Abstract
Biogenesis of the mitochondrial oxidative phosphorylation system, which produces the bulk of ATP for almost all eukaryotic cells, depends on the translation of 13 mtDNA-encoded polypeptides by mitochondria-specific ribosomes in the mitochondrial matrix. These mitoribosomes are dual-origin ribonucleoprotein complexes, which contain mtDNA-encoded rRNAs and tRNAs and ∼80 nucleus-encoded proteins. An increasing number of gene mutations that impair mitoribosomal function and result in multiple OXPHOS deficiencies are being linked to human mitochondrial diseases. Using exome sequencing in two unrelated subjects presenting with sensorineural hearing impairment, mild developmental delay, hypoglycemia, and a combined OXPHOS deficiency, we identified mutations in the gene encoding the mitochondrial ribosomal protein S2, which has not previously been implicated in disease. Characterization of subjects’ fibroblasts revealed a decrease in the steady-state amounts of mutant MRPS2, and this decrease was shown by complexome profiling to prevent the assembly of the small mitoribosomal subunit. In turn, mitochondrial translation was inhibited, resulting in a combined OXPHOS deficiency detectable in subjects’ muscle and liver biopsies as well as in cultured skin fibroblasts. Reintroduction of wild-type MRPS2 restored mitochondrial translation and OXPHOS assembly. The combination of lactic acidemia, hypoglycemia, and sensorineural hearing loss, especially in the presence of a combined OXPHOS deficiency, should raise suspicion for a ribosomal-subunit-related mitochondrial defect, and clinical recognition could allow for a targeted diagnostic approach. The identification of MRPS2 as an additional gene related to mitochondrial disease further expands the genetic and phenotypic spectra of OXPHOS deficiencies caused by impaired mitochondrial translation.
Keywords: mitochondrial ribosomes, mitochondrial translation defect, combined OXPHOS complex deficiencies, hearing loss, wrinkly skin, 2-oxoglutaric acid, complexome profiling
Main Text
Mitochondria are essential organelles that harbor the oxidative phosphorylation system (OXPHOS), an ATP-producing system encompassing five multi-subunit enzymatic complexes whose biogenesis is strictly dependent on the coordinated expression of genes encoded by nuclear and mitochondrial DNA (mtDNA). mtDNA encodes 13 subunits that are essential structural components of OXPHOS complexes I, III, IV, and V, in addition to encoding two rRNAs (16S and 12S) and 22 tRNAs required for the translation of the subunits.1 Mitochondrial translation is executed by a dedicated translation machinery that includes many regulatory factors and a mitochondrial-specific ribosome composed of two subunits: the 28S (mt-SSU) and 39S (mt-LSU) ribosomal subunits. These subunits are large ribonucleoprotein complexes containing around 80 nuclear-encoded structural mitoribosomal proteins (MRPs) and mt-DNA-encoded RNAs: 12S rRNA in the mt-SSU and 16S rRNA, in addition to either mt-tRNAVal or mt-tRNAPhe, in the mt-LSU subunit.2, 3, 4, 5 The three-dimensional structures of the mammalian3, 6, 7 and human2 mitoribosomes have only been elucidated recently.
Considering the many factors involved in this process, it is not surprising that an increasing number of gene mutations cause defective mitochondrial translation and are linked to mitochondrial disease.1, 3 Mitochondrial translation defects usually result in a combined OXPHOS complex deficiency, leading to disorders with severe multisystem involvement and often an early lethal outcome. To date, mutations in eight mitoribosomal protein-encoding genes—MRPS7 (MIM: 611974), MRPS16 (MIM: 609204), MRPS22 (MIM: 605810), MRPS23 (MIM: 611985), and MRPS34 (MIM: 611994) from the mt-SSU and MRPL3 (MIM: 607118), MRPL12 (MIM: 602375), and MRPL44 (MIM: 611849) from the mt-LSU—have been linked to mitochondrial disease in a total of 22 subjects (summarized in Table 1).
Table 1.
Overview of the Clinical Features of Subjects Carrying Mutations in Mitoribosomal Subunits
| Reference |
MRPS2 |
MRPS2 |
MRPS7 |
MRPS16 |
MRPS22 |
MRPS22 |
MRPS22 |
MRPS23 |
MPRS34 |
MRPL3 |
MRPL12 |
MRPL44 |
MRPL44 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| this article (S1) | this article (S2) | Menezes et al.16 | Miller et al.11 | Saada et al.13 | Smits et al.9 | Baertling et al.19 | Kohda et al.14 | Lake et al.12 | Galmiche et al.8 | Serre et al.33 | Distelmaier et al.10 | Distelmaier et al.10 | |
| Number of subjects | 1 | 1 | 2 siblings | 1 | 3 siblings | 1 | 1 | 1 | 6 | 4 siblings | 1 | 1 | 1 |
| Dysmorphic features | + | − | − | + | NR | + | NR | NR | + | − | + | − | − |
| Cardiac involvement | − | − | − | + | + | + | + | NR | − | + | NR | + | + |
| Hypotonia | + | + | NR | + | + | + | NR | NR | + | NR | + | + | NR |
| Skin involvement | + | − | − | redundant skin of the neck | − | redundant skin of the neck | − | NR | − | − | NR | − | − |
| Structural brain abnormalities | − | − | NR | + | NR | + | + | NR | + | + | + | NR | NR |
| Hearing impairment | + | + | ++ | NR | NR | − | NR | NR | − | NR | NR | NR | NR |
| Developmental delay | +/− | + | − | NR | NR | ++ | NR | NR | + | ++ | + | − | +/− |
| Growth delay | FTT | − | FTT | SGA | NR | NR | + | NR | + | FTT | SGA, FTT | NR | NR |
| Increased lactate levels | ++ | + | + | + | ++ | + | ++ | NR | + | + | + | + | + |
| Combined OXPHOS deficiency | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Hypoglycemia | + | + | NR | NR | NR | NR | NR | + | NR | NR | NR | NR | NR |
| Age of presentation | infantile | infantile | infantile | neonatal | neonatal | neonatal | neonatal | infantile | neonatal or infantile | infantile | neonatal | infantile | neonatal |
| Age of death (or age at last follow up) | alive at 11 years | alive at 11 years | 14 years; alive at 17 years | 3 days | 2–22 days | alive at 5 years | 3 days | alive at 1 year and 6 months | range: dead at 8.5 months to alive at 17 years | two died at 15 and 17 months; two are alive at 3 years | 2 years | alive at 8 years | alive at 26 years |
Abbreviations are as follows: −, not present; +/−, mildly affected; +, present; ++, severely affected; NR, not reported; FTT, failure to thrive; SGA, small for gestational age.
