Abstract
The multiple morphological abnormalities of the flagella (MMAF) phenotype is among the most severe forms of sperm defects responsible for male infertility. The phenotype is characterized by the presence in the ejaculate of immotile spermatozoa with severe flagellar abnormalities including flagella being short, coiled, absent, and of irregular caliber. Recent studies have demonstrated that MMAF is genetically heterogeneous, and genes thus far associated with MMAF account for only one-third of cases. Here we report the identification of homozygous truncating mutations (one stop-gain and one splicing variant) in CFAP69 of two unrelated individuals by whole-exome sequencing of a cohort of 78 infertile men with MMAF. CFAP69 encodes an evolutionarily conserved protein found at high levels in the testis. Immunostaining experiments in sperm from fertile control individuals showed that CFAP69 localized to the midpiece of the flagellum, and the absence of CFAP69 was confirmed in both individuals carrying CFPA69 mutations. Additionally, we found that sperm from a Cfap69 knockout mouse model recapitulated the MMAF phenotype. Ultrastructural analysis of testicular sperm from the knockout mice showed severe disruption of flagellum structure, but histological analysis of testes from these mice revealed the presence of all stages of the seminiferous epithelium, indicating that the overall progression of spermatogenesis is preserved and that the sperm defects likely arise during spermiogenesis. Together, our data indicate that CFAP69 is necessary for flagellum assembly/stability and that in both humans and mice, biallelic truncating mutations in CFAP69 cause autosomal-recessive MMAF and primary male infertility.
Keywords: male infertility, infertility genetics, MMAF, CFAP69, sperm flagellum, asthenozoospermia, teratozoospermia, whole-exome sequencing, KO mouse model, manchette
Introduction
Human male infertility remains a persistent problem, affecting an estimated 15% of couples.1 Male infertility often manifests as decreased sperm count (oligozoospermia), decreased sperm motility (asthenozoospermia), a higher proportion of morphologically defective sperm in the ejaculate (teratozoospermia), or a combination of the above defects. An estimated 30%–50% of male infertility cases have a genetic component.2 Thus, considerable effort has been made to identify and characterize the large number of genes required for sperm development. The advent of high-throughput sequencing (HTS) technologies has greatly facilitated the identification in infertile males of potentially responsible genes, but detailed studies of the functions of these genes in spermatogenesis is generally not possible in humans, requiring animal models instead.3, 4 Indeed, many mutant mouse models with defective spermatogenesis have been characterized, and molecular and genetic studies of sperm development in these models have yielded a deeper understanding of spermatogenesis.5
Multiple morphological abnormalities of the flagella (MMAF) phenotype is one of the most severe forms of qualitative sperm defects responsible for male infertility.6 This peculiar phenotype is characterized by the presence in the ejaculate of immotile spermatozoa presenting with several severe abnormalities of the sperm flagellum including being short, coiled, absent, and of irregular caliber.7 Whole-exome sequencing (WES) analysis revealed that mutations in DNAH1 (MIM: 603332), CFAP43 (MIM: 617558), and CFAP44 (MIM: 617559) are frequently found in MMAF-affected individuals and account for about one-third of MMAF cases.8, 9, 10, 11 These results established a strong genetic component for MMAF. They also demonstrated that MMAF is genetically heterogeneous and that other relevant genes still await identification. In a large cohort of 78 infertile male individuals with MMAF, we have now identified two unrelated individuals harboring homozygous truncating mutations in CFAP69 in addition to identifying individuals with mutations in the known MMAF genes, indicating that this gene is likely to be important for sperm flagellum morphogenesis and male fertility. In parallel, we characterized a Cfap69 knockout mouse model, which we found to recapitulate the MMAF phenotype. Overall, our work demonstrates that CFAP69 is required for sperm flagellum assembly/stability and that truncating mutations of CFAP69 induce autosomal-recessive MMAF and primary male infertility.
Subjects and Methods
Human Case and Control Subjects
WES was performed for a large cohort of 78 MMAF-affected individuals as previously described.12 All individuals presented with a typical MMAF phenotype characterized by severe asthenozoospermia (total sperm motility below 10%) with >5% of sperm having at least three flagellar abnormalities (absent, short, coiled, bent, or irregular flagella).
Individual CFAP69_1 is from Iran and was treated in Tehran at the Royan Institute for primary infertility from 2008 to 2015. Individual CFAP69_2 is from North Africa and consulted for primary infertility at the Cochin University Hospital in Paris (France) from 2015 to the present. Both individuals were born to first-cousin parents, had normal somatic karyotypes (46,XY), and were negative for Y chromosome Azoospermia Factor (AZF) microdeletions.
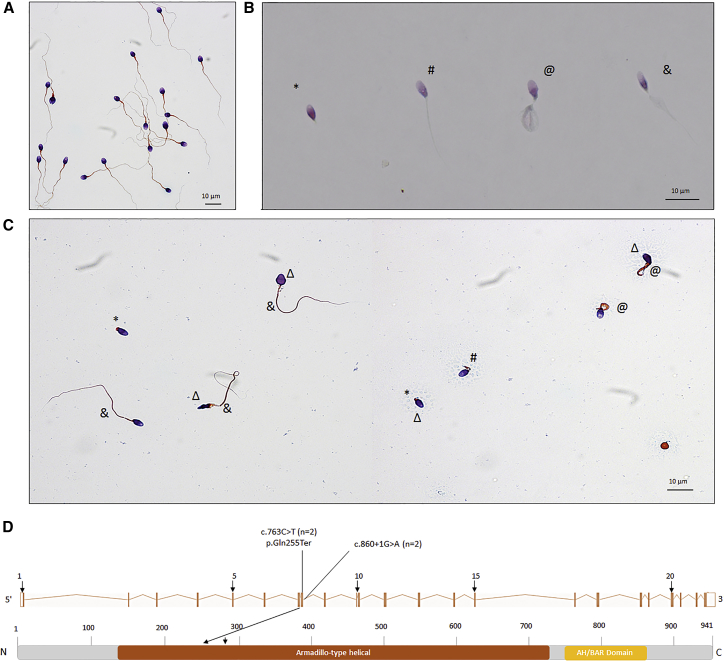
Sperm analysis was carried out in the source laboratories during routine biological examination of the individuals according to World Health Organization (WHO) guidelines.13 The morphology of the individuals’ sperm was assessed with Papanicolaou staining (Figures 1A–1C). The detailed semen parameters of the two CFAP69 individuals are presented in Table 1, and the average semen parameters of the studied MMAF cohort according to genotype are described in Table S1.
