Abstract
The major diseases affecting the thoracic aorta are aneurysms and acute dissections, and pathogenic variants in 11 genes are confirmed to lead to heritable thoracic aortic disease. However, many families in which multiple members have thoracic aortic disease do not have alterations in the known aortopathy genes. Genes highly expressed in the aorta were assessed for rare variants in exome sequencing data from such families, and compound rare heterozygous variants (p.Pro45Argfs∗25 and p.Glu750∗) in LTBP3 were identified in affected members of one family. A homozygous variant (p.Asn678_Gly681delinsThrCys) that introduces an additional cysteine into an epidermal growth factor (EGF)-like domain in the corresponding protein, latent TGF-β binding protein (LTBP-3), was identified in a second family. Individuals with compound heterozygous or homozygous variants in these families have aneurysms and dissections of the thoracic aorta, as well as aneurysms of the abdominal aorta and other arteries, along with dental abnormalities and short stature. Heterozygous carriers of the p.Asn678_Gly681delinsThrCys variant have later onset of thoracic aortic disease, as well as dental abnormalities. In these families, LTBP3 variants segregated with thoracic aortic disease with a combined LOD score of 3.9. Additionally, heterozygous rare LTBP3 variants were found in individuals with early onset of acute aortic dissections, and some of these variants disrupted LTBP-3 levels or EGF-like domains. When compared to wild-type mice, Ltbp3−/− mice have enlarged aortic roots and ascending aortas. In summary, homozygous LTBP3 pathogenic variants predispose individuals to thoracic aortic aneurysms and dissections, along with the previously described skeletal and dental abnormalities.
Keywords: amelogenesis imperfecta, aortic dissections, heritable thoracic aortic diseases, LTBP3
Main Text
Thoracic aortic aneurysms involving the aortic root and ascending aorta are typically asymptomatic as they progressively enlarge and eventually lead to acute ascending aortic dissections in the absence of surgical repair of the aortic aneurysm. These thoracic aortic diseases can be triggered by mutations in single genes, either in association with syndromic features (e.g., in Marfan syndrome [MFS, MIM: 154700]) or in the absence of syndromic features. Heritable thoracic aortic disease (HTAD) is a genetically heterogeneous condition typically inherited in an autosomal-dominant manner, and there is sufficient evidence to classify mutations in 11 genes as causative.1, 2 These HTAD genes encode proteins involved in aortic smooth muscle cell (SMC) contraction, the extracellular matrix (ECM), and transforming growth factor-beta (TGF-β) signaling.3, 4, 5, 6, 7, 8, 9, 10, 11, 12 Despite identification of these HTAD genes, the causative genes in the majority of families with thoracic aortic disease have not been identified to date.
TGF-β signaling is initiated when the cytokine binds to the cell-surface TGF-β type II receptor, which then recruits and phosphorylates the TGF-β type I receptor. The type I receptor phosphorylates SMAD2 and SMAD3 (mothers against decapentaplegic homolog 2 and 3 (MIM: 601366 and MIM: 603109, respectively), which form a complex with SMAD4 and translocate to the nucleus to alter gene transcription. Mutated HTAD genes in this pathway include genes encoding one of the three TGF-β ligand family members, TGFB2 (TGF-β2; MIM: 190220), the TGF-β cellular receptors, TGFBR2 (MIM: 190182), TGFBR1 (MIM: 19018), and SMAD3. Pathogenic variants in these genes have been shown or are predicted to decrease cellular TGF-β signaling as a result of haploinsufficiency (e.g., nonsense or frameshift mutations) or missense variants that disrupt the protein function (e.g., decrease the intracellular kinase activity of the TGF-β receptors).13, 14
To identify pathogenic variants in novel genes predisposing to HTAD, we obtained whole-exome sequencing data on affected probands and family members from 271 unrelated HTAD families in which a HTAD gene mutation had not been identified (dbGap: phs000693.v5.p1). We collected blood or saliva samples from both affected and unaffected family members after obtaining approval from the institutional review board at the University of Texas Health Science Center at Houston. All participants provided informed consent. Gene variants identified by whole-exome sequencing were filtered for further analyses on the basis of the following criteria: (1) variants that altered amino acids in proteins, including nonsynonymous, stop-loss, stop-gain, coding indel, frameshift, or splice-site variants; (2) variants with a minor-allele frequency less than 0.5% in the Genome Aggregation Database (gnomAD); (3) variants that were heterozygous, homozygous, or compound heterozygous in affected family members; and (4) variants that segregated with thoracic aortic disease in families.