Most subjects presented in the neonatal period, and about one-third of the subjects died before the age of 1 year. However, besides the common feature of severe lactic acidosis, these disorders show a large variety in clinical presentation. Some have specific clinical features such as early-onset cardiomyopathy,8, 9, 10 developmental abnormalities,10, 11, 12 corpus callosum agenesis,13 hypoglycemia,14 Leigh syndrome,12 or short stature and dysmorphic features.9, 12, 13 Wrinkled or redundant skin has been observed in mitochondrial translation defects caused by mutations in MRPS229, 13 and MRPS16.11
Here, we report two unrelated subjects presenting with sensorineural hearing impairment, developmental delay, hypoglycemia, lactic acidemia, and a combined OXPHOS deficiency. Using exome sequencing, we identified bi-allelic mutations in MRPS2 (MIM: 611971), which encodes the mitochondrial ribosomal protein S2 and has not been implicated in disease until now.
Our study adhered to the Declaration of Helsinki and was approved by the institutional review boards at each research site. Written informed consent was obtained from the parents of the subjects.
Subject 1, a girl, was born at term after an uneventful pregnancy as the third child of non-consanguineous healthy parents of Austrian origin (Figure 1). Birth parameters were within normal limits. She had minor dysmorphic features, including low-set ears and slightly up-slanting palpebral fissures. Skin wrinkling, most pronounced on the abdomen and hands, was apparent from birth. During the first year of life she developed a failure to thrive. She had a psychomotor developmental delay in which the motor delay was most pronounced. She had an intermittent divergent strabismus of the left eye. At the age of 3 years, she had developed progressive sensorineural hearing loss, which required the use of a hearing aid and was corrected with bilateral cochlea implants. After this correction, both her speech and development improved markedly. Her last formal developmental assessment at the age of 5 years and 10 months showed an average developmental state of 2 years and 6 months of age. A cerebral MRI at the age of 3 years did not reveal any structural anomalies. She was prone to developing hypoglycemia. Biochemical evaluation revealed elevated liver enzymes (3- to 4-fold elevation in aspartate-amino transferase and alanine-amino transferase, repetitive elevated lactate levels (>8 mmol/L; reference values < 2 mmol/L), elevated serum alanine (up to 850 μmol/L; reference values = 99–350 μmol/L), and increased excretion of Krebs cycle intermediates (2-oxoglutaric acid between 50–220 μmol/mmol creatinine; reference values = 0–50 μmol/mmol creatinine and trace elevation of succinic acid). Glycosylation screening (transferrin and apolipoprotein-CIII isoelectric focusing) was unremarkable. Normal serum creatine kinase (CK) concentrations were found. Measurements of OXPHOS complex activities in liver, muscle, and fibroblasts showed a decrease in multiple enzyme complexes (Table 2).
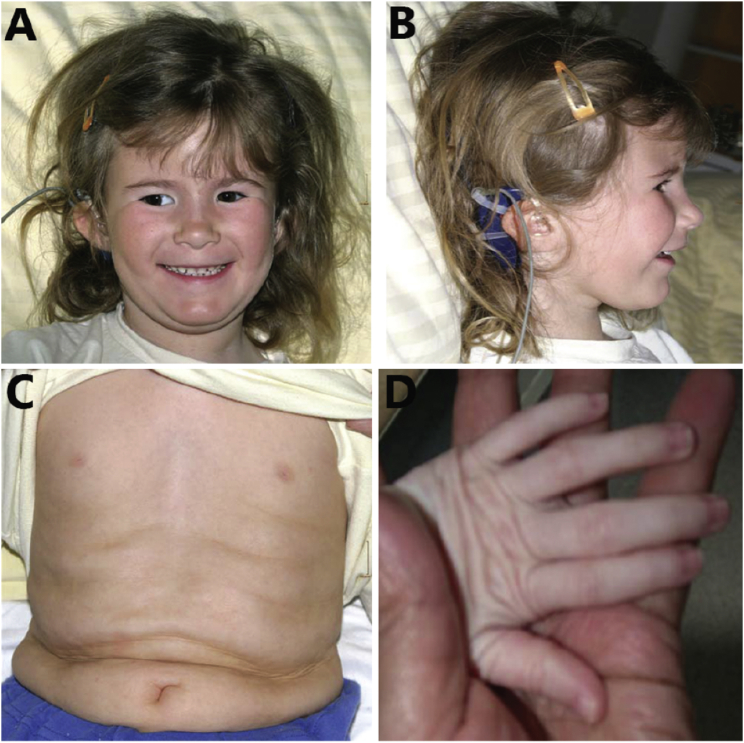
Figure 1.
Clinical Characteristics of Subject 1 at the Age of 5 Years
Clinical features include (A) slightly up-slanting palpebral fissures and left-eye strabismus, (B) low-set ears, and redundant skin on the (C) abdomen and (D) hands.
Table 2.
OXPHOS Complex Enzyme Activities in Muscle, Liver, and Fibroblasts
|
Complex Activity in Muscle |
Complex Activity in Liver |
Complex Activity in Fibroblasts |
||||
|---|---|---|---|---|---|---|
| Subject 1 | Subject 2 | Subject 1 | Subject 2 | Subject 1 | Subject 2 | |
| Complex I | 0. 14 (0.14–0.35) | 0.20 (0.13–0.29) | 0.02 ↓ (0.24; 0.59) | 0.02 ↓ (0.35–0.50) | 0.04 (0.04–0.12) | 0.16 (0.13–0.26) |
| Complex II | 0.22 ↓ (0.23–0.41) | 0.19 ↑(0.09–0.15) | 1.43 (0.85; 1.80) | 0.9 ↓ (1.70–2.50) | 0.23 (0.18–0.43) | 0.36 ↑(0.25–0.34) |
| Complex III | 0.81 ↓ (1.45–3.76) | NP | 0.34 ↓ (2.18; 3.18) | 0.23 ↓ (1.00–1.40) | 0.96 (0.72–2.23) | 2.16 ↓ (2.23–2.98) |
| Complex IV | 0.51 ↓ (0.82–2.04) | 0.42 ↓ (0.45–0.75) | 0.09 ↓ (1.44; 1.66) | 0.12 ↓ (1.00–1.40) | 0.24 ↓ (0.90–1.79) | 0.99 ↓ (1.14–1.54) |
| Complex V | 0.67 (0.42–1.26) | NP | 0.43 (0.31; 1.21) | NP | 0.39 (0.39–0.79) | 0.48 (0.23–0.33) |
Laboratory reference values were available for muscle, fibroblasts, and liver and are shown in parentheses. Complex activities in liver for subject 1 are compared with those in two control samples that were used in the same experiment. These values are reported in parentheses. The following abbreviation is used: NP, not performed.
