately in each ROA class, a number of interesting patterns emerge. First, rates of gain of damaging homozygotes with increasing ROA coverage in OMIM recessive (Figure S8) and ClinVar (Figure S9) genes significantly outpace those in non-OMIM and non-ClinVar genes, respectively, with all ROA classes (Figure 8A, Table S4). This pattern generally persists at the population level with longer class 3 to 5 ROAs that arise via more recent parental relatedness, but not with shorter class 1 and 2 ROAs that arise via more distant background parental relatedness (Figures 9B and 9C, respectively; Tables S5 and S6). Second, at the worldwide scale, OMIM dominant genes are observed to have a significant depletion of damaging homozygotes in long class 5 and short class 1 ROAs relative to non-OMIM genes, while small gains are instead observed with class 2 and 3 ROAs (Figures 8A and S8, Table S4). However, at the population-level, the rates for OMIM dominant genes are rarely different from those of non-OMIM genes (Figure 9A, Table S7), suggesting that worldwide patterns are driven primarily by population differences.
Figure 8.
Differences in the Rate of Gain of Damaging Homozygotes in Disease and Non-Disease Gene Sets
A bar plots depicting for each disease gene set and ROA class the magnitude of β3 from regressions comparing the rates of gain of damaging nonreference homozygotes in disease-associated and non-disease-associated gene sets with increasing genomic ROA coverage at the worldwide scale.
(A) Genes included in the OMIM and ClinVar databases and encoding FDA-approved drug targets, and genes located nearest to reported GWAS signals.
(B) Eight complex diseases and traits for which ROA load has been identified as a genetic risk factor.
Bars depicting β3 with p < 0.05 are shown in color, while those with p ≥ 0.05 are shown in gray. β3 values and their associated p value can be found in Tables S4 and S10 for (A) and (B), respectively.
Figure 9.
Differences in the Rate of Gain of Damaging Homozygotes in Disease and Non-Disease Gene Sets in Each Population
Bar plots showing for each population and ROA class the magnitude of β3 from regressions comparing the rates of gain of damaging nonreference homozygotes in disease-associated and non-disease-associated gene sets with increasing genomic ROA coverage.
(A) OMIM dominant genes.
(B) OMIM recessive genes.
(C) ClinVar genes.
(D) FDA-approved drug target genes.
(E) Genes located nearest to reported GWAS signals.
Bars depicting β3 with p < 0.05 are shown in color, while those with p ≥ 0.05 are shown in gray. β3 values and their associated p value can be found in Tables S5–S9.
Third, rates of gain of damaging homozygotes in FDA-approved drug target genes significantly outpace those of genes that do not encode drug target genes with all ROA classes. At the worldwide scale, the magnitude of the difference generally increases with increasing ROA class (Figures 8A and S10, Table S4), while at the population level, differences are observed most often with the shortest and longest ROA classes (Figure 9D, Table S8). This suggests that both weaker damaging alleles that persist on older haplotypes segregating in the general population as well as stronger damaging alleles that arise on younger haplotypes that arise via recent inbreeding contribute to damaging homozygote loads present in drug target genes. Notably, we find much weaker support for a difference in rates of gain in damaging homozygotes between FDA-approved drug target genes and non-drug-target genes in African Europeans as well as the Esan (ESN) and Sri Lankan Tamil (STU) populations.
Fourth, while rates of gain of damaging homozygotes in genes nearest to reported GWAS hits are significantly higher than those of all other genes with class 3 and 4 ROAs at the worldwide scale, they are significantly lower for short class 1 and long class 5 ROAs (Figures 8A and S11, Table S4). In agreement with the generally weak patterns observed at a worldwide scale, at the population level, significant differences in rates of gain for damaging homozygotes in genes nearest to reported GWAS hits and in all other genes are only frequently observed with long class 5 ROAs that arise through recent parental relatedness (Figure 9E, Table S9), with 12 of the 26 populations showing a significant depletion, and two populations exhibiting an enrichment. Intriguingly, depletion of damaging homozygotes is observed in most African and South Asian populations in addition to the Iberian (IBS), CHS, Colombian (CLM), and Puerto Rican (PUR) populations, while enrichment is observed in Utah Mormons with Northern and Western European ancestry (CEU) and the Toscani (TSI). These findings highlight the potential confounding effects of evolutionary and cultural histories, which may be shared among populations from the same country or geographic area, on the relationship between damaging homozygote load and ROAs that can lead to differences in their contributions to general genetic risk for complex disease across populations.