Figure 1.
Morphology of Normal and CFAP69 Mutant Spermatozoa, and the Mutations Identified in CFAP69-Mutant Individuals
(A–C) Morphology of spermatozoa from fertile control subjects (A), individual CFAP69_1 (B), and individual CFAP69_2 (C). Most spermatozoa from CFAP69 individuals have flagella that are short (#), absent (∗), of irregular caliber (&), or coiled (@). Head malformations were also observed (Δ).
(D) Location and nature of CFAP69 mutations in CFAP69 and protein. Colored squares stand for Armadillo-like helical (orange) repeats and AH/BAR domain (yellow) as predicted by InterPro server. Mutations are annotated in accordance to the HGVS’s recommendations. n = number of alleles.
Table 1.
Detailed Semen Parameters in the Two MMAF-Affected Individuals Harboring a CFAP69 Mutation
|
CFAP69-Mutated Individuals |
Semen Parameters |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Individuals | Age (years) | CFAP69 Mutation | Sperm Volume (mL) | Sperm Concentration (106/mL) | Total Motility 1 hr | Vitality | Normal Spermatozoa | Absent Flagella | Short Flagella | Coiled Flagella | Bent Flagella | Flagella of Irregular Caliber | Tapered Head | Thin Head | Micro-cephalic | Macro-cephalic | Multiple Heads | Abnormal Base | Abnormal Acrosomal Region |
| CFAP69_1 | 42 | c.860+1G>A | 5 | 4 | 1 | 13 | 1 | 1 | 79 | 1 | 0 | 1 | 20 | 14 | 0 | 0 | 0 | 15 | 64 |
| CFAP69_2 | 51 | c.763C>T | 4 | 6 | 10 | 62 | 12 | 12 | 13 | 7 | 5 | 7 | 1 | 37 | 1 | 0 | 0 | 27 | 70 |
| Reference Limitsa | 1.5 (1.4–1.7) | 15 (12–16) | 40 (38–42) | 58 (55–63) | 23 (20–26) | 5 (4–6) | 1 (0–2) | 17 (15–19) | 13 (11–15) | 2 (1–3) | 3 (2–4) | 14 (12–16) | 7 (5–9) | 1 (0–2) | 2 (1–3) | 42 (39–45) | 60 (57–63) | ||
Informed consent was obtained from all the subjects participating in the study according to local protocols and the principles of the Declaration of Helsinki. Sperm samples were obtained following informed consent from both individuals CFAP69_1 and CFAP69_2. The study was approved by local ethics committees, and the samples were then stored in the CRB Germetheque (certification under ISO-9001 and NF-S 96-900) following a standardized procedure. Sperm samples from fertile individuals with normal spermograms were obtained from CRB Germetheque. Consent for CRB storage was approved by the CPCP Sud-Ouest of Toulouse (coordinator of the multi-site CRB Germetheque).
Whole-Exome Sequencing (WES) and Bioinformatic Analysis
WES and bioinformatic analysis were performed according to our previously described protocol.12 For details, see Supplemental Material and Methods.
Sanger Sequencing
CFAP69 mutations identified by WES were validated by Sanger sequencing. PCR primers and protocols used for each individual are listed in the Table S2. Sequencing reactions were carried out with BigDye Terminator v3.1 (Applied Biosystems). Sequencing was carried out on ABI 3130XL (Applied Biosystems). Sequences were analyzed using SeqScape software (Applied Biosystems).
Quantitative Real-Time RT-PCR (RT-qPCR) Analysis
RT-qPCR was performed with cDNAs from various human tissues purchased from Life Technologies. A panel of seven organs was used for experiments: testis, brain, lung, kidney, liver, intestine, and heart. Each sample was assayed in triplicate for each gene on a StepOnePlus (LifeTechnologies) with Power SYBR Green PCR Master Mix (Life Technologies). The PCR cycle was as follows: 10 min at 95°C, 1 cycle for enzyme activation; 15 s at 95°C, 60 s at 60°C with fluorescence acquisition, 40 cycles for the PCR. RT-qPCR data were normalized using the reference housekeeping gene ACTB for human with the -ΔΔCt method.14 The 2-ΔΔCt value was set at 0 in brain cells, resulting in an arbitrary expression of 1. Primer sequences and RT-qPCR conditions are indicated in Table S3. The efficacy of primers was checked using a standard curve. Melting curve analysis was used to confirm the presence of a single PCR product. Statistics were performed using a two-tailed t test on Prism 4.0 software (GraphPad) to compare the relative expression of CFAP69 transcripts in several organs. Statistical tests with a two-tailed p value ≤ 0.05 were considered significant.