Subject 2, a boy, presented with fasting hypoglycemia at the age of 6 years. He was the first child of healthy unrelated parents of Tunisian origin and was born at term after an uneventful pregnancy. Birth parameters were within normal limits. Medical history was marked by several acute episodes of hypoglycemia after an overnight or a 12-hr fasting since the age of 18 months, especially when these episodes coincided with illness and poor oral intake, which associated with lactic acidosis. He could walk at 22 months. Speech development was delayed as a result of severe sensorineural deafness at the age of 2 years, necessitating the use of a hearing aid. Speech development improved after the hearing loss was corrected. At the age of 11 years he had a normal physical appearance, with normal growth parameters, but a moderate intellectual disability, frequent headache episodes, and muscular weakness of the lower limbs. He suffered from exercise intolerance marked by myalgia after walking. His brain MRI was normal. Metabolic investigations showed repetitive mildly increased lactate levels in plasma (2.2–3.8 mmol/L; reference values < 2 mmol/L) and urine (292 μmol/mmol creatinine; reference values = 25–100 μmol/mmol creatinine). A clinical fasting test showed hypoglycemia with hyperlactatemia (5 mmol/L) and slightly increased excretion of 2-oxoglutarate (37 μmol/mmol creatinine; control range < 27 μmol/mmol creatinine) in urine. Measurements of OXPHOS complex activities showed a decrease in multiple complexes in liver and fibroblasts and a complex IV deficiency in muscle. This subject had two unaffected siblings. A third sibling died during pregnancy (Figure 2B). The cause of her death is unknown, so it is unclear whether it is related to mitochondrial disease.
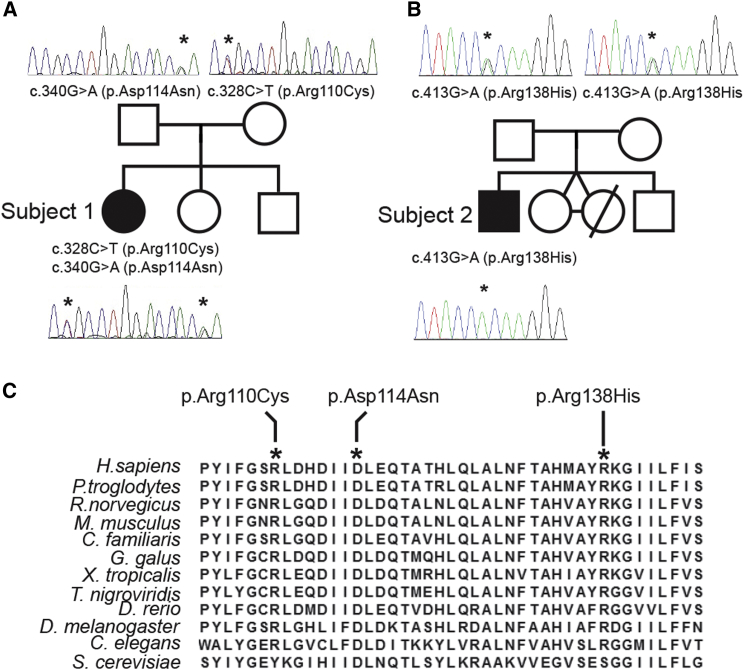
Figure 2.
MRPS2 Mutation Analysis and Evolutionary Conservation of the Affected Amino Acid Residues
(A and B) Pedigree and sequencing chromatograms for (A) S1 and (B) S2 and their families, depicting the segregation of the identified recessive mutations.
(C) Interspecies alignment of the MRPS2 region containing the amino acid residues altered (depicted in bold and italics) in S1 and S2.
Exome sequencing was performed to identify pathogenic variants underlying the disease in both subjects (for methods, see Supplemental Data). For subject 1, a series of filter steps, comparable to those described by Gilissen et al.,15 was applied to the variant data for the creation of a candidate gene list. Non-genic, intronic (except for splice sites), synonymous changes and common variants were excluded by comparison with dbSNPv132 and our in-house variant database (cutoff > 1%). Variants were further prioritized on the basis of PhyloP scores (cutoff > 3) as well as molecular pathways and segregation analysis. This resulted in the identification of a single candidate gene, mitochondrial ribosomal protein S2 (MRPS2; GenBank: NM_016034.3), carrying two heterozygous sequence variants, c.328C>T (p.Arg110Cys) and c.340G>A (p.Asp114Asn) (Figure 2A). Segregation analysis confirmed that both parents are heterozygous for one of these variants. One healthy sibling was homozygous for both wild-type alleles, whereas the second was heterozygous for the c.328C>T variant (data not shown).
For subject 2, the pathogenic variant was selected according to similar criteria: exclusion of known SNPs reported with a frequency > 0.1% in dbSNP, 1000 Genomes, Exome Variant Server, or our in-house database; exclusion of non-coding variants; and selection of variants that were predicted to be possibly damaging by PolyPhen and SIFT. This filtering identified a homozygous variation (c.413G>A [p.Arg138His]) in only one gene: MRPS2, which encodes a known mitochondrial protein. Segregation analysis confirmed that both parents are heterozygous for this variant (Figure 2B).
All identified variants affect highly conserved amino acids (Figure 2C) and were predicted to be pathogenic by at least two of the three in silico prediction programs that were used. These variants are reported in the GnomAD database only as heterozygous variants with very low minor allele frequencies (Table S1).
Given that MRPS2 is a mitochondrial ribosomal protein, we proceeded to characterize the effects of the identified variants on mitochondrial physiology by using fibroblasts obtained from skin biopsies of both subjects (S1 and S2). A previously characterized fibroblast cell line (designated S3) carrying disease-causing mutations in MRPS22 was used as a positive control for mitoribosomal dysfunction.9
SDS-PAGE analysis of mitochondrial extracts (for methods, see Supplemental Data) from these fibroblasts showed decreased steady-state amounts of the protein MRPS2 in both S1 and S2 fibroblasts (Figure 3A). Similarly, the mt-SSU proteins MRPS5, MRPS18B, and MRPS28 were less abundant in subject fibroblasts. Amounts of the mt-LSU proteins MRPL37 and MRPL44 remained unchanged. Stability of newly imported mitoribosomal proteins and nascent 12S rRNA depends on their coordinated assembly into ribonucleoprotein complexes. Loss of individual MRPS proteins has been previously shown to result in decreased 12S rRNA steady-state abundance.9, 11, 13, 16 Consistent with this, steady-state abundance of 12S rRNA, but not that of 16S rRNA, was specifically decreased in S1 and S2 fibroblasts; a similar decrease was seen in S3 fibroblasts (Figure 3B) on northern blot analysis (for methods, see Supplemental Data).