Relationship between Damaging Homozygotes and ROAs in GWAS Gene Sets
Our findings, which used genes nearest to reported GWAS hits as a surrogate for those that truly influence complex disease risk, are potentially confounded by variability in the proportion of genetic risk attributable to ROA levels for different diseases, as suggested by past studies investigating ROA-phenotype associations in disease cohorts. Therefore, we next used the GWAS catalog49 to create lists of genes associated via GWAS with increased risk for eight complex diseases and traits for which ROA levels have been identified as a risk factor: standing height (568 genes), CAD (327), ALS (262), Alzheimer disease (375), Parkinson disease (186), schizophrenia (1,104), colorectal cancer (181), and thyroid cancer (25). For each GWAS gene set, we compared it to a set of genes not currently associated with that disease in the GWAS catalog.
ROA Coverage
Intriguingly, while we find genes associated with ALS (Figure S12C) and thyroid cancer (Figure S12G) to have significantly lower overall ROA coverage than genes not associated with these diseases (p < 10−16 in both comparisons, Wilcoxon signed-rank test), genes associated with standing height (Figure S12A), CAD (Figure S12B), Alzheimer disease (Figure S12D), Parkinson disease (Figure S12E), and schizophrenia (Figure S12F) instead had significantly higher overall ROA coverage (p < 10−16 in all comparisons); genes associated with colorectal cancer had similar overall ROA coverage to genes not associated with colorectal cancer (p = 0.775; Figure S12H). The difference between disease and non-disease gene sets was most apparent for schizophrenia, but overall differences between GWAS and non-GWAS gene sets are notably smaller than those observed in the OMIM, ClinVar, and FDA-approved drug target comparisons (Figure 6), consistent with the expectation that purifying selection acting on mutations in genes associated with largely single-gene disorders will exert a larger effect on genetic diversity patterns than those in genes associated with polygenic diseases and traits.
Rates of Gain in Damaging Homozygotes
Despite differences in overall ROA coverage levels among GWAS gene sets, with the exception of thyroid cancer, all were found to gain damaging homozygotes inside ROAs at rates exceeding those of their comparative gene set per unit increase in ROA coverage (Figure 8B, Table S10). However, differences were present in the rates of gain in each ROA class across diseases and traits. The greatest differences were observed with genes associated with colorectal cancer, where the magnitude of the difference from the non-colorectal-cancer gene set increased as a function of ROA class (Figures 8B and S13, Table S10). The rate of gain of damaging homozygotes inside ROAs for Alzheimer disease (Figure S14), Parkinson disease (Figure S15), and schizophrenia (Figure S16) were significantly higher than their comparative gene sets with class 1 to 4 ROAs that arise through background relatedness that is due to evolutionary history within a population (Figure 8B, Table S10), while rates of gain in class 5 ROAs that arise through recent parental relatedness were instead generally significantly lower. Rates of gain of damaging homozygotes inside ROAs in genes associated with standing height (Figure S17) and CAD (Figure S18) were significantly higher than in non-height- and non-CAD-associated gene sets, respectively, for class 2 to 4 ROAs, while rates in class 1 and 5 ROAs were generally significantly lower (Figure 8B, Table S10). The ALS gene set experienced a significantly higher rate of gain of damaging homozygotes in class 1, 4, and 5 ROAs than the non-ALS gene set (Figure S19), while a significantly lower rate was observed for class 2 ROAs (Figure 8B, Table S10). Interestingly, the thyroid cancer gene set was observed to have significantly decreased rates of gain of damaging homozygotes in shorter class 1 to 3 ROAs formed by older haplotypes underlying LD patterns relative to the non-thyroid-cancer gene set (Figures 8A and S20, Table S10); rates were not significantly different with longer class 4 and 5 ROAs that arise due to parental relatedness.
In agreement with patterns observed at the worldwide scale (Figure 8B, Table S10), at the population level (Figure 10, Tables S11–S18) genes associated with colorectal cancer exhibited the greatest consistency across populations, with rates of gain of damaging homozygotes inside longer class 3 to 5 ROAs significantly greater in disease-associated than non-disease-associated genes in all populations, while significant differences in rates with shorter class 1 and 2 ROAs are less common (Figure 10G, Table S11). Similarly, we find that rates of gain in genes associated with thyroid cancer are rarely significantly different from those for genes not associated with thyroid cancer across ROA classes and populations (Figure 10H, Table S12), with some ROA classes occasionally showing significant increases while others show significant decreases across populations. Patterns observed with standing height (Figure 10A, Table S13) and schizophrenia (Figure 10F, Table S14) generally accord with those at the worldwide scale; however, with long class 5 ROAs that arise via recent parental relatedness, significantly higher rates of gain in damaging homozygotes in GWAS gene sets relative to non-GWAS gene sets are observed in five (CEU, TSI, BEB, CHB, and CDX) and three (CEU, TSI, and CDX) of the 26 populations, respectively, while significantly lower rates are observed in 15 and 14 populations, respectively. These observations highlight how the relationship between ROA levels and damaging variation in sets of genes associated with complex diseases and traits can vary appreciably across populations and phenotypes, providing a potential explanation for observed inconsistencies among studied GWAS cohorts and phenotypes in the association between ROA levels and disease risk or trait variability.