The significant genetic heterogeneity for HTAD complicates the identification of novel genes for this condition. Therefore, candidate genes for HTAD were also prioritized on the basis of tissue-specific expression. Using the data from the Genotype-Tissue Expression (GTEx) project, we identified genes that were highly expressed in the aorta but had relatively low expression in other tissues and assessed these genes for rare variants in the HTAD exome data. LTBP3 (GenBank: NM_001130144) (MIM: 602090) was identified through these analyses, and LTBP3 rare variants were identified in two HTAD probands: compound heterozygous variants, c.132delG (p.Pro45Argfs∗25) and c.2248G>T (p.Glu750∗), were present in family TAA909, and a homozygous insertion/deletion variant, c.2033_2041delinsCTT (p.Asn678_Gly681delinsThrCys), was present in family TAA376 (Figure 1). These variants are not in the gnomAD database. In this database, there are only 44 heterozygous loss-of-function (LoF) LTBP3 variants but no homozygous LoF variants. This gene has a high probability of being intolerant to LoF variants (pLI = 1.0).
Figure 1.
Identification of Homozygous LTBP3 Mutations in HTAD
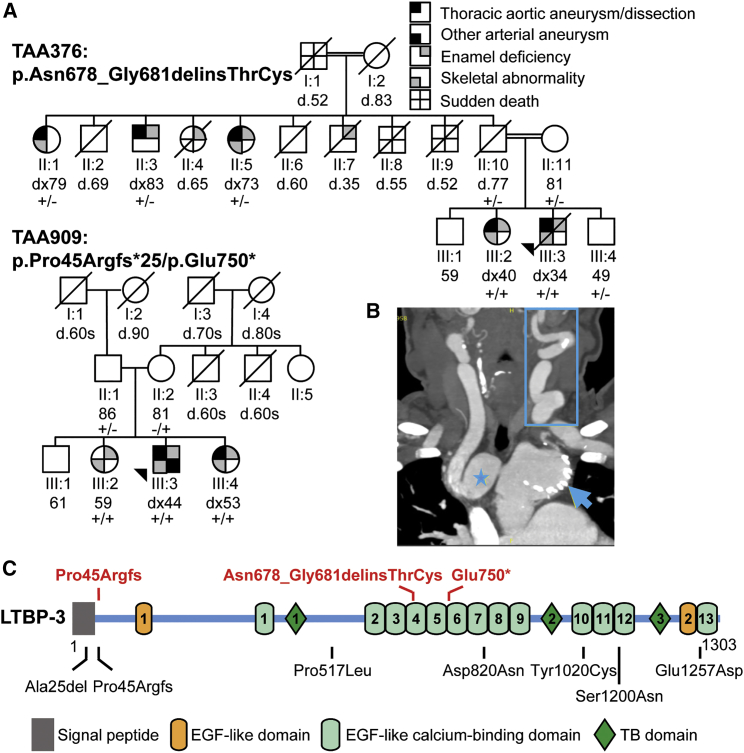
(A) Pedigrees of families showing segregation of LTBP3 variants with DASS and aortic disease. The age at diagnosis (dx) or death (d) in years is shown below the individual symbols. The legend shows the symbols representing the phenotypic features in the family members.
(B) A coronal computed tomography angiogram (CTA) of the neck and chest illustrates innominate artery aneurysm (star), aortic arch aneurysm with stent (arrowhead), and pronounced tortuosity of the left internal carotid artery (rectangle) in individual III:2 of family TAA376.
(C) Schematic diagram of the LTBP-3 domain structure and mutation. The pathogenic variants associated with HTAD are shown in red letters above the protein diagram, and the rare variants identified in cases with early-onset thoracic aortic dissection are shown in black letters below the protein diagram.
LTBP3 homozygous loss-of-function variants have been reported to cause dental abnormalities and short stature (DASS) in affected individuals between the ages of 2 and 28 years, but thoracic aortic disease has not been reported.15, 16, 17, 18 The proband of family TAA909 has DASS and also had multiple aortic and arterial aneurysms, including abdominal aortic aneurysm requiring surgical repair at the age of 44, aortic root dilation, and multiple visceral and peripheral arterial aneurysms at the age of 54 years (Table 1). His sisters inherited both LTBP3 variants and have DASS. Individual III:2 has no aortic abnormalities on echocardiogram and imaging of the abdominal aorta but has mild mitral valve prolapse and regurgitation. Individual III:4 has mild aortic root dilation (z-score = 2.1) and mild mitral valve prolapse. Their parents, both heterozygous for a LTBP3 variant, did not have DASS or aortic disease.
Table 1.