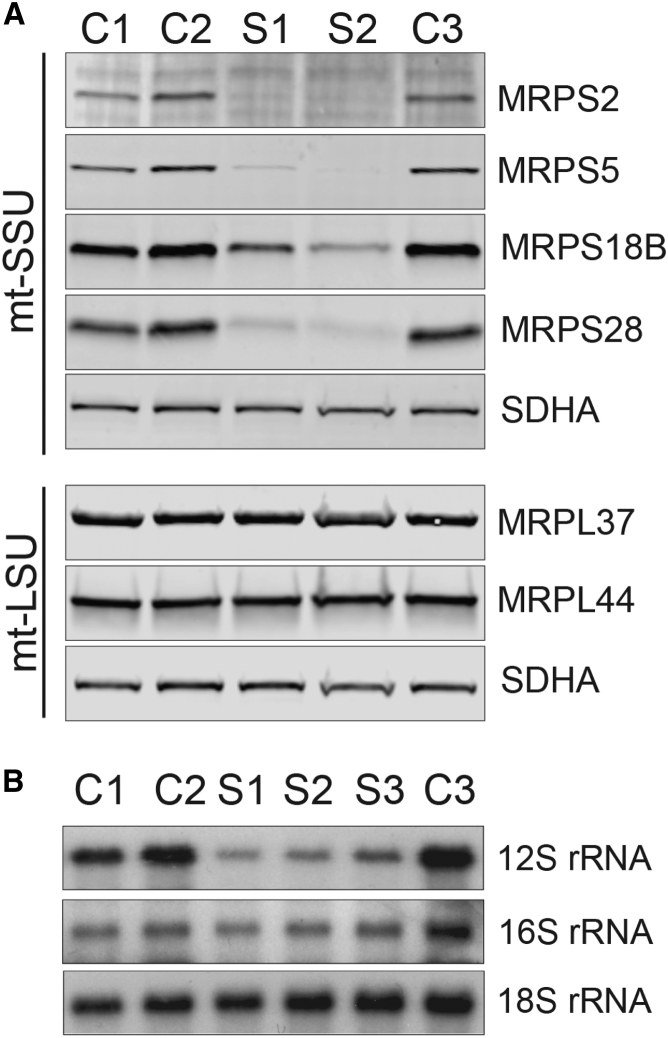
Figure 3.
Abundance of mtDNA-Encoded rRNAs and Mitoribosomal Subunits
(A) SDS-PAGE analysis of proteins from the small (MRPS2, MRPS5, MRPS18B, and MRPS28) and large (MRPL37 and MRPL44) mitoribosomal subunits in both subjects and control cells revealed decreased steady-state levels of MRPS2 and the other mt-SSU proteins. Steady-state levels of the mt-LSU proteins are unaffected. OXPHOS complex II protein SDHA was used as a loading control.
(B) Northern blot analysis of the steady-state levels of 12S and 16S rRNA in fibroblasts from subjects and controls demonstrates that 12S rRNA, but not 16S rRNA, is decreased in fibroblasts carrying mutations in MRPS2 (S1 and S2) or MRPS22 (S3). 18S rRNA from the cytosolic ribosome was used as a loading control.
To further characterize the detrimental effects of MRPS2 mutations on mitoribosomal biogenesis, we analyzed the assembly and abundance of mt-SSU and mt-LSU particles with complexome profiling of mitochondrial extracts from control, S1, and S3 fibroblasts (for methods, see Supplemental Data). Complexome profiling enables the investigation of the abundance and composition of macromolecular protein complexes.17 Fully assembled mt-SSU particles were hardly detectable in both S1 and S3 fibroblasts, whereas mt-LSU particles were assembled at a normal level (Figure 4). A previously reported, ∼300 kDa mt-SSU subassembly containing MRPS16, MRPS17, MRPS18B, MRPS22, MRPS26, MRPS27, and MRPS3418 was detectable in control fibroblasts and was also preserved, albeit at reduced levels, in S1 mitochondria, suggesting that MRPS2 is unlikely to function in the early assembly pathway. Cumulatively, these data indicate that the identified mutations in MRPS2 destabilize the protein and thereby impair mt-SSU assembly in subjects 1 and 2.
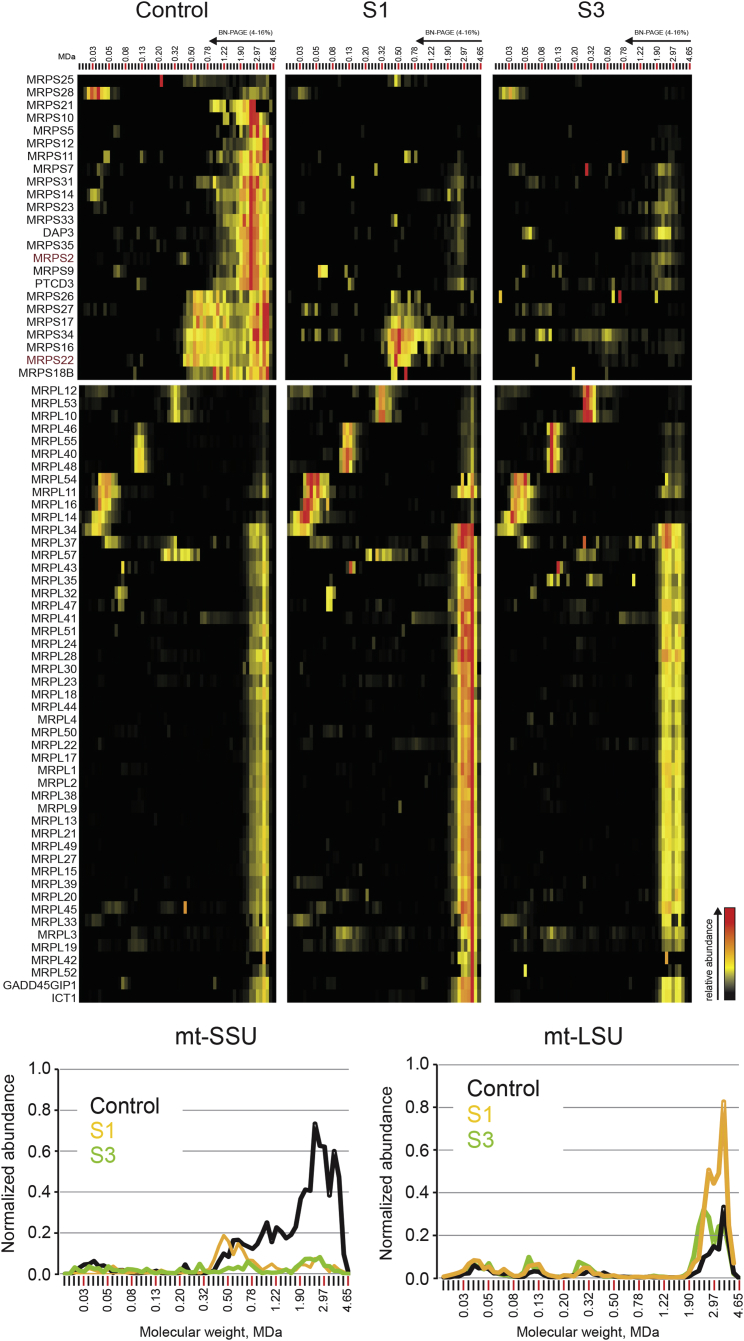
Figure 4.
Complexome Profiling
Heatmap representation (top) of the migration profiles of mt-SSU and mt-LSU in control, S1, and S3 fibroblasts. Interaction heatmaps were created by hierarchical clustering, including the manual addition of known ribosomal components that were not grouped together by the algorithm. Top-center and top-right panels show decreased amounts of fully assembled mt-SSU in both S1 and S3 fibroblasts and a subassembly of eight subunits present in control and S1 cells but absent in S3 cells. The abundance of mt-LSU is unaffected in S1 and S3. Migration profiles (bottom) of both mitoribosomal subunits show the relative abundance of proteins plotted against their apparent molecular mass and reveal decreased abundance of mt-SSU and unaffected abundance of mt-LSU in both subjects. The relative abundance was calculated as the average of the normalized iBAQ values for all mt-SSU or mt-LSU proteins.