RT-PCR Analysis
Total RNA from whole blood from individual CFAP69_1 was extracted using the mirVana PARIS Kit (Life Technologies) according to the manufacturer’s protocol. Reverse transcription was carried out for individual CFAP69_1 and two healthy control subjects (C1 and C2) with 5 μL of extracted RNA (approximately 500 ng). Hybridization of the oligo-dT was performed by incubation for 5 min at 65°C and quenching on ice in the following mix: 5 μL of RNA, 3 μL of poly T oligo primers (dT)12–18 (10 mM, Pharmacia), 3 μL of the four dNTPs (0.5 mM, Roche Diagnostics), and 2.2 μL of H2O. Reverse transcription was then carried out for 30 min at 55°C after the addition of 4 μL of 5× buffer, 0.5 μL RNase inhibitor, and 0.5 μL of Transcriptor Reverse transcriptase (Roche Diagnostics). 2 μL of the obtained cDNA mix was used for subsequent PCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a housekeeping gene (internal control). CFAP69 primer sequences and RT-PCR conditions are indicated in the Table S4.
Immunostaining in Human Sperm Cells
Immunofluorescence (IF) experiments were performed using sperm cells from control individuals and from one or both individuals carrying CFAP69 mutations. Sperm cells were fixed in phosphate-buffered saline (PBS)/4% paraformaldehyde for 1 min at room temperature. After washing in 1 mL PBS, the sperm suspension was spotted onto 0.1% poly L-lysine pre-coated slides (Thermo Scientific). After attachment, sperm were permeabilized with 0.1% (v/v) Triton X-100 –DPBS (Triton X-100; Sigma-Aldrich) for 5 min at RT. Slides were then blocked in 5% normal serum-DPBS (normal goat or donkey serum; GIBCO, Invitrogen) and incubated overnight at 4°C with the following primary antibodies: rabbit polyclonal anti-CFAP69 (ab171156, Abcam, 1:200), mouse monoclonal anti-Hsp60 (ab13532, Abcam, 1:500), rabbit polyclonal anti-SPAG6 (HPA038440, Sigma-Aldrich, 1:500), rabbit polyclonal anti-SPEF2 (HPA040343, Sigma-Aldrich, 1:500), rabbit polyclonal anti-DNAH5 (HPA037470, Sigma-Aldrich, 1:100), rabbit polyclonal anti-DNALI1 (HPA028305, Sigma-Aldrich, 1:100), and monoclonal mouse anti-acetylated-α-tubulin (T7451, Sigma-Aldrich, 1:2,000). Washes were performed with 0.1% (v/v) Tween 20-DPBS, followed by 1 hr incubation at room temperature with secondary antibodies. Highly cross-adsorbed secondary antibodies (Dylight 488 and Dylight 549, 1:1,000) were from Jackson Immunoresearch. Appropriate controls were performed, omitting the primary antibodies. Samples were counterstained with 5 mg/mL Hoechst 33342 (Sigma-Aldrich) and mounted with DAKO mounting media (Life Technologies). Fluorescence images were captured with a confocal microscope (Zeiss LSM 710).
Animals
For all experiments involving mice, animals were handled and euthanized in accordance with methods approved by the Animal Care and Use Committees of each applicable institution. All mice used were adult (6 weeks or older) male mice.
Generation of Cfap69tm1b/tm1b Mice
We obtained Cfap69tm1a mice as previously described.15 The Cfap69tm1a mouse line carries a Cfap69 knockout first allele,16 in which a promoterless, gene-trapping cassette including the LacZ and neo genes was inserted in introns 4–5 of Cfap69, resulting in a floxed exon 5. To generate the Cfap69tm1b allele, in which exon 5 and neo are excised, resulting in truncation of CFAP69, but LacZ remains as a reporter, Cfap69tm1a mice were crossed with the E2a-cre mouse line (B6.FVB-Tg(EIIa-cre)C5379Lmgd/J, The Jackson Laboratory), which has early embryonic expression of cre recombinase leading to ubiquitous recombination. Genotyping was performed with two primer pairs: for the Cfap69tm1b allele, CTCCAGTGAAAGCCCACCT (forward) and CGGTCGCTACCATTACCAGT (reverse) with an expected product size of ∼450 bp; for the WT Cfap69 allele, ATGACCTAGGATTCATAAGCTTGATCT (forward) and GCGCTGCAACTGGAATCAGA (reverse) with an expected product size of ∼300 bp.
X-gal Staining and Imaging
To collect tissue, mice were deeply anesthetized with Avertin (2,2,2-tribromoethyl alcohol in 2-methyl-2-butanol) and then perfused transcardially first with PBS (pH 7.4) followed by 4% paraformaldehyde (PFA) in PBS. Testes and epididymides were dissected out, and the tunica albigunea of the testis was punctured several times with a needle at each pole. Tissues were then immersed in 4% PFA for 2 hr at 4°C. For whole-mount X-gal staining, testes and epididymides were washed in PBS and directly immersed in X-gal staining solution (1 mg/mL X-gal, 2 mM MgCl2, 0.01% sodium deoxycholate, 0.02% NP-40, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide) for 48 hr at 37°C. To prepare tissue cryosections, testes and epididymides were washed in PBS before being cryoprotected in 30% (w/v) sucrose in PBS for 48 hr. The tissues were then embedded in Tissue-Tek O.C.T. (Sakura, 4583) and cut into 12 μM sections, which were adhered to glass slides. Sections were fixed for 5 min in 4% PFA at room temperature, washed in PBS, and immersed in X-gal staining solution for 24 hr at 37°C. Sections were then fixed in 4% PFA at room temperature for 10 min, washed in PBS, and blocked in 5% normal goat serum and 0.1% Triton X-100 in PBS for 1 hr at room temperature. Alexa Fluor 488-conjugated peanut agglutinin (PNA) (Thermo Fisher Scientific, L32458) (1:200) was diluted in blocking solution and applied to sections overnight at 4°C. Sections were washed in PBS with 0.1% Tween-20, counterstained in DAPI (1 μg/mL in PBS) for 5 min, and mounted in Fluoromount (Sigma-Aldrich, F4680).