Clinical Characteristics of Individuals with LTBP3 Variants
| Family ID | Individual ID | Age/ Gendera | LTBP3 Genotype | Aortic Diseaseb | Other Cardiovascular Diseaseb | Other Clinical Featuresb | Height (cm) |
|---|---|---|---|---|---|---|---|
| TAA909 | III:3 | 54/M | +/+ | abdominal aortic aneurysm (44), aortic root dilation (54) | arterial aneurysms: axillary, iliac, hepatic, celiac axis (54) | amelogenesis imperfecta; cataracts | 160∗ |
| III:4 | 55/F | +/+ | mild aortic root dilation (53) | mild mitral valve prolapse (53) | amelogenesis imperfecta | 152∗ | |
| III:2 | 59/F | +/+ | none | mild mitral valve prolapse (54), mild mitral valve, tricuspid valve regurgitation | amelogenesis imperfecta; diffuse sclerosis of thoracic spine (58), osteoporosis, femur fracture (54) | 147∗ | |
| II:1 | 86/M | +/− | none | none | 163 | ||
| II:2 | 81/F | -/+ | none | none | 158 | ||
| TAA376 | III:3 | d.44/M | +/+ | type A dissection (34) | white-matter changes, small lacunar infarcts, remote injury in left cerebellum | tooth-enamel deficiency | 160∗ |
| III:2 | 58/F | +/+ | ascending aortic aneurysm (34), type A dissection (40), abdominal aortic disease (47) | pulmonary artery dilation (54), arterial aneurysms (innominate, common carotid; 58) cervical artery tortuosity, white-matter changes, mitral valve disease s/p repair (34), atrial fibrillation (58), mild pulmonary hypertension | tooth-enamel deficiency; scoliosis; osteopenia; inguinal hernia; diverticulosis | 150∗ | |
| III:4 | 50/M | +/− | none | none | none | 175 | |
| II:1 | 88/F | +/− | type B dissection (79), ascending aortic aneurysm (79) | congestive heart failure | 155 | ||
| II:3 | 84/M | +/− | aortic root and ascending aortic aneurysm (84) | atrial fibrillation (83); non-obstructive coronary artery disease | dental anomalies | 185 | |
| II:5 | 77/F | +/− | ascending aortic aneurysm (74) | non-obstructive coronary artery disease, mild pulmonary hypertension | dental anomalies | 160 | |
| II:10 | d.77/M | +/− | none | none | none | 173 | |
| II:11 | 81/F | +/− | none | none | none | 165 |
Symbols: (+/+) – homozygous or heterozygous, (+/−) – heterozygous, (∗) – less than the 5th percentile of height in the United States population of the same age group and gender.
Age at last follow-up or death (d.)
Age at diagnosis is shown in parenthesis.
The proband of family TAA376 was born to consanguineous parents belonging to a known inbred community. He had DASS and presented with a Stanford type A aortic dissection at the age of 34 years and underwent replacement of his ascending aorta and aortic valve. At the age of 40, he required re-operation of his ascending aorta graft and replacement of his aortic arch and descending aorta. His sister (III:2) also has the homozygous LTBP3 variant and DASS. She had an ascending aortic aneurysm at the age of 34 and Stanford type A aortic dissection at the age of 40, and she underwent composite graft of the aortic valve and ascending aorta. She further developed aortic arch, descending and abdominal aortic disease requiring replacement of her aortic arch, endovascular stent graft of her descending thoracic aorta, and open surgical repair of her abdominal aorta. She also has pulmonary artery dilation, other arterial aneurysms, cervical artery tortuosity (Figure 1B), and mitral valve disease, which required repair at the age of 34 years; additionally, she has scoliosis. A brother (III:4) is heterozygous for the LTBP3 variant and does not have DASS or aortic disease. Three paternal relatives with thoracic aortic aneurysm or dissection at older ages were heterozygous for the LTBP3 variant and had normal stature and variable dental features. Individual II:1 had a descending thoracic aortic dissection requiring endovascular repair at the age of 79, as well as an ascending aortic aneurysm. Individual II:3 underwent repair of an ascending aortic aneurysm at the age of 84 years; he also had dental abnormalities and was fitted for dentures in his 50s. Individual II:5 was diagnosed with a 4.4 cm ascending aortic aneurysm at the age of 74 years and required a full set of dentures at an early age. Three other paternal relatives of this generation died suddenly of unknown causes at 52, 55, and 65 years of age; one of these relatives was individual II:4, who had dental abnormalities and required dentures at a young age. Individual II:7 died at 35 years old from unrelated causes and had dental anomalies. The proband’s parents were of normal height, had no dental anomalies, and had normal echocardiograms in their 70s.