Figure 10.
Differences in the Rate of Gain of Damaging Homozygotes in Complex Disease and Non-Disease Gene Sets in Each Population
Bar plots showing for each population and ROA class the magnitude of β3 from regressions comparing the rates of gain of damaging nonreference homozygotes in disease-associated and non-disease-associated gene sets with increasing genomic ROA coverage.
(A) Standing height.
(B) CAD.
(C) ALS.
(D) Alzheimer disease.
(E) Parkinson disease.
(F) Schizophrenia.
(G) Colorectal cancer.
(H) Thyroid cancer.
Bars depicting β3 with p < 0.05 are shown in color, while those with p ≥ 0.05 are shown in gray. β3 values and their associated p value can be found in Tables S11–S18.
Discussion
Our findings contribute to the emerging picture of the variable roles of population, cultural, and genomic processes in shaping patterns of ROAs and deleterious variation in the human genome.6, 7 They are consistent with processes acting to increase genomic ROA coverage leading to enrichment for homozygotes of alleles that are predicted to have a measurable impact on gene and protein function, particularly in genes associated with phenotypic variation and disease risk. However, they also suggest that their baseline numbers in the genomes of noninbred individuals and their degree of enrichment in the genomes of more inbred individuals may vary greatly across populations as well as among diseases and traits, potentially reflecting differences in the levels and properties of deleterious variation across populations that have arisen due to their distinct histories and cultural norms.56, 57, 58 While natural selection has no doubt contributed to the presence of functionally important alleles on the long haplotypes underlying ROAs, and to observed differences in enrichment of ROAs for such alleles across populations, we have previously shown signals of natural selection to be only weakly correlated with ROA patterns;2, 5 thus, the historical actions of natural selection cannot wholly explain the observed enrichment of ROAs for predicted damaging alleles. Taken altogether, our findings provide important new insights into how parental relatedness at different genealogical depths contributes to the shaping of patterns of harmful variation in individual genomes and populations that can lead to a general decrease in biological fitness, or inbreeding depression,8 and increased genetic risk for the development of Mendelian and complex disease.59
Rates of gain in damaging homozygotes across ROA classes are consistent with recent parental inbreeding, the primary force creating long class 5 ROAs, elevating numbers of damaging homozygotes, and strongly damaging homozygotes in particular, at a rate greater than would be expected based upon nondamaging homozygotes. The lower numbers of damaging homozygotes in class 1 to 4 ROAs, which arise via population and genomic processes, relative to nondamaging homozygotes is consistent with purifying selection acting to remove haplotypes with harmful alleles from the general population. Consequently, damaging alleles are generally present at low frequencies and are most frequently found in homozygous form in long class 5 ROAs that commonly arise through recent inbreeding since purifying selection will not yet have had time to remove these young haplotypes carrying damaging alleles from the gene pool. Nevertheless, rates of gain in homozygotes for damaging alleles in intermediate-length class 3 and 4 ROAs that outpace those for nondamaging alleles are consistent with population processes that decrease the effective size of populations or maintain a small effective size contributing to the enrichment of individual genomes for deleterious variation carried in homozygous form, albeit to a lesser extent than recent inbreeding.