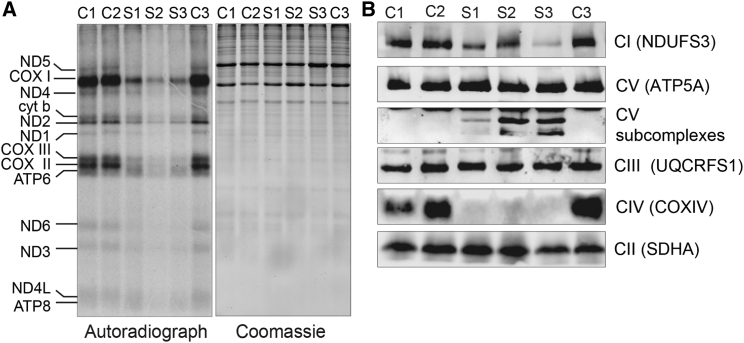
Decreased mt-SSU assembly and the resulting lack of functional mitoribosomes invariably lead to an inhibition of mitochondrial translation and multiple OXPHOS deficiencies. Indeed, in vitro pulse labeling of mitochondrial translation products with radiolabeled methionine and cysteine revealed a profound and generalized translation defect of mtDNA-encoded polypeptides in both S1 and S2 fibroblasts, as well as in the MRPS22-deficient fibroblasts (Figure 5A; for methods, see Supplemental Data). In line with this impaired translation, BN-PAGE analysis of OXPHOS complex assembly revealed decreased amounts of fully assembled OXPHOS complexes I and IV but not of complex III and the exclusively nucleus-encoded complex II (Figure 5B; for methods, see Supplemental Data). Although no differences in abundance of fully assembled complex V were seen in S1 and S2 fibroblasts, BN-PAGE analysis revealed the presence of subcomplexes of complex V, which were absent in the controls (Figure 5B). Interaction profiles of the different OXPHOS complexes extracted from the complexome profiling data for S1 consistently confirmed a decrease in the abundance of fully assembled complex I and IV. Furthermore, complexome profiling showed an accumulation of complex I and complex V subassemblies at lower molecular masses in the S1 fibroblasts than in the control cells (Figure S1).
Figure 5.
Pulse Labeling of Mitochondrial Translation Products and OXPHOS Assembly Analysis
(A) Pulse labeling of mitochondrial translation products in control (C1–C3), S1, S2, and S3 fibroblasts shows decreased mitochondrial protein synthesis in all subjects’ cell lines. Coomassie staining of the gels was used for the assessment of loading.
(B) BN-PAGE analysis of OXPHOS assembly in control, S1, S2, and S3 fibroblasts shows decreased amounts of fully assembled OXPHOS complexes I and IV in the fibroblasts from the MRPS2- and MRPS22-deficient subjects. Subcomplexes of complex V accumulate in subjects’ fibroblasts and are absent in the control cells.
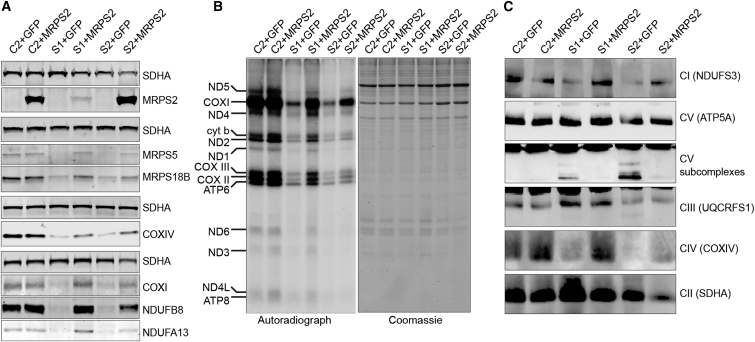
To demonstrate the disease-causing nature of MRPS2 mutations, we carried out functional complementation experiments by generating control, S1, and S2 cell lines stably expressing either green fluorescent protein (GFP) as a negative control or wild-type MRPS2 (for methods, see Supplemental Data). SDS-PAGE analysis and immunoblotting of whole-cell extracts from these cell lines revealed that despite the markedly different abundance of MRPS2 (Figure 6A), the amounts of steady-state MRPS5 and MRPS18B were higher in S1 and S2 cells complemented with MRPS2 than in the same cells expressing GFP. Likewise, the OXPHOS proteins NDUFB8 and NDUFA13 from complex I as well as COXI and COXIV from complex IV accumulated in MRPS2-complemented fibroblasts, suggesting restoration of OXPHOS biogenesis. Consistent with these observations, mitochondrial translation was partially restored in both subject cell lines complemented with wild-type MRPS2 (Figure 6B), as was the assembly of OXPHOS complexes I and IV (Figure 6C). We also noted fewer complex V subassemblies in MRPS2-expressing subjects’ fibroblasts than in the negative control cells. Overexpression of MRPS2 did not exhibit any negative effect on mitochondrial translation or OXPHOS assembly, as evidenced by the fact that these processes were unaffected in control cells overexpressing MRPS2. Cumulatively, our complementation experiments confirm the pathogenicity of MRPS2 mutations.
Figure 6.
Amounts of Wild-Type MRPS2 in S1 and S2 Fibroblasts
(A) Immunoblot analysis of whole-cell extracts from control (C), S1, and S2 cell lines transduced with a lentivirus expressing either wild-type MRPS2 (+MRPS2) or green fluorescent protein (+GFP) as a negative control. The presence of wild-type MRPS2 leads to increased amounts of MRPS5 and MRPS18B, as well as NDUFB8 and NDUFA13 from complex I and COXI and COXIV from complex IV. MRPS2 was detected with a specific antiserum that does not detect the endogenous MRPS2 in whole-cell extracts. SDHA was used as a loading control for each separate gel.
(B) Pulse labeling of mitochondrial translation products demonstrates that the presence of wild-type MRPS2 partially restores mitochondrial translation in S1 and S2 fibroblasts. The gel was stained with Coomassie colloidal dye for the assessment of loading.
(C) BN-PAGE analysis of OXPHOS assembly in C, S1, and S2 fibroblasts transfected with either GFP- or wild-type-MRPS2-expressing lentivirus shows partial restoration of OXPHOS complex assembly in subject cells expressing MRPS2.
The cases presented here extend the genotypic and phenotypic spectra of mitochondrial translation deficiencies. Although all reported MRP variations invariably lead to combined OXPHOS deficiencies, their clinical presentations and prognoses are in some cases markedly different (Table 1). Defects in three proteins of mt-SSU, MRPS16,11 MRPS22,9, 13, 19 and MRPS34,12 are reported to result in a phenotype with early lethality: four of the six subjects died in the first month of life and had a severe lactate acidemia, cardiomyopathy (except for one MRPS22-deficient subject9), and multi-organ failure.
