Abstract
Hypomyelinating leukodystrophies are genetic disorders characterized by insufficient myelin deposition during development. They are diagnosed on the basis of both clinical and MRI features followed by genetic confirmation. Here, we report on four unrelated affected individuals with hypomyelination and bi-allelic pathogenic variants in EPRS, the gene encoding cytoplasmic glutamyl-prolyl-aminoacyl-tRNA synthetase. EPRS is a bifunctional aminoacyl-tRNA synthetase that catalyzes the aminoacylation of glutamic acid and proline tRNA species. It is a subunit of a large multisynthetase complex composed of eight aminoacyl-tRNA synthetases and its three interacting proteins. In total, five different EPRS mutations were identified. The p.Pro1115Arg variation did not affect the assembly of the multisynthetase complex (MSC) as monitored by affinity purification-mass spectrometry. However, immunoblot analyses on protein extracts from fibroblasts of the two affected individuals sharing the p.Pro1115Arg variant showed reduced EPRS amounts. EPRS activity was reduced in one affected individual’s lymphoblasts and in a purified recombinant protein model. Interestingly, two other cytoplasmic aminoacyl-tRNA synthetases have previously been implicated in hypomyelinating leukodystrophies bearing clinical and radiological similarities to those in the individuals we studied. We therefore hypothesized that leukodystrophies caused by mutations in genes encoding cytoplasmic aminoacyl-tRNA synthetases share a common underlying mechanism, such as reduced protein availability, abnormal assembly of the multisynthetase complex, and/or abnormal aminoacylation, all resulting in reduced translation capacity and insufficient myelin deposition in the developing brain.
Keywords: hypomyelinating leukodystrophy, tRNA-synthetase, EPRS
Main Text
Leukodystrophies are genetically determined disorders characterized by abnormal white matter on brain imaging.1, 2 These disorders can be classified into hypomyelinating leukodystrophies and leukodystrophies with other white matter pathology on the basis of their MRI characteristics2 and on whether they arise from deficient myelin deposition during development or other types of white-matter involvement, respectively. With advances in next-generation sequencing, several novel genes causing leukodystrophies have been identified, allowing a reduction in the number of molecularly undiagnosed affected individuals from about 50% to 20%–30%.3, 4, 5
In recent literature, pathogenic variants in genes encoding for aminoacyl-tRNA synthetases (ARSs) have been implicated in several central nervous system disorders, including hypomyelinating leukodystrophies6, 7 (Table 1). ARSs are responsible for catalyzing esterification reactions that link amino acids with their cognate tRNAs; these reactions are a critical step in the faithful transfer of genetic information from mRNA to protein.8, 9 Separate sets of ARSs are present in the cytoplasm for the translation of nuclear genes and in the mitochondria for the translation of mitochondrial genes. Cytoplasmic ARSs form a large 11-subunit multisynthetase complex (MSC) that is critical for the aminoacylation function of ARS subunits and for channeling loaded tRNAs directly to the ribosome.8, 9, 10, 11 EPRS is a bifunctional aminoacyl tRNA synthetase that catalyzes the aminoacylation of both glutamic acid and proline tRNA species. It is also part of the GAIT (gamma-interferon-activated inhibitor of translation) complex, which is involved in selective translational silencing of more than 30 known targets. The GAIT complex consists of the ribosomal protein L13a (RPL13A), EPRS, synaptotagmin-binding cytoplasmic RNA-interacting protein (SYNCRIP, also known as NSAP1), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH).12
Table 1.
Bi-allelic Variants in EPRS Cause Hypomyelinating Leukodystrophy
| Subject | Ethnicity | Consanguinity | Genomic Position (NM_004446.2; GRCh37/hg19) | Exon | cDNA | Protein | Inheritance | In Silico Prediction |
|---|---|---|---|---|---|---|---|---|
| 1 | W | yes | chr1: 220,156,143 | 23 | c.3344C>G | p.Pro1115Arg | HMZ | damaging |
| 2 | W | no | chr1: 220,195,789 | 9 | c.1015C>T | p.Arg339∗ | M | NA |
| chr1: 220,156,143 | 23 | c.3344C>G | p.Pro1115Arg | P | damaging | |||
| 3 | W | no | chr1: 220,154,175 | 24 | c.3478C>T | p.Pro1160Ser | P | damaging |
| chr1: 220,153,471 | 26 | c.3667delA | p.Thr1223Leufs3∗ | M | NA | |||
| 4 | W | yes | chr1: 220,154,796 | 24 | c.3377T>C | p.Met1126Thr | HMZ | damaging |
Abbreviations are as follows: HMZ, homozygous; M, maternal; NA, not applicable; P, paternal; W, white.
The EPRS linker region contains three helix-turn-helix WHEP domains, the first two of which bind the RNA element, whereas the second overlapping pair binds the other three GAIT proteins. WHEP domains are present in five ARSs, WARS, HARS, EPRS (from which the domain is named), and also GARS and MARS, but not in any non-ARS protein.13 EPRS joins other WHEP-domain-containing aminoacyl tRNA synthetases with noncanonical functions.12 It is inducibly phosphorylated by mTORC1–S6K1 at Ser999 in the linker region and released from the MSC to perform GAIT-mediated translational control function. Indeed, EPRS has a special role in this process; it is solely responsible for recognition and interaction with GAIT elements in target mRNAs.14
In this study, we describe four unrelated affected individuals with hypomyelinating leukodystrophy caused by bi-allelic pathogenic variants in EPRS.
Four affected individuals with hypomyelinating leukodystrophy were selected from two cohorts of molecularly unsolved hypomyelinating leukodystrophy cases on the basis of their clinical and MRI characteristics.15 The project was approved by the Montreal Children’s Hospital Research Ethics Board [11-105-PED] and the institutional review board of the VU Medical Center. Informed consent was obtained from all participating families. Charts and brain magnetic resonance images of all four affected individuals were reviewed.