Patterns in ROA coverage and rates of gain in damaging homozygotes in different sets of genes associated with Mendelian and complex disease are consistent with the expectation that mutations in OMIM and ClinVar genes, which commonly cause disease in a Mendelian manner, have a greater chance of being damaging than mutations in genes indirectly implicated in complex disease through their proximity to GWAS signals. This leads to a more rapid increase in numbers of damaging homozygotes observed per unit increase in ROA coverage in OMIM and ClinVar genes than in genes nearest reported GWAS hits. We would also expect haplotypes harboring one or more damaging alleles to experience stronger negative selection at OMIM and ClinVar genes than at genes located near to GWAS signals due to their greater potential to significantly limit the survival and reproductive potential of an individual. Since damaging recessive alleles are removed from the gene pool much less effectively than dominant damaging alleles (because purifying selection acts only on their homozygous form), they are able to reach nontrivial frequencies in the general population, enabling them to occur in homozygous form through both population processes (class 1–4 ROAs) and recent inbreeding (class 5 ROAs) more frequently than damaging dominant alleles, which given their high deleteriousness are rarely observed in homozygous form in the extant population. Therefore, for genes where damaging recessive alleles play a significant role in determining disease risk, we would expect to observe a greater decrease in ROA levels relative to genes that do not contribute to disease risk or where damaging dominant alleles are more common since purifying selection will act to remove damaging recessive homozygotes much more frequently. Compatible with this view, we observe a greater decrease in ROA levels in OMIM recessive than in OMIM dominant genes relative to non-OMIM genes. Moreover, while OMIM recessive genes show enrichment for damaging homozygotes in longer class 4 and 5 ROAs relative to non-OMIM genes, OMIM dominant genes instead show a depletion of damaging homozygotes in long class 5 ROAs and no appreciable difference in intermediate-length class 4 ROAs. This is consistent with the expectation that strongly damaging dominant alleles will be rarer in the extant population due to more efficient removal by purifying selection.
The decreased ROA coverage and increased rate of gain of damaging homozygotes observed in genes encoding the targets of FDA-approved drugs relative to genes that do not is compatible with an intriguing scenario. Some of these damaging alleles may contribute meaningfully to development of phenotypes that are severe enough to elicit a purifying selection response when formed into homozygotes in ROAs generated through both recent and more distant inbreeding. These findings are consistent with the expectation that the targets of drugs used in the treatment of disease most likely function in pathways that contribute to disease development and progression, and thus genetic changes that alter their structure or function have a high potential to contribute to the onset of disease. However, the accumulation of mildly damaging genetic variants, as might be expected with increasing ROA coverage, could lead to subtle changes to protein structure that impact the interaction of a drug with its protein target without leading to disease. These findings highlight the need for pharmacogenomic investigations into the possible consequences of elevated damaging allele loads carried in ROAs on drug efficacy in populations where demographic and cultural processes that elevate ROA levels are known to exist.
We have uncovered differences in overall ROA coverage and in rates of gain of damaging homozygotes with increasing ROA coverage, between sets of genes associated and not-associated with eight complex diseases and traits for which ROA levels have been identified as a risk factor, as well as variable patterns among diseases and traits. These observations highlight potential differences in the contribution of ROAs and the damaging homozygotes they harbor to the determination of genetic risk across complex diseases and traits. This is consistent with the expectation that genetic determinants contributing to risk for polygenic diseases and traits will be highly variable, and the relative roles of dominant and recessive alleles in determining overall risk will be decided by the genes involved and their degree of influence on the phenotype in question. Patterns in damaging homozygotes across ROA classes are consistent with the idea that the primary factors elevating genetic risk with increasing genomic ROA load for complex diseases and traits are weakly damaging alleles segregating on older haplotypes. Such older haplotypes are most commonly paired IBD through population processes that shape levels of background relatedness within a population. They form class 1 to 4 ROAs that we observe to be frequently enriched for damaging homozygotes located in genes associated with standing height, CAD, Alzheimer disease, Parkinson disease, schizophrenia, and colorectal cancer. Interestingly, while long class 5 ROAs that arise most frequently through recent inbreeding are enriched for damaging homozygotes located in genes associated with ALS and colorectal cancer, a depletion was instead observed for genes associated with standing height, Parkinson disease, and schizophrenia. These observations underscore the variable contributions of recent inbreeding to the determination of genetic risk for complex diseases and traits, which is in contrast to genes causing monogenic disorders for which class 5 ROAs were frequently found to be significantly enriched for damaging homozygotes.
For the gene sets associated with standing height, CAD, Alzheimer disease, Parkinson disease, and schizophrenia, their higher overall ROA coverage and higher rates of gain of damaging homozygotes in class 2 to 4 ROAs per unit increase in ROA coverage than genes not associated with these phenotypes would be compatible with a subset of alleles classed as damaging by the CADD method being instead beneficial. As such, CADD scores reflect the probability that a given nonreference allele will have a measurable impact on gene or protein function, which is commonly assumed will be harmful in nature. However, it is possible that such changes will in some instances be beneficial. In this scenario, increased overall ROA coverage is observed through the actions of positive rather than purifying selection acting in numerous trait-associated genes on beneficial alleles that act in a recessive manner. Conversely, patterns with ALS and thyroid cancer are consistent with homozygosity for damaging alleles being a bigger driver of ROA patterns in genomic regions harboring genes contributing to these diseases.