Two-point linkage analysis of thoracic aortic disease with the LTBP3 variants in pedigree TAA909 yielded a maximum LOD score of 0.6 (θ = 0) for a fully penetrant autosomal-recessive model. For pedigree TAA376, it was assumed that the variants could cause a milder disease under an autosomal-dominant model and a more severe disease phenotype under an autosomal-recessive model. Two-point linkage analysis produced a LOD score of 3.3 (θ = 0.). For both families, a disease and a variant allele frequency of 0.0001 were used. The combined LOD score for the two families is 3.9.
To further investigate whether LTBP3 rare variants predispose individuals to thoracic aortic dissections, we evaluated exome sequencing data of 338 individuals who had thoracic aortic dissections at < 56 years of age but no family history, syndromic features, or known HTAD gene mutation.19 Nine individuals had heterozygous LTBP3 rare variants, including a frameshift variant, coding deletion, nonsense variant, and five missense substitutions (Table S1). One of these individuals also had a pathogenic variant in COL3A1 (MIM: 120180). LTBP3 is structurally similar to FBN1 (MIM: 134797) in that both proteins have multiple EGF-like repeats and unique 8-cys domains.20 Heterozygous FBN1 missense mutations that disrupt an EGF-like domain, through either a deletion or an insertion of a cysteine or through alteration of amino acids important in calcium binding, are a common cause of MFS.21 Two LTBP3 rare variants in the individuals who had undergone dissections similarly disrupt EGF-like domains by inserting a cysteine (p.Tyr1020Cys) into domain 10 or altering an amino acid important for calcium binding (p.Glu1257Asp) in domain 13 (Figure 1C). The p.Tyr1020Cys and p.Glu1257Asp variants were found in a 52-year-old male with a type B aortic dissection and a 48-year-old male with type A aortic dissection, respectively. Interestingly, the homozygous LTBP3 indel variant in TAA376 also disrupts an EGF domain by inserting an additional cysteine (Figure 1C). An individual presenting with a type B aortic dissection at 50 years of age and mild dilation of the aortic arch is heterozygous for the same LTBP3 frameshift variant (p.Pro45Argfs∗25) identified in TAA909. None of these individuals with rare, heterozygous LTBP3 variants were reported to have DASS.
LTBP3 encodes latent TGF-β binding protein-3 (LTBP-3), which belongs to a family of proteins that regulate TGF-β activity by enabling its secretion, directing it to specific sites in the ECM, and participating in its activation.21 TGF-β is secreted from cells bound to a complex that includes its dimeric pro-peptide (termed latency-associated peptide, or LAP) and one of three LTBPs. LTBP-3 binds to all three TGF-β ligands, and the secretion of LTBP-3 from cells requires LTBP-3 to be complexed with TGF-β and LAP.22 Once secreted, LTBP-3, with LAP and inactive TGF-β, associates with fibrillin-1 in the ECM. Loss of fibrillin-1 in cells or aortic tissues diminishes LTBP-3 immunofluorescence in the ECM.23 Thus, loss of LTBP-3 would be predicted to decrease both TGF-β secretion from cells and the amount of LTBP-3 incorporated into fibrillin-1-containing microfibrils.