Fluorescent X-gal imaging using confocal microscopy was performed as previously described.17 Imaging was performed on a Zeiss LSM 700 confocal microscope. X-gal inclusions were excited with a 637 nm laser, and emitted fluorescent signal between 650 and 770 nm was recorded. Transmitted light signal was also recorded. PNA and DAPI were simultaneously imaged using standard methods. To produce the final fluorescent image of X-gal staining, the transmitted light image was subtracted from the emitted light image using the “Image Calculator” function in ImageJ.
Western Blotting
Whole testes were homogenized in 2× Laemmli buffer (65.8 mM Tris-HCl, 26.3% glycerol, 2.1% SDS, 0.01% bromophenol blue, 10% 2-mercaptoethanol [pH 6.8]) using a dounce homogenizer and then heated at 95°C for 10 min. Lysates were fractionated by SDS-PAGE on 8% polyacrylamide gels and transferred to polyvinylidene fluoride membranes by wet transfer in Towbin Buffer without methanol (25 mM Tris base, 192 mM glycine, 0.025% SDS). Membranes were blocked in 3% BSA in tris-buffered saline with Tween-20 (TBST) for 1 hr at room temperature. Custom anti-CFAP69 antibody15 was diluted 1:1,000 in blocking solution and incubated with the membrane overnight at 4°C. The membrane was then washed in TBST and incubated with HRP-conjugated anti-Rabbit IgG antibody (GE Healthcare, NA934) at a 1:2,000 dilution in blocking solution for 1 hr at room temperature. After washing, Western Lightning Plus-ECL reagent (PerkinElmer NEL103001) was applied to the membrane according to manufacturer instructions. The membranes were then exposed to autoradiography film.
Mouse Epididymal Sperm Collection
Male mice were deeply anesthetized with Avertin. To collect sperm, the cauda epididymides were dissected out and minced in PBS. The tissue was incubated at 37°C for 30 min before the sperm-containing supernatant was collected.
Scanning Electron Microscopy
Sperm were deposited on poly-L-lysine coated coverslips, fixed in 2.5% glutaraldehyde, and post-fixed in osmium tetroxide. Coverslips were washed in distilled water and dehydrated through cold 50%, 70%, 95%, and 100% ethanol. Coverslips were then dried at critical point in a Tousimis Autosamdri-810 Critical Point Dryer, mounted onto specimen stubs, and sputter-coated with palladium before being viewed with a FEI Quanta ESEM 200 scanning electron microscope.
Transmission Electron Microscopy
Tissue was prepared for transmission electron microscopy (TEM) according to standard protocols. Briefly, mice were deeply anesthetized with Avertin and perfused transcardially with PBS and then 5% glutaraldehyde (in 0.1 M phosphate buffer [pH 7.4]). Testes were dissected out and cut into pieces no more than 2 mm thick in any dimension before being immersed in 5% glutaraldehyde overnight at 4°C. Samples were then washed in phosphate buffer, post-fixed in osmium tetroxide for 1 hr at 4°C, washed again, and dehydrated through 1 change each of cold 50%, 70%, 95% ethanol, and 3 changes of 100% ethanol for 15 min each. Tissue fragments were then immersed in 2 changes of propylene oxide, 15 min each, before being placed in a 1:1 propyelene oxide:EMbed812 (Electron Microscopy Sciences, 14120) mixture overnight under vacuum. Samples were further infiltrated with EMbed812 for 6 hr under vacuum before being placed in fresh EMbed812. Blocks were polymerized at 60°C for 48 hr. Pale gold thin sections were cut, collected on copper grids, and stained with uranyl acetate and lead citrate. Images were obtained using a Phillips EM 420 transmission electron microscope.
Histology
Tissue was prepared the same way as for TEM analysis. Semi-thin sections 1–2 μM in thickness were cut from EMbed812-embedded testis blocks with glass knives and placed on drops of water on glass slides. The slides were then placed on a hot plate at low heat for about 30 s to dry the water and adhere the sections to the slides. The sections were then covered in a 1% solution of toluidine blue in 2% sodium tetraborate for an additional 30 s. The slides were then removed from the hot plate, rinsed in distilled water, and mounted in EMbed812.
Results
Whole-Exome Sequencing (WES) Identifies Homozygous Truncating Mutations in CFAP69 in MMAF Individuals
In the cohort of 78 MMAF-affected individuals we analyzed, 22 individuals were identified with harmful mutations in the known MMAF-related genes DNAH1, CFAP43, or CFAP44.12 After applying stringent filters and reanalysis of the remaining exomes, we identified two individuals with truncating mutations in CFAP69, which accounts for 2.5% of our cohort (Figure 1D, Table S5). CFAP69 was reported in public databases to be strongly expressed in the testis and connected with cilia or the flagellum.19 We confirmed these data by RT-qPCR experiments in human tissue panels, which indicate that expression of CFAP69 mRNA in testis is predominant and is significantly higher than in other tested tissues (Figure S1). For both individuals, no variants with low frequency in control databases were identified in other genes reported to be associated with cilia, flagella, or male fertility. We therefore focused on CFAP69, which appeared to be an excellent MMAF candidate.
CFAP69 (formerly known as c7orf63; GenBank: NM_001039706) is located on chromosome 7 and contains 23 exons encoding a predicted 941-amino acid protein (A5D8W1). The two CFAP69 variants were found in two unrelated individuals. These two variants were absent from control sequence databases (dbSNP, 1000 Genomes Project, NHLBI Exome Variant Server, gnomAD, and in-house database). The variant identified in individual CFAP69_1 is a splicing variant c.860+1G>A, altering a consensus splice donor site of CFAP69 exon 8. The variant identified in individual CFAP69_2 was a stop-gain mutation c.763C>T (p.Gln255Ter) located in exon 8 (Figure 1D, Table S5). The presence of the two variants was confirmed by Sanger sequencing of CFAP69 exon 8 in both individuals (Figure S2). According to the Human Splicing Finder, the c.860+1G>A mutation abrogates the consensus donor site, leading to altered splicing and subsequent frameshift. No Cfap69 mRNA could be detected by RT-PCR in individual CFAP69_1 (Figure S3), possibly indicating a frameshift and premature translation termination leading to nonsense-mediated decay.