Two (out of six) of the subjects with mutations in MRPS34 did not have cardiac involvement but died in infancy from respiratory failure. The four siblings with MRPL3 mutations also have a cardiomyopathy, but this defect seems to have a better prognosis: two siblings have survived into the second year of life, and the other two are alive at the age of 3.8 In one of the subjects with a MRPL44 defect the cardiomyopathy was diagnosed at the age of 3 years. At 8 years old, she has a moderate non-obstructive hypertrophic cardiomyopathy.10
The subjects presented in this paper are at the less severe end of the clinical spectrum, and at the age of 11 years, neither shows progressive disease, despite the profound decrease in mitoribosomal assembly and OXPHOS enzyme activity detected in fibroblast cultures from these subjects. These divergent clinical presentations are inconsistent with the ubiquitous expression and seemingly equivalent roles of the mutated proteins in mitoribosomal function.
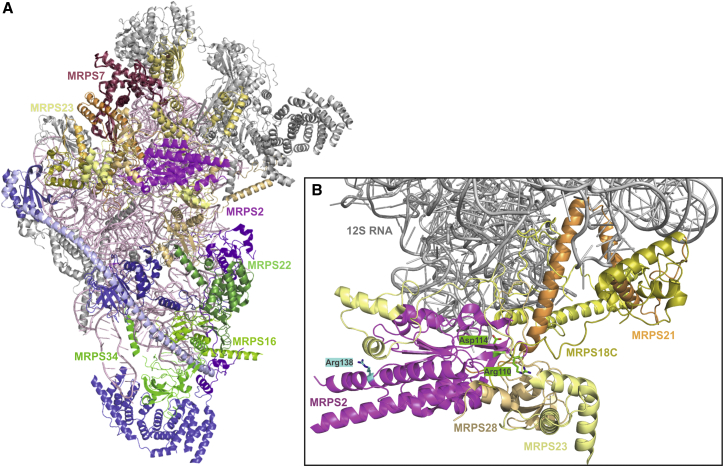
The explanation for the clinical variability in subjects with mutations in a mitochondrial ribosomal protein is likely to reside at the intersection between the different effect of each mutation on the stability or activity of the affected protein and the relative role of the different subunits in the mitoribosomal structure and assembly process. Examination of the structure of the human mt-SSU6, 7 revealed that, indeed, the three proteins for which variants lead to early lethality and that are components of the stable ∼300 kDa subassembly (MRPS16, MRPS22, and MRPS34) are in close contact with each other and form a common subdomain. This subdomain is clearly separated from the other disease-implicated MRPS proteins (Figure 7A). Part of the explanation of the diverse clinical phenotypes seen in subjects with MRPS or MRPL mutations could also lie in the effects different mutations have on the stability, activity, or interactions of the mutant proteins. For example, the mutations identified in the MRPS1611-deficient subjects are nonsense mutations, resulting in the loss of MRPS16’s mitochondrial-specific elongation extension that connects the protein to MRPS26 and MRPS307 and thereby disturbing its putative assembly function.6, 7 To date, almost all identified MRPS or MRPL mutations have been found to lead to moderate or severe depletion of the affected proteins. Because most mutations in mitoribosomal subunits are not complete loss-of-function alleles, they will have different effects on protein folding, stability, or activity, which might result in different residual levels of (semi)functional mitoribosomes in different tissues. Because of the different thresholds at which individual tissues are affected by diminished energy supply, this will lead to differences in tissue involvement between different mutations in mitochondrial ribosomal protein-encoding genes.
Figure 7.
Structural Model of the Human mt-SSU Shows the Positions of Affected Proteins and Residues
(A) The human 28S mt-SSU (PDB: 3J9M7) was drawn in cartoon representation with PyMol version 1.7.4.0. The subunits known to be implicated in disease are shown with color-matched labels: magenta, MRPS2; raspberry, MRPS7; pale yellow, MRPS23; and shades of green, MPRS16, MRPS22, and MRPS34. Subunits interacting with MRPS2 are shown in shades of yellow, and constituents of the ∼300 kDa subassembly are shown in shades of blue and green. Other subunits are shown in gray, and 12S RNA is shown in light pink.
(B) Partial model of (A) shows only the 12S RNA (gray), MRPS2 (magenta), and surrounding proteins MRPS18C, MRPS21, and MRPS28 (shades of yellow). The positions of the residues of MRPS2 altered in S1 (Arg110 and Asp114) and in S3 (Arg138) are shown in green and cyan, respectively, as stick models.
Another factor that might contribute to the differences in clinical presentation of subjects with mutations in MRPL or MRPS might be the hypothesis that destabilizing variants in subunits that join the mt-SSU or mt-LSU at an early assembly stage (as opposed to mutations in proteins that assemble later during mitoribosomal biogenesis) might lead to a more profound metabolic disturbance. Despite recent advances in elucidating the structure of the mammalian mitoribosome,3 the order of steps in the assembly process of the mitochondrial ribosome remains elusive.
MRPS2 is an evolutionary conserved protein, which suggests that it plays an important but currently unclear role in mitoribosomal biogenesis or function. The bacterial ortholog of MRPS2, bS2, is a dual-function protein. It is important for the translation of mRNAs that contain a Shine-Dalgarno sequence, a purine-rich sequence upstream of the start codon that plays an important role in the binding of the mRNA to the ribosomal RNA. BS2 recruits the bS1 ribosomal protein to bacterial SSU and stabilizes the mRNA’s Shine-Dalgarno sequence during translation initiation.20, 21 In addition, bS2 plays a role in the autogenous control of bacterial ribosomal assembly as a regulator of the S2-EF-Ts operon (rpsB-tsf).22
BS2 is among the last proteins to assemble in the SSU particle, and because it is only loosely associated with the particle, it can be easily exchanged between subunits.23 Mammalian MRPS2 shares a central region of orthology with bS2, but in addition it has evolved mitochondria-specific N- and C-terminal extensions, as have many other mitoribosomal proteins that are conserved in bacteria.
MRPS2 spans all three domains of the mt-SSU6, 7 and interacts with seven other mt-SSU proteins: MRPS5, MRPS9, MRPS23, MRPS28 (bS1m), MRPS18C, MRPS21, and MRPS372 (Figure 7A). This extended range of interactors suggests that MRPS2 has adopted a primarily structural role, which is consistent with our findings that its loss is associated with impaired mt-SSU assembly.
Although MRPS2 interacts with MRPS5 and MRPS28, which are involved in mRNA recruitment and exit from the mitoribosome, respectively, the role of MRPS2 in translation has most likely been lost in evolution. Mitochondrial mRNAs do not have Shine-Dalgarno sequences. Bacterial bS2 is dispensable for the translation of mRNAs without such a sequence, and this could be the case for MRPS2 in mitochondria as well. In summary, our data support a structural role for MRPS2 in the connection of different domains of the mitochondrial ribosome.
We propose that MRPS2, like bS2 and consistent with our complexome profiling data (Figure 4) and the structure of mtSSU (Figure 7A), most likely assembles late in mitoribosomal biogenesis and that this assembly is probably independent of the set of core proteins, which include MRPS22 and MRPS16, forming the ∼300 kDa subassembly. Notably, after depletion of these subunits in subjects with mutations in the encoding genes, large amounts of MRPS2 and mt-LSU were still detectable.24 This late assembly might explain the less severe phenotype in our subjects.