The first individual (P1), a boy, was born to a consanguineous white couple. His initial psychomotor development was normal except for subtle personality changes starting around the age of 3 years. At age 7 years, he developed amblyopia with marked prosopagnosia, progressive ataxia, dysarthria, spasticity, dystonia, and severe intention tremor, as well as cognitive regression. Hypermetropia was also noted. He never showed signs of posterior column involvement. Presently, at the age of 16 years, he is nearly blind from bilateral optic atrophy, has lost ambulation, and has developed mild dysphagia. Of note, he has always been prone to cavities, but he is without other dental abnormalities or endocrine features.
The second affected individual (P2), a girl, was born to a non-consanguineous white couple. She presented at 14 years of age with deteriorating school performance. By age 18, she had developed spasticity and dystonia. She exhibited prominent cerebellar features, including severe intention tremor, ataxia, nystagmus, and dysarthria. Ophthalmologic evaluation revealed optic atrophy, restriction of visual fields, and significant vision loss. In addition, she lacked seven teeth, and her remaining teeth were unusually small. She did not have hormonal abnormalities. Epilepsy started at age 23 years. She experienced cognitive and motor regression but remained ambulatory.
The third individual (P3), another female, was born to a non-consanguineous white couple. She presented in the first months of life with failure to thrive, microcephaly, and delayed motor development. She achieved unsupported sitting, but she could never walk. Around 18 months of age, she developed motor regression, with ataxia, dystonia, and pyramidal signs. Dysphagia led to insertion of a gastrostomy tube at age 2 years; despite tube feeding, her growth lagged. Her dentition was normal, and she did not exhibit ophthalmologic abnormalities. She passed away at age 9 years due to pneumonia.
The fourth affected individual (P4) is a girl born to a consanguineous white couple. She presented at 12 months of age with microcephaly, nystagmus, and developmental delay. She never achieved unsupported walking. At the age of 2 years, she displayed motor regression, with signs of upper-motor-neuron involvement, as well as ataxia, dystonia, and athetoid movements. Parents reported episodic deterioration with febrile illnesses. She later developed cognitive regression and sensorineural hearing loss. She also had short stature and severe dysphagia. She required insertion of a gastrostomy tube at 5 years of age. Ophthalmologic assessment revealed optic atrophy. An equally affected older sister had died at age 3 years.
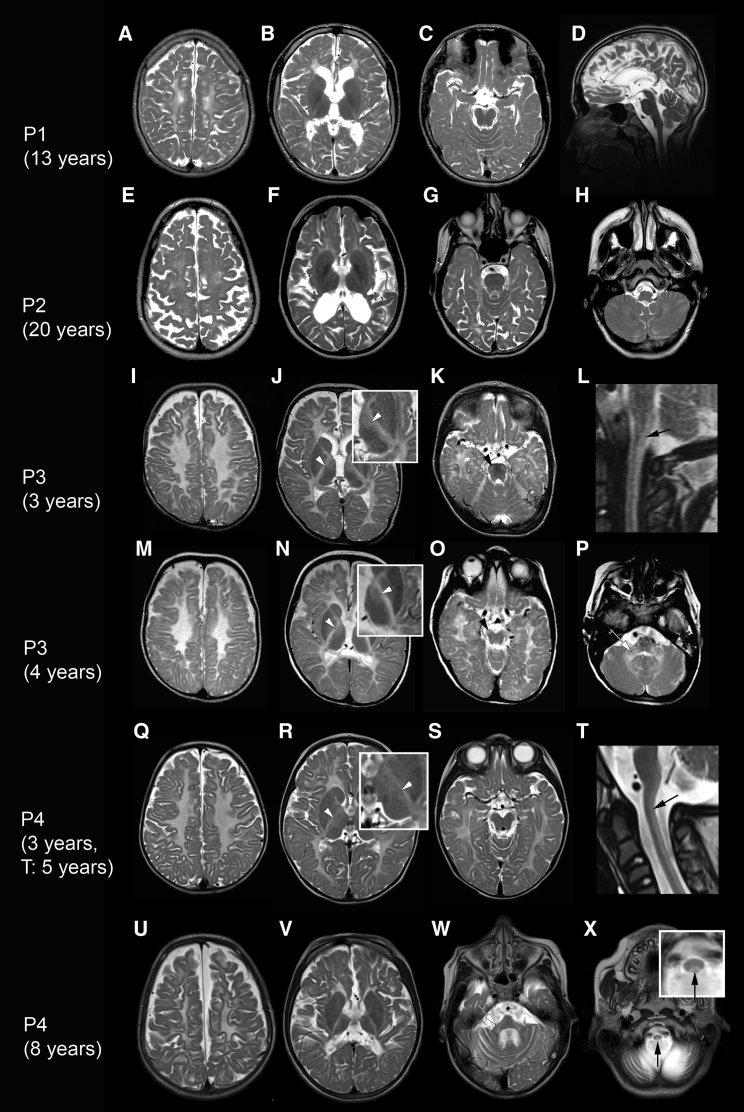
Brain MRI demonstrated a hypomyelinating leukodystrophy with thinning of the corpus callosum in all four affected individuals (Figure 1, Table S1). Supratentorial atrophy was severe in the older individuals (P1 and P2, aged 13 and 20 years, respectively, at the time of the MRI). If present, cerebellar atrophy was mild. Affected individuals P3 and P4 had involvement of the thalami and of specific tracts in the brainstem and spinal cord.
Figure 1.