We compared semen parameters of individuals carrying CFAP69 mutations with those of individuals carrying mutations in previously described MMAF-related genes in order to test for potential phenotype-genotype correlation (Table S5). Although there were no significant statistical differences between the semen parameters of the CFAP69 individuals and MMAF-affected individuals with mutations in DNAH1, CFAP43, or CFAP44, we observe very low sperm concentrations and total sperm counts in the ejaculates from both CFAP69-mutant individuals. Compared to sperm from control samples (Figure 1A), sperm from both individuals showed severe defects characteristic of MMAF (Figures 1B and 1C). A high rate of head malformations, in particular thin heads and an abnormal acrosomal region, were also observed (Figure 1C, Tables 1 and S1). Interestingly, when selecting sperm for intracytoplasmic sperm injection by density gradient centrifugation from individual CFAP69_2, numerous isolated sperm heads were recovered, suggesting a fragility of the head-flagellum connection.
CFAP69 Is Located in the Midpiece of the Human Sperm Flagellum, and Its Absence Is Associated with Axonemal Defects
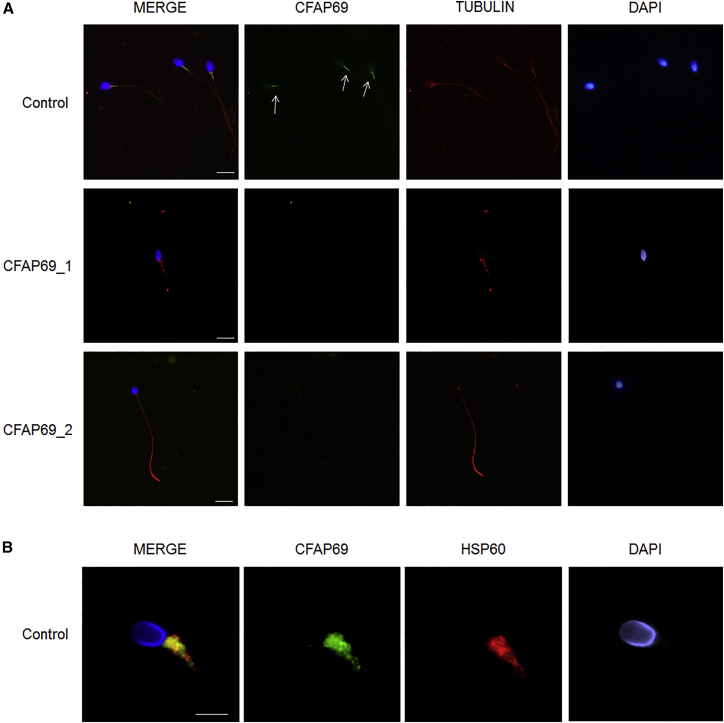
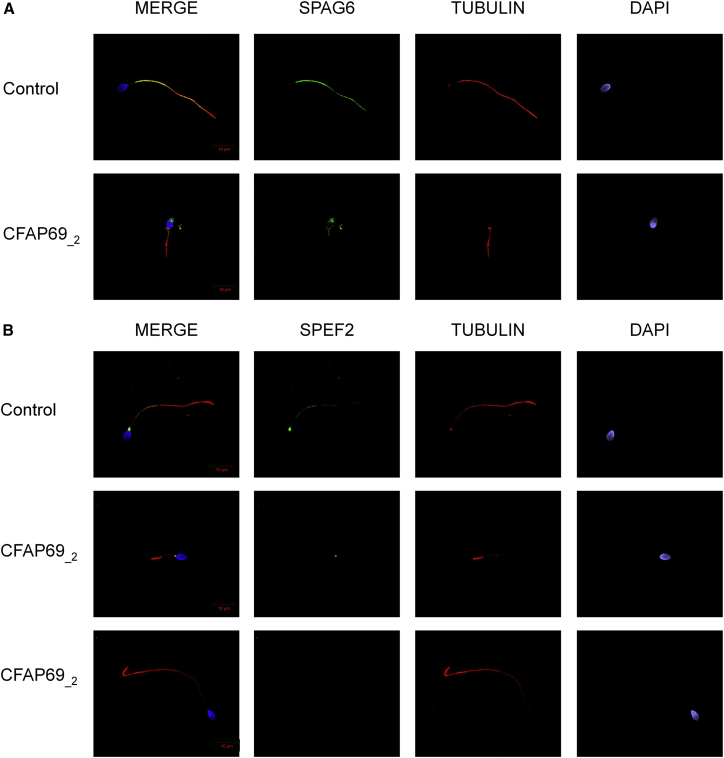
To further investigate the pathogenicity of the CFAP69 variants identified, we examined the distribution of CFAP69 in sperm cells from control and CFAP69-mutant individuals by immunofluorescence staining. In sperm from control individuals, we observed CFAP69 concentrated in the midpiece of the sperm flagellum (Figure 2A). This localization was confirmed by co-staining of the sperm mitochondrial protein HSP60 (Figure 2B), which visualizes the mitochondrial sheath in the midpiece.20 Importantly, CFAP69 was absent from all sperm cells from both individuals CFAP69_1 and CFAP69_2 (Figure 2A), consistent with the absence of CFAP69 transcript observed in individual CFAP69_1 (Figure S3). We next investigated the flagellar and axonemal defects of CFAP69-mutant individuals. Due to sample availability, these analyses were carried out only for individual CFAP69_2. We observed that in sperm from individual CFAP69_2, staining of SPAG6, an axoneme central pair complex (CPC) protein,21 was absent from the flagellum or displayed abnormal localization in the midpiece and the acrosomal region of the spermatozoa (Figure 3A). In contrast, immunostaining of DNALI1 and DNAH5 in sperm from individual CFAP69_2 was comparable with that observed in control cells, suggesting that outer dynein arms (ODAs) and inner dynein arms (IDAs), respectively, were not directly affected by mutations in CFAP69 (Figures S4A and S4B). We also examined the localization of SPEF2, a protein required for sperm flagellum assembly in the mouse22 and described as a putative partner to CFAP69.23 In control sperm, SPEF2 immunostaining appeared very strongly at the base of the flagellum, likely corresponding to the basal body, and lightly throughout the whole sperm flagellum, but in individual CFAP69_2, SPEF2 staining was weak or completely absent whereas tubulin staining remained detectable in the axoneme (Figure 3B).