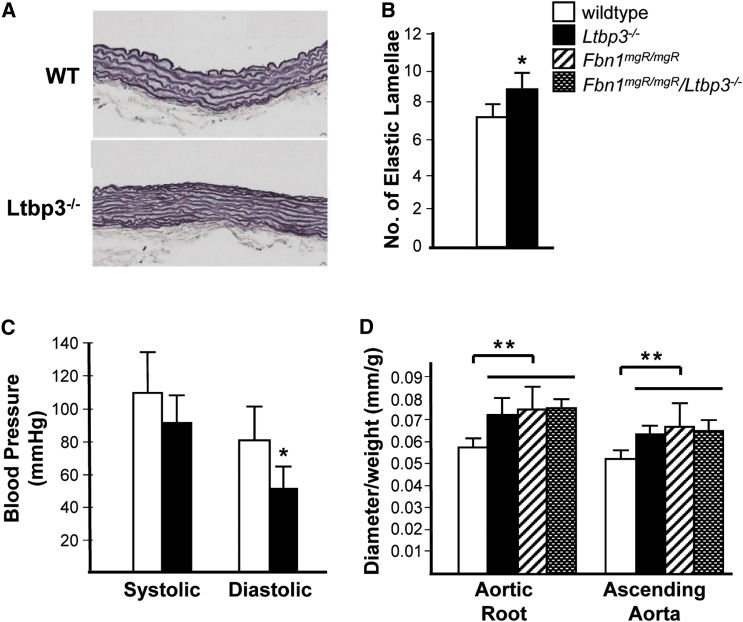
Ltbp3−/− mice have been extensively documented as being smaller than wild-type or heterozygous littermates and as having dental anomalies and defects in skeletal development; such defects include domed skulls, shortened maxilla, anteriorized foramen magnum, and kyphosis, abnormalities that can result from the reduction of TGF-β bioavailability in mice.24, 25 Neither aortic pathology nor aortic disease has been described in these mice. We investigated these mice for cardiovascular pathology and identified significantly increased elastic lamellar units in Ltbp3−/− ascending aortas compared to wild-type aortas, a finding that is also observed in another HTAD mouse model, Acta2−/− mice (Figures 2A and 2B).26, 27 Similar to the Acta2−/− mice, Ltbp3−/− mice had reduced diastolic blood pressures in comparison to those of wild-type mice (Figure 2C).26, 27 Previous studies reported that the diameters of the aortic root and ascending aorta of homozygous Ltbp3−/− mice were similar to those of wild-type mice.26 However, these studies did not correct for the diminished weight of the mutant mice compared to wild-type mice, a variable that is important to control for when one assesses aortic diameter in both humans and rodents.28, 29 When aortic diameters were normalized by body mass, the diameters of both the aortic root and the ascending aorta of Ltbp3−/− mice were significantly larger than those of wild-type mice (Figure 2D). It is notable that approximately 10% of Ltbp3−/− mice died of unknown causes within 30 days of birth.26
Figure 2.
Ltbp3−/− Mice Have Decreased Blood Pressure and Altered Aortic Pathology, Which Lead to Thoracic Aortic Dilatation
(A and B) The average number of elastic lamellae in mouse aortic walls was increased in Ltbp3−/− mice (n = 3) compared to wild-type mice (n = 4).
(C) Average systolic, diastolic, and mean blood pressures in wild-type (n = 6) and Ltbp3−/− (n = 5) mice. Data are presented as the mean ± SD for each mouse genotype.
(D) Average diameters of the aortic root and ascending aorta were measured by echocardiogram of 2-month-old wild-type (n = 8), Ltbp3−/− (n = 5), Fbn1mgR/mgR (n = 9), and Fbn1mgR/mgR/Ltbp3−/− (n = 5) mice; values were normalized to the weight of each mouse. Data are presented as the mean ± SD for each mouse genotype. ∗p < 0.05, ∗∗p < 0.005.
Loss of LTBP-3 has been reported to prevent thoracic aortic disease in the hypomorphic mouse model of MFS (Fbn1mgR/mgR). This mouse model has been used extensively for studying the role of TGF-β signaling in MFS.26 These mice develop impaired pulmonary alveolar septation associated with increased TGF-β signaling, and perinatal administration of a polyclonal TGF-β neutralizing antibody prevents the pulmonary defect.30, 31 In contrast, perinatal administration of a TGF-β neutralizing antibody exacerbated the thoracic aortic disease in the Fbn1mgR/mgR mice, leading to earlier deaths due to aortic rupture. Interestingly, later administration of the neutralizing antibody slightly delayed death in these mice but did not decrease aneurysm growth or medial degeneration. To investigate a role of LTBP-3 in MFS aortic disease, we crossed the Fbn1mgR/mgR mice with the Ltbp3−/− mice.26 Although initial analyses indicated that loss of LTBP-3 prevented aortic aneurysms in this mouse model, when the size of the aorta is corrected for the weight of the mouse, LTBP-3 deficiency does not alter aneurysm growth in Fbn1mgR/mgR mice (Figure 2D). Similar to late treatment with a polyclonal TGF-β neutralizing antibody, loss of LTBP-3 does prevent deaths due to aortic rupture in this mouse model.32 These data indicate that diminished TGF-β levels resulting from neutralizing antibodies or loss of LTBP-3 do not disrupt aneurysm growth in the Fbn1mgR/mgR mice but do decrease late aortic ruptures and deaths. Further studies will determine whether LTBP-3 loss alters TGF-β bioavailability, disrupts the ECM, or both.