MRI Characteristics of Hypomyelinating Leukodystrophy Caused by Mutations in EPRS
Brain MRI findings in the four affected individuals with EPRS pathogenic variants. Axial and sagittal (D, L, and T) T2-weighted images of affected individual P1 at age 13 years (A–D), affected individual P2 at age 20 years (E–H), affected individual P3 (I–L: age 3 years; M–P: age 4 years), and affected individual P4 (Q–S: age 3 years; T: age 5 years; U–X: age 8 years). All affected individuals show diffusely elevated T2 signal of the entire supratentorial white matter. There is no (Q–S) or only mild (I–K) atrophy in the younger affected individuals, whereas in the older affected individuals (A–C, E–G, and U–W) there is pronounced supratentorial atrophy with preserved (D and H) or mildly atrophic cerebellum. Affected individuals P3 and P4 show hyperintense T2 signal in the posterior columns of the spinal cord (L, T, and X; black arrows) and also in the ventrolateral thalami (J, N, and R; white arrowheads). In addition, affected individual P3 has elevated T2 signal of the pyramidal tracts in the midbrain (O, black arrowhead) and pons (K, black arrowhead). The cerebellar white matter (H and P) and the middle cerebellar peduncles (P and W; white arrows) were also hyperintense on T2.
Whole-exome sequencing with either a HiSeq2000 or HiSeq2500 sequencing system (Illumina, San Diego, CA) was performed on the affected individuals P1 and P2 (HiSeq2000) and P3 (HiSeq2500) after informed consent was obtained and with methods as previously published.6, 16 Because individuals P3 and P4 showed similar MRI patterns, EPRS was sequenced directly in individual P4. Variants were filtered to less than 1% minor allele frequency in population databases and then prioritized on the basis of American College of Medical Genetics categorization, OMIM identity, phenotypic assessment, suspected mode of inheritance, and in silico prediction tools. Candidate genes were evaluated in correlation to phenotype, gene function, and expression.
Using primer pairs designed with the Primer3 software, we performed Sanger sequencing and co-segregation analysis on genomic DNA of individuals P1, P2, and P3 to confirm the variants identified by WES (whole exome sequencing). To identify EPRS variants in individual P4, we designed EPRS-specific primers to cover all exons and exon-intron boundaries of human EPRS (NM_004446.2; GRCh37/hg19). PCR amplification was performed with Q5 Hot Start High-Fidelity DNA Polymerase Kit according to manufacturer instructions (New England BioLabs) or HotStart Taq Polymerase (QIAGEN, Germany). PCR products were forward- and reverse-sequenced at the McGill University and Genome Quebec Innovation Centre with a 3730xl DNA Analyzer from Applied Biosystems (ABI) or in the Metabolic Unit in Amsterdam with an ABI 3130 DNA Analyzer. Sequences were analyzed with SeqMan 4.03 (DNAStar) and Mutation Surveyor (Softgenetics).
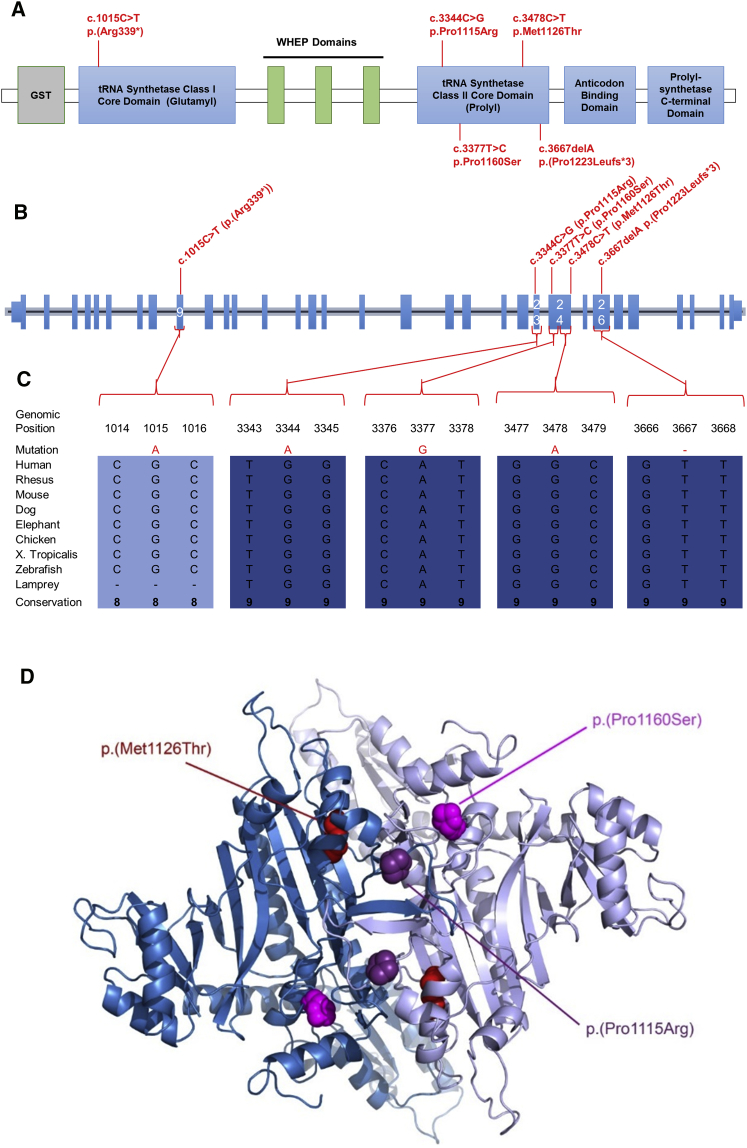
Analysis of WES data with in-house filters in three unrelated affected individuals revealed suggestive diagnostic EPRS genotypes: a homozygous missense variant in individual P1 (c.3344C>G [p.Pro1115Arg]) and compound heterozygous variants in affected individual P2 (c.3344C>G [p.Pro1115Arg] and c.1015C>T [p.Pro339∗]) and individual P3 (c.3478C>T [p.Pro1160Ser] and c.3667delA [p.Thr1223Leufs3∗]). Sanger sequencing of EPRS in individual P4 identified a homozygous missense variant (c.3377T>C [p.Met1126Thr]). Variant interpretation included familial segregation studies, minor allele frequency, conservation, and in silico analysis (Figure 2, Table S2). Apart from the nonsense variant observed once in heterozygous form in the Broad Institute’s ExAC dataset (0.01%, 1/8654 South Asian alleles; 8.237e-06, 1/121404 total alleles), all were private changes predicted to be deleterious (Table S3) and to affect conserved residues among orthologs. All missense variants were in the portion of the protein responsible for the proline tRNA synthesis (Figure 2, Table S3).