Differences were observed in the relationship between ROAs and damaging homozygotes in the Northern (CHB) and Southern (CHS) Han, a surprising finding given their recent shared ancestry.60 This includes a difference in the net change with a 10% increase in ROA coverage, where the CHB showed a positive change and the CHS showed a negative change. It also includes a difference in the rates of gain of damaging and nondamaging homozygotes in long class 5 ROAs that arise through recent parental relatedness, where the rate of gain of damaging homozygotes outpaces that of nondamaging homozygotes in the CHB but not in the CHS. These inconsistencies potentially reflect differences in their frequencies of consanguinity—1.16% in the CHB53, 61, 62 and 3.43% in the CHS53, 62—that have led to elevated ROA levels in the CHS compared with the CHB5 as well as contributed to the development of low but detectable genetic structure between these two groups.36 Importantly, these differences have likely contributed to the development of small but noticeable dissimilarities in their rates of gain of damaging homozygotes in genes that are associated with Mendelian and complex diseases and traits. This includes dissimilarities in rates for long class 5 ROAs with genes associated with standing height and encoding FDA-approved drug targets. Such differences between the closely related CHB and CHS populations, which are represented here by large and similarly sized sets of individuals,5 highlight the complex relationship between ROAs and deleterious variation patterns and their joint contribution to genetic risk for Mendelian and complex disease. Moreover, they suggest that such contributions are sensitive to the effects of population and cultural processes that impact genetic diversity patterns over relatively short timescales.
Future studies in disease cohorts with clearly defined population backgrounds and available WGS data as well as improved methods for predicting the relative effects of observed alleles, both harmful and beneficial, will be required to clarify the role of ROAs and the trait-influencing alleles they contain in the determination of genetic risk for complex disease and variability in complex traits. Our finding that net gains in damaging homozygotes with increasing genomic ROA coverage vary appreciably across populations would suggest that ROA-phenotype associations may be most apparent, and most important, in populations where ROA levels have the greatest impact on damaging homozygote loads, such as admixed Amerindian-European and South Asian populations. Altogether, our findings highlight the need for studies to appropriately control for population background when performing investigations in large cohorts used to investigate ROA-phenotype associations, particularly when undertaken in a meta-analysis framework. Moreover, they draw attention to the need to expand the diversity of populations and diseases examined to disentangle and clarify the variable roles of numerous population and cultural forces shaping ROAs and deleterious variation patterns in the determination of population attributable risk for diseases of major public health concern worldwide.
Acknowledgments
This research was partially supported by an institutional start-up fund from the University of Manitoba (T.J.P.). Partial support to Z.A.S. was provided by the National Human Genome Research Institute of the NIH under award number R01HG007644 (awarded to Ryan D. Hernandez, University of California, San Francisco). The authors thank Noah Rosenberg at Stanford University for helpful comments and discussions.
Published: March 15, 2018
Footnotes
Supplemental Data include 20 figures and 18 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.02.013.
Web Resources
1000 Genomes Project, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/
DrugBank, https://www.drugbank.ca
GWAS Catalog, http://www.ebi.ac.uk/gwas/
McArthur lab gene lists, https://github.com/macarthur-lab/gene_lists
OMIM, http://www.omim.org/
UCSC Genome Browser, http://hgdownload.cse.ucsc.edu/downloads.html#human
Supplemental Data
References