A more detailed inspection of the position of the affected residues within MRPS2 (Figure 7B) showed that the two residues that were altered in S1 are found in a region that faces the 12S RNA and tightly interacts with several other proteins. In contrast, the arginine exchanged in S2 is located near the surface of mt-SSU, suggesting more indirect effects on the stability of the complex. This difference might also explain why the amount of 12S RNA found in S1 was slightly lower than in S2 (Figure 3B).
A distinctive clinical feature in subject 1 was wrinkly skin. To date, dermatological symptoms in subjects with mutations in MRP genes have been limited to redundant skin of the neck, which was previously described in single subjects carrying either MRPS22 or MRPS16 mutations.9, 11, 13 However, disturbed mitochondrial metabolism has been previously linked to wrinkled skin in association with progeria25, 26, 27 and in subjects carrying PYCR1 (MIM: 179035) and ALDH18A1 (also known as P5CS) (involved in de novo proline synthesis) mutations leading to multisystemic disorders with cutis laxa, a typical facial gestalt, hypotonia, and intellectual disability.25, 26, 27, 28 Of note, although metabolic markers (blood lactate and alanine elevation) of mitochondrial disease have been found in one subject with PYCR1 deficiency,25 OXPHOS activities and lactate levels are typically normal in these subjects, which sets them apart from those carrying MRP alterations. Moreover, hearing impairment is very rare in cutis laxa or wrinkly skin syndromes; it has only been described as an incidental finding in studies of P5CS deficiency, which makes this wrinkly-skin syndrome recognizable.29, 30, 31, 32
Together, our data demonstrate that mutations in MRPS2, which encodes a mitoribosomal protein, cause a recognizable phenotype involving sensorineural deafness, hypoglycemia, lactic acidemia, 2-oxoglutaric aciduria, developmental delay, and multiple-OXPHOS-complex dysfunction. Especially in the presence of wrinkled skin, these are discriminative features that, when encountered, should raise suspicion for a MRP-related mitochondrial defect. The clinical recognition of these features should allow a targeted diagnostic approach to MRP deficiencies.
Acknowledgments
We thank the subjects and their parents for their participation. This study was financially supported by the Dutch Metakids foundation (2012, M.M.), the Netherlands Organisation for Scientific Research (2011, NWO project 017.008.052, M.M.), the E-Rare project GENOMIT (01GM1207, A.R.), the National Institutes of Health (HL090648, Z.U.), and the Association Français contre les Myopathies (19876, M.D.M). B.R., A.R., and M.D.M would like to acknowledge the technical assistance with cell culture from Ms. Coralie Zangarelli. Exome sequencing of subject 2 was done in collaboration with the Centre National de Génotypage.
Published: March 22, 2018
Footnotes
Supplemental Data include Supplemental Material and Methods, one figure, and one table and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.02.012.
Contributor Information
Metodi D. Metodiev, Email: metodi.metodiev@inserm.fr.
Eva Morava, Email: emoravakozicz@tulane.edu.
Web Resources
gnomAD Browser, http://gnomad.broadinstitute.org
MITOMAP, http://www.mitomap.org/mitomap
OMIM, http://www.omim.org/
PDB, http://www.rcsb.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org
Supplemental Data
References
- 1.Pearce S., Nezich C.L., Spinazzola A. Mitochondrial diseases: translation matters. Mol. Cell. Neurosci. 2013;55:1–12. doi: 10.1016/j.mcn.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 2.Amunts A., Brown A., Toots J., Scheres S.H.W., Ramakrishnan V. Ribosome. The structure of the human mitochondrial ribosome. Science. 2015;348:95–98. doi: 10.1126/science.aaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greber B.J., Boehringer D., Leibundgut M., Bieri P., Leitner A., Schmitz N., Aebersold R., Ban N. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;515:283–286. doi: 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- 4.Greber B.J., Boehringer D., Leitner A., Bieri P., Voigts-Hoffmann F., Erzberger J.P., Leibundgut M., Aebersold R., Ban N. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;505:515–519. doi: 10.1038/nature12890. [DOI] [PubMed] [Google Scholar]
- 5.Rorbach J., Gao F., Powell C.A., D’Souza A., Lightowlers R.N., Minczuk M., Chrzanowska-Lightowlers Z.M. Human mitochondrial ribosomes can switch their structural RNA composition. Proc. Natl. Acad. Sci. USA. 2016;113:12198–12201. doi: 10.1073/pnas.1609338113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaushal P.S., Sharma M.R., Booth T.M., Haque E.M., Tung C.S., Sanbonmatsu K.Y., Spremulli L.L., Agrawal R.K. Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome. Proc. Natl. Acad. Sci. USA. 2014;111:7284–7289. doi: 10.1073/pnas.1401657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaushal P.S., Sharma M.R., Booth T.M., Haque E.M., Tung C.S., Sanbonmatsu K.Y., Spremulli L.L., Agrawal R.K. Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome. Proc. Natl. Acad. Sci. USA. 2014;111:7284–7289. doi: 10.1073/pnas.1401657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galmiche L., Serre V., Beinat M., Assouline Z., Lebre A.S., Chretien D., Nietschke P., Benes V., Boddaert N., Sidi D. Exome sequencing identifies MRPL3 mutation in mitochondrial cardiomyopathy. Hum. Mutat. 2011;32:1225–1231. doi: 10.1002/humu.21562. [DOI] [PubMed] [Google Scholar]
- 9.Smits P., Saada A., Wortmann S.B., Heister A.J., Brink M., Pfundt R., Miller C., Haas D., Hantschmann R., Rodenburg R.J. Mutation in mitochondrial ribosomal protein MRPS22 leads to Cornelia de Lange-like phenotype, brain abnormalities and hypertrophic cardiomyopathy. Eur. J. Hum. Genet. 2011;19:394–399. doi: 10.1038/ejhg.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Distelmaier F., Haack T.B., Catarino C.B., Gallenmüller C., Rodenburg R.J., Strom T.M., Baertling F., Meitinger T., Mayatepek E., Prokisch H., Klopstock T. MRPL44 mutations cause a slowly progressive multisystem disease with childhood-onset hypertrophic cardiomyopathy. Neurogenetics. 2015;16:319–323. doi: 10.1007/s10048-015-0444-2. [DOI] [PubMed] [Google Scholar]
- 11.Miller C., Saada A., Shaul N., Shabtai N., Ben-Shalom E., Shaag A., Hershkovitz E., Elpeleg O. Defective mitochondrial translation caused by a ribosomal protein (MRPS16) mutation. Ann. Neurol. 2004;56:734–738. doi: 10.1002/ana.20282. [DOI] [PubMed] [Google Scholar]