Figure 2.
EPRS Pathogenic Variants are Found within the Proline tRNA Synthetase Core Domain
(A) The locations of the EPRS pathogenic variants are shown according to their position within the different functional domains.
(B) Genomic organization of EPRS in humans (UCSC Genome Browser hg19): positions of EPRS pathogenic variants (in red) within the EPRS gDNA.
(C) EPRS mutations in individuals with leukodystrophy affect amino acids that are conserved through species.
(D) 3D representations (created with MacPyMOL) of identified EPRS missense variants: point mutations identified in EPRS are displayed according to their equivalent positions in the proline tRNA synthetase domain of the human EPRS (PDB 4K86).
We hypothesized that the variants found in EPRS lead to one or more of the following mechanisms: (1) decreased protein availability, (2) impaired tRNA synthetase function, and/or (3) abnormal assembly of the MSC. To test these hypotheses, we performed immunoblot, protein-activity determination, and mass spectrometry (MS).
Immunoblot was performed with protein extracts from fibroblasts of affected individuals P1 and P2 and age- and sex-matched healthy controls. Affected individuals’ and controls’ fibroblasts were harvested at three consecutive passages, and each sample was used for western blot analysis in triplicate. Visualization of EPRS and GAPDH amounts was achieved with antibodies targeted to the N terminus of EPRS ([AP7565a, Abgent] purified rabbit antibody, dilution 1:500) and GAPDH ([FL-335, Santa Cruz Biotechnology] purified rabbit antibody, dilution 1:2,000), in conjunction with a donkey anti-Rabbit (ECL anti-rabbit IgG, horseradish peroxidase linked whole antibody) secondary antibody, according to previously described methods.17 Band intensity was measured and quantified with ImageQuant TL 8.1 (GE Life Sciences). In order to determine statistical significance, a one-tailed, unpaired t test was performed. Decreased EPRS amount was seen in the tested fibroblasts of individuals P1 and P2 compared to age- and sex-matched controls (Figures 3A and 3B, Figure S1). Of note, fibroblasts from affected individuals P3 and P4 were not available for testing. However, we hypothesized that both the p.Arg339∗ (c.1015C>T) and p.Thr1223Leufs3∗ (c.3667delA) variations, resulting in premature stop codons in exon 9 and exon 26, respectively, lead to transcripts subject to nonsense-mediated RNA decay.
Figure 3.
EPRS Protein Amount and Activity Are Decreased in Affected Individuals with Hypomyelinating Leukodystrophy-Causing Mutations in EPRS
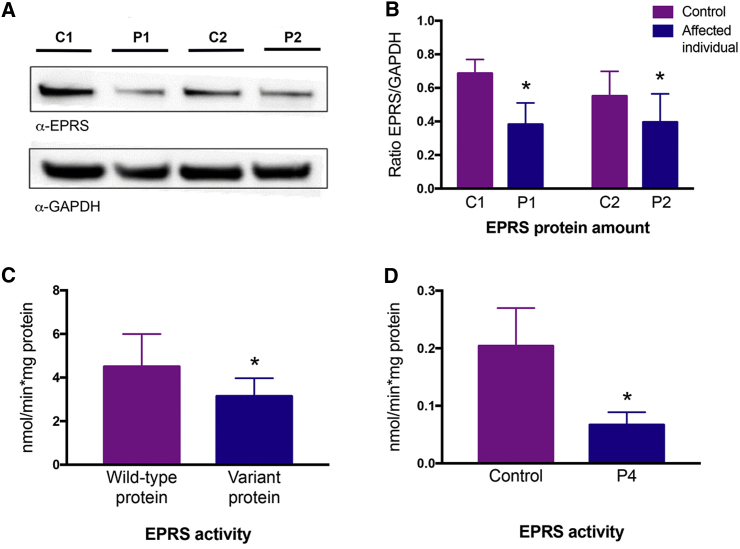
(A) Immunoblot with protein extracted from hypomyelinating leukodystrophy-affected individuals and age- and sex-matched control fibroblasts. Immunoblot analysis with anti-EPRS revealed a reduced abundance of EPRS, ∼60% of the control amount (control 1 [C1], control 2 [C2]), in individuals P1 and P2. Anti-GAPDH was used as a loading control.
(B) Endogenous amounts of EPRS in fibroblasts between affected individuals and age-sex-matched controls were quantified by a ratio of EPRS band intensity over GAPDH with the three immunoblot analysis experiments, one of which is shown in (A). Abundance of EPRS relative to GAPDH was assessed in three independent experiments using three different protein extracts from each sample. Bar chart shows quantification of EPRS protein amounts compared to control in each condition.
(C) Activity of wild-type and mutated recombinant prolyl-tRNA synthetase protein (p.Pro1115Arg) (nmol released Pi/min/mg protein). Assay was performed at least in triplicate.
(D) 5 μg of total protein was used for determining EPRS activity in lymphoblast lysates from affected individual P4.
Error bars show standard deviation, ∗p < 0.05.
In order to evaluate tRNA synthetase activity, we purified human prolyl-tRNA synthetase (hPRS) (wild-type and c.3344C>G mutant). The cDNA encoding the C-terminal region of the hPRS, including PRS from residues 1001 to 1512, was amplified by PCR and cloned into pGEX 4T1 vector (Addgene) according to the manufacturer's instructions. GST-tagged hPRS expression plasmids were generated and transformed in One Shot BL21 Chemically Competent E. coli (Thermofisher) according to standard methods.18 Cell cultures were induced with IPTG, and after an induction process of 4 hr, the cultures were subjected to French press disruption as previously described.18 To purify GST-tagged proteins, we used batch purification with Glutathione-Sepharose 4B (GE Healthcare). The elution products were visualized with SDS-PAGE followed by Coomassie Blue staining to ensure the presence of the target protein (Figure S2), then pooled, concentrated with size exclusion centrifugation, and dialysed according to standard methods.18 All proteins were stored at −80°C until use.