- 1.Kirin M., McQuillan R., Franklin C.S., Campbell H., McKeigue P.M., Wilson J.F. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE. 2010;5:e13996. doi: 10.1371/journal.pone.0013996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pemberton T.J., Absher D., Feldman M.W., Myers R.M., Rosenberg N.A., Li J.Z. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 2012;91:275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karafet T.M., Bulayeva K.B., Bulayev O.A., Gurgenova F., Omarova J., Yepiskoposyan L., Savina O.V., Veeramah K.R., Hammer M.F. Extensive genome-wide autozygosity in the population isolates of Daghestan. Eur. J. Hum. Genet. 2015;23:1405–1412. doi: 10.1038/ejhg.2014.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang J.T.L., Goldberg A., Edge M.D., Behar D.M., Rosenberg N.A. Consanguinity rates predict long runs of homozygosity in Jewish populations. Hum. Hered. 2017;82:87–102. doi: 10.1159/000478897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blant A., Kwong M., Szpiech Z.A., Pemberton T.J. Weighted likelihood inference of genomic autozygosity patterns in dense genotype data. BMC Genomics. 2017;18:928. doi: 10.1186/s12864-017-4312-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szpiech Z.A., Xu J., Pemberton T.J., Peng W., Zöllner S., Rosenberg N.A., Li J.Z. Long runs of homozygosity are enriched for deleterious variation. Am. J. Hum. Genet. 2013;93:90–102. doi: 10.1016/j.ajhg.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mezzavilla M., Vozzi D., Badii R., Alkowari M.K., Abdulhadi K., Girotto G., Gasparini P. Increased rate of deleterious variants in long runs of homozygosity of an inbred population from Qatar. Hum. Hered. 2015;79:14–19. doi: 10.1159/000371387. [DOI] [PubMed] [Google Scholar]
- 8.Charlesworth D., Willis J.H. The genetics of inbreeding depression. Nat. Rev. Genet. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- 9.Wright S. Systems of mating. Genetics. 1921;6:111–123. doi: 10.1093/genetics/6.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudan I., Smolej-Narancic N., Campbell H., Carothers A., Wright A., Janicijevic B., Rudan P. Inbreeding and the genetic complexity of human hypertension. Genetics. 2003;163:1011–1021. doi: 10.1093/genetics/163.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fareed M., Afzal M. Evidence of inbreeding depression on height, weight, and body mass index: a population-based child cohort study. Am. J. Hum. Biol. 2014;26:784–795. doi: 10.1002/ajhb.22599. [DOI] [PubMed] [Google Scholar]
- 12.Fareed M., Afzal M. Increased cardiovascular risks associated with familial inbreeding: a population-based study of adolescent cohort. Ann. Epidemiol. 2016;26:283–292. doi: 10.1016/j.annepidem.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Rudan I. Inbreeding and cancer incidence in human isolates. Hum. Biol. 1999;71:173–187. [PubMed] [Google Scholar]
- 14.Rudan I., Rudan D., Campbell H., Carothers A., Wright A., Smolej-Narancic N., Janicijevic B., Jin L., Chakraborty R., Deka R., Rudan P. Inbreeding and risk of late onset complex disease. J. Med. Genet. 2003;40:925–932. doi: 10.1136/jmg.40.12.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McQuillan R., Eklund N., Pirastu N., Kuningas M., McEvoy B.P., Esko T., Corre T., Davies G., Kaakinen M., Lyytikäinen L.P., ROHgen Consortium Evidence of inbreeding depression on human height. PLoS Genet. 2012;8:e1002655. doi: 10.1371/journal.pgen.1002655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verweij K.J., Abdellaoui A., Veijola J., Sebert S., Koiranen M., Keller M.C., Järvelin M.R., Zietsch B.P. The association of genotype-based inbreeding coefficient with a range of physical and psychological human traits. PLoS ONE. 2014;9:e103102. doi: 10.1371/journal.pone.0103102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joshi P.K., Esko T., Mattsson H., Eklund N., Gandin I., Nutile T., Jackson A.U., Schurmann C., Smith A.V., Zhang W. Directional dominance on stature and cognition in diverse human populations. Nature. 2015;523:459–462. doi: 10.1038/nature14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samuels D.C., Wang J., Ye F., He J., Levinson R.T., Sheng Q., Zhao S., Capra J.A., Shyr Y., Zheng W., Guo Y. Heterozygosity ratio, a robust global genomic measure of autozygosity and its association with height and disease risk. Genetics. 2016;204:893–904. doi: 10.1534/genetics.116.189936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christofidou P., Nelson C.P., Nikpay M., Qu L., Li M., Loley C., Debiec R., Braund P.S., Denniff M., Charchar F.J. Runs of homozygosity: association with coronary artery disease and gene expression in monocytes and macrophages. Am. J. Hum. Genet. 2015;97:228–237. doi: 10.1016/j.ajhg.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomsen H., Chen B., Figlioli G., Elisei R., Romei C., Cipollini M., Cristaudo A., Bambi F., Hoffmann P., Herms S. Runs of homozygosity and inbreeding in thyroid cancer. BMC Cancer. 