In summary, the data presented here demonstrate that mutations in LTBP3 lead to a risk for autosomal-recessive thoracic aortic aneurysms and dissections and other arterial aneurysms, along with previously reported skeletal and dental defects. Individuals with heterozygous mutations in LTBP3 might also be at increased risk for later-onset thoracic aortic disease. All HTAD gene mutations identified thus far have been linked to autosomal-dominant disorders; the one exception is biglcan (BGN, MIM: 301870) mutations, which cause X-linked thoracic aortic disease. Ltbp3 loss in mice also leads to thoracic aortic aneurysms, but additional studies are needed if we are to determine whether the disease is due to LTBP-3′s role in directing TGF-β bioavailability, non-TGF-β-mediated ECM functions, or both.
Acknowledgments
This work was supported by the National Heart, Lung, and Blood Institute (RO1 HL62594 and P01HL110869-01 to D.M.M.), the John Ritter Foundation, NIH UL1 RR024148 (through the Clinical and Translational Science Awards program), the National Human Genome Research Institute (1U54HG006493 to D.A.N., M.J.B., and S.M.L.), and the National Heart, Lung and Blood Institute Resequencing and Genotyping Service (HHSN268201100037C). Marfan Foundation (D.B.R.).
Published: April 5, 2018
Footnotes
Supplemental Data include one table and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.002.
Web Resources
dbNSFP v.2.0 annotation, https://sites.google.com/site/jpopgen/dbNSFP
Depletion, http://cadd.gs.washington.edu/score
Ensembl Genome Browser, http://www.ensembl.org/index.html
GTEx database, https://gtexportal.org/home/
NCBI nucleotide, http://www.ncbi.nlm.nih.gov/nuccore
NHLBI Exome Sequencing Project, http://evs.gs.washington.edu/EVS/
OMIM, https://www.omim.org/
SeattleSeq Annotation 137, http://snp.gs.washington.edu/SeattleSeqAnnotation137/
Supplemental Data
References
- 1.Biddinger A., Rocklin M., Coselli J., Milewicz D.M. Familial thoracic aortic dilatations and dissections: a case control study. J. Vasc. Surg. 1997;25:506–511. doi: 10.1016/s0741-5214(97)70261-1. [DOI] [PubMed] [Google Scholar]
- 2.Albornoz G., Coady M.A., Roberts M., Davies R.R., Tranquilli M., Rizzo J.A., Elefteriades J.A. Familial thoracic aortic aneurysms and dissections--incidence, modes of inheritance, and phenotypic patterns. Ann. Thorac. Surg. 2006;82:1400–1405. doi: 10.1016/j.athoracsur.2006.04.098. [DOI] [PubMed] [Google Scholar]
- 3.Mizuguchi T., Collod-Beroud G., Akiyama T., Abifadel M., Harada N., Morisaki T., Allard D., Varret M., Claustres M., Morisaki H. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat. Genet. 2004;36:855–860. doi: 10.1038/ng1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu L., Vranckx R., Khau Van Kien P., Lalande A., Boisset N., Mathieu F., Wegman M., Glancy L., Gasc J.M., Brunotte F. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat. Genet. 2006;38:343–349. doi: 10.1038/ng1721. [DOI] [PubMed] [Google Scholar]
- 5.Guo D.C., Pannu H., Tran-Fadulu V., Papke C.L., Yu R.K., Avidan N., Bourgeois S., Estrera A.L., Safi H.J., Sparks E. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat. Genet. 2007;39:1488–1493. doi: 10.1038/ng.2007.6. [DOI] [PubMed] [Google Scholar]
- 6.Wang L., Guo D.C., Cao J., Gong L., Kamm K.E., Regalado E., Li L., Shete S., He W.Q., Zhu M.S. Mutations in myosin light chain kinase cause familial aortic dissections. Am. J. Hum. Genet. 2010;87:701–707. doi: 10.1016/j.ajhg.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo D.C., Regalado E., Casteel D.E., Santos-Cortez R.L., Gong L., Kim J.J., Dyack S., Horne S.G., Chang G., Jondeau G., GenTAC Registry Consortium. National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project Recurrent gain-of-function mutation in PRKG1 causes thoracic aortic aneurysms and acute aortic dissections. Am. J. Hum. Genet. 2013;93:398–404. doi: 10.1016/j.ajhg.