In purified protein, we measured the reaction between ATP and proline by quantifying the amount of phosphate (pi) released in the conversion of ATP to AMP.19 The assay was performed with approximately 1 μg of pure protein (wild-type or mutant) in a reaction buffer (50 mM Tris buffer [pH 7.5], 12 mM MgCl2, 25 mM KCl) containing 1 mM ATP and 0.3 mM proline. Pyrophosphatase (0.1U) was added to convert pyrophosphate (ppi) to pi. After 30 min of incubation at 22°C, the released pi was quantified with the BioMol Green kit (Enzo) according to the manufacturer specifications. Statistical significance was determined with a one-tailed, unpaired t test. The purified hPRS with the p.Pro1115Arg variant detected in affected individuals P1 and P2 had less activity than the wild type hPRS (Figure 3C).
Steady state aminoacylation assays were performed in triplicate in lymphoblast lysates of affected individual P4, homozygous for p.Met1126Thr, at 37°C for 10 min in a reaction buffer (50 mM Tris buffer [pH 7.5], 12 mM MgCl2, 25 mM KCl) containing 1 mg/mL bovine serum albumin, 0.5 mM spermine, 1 mM ATP, 0.2 mM yeast total tRNA, 1 mM DTT, and 0.3 mM [13C4,15N] proline. The reaction was terminated with trichloroacetic acid (TCA). After samples were washed with TCA, ammonia was added so that [13C4,15N] proline would be released from the tRNAs [13C2,15N], and glycine was added as an internal standard. Labeled amino acids were quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Intra-assay variation was <15%. Statistical significance was determined with a one-tailed, unpaired t test. The prolyl-tRNA synthetase activity was significantly decreased in affected-individual-derived samples relative to control samples (Figure 3D).
We performed affinity purification and MS experiments to assess the integrity of the MSC with the c.3344C>G mutation. FLAG-tagged EPRS (wild-type or mutant) was produced in HEK293T cells for 24 hr, and affinity purification from the soluble fractions was performed with standard procedures.20, 21 The eluates were digested with trypsin, and the resulting tryptic peptides were purified and identified via LC-MS/MS with a microcapillary reversed-phase, high-pressure LC-coupled LTQ-Orbitrap (ThermoElectron) quadrupole ion trap mass spectrometer with a nanospray interface, as recently described.22 Protein database searching was performed with Mascot 2.3 (Matrix Science) against the human NCBInr protein database (version July 18, 2014).23 Known protein contaminants such as keratins, which are not produced in HEK293T cells, were excluded from the dataset. In cases where multiple gene products were identified from the same peptide set, all were unambiguously removed from the dataset. When multiple isoforms were identified for a unique gene, only the isoform with the best sequence coverage was reported. Proteins identified on the basis of a single spectrum were also discarded. For each LC-MS/MS analysis, a set of high-confidence interactors were assigned for EPRS (wild-type and mutant) in order to assess the reliability of the data obtained from AP-MS experiments. By comparing the spectral counts of the interactors obtained from the purifications of the paired LC-MS/MS of the wild-type or p.Pro1115Arg proteins to those with an empty vector (EV) of the FLAG tag (non-specific interactions), we identified high-confidence interactors. A protein was labeled as a high-confidence interactor if it was identified and quantified in all three replicates of EPRS (wild-type and Pro1115Arg) and if the ratio of the average spectral counts across the three replicates (WT/EV or MUT/EV) was greater than 5. These stringent criteria allowed us to eliminate the vast majority of non-specific interactors of EPRS.16 The spectral counts of various forms of EPRS (wild-type and p.Pro1115Arg) were equivalent and comparable (Table S4), and the spectral counts were normalized to the amount of the bait in each of the purifications. Stringent criteria were used to obtain high-confidence interactors for both wild-type EPRS and p.Pro1115Arg protein. Both wild-type EPRS and the p.Pro1115Arg variant strongly interacted with the remaining ten MSC subunits (Figure S3).
Using clinical phenotyping combined with MRI pattern recognition and next-generation sequencing, we identified a hypomyelinating leukodystrophy caused by bi-allelic pathogenic variants in EPRS. In total, we identified five pathogenic variants, either nonsense or missense, located in the protein domain responsible for prolyl-tRNA synthesis. Affected individuals with this newly recognized hypomyelinating leukodystrophy present similarities with individuals who have other tRNA-synthetase-related disorders; the two younger and most severely affected individuals showed involvement of selected tracts in the brainstem (pyramidal tracts, middle cerebellar peduncles) and spinal cord (posterior columns). These tracts are also involved in two other tRNA-synthetase-related leukodystrophies, LBSL (leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation) (caused by mutations in DARS2) and HBSL (hypomyelination with brainstem and spinal cord involvement and leg spasticity) (caused by mutations in DARS),7, 24 although both HBSL and EPRS-related hypomyelination do not share all tract involvement with LBSL. In the two more mildly affected and older individuals, involvement of the brainstem and spinal-cord tracts was absent, perhaps because they represent the milder end of the spectrum, but it might also be possible that the specific tract involvement is not visible because of the prominent degenerative changes with pronounced atrophy. Interestingly, individual P2 also had clinical similarities (hypodontia) with POLR3-related leukodystrophy. Highlighting the importance of protein production for myelination, these observations suggest that the clinical and MRI manifestations of EPRS-related leukodystrophy overlap with those of both POLR3-related and tRNA-synthetase-related leukodystrophies, which are disorders of transcription and translation, respectively.