2016;16:227. doi: 10.1186/s12885-016-2264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bacolod M.D., Schemmann G.S., Wang S., Shattock R., Giardina S.F., Zeng Z., Shia J., Stengel R.F., Gerry N., Hoh J. The signatures of autozygosity among patients with colorectal cancer. Cancer Res. 2008;68:2610–2621. doi: 10.1158/0008-5472.CAN-07-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghani M., Sato C., Lee J.H., Reitz C., Moreno D., Mayeux R., St George-Hyslop P., Rogaeva E. Evidence of recessive Alzheimer disease loci in a Caribbean Hispanic data set: genome-wide survey of runs of homozygosity. JAMA Neurol. 2013;70:1261–1267. doi: 10.1001/jamaneurol.2013.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghani M., Reitz C., Cheng R., Vardarajan B.N., Jun G., Sato C., Naj A., Rajbhandary R., Wang L.S., Valladares O., Alzheimer’s Disease Genetics Consortium Association of long runs of homozygosity with Alzheimer disease among African American individuals. JAMA Neurol. 2015;72:1313–1323. doi: 10.1001/jamaneurol.2015.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simón-Sánchez J., Kilarski L.L., Nalls M.A., Martinez M., Schulte C., Holmans P., Gasser T., Hardy J., Singleton A.B., Wood N.W., International Parkinson’s Disease Genomics Consortium. Wellcome Trust Case Control Consortium Cooperative genome-wide analysis shows increased homozygosity in early onset Parkinson’s disease. PLoS ONE. 2012;7:e28787. doi: 10.1371/journal.pone.0028787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaughlin R.L., Kenna K.P., Vajda A., Heverin M., Byrne S., Donaghy C.G., Cronin S., Bradley D.G., Hardiman O. Homozygosity mapping in an Irish ALS case-control cohort describes local demographic phenomena and points towards potential recessive risk loci. Genomics. 2015;105:237–241. doi: 10.1016/j.ygeno.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Carlson C.S., Eberle M.A., Kruglyak L., Nickerson D.A. Mapping complex disease loci in whole-genome association studies. Nature. 2004;429:446–452. doi: 10.1038/nature02623. [DOI] [PubMed] [Google Scholar]
- 27.Freimer N., Sabatti C. The use of pedigree, sib-pair and association studies of common diseases for genetic mapping and epidemiology. Nat. Genet. 2004;36:1045–1051. doi: 10.1038/ng1433. [DOI] [PubMed] [Google Scholar]
- 28.Wright A., Charlesworth B., Rudan I., Carothers A., Campbell H. A polygenic basis for late-onset disease. Trends Genet. 2003;19:97–106. doi: 10.1016/s0168-9525(02)00033-1. [DOI] [PubMed] [Google Scholar]
- 29.McQuillan R., Leutenegger A.L., Abdel-Rahman R., Franklin C.S., Pericic M., Barac-Lauc L., Smolej-Narancic N., Janicijevic B., Polasek O., Tenesa A. Runs of homozygosity in European populations. Am. J. Hum. Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reich D.E., Lander E.S. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 31.Pemberton T.J., Rosenberg N.A. Population-genetic influences on genomic estimates of the inbreeding coefficient: a global perspective. Hum. Hered. 2014;77:37–48. doi: 10.1159/000362878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keller M.C., Simonson M.A., Ripke S., Neale B.M., Gejman P.V., Howrigan D.P., Lee S.H., Lencz T., Levinson D.F., Sullivan P.F., Schizophrenia Psychiatric Genome-Wide Association Study Consortium Runs of homozygosity implicate autozygosity as a schizophrenia risk factor. PLoS Genet. 2012;8:e1002656. doi: 10.1371/journal.pgen.1002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson E.C., Bjelland D.W., Howrigan D.P., Abdellaoui A., Breen G., Borglum A., Cichon S., Degenhardt F., Forstner A.J., Frank J., Schizophrenia Working Group of the Psychiatric Genomics Consortium No reliable association between runs of homozygosity and schizophrenia in a well-powered replication study. PLoS Genet. 2016;12:e1006343. doi: 10.1371/journal.pgen.1006343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howrigan D.P., Simonson M.A., Davies G., Harris S.E., Tenesa A., Starr J.M., Liewald D.C., Deary I.J., McRae A., Wright M.J. Genome-wide autozygosity is associated with lower general cognitive ability. Mol. Psychiatry. 2016;21:837–843. doi: 10.1038/mp.2015.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Power R.A., Nagoshi C., DeFries J.C., Plomin R., Wellcome Trust Case Control Consortium 2 Genome-wide estimates of inbreeding in unrelated individuals and their association with cognitive ability. Eur. J. Hum. Genet. 2014;22:386–390. doi: 10.1038/ejhg.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White S.J., Laros J.F.J., Bakker E., Cambon-Thomsen A., Eden M., Leonard S., Lochmüller H., Matthijs G., Mattocks C., Patton S. Critical points for an accurate human genome analysis. Hum. Mutat. 2017;38:912–921. doi: 10.1002/humu.23238. [DOI] [PubMed] [Google Scholar]