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo D.C., Regalado E.S., Gong L., Duan X., Santos-Cortez R.L., Arnaud P., Ren Z., Cai B., Hostetler E.M., Moran R., University of Washington Center for Mendelian Genomics LOX Mutations Predispose to Thoracic Aortic Aneurysms and Dissections. Circ. Res. 2016;118:928–934. doi: 10.1161/CIRCRESAHA.115.307130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boileau C., Guo D.C., Hanna N., Regalado E.S., Detaint D., Gong L., Varret M., Prakash S.K., Li A.H., d’Indy H., National Heart, Lung, and Blood Institute (NHLBI) Go Exome Sequencing Project TGFB2 mutations cause familial thoracic aortic aneurysms and dissections associated with mild systemic features of Marfan syndrome. Nat. Genet. 2012;44:916–921. doi: 10.1038/ng.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbier M., Gross M.S., Aubart M., Hanna N., Kessler K., Guo D.C., Tosolini L., Ho-Tin-Noe B., Regalado E., Varret M. MFAP5 loss-of-function mutations underscore the involvement of matrix alteration in the pathogenesis of familial thoracic aortic aneurysms and dissections. Am. J. Hum. Genet. 2014;95:736–743. doi: 10.1016/j.ajhg.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van de Laar I.M., Oldenburg R.A., Pals G., Roos-Hesselink J.W., de Graaf B.M., Verhagen J.M., Hoedemaekers Y.M., Willemsen R., Severijnen L.A., Venselaar H. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat. Genet. 2011;43:121–126. doi: 10.1038/ng.744. [DOI] [PubMed] [Google Scholar]
- 12.Regalado E.S., Guo D.C., Villamizar C., Avidan N., Gilchrist D., McGillivray B., Clarke L., Bernier F., Santos-Cortez R.L., Leal S.M., NHLBI GO Exome Sequencing Project Exome sequencing identifies SMAD3 mutations as a cause of familial thoracic aortic aneurysm and dissection with intracranial and other arterial aneurysms. Circ. Res. 2011;109:680–686. doi: 10.1161/CIRCRESAHA.111.248161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inamoto S., Kwartler C.S., Lafont A.L., Liang Y.Y., Fadulu V.T., Duraisamy S., Willing M., Estrera A., Safi H., Hannibal M.C. TGFBR2 mutations alter smooth muscle cell phenotype and predispose to thoracic aortic aneurysms and dissections. Cardiovasc. Res. 2010;88:520–529. doi: 10.1093/cvr/cvq230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horbelt D., Guo G., Robinson P.N., Knaus P. Quantitative analysis of TGFBR2 mutations in Marfan-syndrome-related disorders suggests a correlation between phenotypic severity and Smad signaling activity. J. Cell Sci. 2010;123:4340–4350. doi: 10.1242/jcs.074773. [DOI] [PubMed] [Google Scholar]
- 15.Noor A., Windpassinger C., Vitcu I., Orlic M., Rafiq M.A., Khalid M., Malik M.N., Ayub M., Alman B., Vincent J.B. Oligodontia is caused by mutation in LTBP3, the gene encoding latent TGF-beta binding protein 3. Am. J. Hum. Genet. 2009;84:519–523. doi: 10.1016/j.ajhg.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dugan S.L., Temme R.T., Olson R.A., Mikhailov A., Law R., Mahmood H., Noor A., Vincent J.B. New recessive truncating mutation in LTBP3 in a family with oligodontia, short stature, and mitral valve prolapse. Am. J. Med. Genet. A. 2015;167:1396–1399. doi: 10.1002/ajmg.a.37049. [DOI] [PubMed] [Google Scholar]
- 17.Huckert M., Stoetzel C., Morkmued S., Laugel-Haushalter V., Geoffroy V., Muller J., Clauss F., Prasad M.K., Obry F., Raymond J.L. Mutations in the latent TGF-beta binding protein 3 (LTBP3) gene cause brachyolmia with amelogenesis imperfecta. Hum. Mol. Genet. 2015;24:3038–3049. doi: 10.1093/hmg/ddv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McInerney-Leo A.M., Le Goff C., Leo P.J., Kenna T.J., Keith P., Harris J.E., Steer R., Bole-Feysot C., Nitschke P., Kielty C. Mutations in LTBP3 cause acromicric dysplasia and geleophysic dysplasia. J. Med. Genet. 2016;53:457–464. doi: 10.1136/jmedgenet-2015-103647. [DOI] [PubMed] [Google Scholar]
- 19.Guo D.C., Hostetler E.M., Fan Y., Kulmacz R.J., Zhang D., Nickerson D.A., Leal S.M., LeMaire S.A., Regalado E.S., Milewicz D.M., GenTAC Investigators Heritable thoracic aortic disease genes in sporadic aortic dissection. J. Am. Coll. Cardiol. 2017;70:2728–2730. doi: 10.1016/j.jacc.2017.09.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson I., Jensen S., Handford P. TB domain proteins: evolutionary insights into the multifaceted roles of fibrillins and LTBPs. Biochem. J. 2011;433:263–276. doi: 10.1042/BJ20101320. [DOI] [PubMed] [Google Scholar]