Emerging literature has shown that pathogenic variants in genes encoding both cytoplasmic and mitochondrial ARSs can cause a wide range of central and peripheral nervous system diseases (Table 2). Specifically, hypomyelinating leukodystrophies can be caused by biallelic pathogenic variants in genes encoding two other cytoplasmic aminoacyl-tRNA synthetases: RARS6 (MIM: 616140), encoding arginyl-tRNA synthetase, and DARS, encoding aspartyl-tRNA synthetase (HBSL; MIM: 615281).7 EPRS, RARS, and DARS are all subunits of the MSC. The MSC is composed of eight ARSs proteins and three aminoacyl-tRNA synthetase interacting multifunctional proteins (AIMP1, 2, and 3).8, 9, 10, 11 Mutations in AIMP1 have been shown to cause hypomyelination secondary to a primary neuronal involvement (MIM: 260600).25
Table 2.
Central and Peripheral Nervous System Diseases Caused by Pathogenic Variants in Genes Encoding Aminoacyl-tRNA Synthetases and Aminoacyl-tRNA Synthetase Interacting Multifunctional Proteins
| Cytoplasmic ARS | |
|---|---|
| AARS | epileptic encephalopathy with persistent myelination defect (MIM: 616339)26 |
| DARS | hypomyelinating leukodystrophy with brainstem involvement and leg spasticity (MIM: 615281)7 |
| EPRS | hypomyelinating leukodystrophy |
| GARS | Charcot-Marie tooth disease (CMT) type 2D (MIM 601472), distal hereditary motor neuropathy 5A (MIM: 600794)27 |
| HARS | CMT type 2W (MIM: 616625)28 |
| KARS | AR deafness 89, recessive intermediate B (MIM: 613916),28 and Charcot Marie Tooth (CMT) Disease Type 2 (MIM: 613641)29 |
| MARS | late-onset CMT type 2U (MIM: 616280)30 |
| QARS | progressive microcephaly, intractable seizures and cerebral and cerebellar atrophy (MIM: 615760)31 |
| RARS | hypomyelinating leukodystrophy6 (MIM: 616140) |
| YARS | CMT dominant intermediate C (MIM: 6088323)32 |
| Mitochondrial ARS | |
| AARS2 | (ovario) leukodystrophy and combined oxidative phosphorylation deficiency (MIM: 615889)33 |
| DARS2 | leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation (MIM: 611105)34 |
| EARS2 | leukoencephalopathy with thalamus and brainstem involvement with high lactate (MIM: 614924)35 |
| FARS2 | alpers encephalopathy (MIM: 614946)36 |
| HARS2 | perrault syndrome (MIM: 614926)37 |
| MARS2 | autosomal recessive spastic ataxia with leukoencephalopathy (MIM: 611390)38 |
| RARS2 | pontocerebellar hypoplasia type 6 (MIM: 611523)39 |
| TARS2 | mitochondrial encephalopathy (combined oxidative phosphorylation deficiency 21) (MIM: 615918)40 |
| VARS2 | mitochondrial encephalopathy (combined oxidative phosphorylation deficiency 20) (MIM: 615917)40 |
| tRNA Synthetase Cofactor Gene | |
| AIMP1 | hypomyelination secondary to neuronal disorder (MIM: 260600)25 |
| AIMP2 | hypomyelination secondary to neuronal disorder41 |
Underlined genes are the genes associated with hypomyelination.
We speculated that pathogenic variants in EPRS result in reduced protein availability, impaired tRNA synthetase function, and/or abnormal assembly of the MSC and thereby lead to decreased translation and thus decreased protein production at a crucial time during brain development, ultimately resulting in insufficient myelination. In the case of EPRS-related leukodystrophy due to the pathogenic variants studied here, we demonstrated that the mechanism is related to abnormal protein production with or without abnormal aminoacylation.
Little is known about the mechanisms underlying DARS- and RARS-related hypomyelination. Because of the clinical and MRI similarities between the three hypomyelinating leukodystrophies caused by mutations in EPRS, DARS, and RARS, we hypothesize that the same potential three mechanisms are at play and that they ultimately lead to impaired canonical functions of these ARSs. Whether non-canonical functions contribute to these pathologies is unknown but appears unlikely. We speculate that alterations at different steps of the tRNA-synthetase pathway lead to impaired canonical functions and therefore abnormal translation. The result might be insufficient production of certain proteins, including myelin proteolipid protein and myelin basic protein, the most abundant CNS myelin proteins, during a crucial developmental period. Additional research will further elucidate the underlying molecular mechanisms of tRNA-synthetase-related hypomyelinating leukodystrophies and shed light on the implication of tRNA synthetases in normal myelination.
Acknowledgments
The authors wish to thank the patients and their families for their participation. This study was supported by grants from the Canadian Institutes of Health Research (201610PJT- 377869 and MOP-G2-341146-159133-BRIDG), the Fondation du Grand Défi Pierre Lavoie, the Fondation les Amis d’Eliott, the Fondation Lueur d’Espoir pour Ayden, and the Réseau de Médecine Génétique Appliquée. Part of this study received financial support from ZonMw TOP (grant 91211005). This research was enabled in part by support provided by Compute Canada (www.computecanada.ca). We also wish to acknowledge the McGill University and Génome Québec Innovation Centre, where Sanger sequencing of affected individuals P1 and P2 was performed. M.G.S. has received the 2016–2017 McGill University Faculty of Medicine Internal Studentship Award. G.B. has received a Research Scholar Junior 1 award from the Fonds de Recherche du Québec en Santé (2012–2016) and the New Investigator Salary Award from the Canadian Institutes of Health Research (201512MSH-360766-171036, 2017–2022).
Published: March 22, 2018
Footnotes
Supplemental Data include three figures and four tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.02.011.
Accession Numbers
The accession numbers for the variants reported in this paper are dbSNP: ss3646217899, ss3646217900, ss3646217901, ss3646217902, and ss3646217903.