Figure 2.
CFAP69 Immunostaining in Human Spermatozoa from Control Subjects and CFAP69 Mutant Individuals
(A) Sperm cells from a fertile control individual and the two CFAP69-mutant individuals stained with anti-CFAP69 (green) and anti-acetylated tubulin (red) antibodies. DNA was counterstained with Hoechst 33342. In the fertile control, the CFAP69 immunostaining (green) is concentrated in the midpiece of the spermatozoa (white arrows) and is not detectable in the principle piece. CFAP69 staining is absent in sperm flagellum from individual CFAP69_1 and individual CFAP69_2. Scale bars: 10 μm.
(B) Sperm cell from a fertile control individual stained with anti-HSP60 (red), which detects a mitochondrial protein located in the midpiece, and anti-CFAP69 (green) antibodies. The merged image shows that in control sperm, CFAP69 and HSP60 staining superimpose. Scale bars: 5 μm.
Figure 3.
SPAG6 and SPEF2 Immunostainings Are Affected by Mutations in CFAP69
(A) Sperm cells from a fertile control subject and CFAP69_2 stained with anti-SPAG6 (green), which detects a protein located in the CPC, and anti-acetylated tubulin (red) antibodies. DNA was counterstained with Hoechst 33342. SPAG6 staining uniformly decorates the full-length flagellum in the fertile control subject whereas it is absent from the flagellum of sperm from individual CFAP69_2. SPAG6 also shows atypical localization in the midpiece and in the acrosomal region of these spermatozoa.
(B) Sperm cells from a fertile control and CFAP69_2 stained with anti-SPEF2 (green) and anti-acetylated tubulin (red) antibodies. DNA was counterstained with Hoechst 33342. In sperm from the fertile control, SPEF2 staining appears mainly located in the basal body and lightly decorates the sperm flagellum. In sperm cells from the individual CFAP69_2, SPEF2 staining is strongly reduced or totally absent. Scale bars: 10 μm.
CFAP69 Is Found in the Mouse Testis and Is Required for Male Fertility
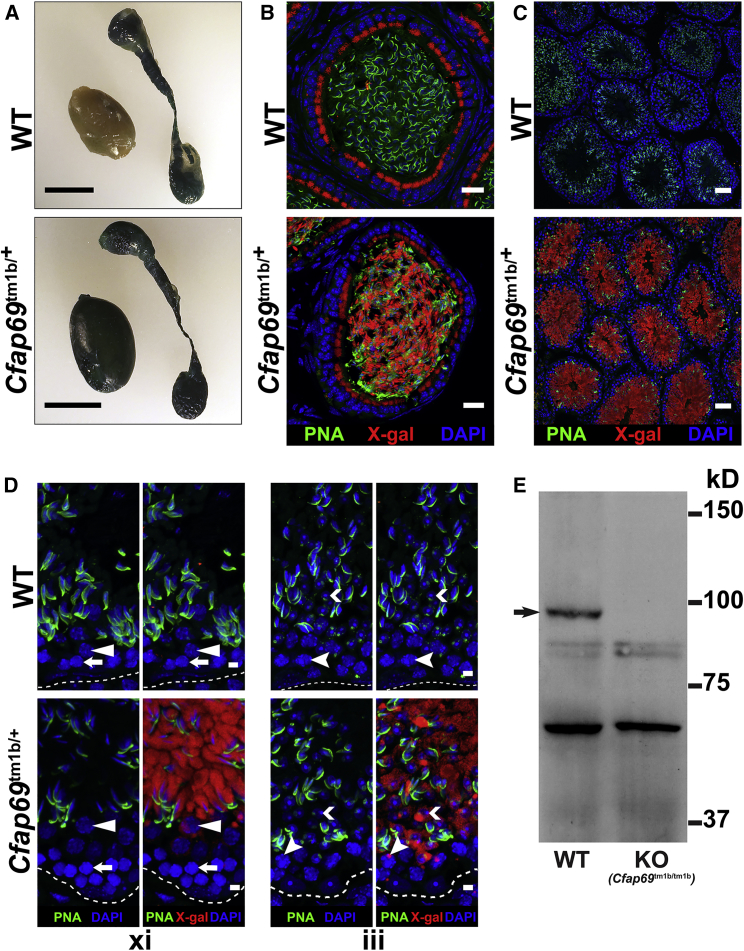
Various expression24 and RNA-seq25, 26 datasets indicate that a Cfap69 transcript is highly expressed in the mouse testis, and the availability of a Cfap69 knockout mouse15 allowed us to investigate the function of CFAP69 in male mouse reproduction. We investigated Cfap69 expression in the male mouse reproductive system by X-gal staining in mice heterozygous for a Cfap69 knockout-reporter allele (Cfap69tm1b/+ mice; see Subjects and Methods). In whole-mount preparations from male Cfap69tm1b/+, but not wild-type, mice, strong X-gal staining was observed in the testes (Figure 4A). The epididymis is known to express endogenous β-galactosidase,27, 28 and accordingly, we found X-gal staining in whole-mount epididymis (Figure 4A) and in the epithelial cells of cauda epididymis from both wild-type and Cfap69tm1b/+ mice (Figure 4B). This suggests that the X-gal staining observed in the Cfap69tm1b/+ epididymal cells is most likely due to endogenous β-galactosidase expression and not Cfap69 expression. However, sperm cells within Cfap69tm1b/+, but not in wild-type, epididymides were positive for X-gal staining (Figure 4B), suggesting that sperm cells or their precursors express the reporter gene. In testis cryosections, X-gal staining was observed in all seminiferous tubules of Cfap69tm1b/+ testes, but not in wild-type testes (Figure 4C). The staining first appeared in pachytene spermatocytes and increased in intensity in early and late spermatids (Figure 4D).