- 38.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng P.C., Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szpiech Z.A., Blant A., Pemberton T.J. GARLIC: Genomic Autozygosity Regions Likelihood-based Inference and Classification. Bioinformatics. 2017;33:2059–2062. doi: 10.1093/bioinformatics/btx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahmood K., Jung C.H., Philip G., Georgeson P., Chung J., Pope B.J., Park D.J. Variant effect prediction tools assessed using independent, functional assay-based datasets: implications for discovery and diagnostics. Hum. Genomics. 2017;11:10. doi: 10.1186/s40246-017-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., Maciejewski A., Arndt D., Wilson M., Neveu V. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blekhman R., Man O., Herrmann L., Boyko A.R., Indap A., Kosiol C., Bustamante C.D., Teshima K.M., Przeworski M. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berg J.S., Adams M., Nassar N., Bizon C., Lee K., Schmitt C.P., Wilhelmsen K.C., Evans J.P. An informatics approach to analyzing the incidentalome. Genet. Med. 2013;15:36–44. doi: 10.1038/gim.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen Z., Duan L., Yang H., Yuan L., Huang Y., Li L., Xu B. Genetic variation of 17 STR loci in Dai population in mainland China. Forensic Sci. Int. Genet. 2015;19:37–38. doi: 10.1016/j.fsigen.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 51.Sun H., Zhou C., Huang X., Lin K., Shi L., Yu L., Liu S., Chu J., Yang Z. Autosomal STRs provide genetic evidence for the hypothesis that Tai people originate from southern China. PLoS ONE. 2013;8:e60822. doi: 10.1371/journal.pone.0060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y.C., Huang W., Tian J.Y., Chen X.Q., Kong Q.P. Exploring the maternal history of the Tai people. J. Hum. Genet. 2016;61:721–729. doi: 10.1038/jhg.2016.36. [DOI] [PubMed] [Google Scholar]
- 53.Wu L. Investigation of consanguineous marriages among 30 Chinese ethnic groups. Hered. Dis. 1987;4:163–166. [Google Scholar]
- 54.Thomas A., Skolnick M.H., Lewis C.M. Genomic mismatch scanning in pedigrees. IMA J. Math. Appl. Med. Biol. 1994;11:1–16. doi: 10.1093/imammb/11.1.1. [DOI] [PubMed] [Google Scholar]
- 55.Thomas A., Camp N.J., Farnham J.M., Allen-Brady K., Cannon-Albright L.A. Shared genomic segment analysis. Mapping disease predisposition genes in extended pedigrees using SNP genotype assays. Ann. Hum. Genet. 2008;72:279–287. doi: 10.1111/j.1469-1809.2007.00406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lohmueller K.E. The impact of population demography and selection on the genetic architecture of complex traits. PLoS Genet. 2014;10:e1004379. doi: 10.1371/journal.pgen.1004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lohmueller K.E. The distribution of deleterious genetic variation in human populations. Curr. Opin. Genet. Dev. 2014;29:139–146. doi: 10.1016/j.gde.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 58.Pedersen C.T., Lohmueller K.E., Grarup N., Bjerregaard P., Hansen T., Siegismund H.R., Moltke I., Albrechtsen A. The effect of an extreme and prolonged population bottleneck on patterns of deleterious variation: insights from the Greenlandic Inuit. Genetics. 2017;205:787–801. doi: 10.1534/genetics.116.193821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bittles A.H., Black M.L. Evolution in health and medicine Sackler colloquium: Consanguinity, human evolution, and complex diseases. Proc. Natl. Acad. Sci. USA. 2010;107(Suppl 1):1779–1786. doi: 10.1073/pnas.0906079106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wen B., Li H., Lu D., Song X., Zhang F., He Y., Li F., Gao Y., Mao X., Zhang L. Genetic evidence supports demic diffusion of Han culture. Nature. 2004;431:302–305. doi: 10.1038/nature02878. [DOI] [PubMed] [Google Scholar]
- 61.Du R.F. [Percentages and types of consanguineous marriage in different nationalities of China (author’s transl)] Zhonghua Yi Xue Za Zhi. 1981;61:723–728. [PubMed] [Google Scholar]
- 62.Zhang J.X. [Effects of consanguineous marriages on hereditary diseases: a study of the Han ethnic group in different geographic districts of Zhejiang Province] Zhonghua Yi Xue Za Zhi. 1992;72:674–676, 703. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.