Web Resources
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Schiffmann R., van der Knaap M.S. The latest on leukodystrophies. Curr. Opin. Neurol. 2004;17:187–192. doi: 10.1097/00019052-200404000-00017. [DOI] [PubMed] [Google Scholar]
- 2.Schiffmann R., van der Knaap M.S. Invited article: an MRI-based approach to the diagnosis of white matter disorders. Neurology. 2009;72:750–759. doi: 10.1212/01.wnl.0000343049.00540.c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kevelam S.H., Steenweg M.E., Srivastava S., Helman G., Naidu S., Schiffmann R., Blaser S., Vanderver A., Wolf N.I., van der Knaap M.S. Update on Leukodystrophies: A Historical Perspective and Adapted Definition. Neuropediatrics. 2016;47:349–354. doi: 10.1055/s-0036-1588020. [DOI] [PubMed] [Google Scholar]
- 4.Parikh S., Bernard G., Leventer R.J., van der Knaap M.S., van Hove J., Pizzino A., McNeill N.H., Helman G., Simons C., Schmidt J.L., GLIA Consortium A clinical approach to the diagnosis of patients with leukodystrophies and genetic leukoencephelopathies. Mol. Genet. Metab. 2015;114:501–515. doi: 10.1016/j.ymgme.2014.12.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanderver A., Simons C., Helman G., Crawford J., Wolf N.I., Bernard G., Pizzino A., Schmidt J.L., Takanohashi A., Miller D., Leukodystrophy Study Group Whole exome sequencing in patients with white matter abnormalities. Ann. Neurol. 2016;79:1031–1037. doi: 10.1002/ana.24650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolf N.I., Salomons G.S., Rodenburg R.J., Pouwels P.J.W., Schieving J.H., Derks T.G.J., Fock J.M., Rump P., van Beek D.M., van der Knaap M.S., Waisfisz Q. Mutations in RARS cause hypomyelination. Ann. Neurol. 2014;76:134–139. doi: 10.1002/ana.24167. [DOI] [PubMed] [Google Scholar]
- 7.Taft R.J., Vanderver A., Leventer R.J., Damiani S.A., Simons C., Grimmond S.M., Miller D., Schmidt J., Lockhart P.J., Pope K. Mutations in DARS cause hypomyelination with brain stem and spinal cord involvement and leg spasticity. Am. J. Hum. Genet. 2013;92:774–780. doi: 10.1016/j.ajhg.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson J.C., Kerjan P., Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: quantitative analysis of protein-protein interactions and mechanism of complex assembly. J. Mol. Biol. 2000;304:983–994. doi: 10.1006/jmbi.2000.4242. [DOI] [PubMed] [Google Scholar]
- 9.Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu. Rev. Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- 10.Kyriacou S.V., Deutscher M.P. An important role for the multienzyme aminoacyl-tRNA synthetase complex in mammalian translation and cell growth. Mol. Cell. 2008;29:419–427. doi: 10.1016/j.molcel.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cestari I., Kalidas S., Monnerat S., Anupama A., Phillips M.A., Stuart K. A multiple aminoacyl-tRNA synthetase complex that enhances tRNA-aminoacylation in African trypanosomes. Mol. Cell. Biol. 2013;33:4872–4888. doi: 10.1128/MCB.00711-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sampath P., Mazumder B., Seshadri V., Gerber C.A., Chavatte L., Kinter M., Ting S.M., Dignam J.D., Kim S., Driscoll D.M., Fox P.L. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell. 2004;119:195–208. doi: 10.1016/j.cell.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 13.Jia J., Arif A., Ray P.S., Fox P.L. WHEP domains direct noncanonical function of glutamyl-Prolyl tRNA synthetase in translational control of gene expression. Mol. Cell. 2008;29:679–690. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao P., Potdar A.A., Arif A., Ray P.S., Mukhopadhyay R., Willard B., Xu Y., Yan J., Saidel G.M., Fox P.L. Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell. 2012;149:88–100. doi: 10.1016/j.cell.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steenweg M.E., Vanderver A., Blaser S., Bizzi A., de Koning T.J., Mancini G.M., van Wieringen W.N., Barkhof F., Wolf N.I., van der Knaap M.S. Magnetic resonance imaging pattern recognition in hypomyelinating disorders. Brain. 2010;133:2971–2982. doi: 10.1093/brain/awq257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiffault I., Wolf N.I., Forget D., Guerrero K., Tran L.T., Choquet K., Lavallée-Adam M., Poitras C., Brais B., Yoon G. Recessive mutations in POLR1C cause a leukodystrophy by impairing biogenesis of RNA polymerase III. Nat. Commun. 2015;6:7623–7631. doi: 10.1038/ncomms8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cloutier P., Poitras C., Durand M., Hekmat O., Fiola-Masson É., Bouchard A., Faubert D., Chabot B., Coulombe B. R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat. Commun. 2017;8:15615. doi: 10.1038/ncomms15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cloutier P., Lavallée-Adam M., Faubert D., Blanchette M., Coulombe B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 2013;9:e1003210. doi: 10.1371/journal.pgen.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harder K.W., Owen P., Wong L.K., Aebersold R., Clark-Lewis I., Jirik F.R. Characterization and kinetic analysis of the intracellular domain of human protein tyrosine phosphatase beta (HPTP beta) using synthetic phosphopeptides. Biochem. J. 1994;298:395–401. doi: 10.1042/bj2980395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen G.I., Gingras A.C. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods. 2007;42:298–305. doi: 10.1016/j.ymeth.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 22.Lavallée-Adam M., Rousseau J., Domecq C., Bouchard A., Forget D., Faubert D., Blanchette M., Coulombe B. Discovery of cell compartment specific protein-protein interactions using affinity purification combined with tandem mass spectrometry. J. Proteome Res. 2013;12:272–281. doi: 10.1021/pr300778b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Scheper G.C., van der Klok T., van Andel R.J., van Berkel C.G., Sissler M., Smet J., Muravina T.I., Serkov S.V., Uziel G., Bugiani M. Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat. Genet. 2007;39:534–539. doi: 10.1038/ng2013. [DOI] [PubMed] [Google Scholar]