Figure 4.
Expression of CFAP69 in the Mouse Testis
(A) X-gal staining of whole-mount preparations of testis and epididymis from wild-type and Cfap69tm1b/+ mice.
(B–D) X-gal staining of cryosections of cauda epididymis (B) and testis (C and D) from wild-type and Cfap69tm1b/+ mice. X-gal staining is shown in red, acrosomes are labeled by peanut agglutinin (PNA, green), and nuclei are stained with DAPI (blue).
(B) X-gal staining is found in epithelial cells of both wild-type and Cfap69tm1b/+ epididymides. However, X-gal staining is found in the sperm of Cfap69tm1b/+ mice only and not in those of wild-type mice.
(C) X-gal staining is found in all seminiferous tubules of Cfap69tm1b/+ testes but not in those of wild-type testes.
(D) In xi, white arrows indicate zygotene spermatocytes, and white solid arrowheads indicate diplotene spermatocytes within stage xi seminiferous epithelia. In Cfap69tm1b/+ testes, strong X-gal staining is found in elongating spermatids, weak X-gal staining, often appearing as a dot, is found in diplotene spermatocytes, and no X-gal staining is found in zygotene spermatocytes. In iii, white chevrons indicate round spermatids, and white cleft arrowheads indicate pachytene spermatocytes within stage iii seminiferous epithelia. In Cfap69tm1b/+ mice, weak X-gal staining appearing as dots is found in pachytene spermatocytes, and stronger X-gal staining in round spermatids. Dashed white lines outline the basement membrane.
(E) Western blotting analysis of whole testis lysates from WT and Cfap69tm1b/tm1b (Cfap69 KO) mice. The arrow points to the band representing CFAP69.
Scale bars: (A) 0.5 cm; (B) 20 μm; (C) 50 μm; (D) 5 μm.
Western blotting analysis of wild-type adult mouse testis lysates using a custom antibody against CFAP6915 detects a band around 100 kD (Figure 4E), consistent with the predicted molecular weight of the protein encoded by the testis Cfap69 transcript. The corresponding band was absent from testes of Cfap69 knockout mice (Cfap69tm1b/tm1b; see Subjects and Methods) (Figure 4E). (Note: for unknown reasons, this antibody gives no staining when used in immunostaining in sections of reproductive tissues.)
Altogether, these results indicate that CFAP69 is found in germ cells of adult mouse testis, possibly beginning in pachytene spermatocytes and at higher levels after meiosis.
We then assessed fertility and reproductive behavior of Cfap69 knockout mice. When male Cfap69 knockout mice 6 weeks of age or older were mated to similarly aged wild-type females, they show normal mounting and produce copulatory plugs after mating, indicating that mating behavior is unimpaired in the absence of CFAP69. However, Cfap69 knockout male mice failed to produce any offspring over 3 months of breeding (8 Cfap69 mutant males mated with 2 wild-type females each, no offspring produced over 3 months), whereas wild-type males routinely produced offspring (8 wild-type males mated with 2 wild-type females each, 3–4 litters produced per female over 3 months, 6.93 ± 0.13 pups per litter). Cfap69 knockout female mice produced offspring at a rate similar to that of wild-type female mice when mated with wild-type males (data not shown). These results indicate that CFAP69 is required for male fertility in the mouse, consistent with phenotype analysis by The International Mouse Phenotyping Consortium.
Sperm of Cfap69 Knockout Mice Have Profound Flagellum Morphology Defects
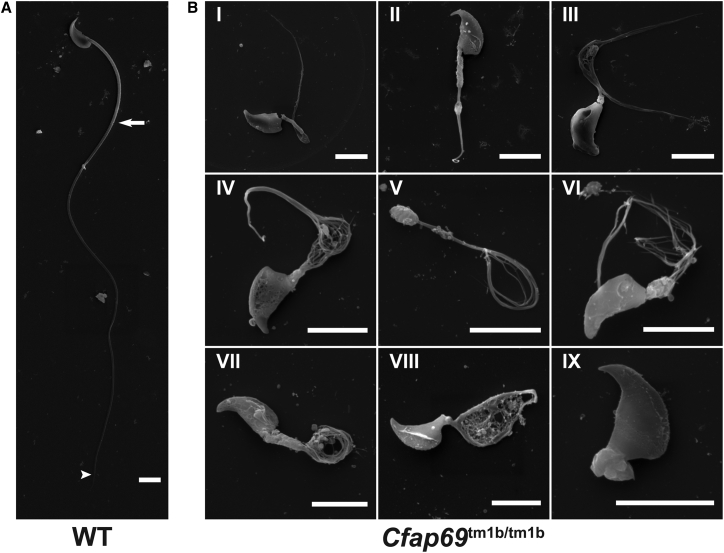
We next examined Cfap69 knockout sperm by collecting sperm from the cauda epididymides of both wild-type and Cfap69 knockout mice. Samples from Cfap69 knockout mice showed a complete lack of normal-looking sperm, and whereas we could observe sperm swimming in samples collected from wild-type mice, no sperm were observed to be motile in Cfap69 mutant samples. When observed by scanning electron microscopy (SEM), wild-type mouse sperm showed a hook-shaped head and a long flagellum with a clearly defined midpiece, principal piece, and end piece (Figure 5A), but all Cfap69 mutant sperm had severe morphology defects of the flagellum (Figure 5B). These defects are highly varied and affect all parts of the flagellum. In general, Cfap69 mutant sperm display shortening of both midpiece and principal piece, leading to overall shorter flagella. The flagella frequently show splaying or perforation or are missing. Additionally, head morphology defects ranging from mild to severe are observed (Figure 5B, III and V), though only a subset of sperm is affected. These defects likely account for the immotility of Cfap69 mutant sperm and the infertility of Cfap69 mutant mice and recapitulate the MMAF phenotype.