- 21.Robertson I.B., Horiguchi M., Zilberberg L., Dabovic B., Hadjiolova K., Rifkin D.B. Latent TGF-β-binding proteins. Matrix Biol. 2015;47:44–53. doi: 10.1016/j.matbio.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y., Dabovic B., Annes J.P., Rifkin D.B. Latent TGF-beta binding protein-3 (LTBP-3) requires binding to TGF-beta for secretion. FEBS Lett. 2002;517:277–280. doi: 10.1016/s0014-5793(02)02648-0. [DOI] [PubMed] [Google Scholar]
- 23.Zilberberg L., Todorovic V., Dabovic B., Horiguchi M., Couroussé T., Sakai L.Y., Rifkin D.B. Specificity of latent TGF-β binding protein (LTBP) incorporation into matrix: role of fibrillins and fibronectin. J. Cell. Physiol. 2012;227:3828–3836. doi: 10.1002/jcp.24094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dabovic B., Chen Y., Colarossi C., Obata H., Zambuto L., Perle M.A., Rifkin D.B. Bone abnormalities in latent TGF-[beta] binding protein (Ltbp)-3-null mice indicate a role for Ltbp-3 in modulating TGF-[beta] bioavailability. J. Cell Biol. 2002;156:227–232. doi: 10.1083/jcb.200111080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morkmued S., Hemmerle J., Mathieu E., Laugel-Haushalter V., Dabovic B., Rifkin D.B., Dollé P., Niederreither K., Bloch-Zupan A. Enamel and dental anomalies in latent-transforming growth factor beta-binding protein 3 mutant mice. Eur. J. Oral Sci. 2017;125:8–17. doi: 10.1111/eos.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zilberberg L., Phoon C.K., Robertson I., Dabovic B., Ramirez F., Rifkin D.B. Genetic analysis of the contribution of LTBP-3 to thoracic aneurysm in Marfan syndrome. Proc. Natl. Acad. Sci. USA. 2015;112:14012–14017. doi: 10.1073/pnas.1507652112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J., Peters A., Papke C.L., Villamizar C., Ringuette L.J., Cao J., Wang S., Ma S., Gong L., Byanova K.L. Loss of smooth muscle α-actin leads to NF-κB-dependent increased sensitivity to angiotensin II in smooth muscle cells and aortic enlargement. Circ. Res. 2017;120:1903–1915. doi: 10.1161/CIRCRESAHA.117.310563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiratzka L.F., Bakris G.L., Beckman J.A., Bersin R.M., Carr V.F., Casey D.E., Jr., Eagle K.A., Hermann L.K., Isselbacher E.M., Kazerooni E.A., American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. American Association for Thoracic Surgery. American College of Radiology. American Stroke Association. Society of Cardiovascular Anesthesiologists. Society for Cardiovascular Angiography and Interventions. Society of Interventional Radiology. Society of Thoracic Surgeons. Society for Vascular Medicine 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with thoracic aortic disease. A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology,American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons,and Society for Vascular Medicine. J. Am. Coll. Cardiol. 2010;55:e27–e129. doi: 10.1016/j.jacc.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 29.Knipp B.S., Ailawadi G., Sullivan V.V., Roelofs K.J., Henke P.K., Stanley J.C., Upchurch G.R., Jr. Ultrasound measurement of aortic diameters in rodent models of aneurysm disease. J. Surg. Res. 2003;112:97–101. doi: 10.1016/s0022-4804(03)00114-8. [DOI] [PubMed] [Google Scholar]
- 30.Nistala H., Lee-Arteaga S., Carta L., Cook J.R., Smaldone S., Siciliano G., Rifkin A.N., Dietz H.C., Rifkin D.B., Ramirez F. Differential effects of alendronate and losartan therapy on osteopenia and aortic aneurysm in mice with severe Marfan syndrome. Hum. Mol. Genet. 2010;19:4790–4798. doi: 10.1093/hmg/ddq409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neptune E.R., Frischmeyer P.A., Arking D.E., Myers L., Bunton T.E., Gayraud B., Ramirez F., Sakai L.Y., Dietz H.C. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat. Genet. 2003;33:407–411. doi: 10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 32.Cohn R.D., van Erp C., Habashi J.P., Soleimani A.A., Klein E.C., Lisi M.T., Gamradt M., ap Rhys C.M., Holm T.M., Loeys B.L. Angiotensin II type 1 receptor blockade attenuates TGF-beta-induced failure of muscle regeneration in multiple myopathic states. Nat. Med. 2007;13:204–210. doi: 10.1038/nm1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.