- 25.Feinstein M., Markus B., Noyman I., Shalev H., Flusser H., Shelef I., Liani-Leibson K., Shorer Z., Cohen I., Khateeb S. Pelizaeus-Merzbacher-like disease caused by AIMP1/p43 homozygous mutation. Am. J. Hum. Genet. 2010;87:820–828. doi: 10.1016/j.ajhg.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simons C., Griffin L.B., Helman G., Golas G., Pizzino A., Bloom M., Murphy J.L., Crawford J., Evans S.H., Topper S. Loss-of-function alanyl-tRNA synthetase mutations cause an autosomal-recessive early-onset epileptic encephalopathy with persistent myelination defect. Am. J. Hum. Genet. 2015;96:675–681. doi: 10.1016/j.ajhg.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antonellis A., Ellsworth R.E., Sambuughin N., Puls I., Abel A., Lee-Lin S.Q., Jordanova A., Kremensky I., Christodoulou K., Middleton L.T. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Safka Brozkova D., Deconinck T., Griffin L.B., Ferbert A., Haberlova J., Mazanec R., Lassuthova P., Roth C., Pilunthanakul T., Rautenstrauss B. Loss of function mutations in HARS cause a spectrum of inherited peripheral neuropathies. Brain. 2015;138:2161–2172. doi: 10.1093/brain/awv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLaughlin H.M., Sakaguchi R., Liu C., Igarashi T., Pehlivan D., Chu K., Iyer R., Cruz P., Cherukuri P.F., Hansen N.F., NISC Comparative Sequencing Program Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am. J. Hum. Genet. 2010;87:560–566. doi: 10.1016/j.ajhg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonzalez M., McLaughlin H., Houlden H., Guo M., Yo-Tsen L., Hadjivassilious M., Speziani F., Yang X.L., Antonellis A., Reilly M.M., Züchner S., Inherited Neuropathy Consortium Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J. Neurol. Neurosurg. Psychiatry. 2013;84:1247–1249. doi: 10.1136/jnnp-2013-305049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X., Ling J., Barcia G., Jing L., Wu J., Barry B.J., Mochida G.H., Hill R.S., Weimer J.M., Stein Q. Mutations in QARS, encoding glutaminyl-tRNA synthetase, cause progressive microcephaly, cerebral-cerebellar atrophy, and intractable seizures. Am. J. Hum. Genet. 2014;94:547–558. doi: 10.1016/j.ajhg.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyun Y.S., Park H.J., Heo S.H., Yoon B.R., Nam S.H., Kim S.B., Park C.I., Choi B.O., Chung K.W. Rare variants in methionyl- and tyrosyl-tRNA synthetase genes in late-onset autosomal dominant Charcot-Marie-Tooth neuropathy. Clin. Genet. 2014;86:592–594. doi: 10.1111/cge.12327. [DOI] [PubMed] [Google Scholar]
- 33.Dallabona C., Diodato D., Kevelam S.H., Haack T.B., Wong L.J., Salomons G.S., Baruffini E., Melchionda L., Mariotti C., Strom T.M. Novel (ovario) leukodystrophy related to AARS2 mutations. Neurology. 2014;82:2063–2071. doi: 10.1212/WNL.0000000000000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyake N., Yamashita S., Kurosawa K., Miyatake S., Tsurusaki Y., Doi H., Saitsu H., Matsumoto N. A novel homozygous mutation of DARS2 may cause a severe LBSL variant. Clin. Genet. 2011;80:293–296. doi: 10.1111/j.1399-0004.2011.01644.x. [DOI] [PubMed] [Google Scholar]
- 35.Steenweg M.E., Ghezzi D., Haack T., Abbink T.E., Martinelli D., van Berkel C.G., Bley A., Diogo L., Grillo E., Te Water Naudé J. Leukoencephalopathy with thalamus and brainstem involvement and high lactate ‘LTBL’ caused by EARS2 mutations. Brain. 2012;135:1387–1394. doi: 10.1093/brain/aws070. [DOI] [PubMed] [Google Scholar]
- 36.Elo J.M., Yadavalli S.S., Euro L., Isohanni P., Götz A., Carroll C.J., Valanne L., Alkuraya F.S., Uusimaa J., Paetau A. Mitochondrial phenylalanyl-tRNA synthetase mutations underlie fatal infantile Alpers encephalopathy. Hum. Mol. Genet. 2012;21:4521–4529. doi: 10.1093/hmg/dds294. [DOI] [PubMed] [Google Scholar]
- 37.Pierce S.B., Chisholm K.M., Lynch E.D., Lee M.K., Walsh T., Opitz J.M., Li W., Klevit R.E., King M.C. Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc. Natl. Acad. Sci. USA. 2011;108:6543–6548. doi: 10.1073/pnas.1103471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayat V., Thiffault I., Jaiswal M., Tétreault M., Donti T., Sasarman F., Bernard G., Demers-Lamarche J., Dicaire M.J., Mathieu J. Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol. 2012;10:e1001288. doi: 10.1371/journal.pbio.1001288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edvardson S., Shaag A., Kolesnikova O., Gomori J.M., Tarassov I., Einbinder T., Saada A., Elpeleg O. Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am. J. Hum. Genet. 2007;81:857–862. doi: 10.1086/521227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diodato D., Melchionda L., Haack T.B., Dallabona C., Baruffini E., Donnini C., Granata T., Ragona F., Balestri P., Margollicci M. VARS2 and TARS2 mutations in patients with mitochondrial encephalomyopathies. Hum. Mutat. 2014;35:983–989. doi: 10.1002/humu.22590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shukla A., Das Bhowmik A., Hebbar M., Rajagopal K.V., Girisha K.M., Gupta N., Dalal A. Homozygosity for a nonsense variant in AIMP2 is associated with a progressive neurodevelopmental disorder with microcephaly, seizures, and spastic quadriparesis. J. Hum. Genet. 2018;63:19–25. doi: 10.1038/s10038-017-0363-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.