- 12.Lake N.J., Webb B.D., Stroud D.A., Richman T.R., Ruzzenente B., Compton A.G., Mountford H.S., Pulman J., Zangarelli C., Rio M. Biallelic Mutations in MRPS34 Lead to Instability of the Small Mitoribosomal Subunit and Leigh Syndrome. Am. J. Hum. Genet. 2017;101:239–254. doi: 10.1016/j.ajhg.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saada A., Shaag A., Arnon S., Dolfin T., Miller C., Fuchs-Telem D., Lombes A., Elpeleg O. Antenatal mitochondrial disease caused by mitochondrial ribosomal protein (MRPS22) mutation. J. Med. Genet. 2007;44:784–786. doi: 10.1136/jmg.2007.053116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohda M., Tokuzawa Y., Kishita Y., Nyuzuki H., Moriyama Y., Mizuno Y., Hirata T., Yatsuka Y., Yamashita-Sugahara Y., Nakachi Y. A Comprehensive Genomic Analysis Reveals the Genetic Landscape of Mitochondrial Respiratory Chain Complex Deficiencies. PLoS Genet. 2016;12:e1005679. doi: 10.1371/journal.pgen.1005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilissen C., Arts H.H., Hoischen A., Spruijt L., Mans D.A., Arts P., van Lier B., Steehouwer M., van Reeuwijk J., Kant S.G. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am. J. Hum. Genet. 2010;87:418–423. doi: 10.1016/j.ajhg.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menezes M.J., Guo Y., Zhang J., Riley L.G., Cooper S.T., Thorburn D.R., Li J., Dong D., Li Z., Glessner J. Mutation in mitochondrial ribosomal protein S7 (MRPS7) causes congenital sensorineural deafness, progressive hepatic and renal failure and lactic acidemia. Hum. Mol. Genet. 2015;24:2297–2307. doi: 10.1093/hmg/ddu747. [DOI] [PubMed] [Google Scholar]
- 17.Heide H., Bleier L., Steger M., Ackermann J., Dröse S., Schwamb B., Zörnig M., Reichert A.S., Koch I., Wittig I., Brandt U. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 2012;16:538–549. doi: 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Wessels H.J., Vogel R.O., Lightowlers R.N., Spelbrink J.N., Rodenburg R.J., van den Heuvel L.P., van Gool A.J., Gloerich J., Smeitink J.A., Nijtmans L.G. Analysis of 953 human proteins from a mitochondrial HEK293 fraction by complexome profiling. PLoS ONE. 2013;8:e68340. doi: 10.1371/journal.pone.0068340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baertling F., Haack T.B., Rodenburg R.J., Schaper J., Seibt A., Strom T.M., Meitinger T., Mayatepek E., Hadzik B., Selcan G. MRPS22 mutation causes fatal neonatal lactic acidosis with brain and heart abnormalities. Neurogenetics. 2015;16:237–240. doi: 10.1007/s10048-015-0440-6. [DOI] [PubMed] [Google Scholar]
- 20.Kaminishi T., Wilson D.N., Takemoto C., Harms J.M., Kawazoe M., Schluenzen F., Hanawa-Suetsugu K., Shirouzu M., Fucini P., Yokoyama S. A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction. Structure. 2007;15:289–297. doi: 10.1016/j.str.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Laughrea M., Moore P.B. Ribosomal components required for binding protein S1 to the 30 S subunit of Escherichia coli. J. Mol. Biol. 1978;122:109–112. doi: 10.1016/0022-2836(78)90111-0. [DOI] [PubMed] [Google Scholar]
- 22.Aseev L.V., Levandovskaya A.A., Tchufistova L.S., Scaptsova N.V., Boni I.V. A new regulatory circuit in ribosomal protein operons: S2-mediated control of the rpsB-tsf expression in vivo. RNA. 2008;14:1882–1894. doi: 10.1261/rna.1099108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brodersen D.E., Clemons W.M., Jr., Carter A.P., Wimberly B.T., Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 24.Emdadul Haque M., Grasso D., Miller C., Spremulli L.L., Saada A. The effect of mutated mitochondrial ribosomal proteins S16 and S22 on the assembly of the small and large ribosomal subunits in human mitochondria. Mitochondrion. 2008;8:254–261. doi: 10.1016/j.mito.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimopoulou A., Fischer B., Gardeitchik T., Schröter P., Kayserili H., Schlack C., Li Y., Brum J.M., Barisic I., Castori M. Genotype-phenotype spectrum of PYCR1-related autosomal recessive cutis laxa. Mol. Genet. Metab. 2013;110:352–361. doi: 10.1016/j.ymgme.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Reversade B., Escande-Beillard N., Dimopoulou A., Fischer B., Chng S.C., Li Y., Shboul M., Tham P.Y., Kayserili H., Al-Gazali L. Mutations in PYCR1 cause cutis laxa with progeroid features. Nat. Genet. 2009;41:1016–1021. doi: 10.1038/ng.413. [DOI] [PubMed] [Google Scholar]
- 27.Bicknell L.S., Pitt J., Aftimos S., Ramadas R., Maw M.A., Robertson S.P. A missense mutation in ALDH18A1, encoding Delta1-pyrroline-5-carboxylate synthase (P5CS), causes an autosomal recessive neurocutaneous syndrome. Eur. J. Hum. Genet. 2008;16:1176–1186. doi: 10.1038/ejhg.2008.91. [DOI] [PubMed] [Google Scholar]
- 28.Wolthuis D.F., van Asbeck E., Mohamed M., Gardeitchik T., Lim-Melia E.R., Wevers R.A., Morava E. Cutis laxa, fat pads and retinopathy due to ALDH18A1 mutation and review of the literature. European journal of paediatric neurology. 2014;18:511–515. doi: 10.1016/j.ejpn.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Berk D.R., Bentley D.D., Bayliss S.J., Lind A., Urban Z. Cutis laxa: a review. J. Am. Acad. Dermatol. 2012;66:1–17. doi: 10.1016/j.jaad.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 30.Morava E., Guillard M., Lefeber D.J., Wevers R.A. Autosomal recessive cutis laxa syndrome revisited. Eur. J. Hum. Genet. 2009;17:1099–1110. doi: 10.1038/ejhg.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gardeitchik T., Mohamed M., Fischer B., Lammens M., Lefeber D., Lace B., Parker M., Kim K.J., Lim B.C., Häberle J. Clinical and biochemical features guiding the diagnostics in neurometabolic cutis laxa. Eur. J. Hum. Genet. 2014;22:888–895. doi: 10.1038/ejhg.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohamed M., Kouwenberg D., Gardeitchik T., Kornak U., Wevers R.A., Morava E. Metabolic cutis laxa syndromes. J. Inherit. Metab. Dis. 2011;34:907–916. doi: 10.1007/s10545-011-9305-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Serre V., Rozanska A., Beinat M., Chretien D., Boddaert N., Munnich A., Rötig A., Chrzanowska-Lightowlers Z.M. Mutations in mitochondrial ribosomal protein MRPL12 leads to growth retardation, neurological deterioration and mitochondrial translation deficiency. Biochim. Biophys. Acta. 2013;1832:1304–1312. doi: 10.1016/j.bbadis.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.