Figure 5.
Scanning Electron Microscopy Analysis of Sperm from Epididymides of Wild-Type and Cfap69 KO Mice
(A) Sperm from wild-type mice. White arrow indicates junction between midpiece and principal piece, and white arrowhead indicates junction between principal piece and end piece.
(B) Sperm from Cfap69 KO mice show severe morphology defects. The thickness and length of the mutant midpiece is variable (compare II, III, VI, VII). The flagella also frequently show splaying into many thin filaments along the entire length of the principal piece (III, VI) or in certain regions (IV, V). These filaments adopt a variety of conformations, including coiled, looped, and tangled. Some sperm essentially lack a flagellum (IX), while others have unusual structures perforated with thin filaments in place of their flagellum (VIII). Head morphology defects ranging from mild (III) to severe (V) are observed, though only a subset of sperm is affected. Scale bar: 5 μm.
Sperm Flagellum Development during Spermiogenesis Requires CFAP69
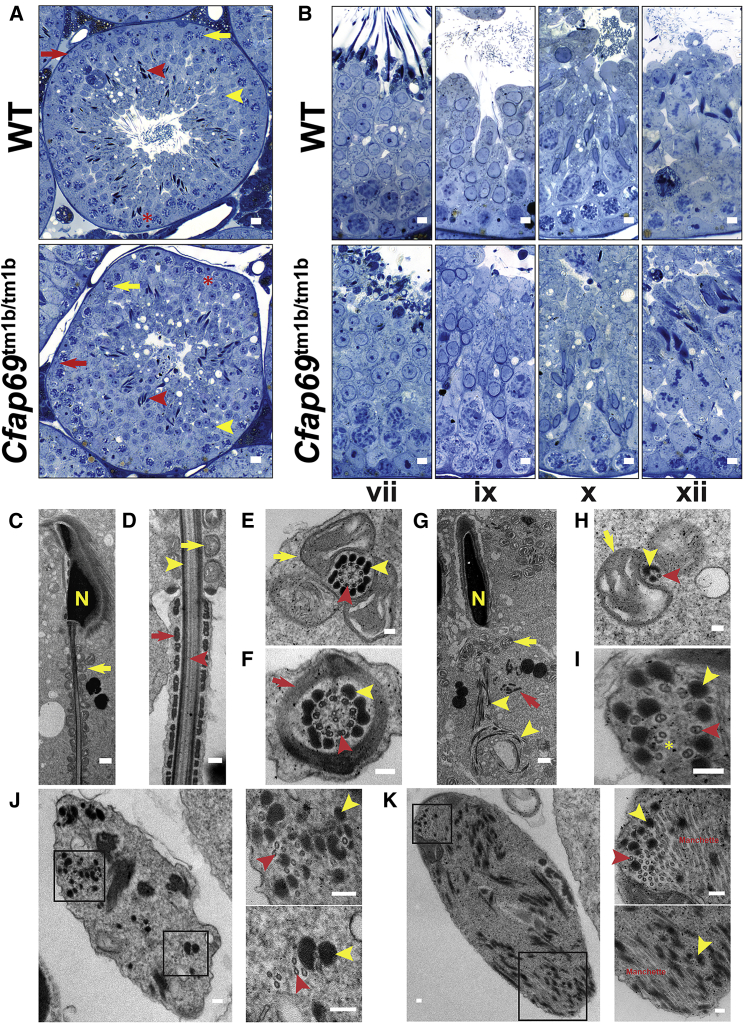
Given the presence of CFAP69 in sperm precursor cells (Figure 4), we sought to gain an understanding of when and how CFAP69 functions during sperm development by examining the histology of Cfap69 knockout testes. Toluidine blue staining of semi-thin testis sections indicates that in knockout mice, the cell composition of the seminiferous epithelium is unaltered as compared to wild-type: all types of germ cells could be observed, including spermatogonia, spermatocytes, spermatids, and spermatozoa (Figure 6A). The somatic Sertoli cells are also present. Staging of seminiferous epithelia by acrosome and nuclear morphology as well as by cell composition reveals that all 12 stages of the mouse seminiferous epithelium are present in Cfap69 knockout testes (Figure 6B). These findings indicate that the overall progression of spermatogenesis is preserved and that many spermatogenesis and spermiogenesis processes are nominally intact, including meiosis (Figure 6B, xii), acrosome development, and nuclear condensation and elongation (Figures 6B and S5). However, the absence of long flagella in the lumens of Cfap69 knockout seminiferous tubules is conspicuous (Figures 6A and 6B). Additionally, sperm with abnormal flagella can be observed in the lumen of Cfap69 knockout seminiferous tubules (Figures 6A and 6B). Thus, CFAP69 appears to be required during spermiogenesis for sperm flagellum development.
Figure 6.
The Progression of Spermatogenesis in Cfap69 KO Mice Is Preserved, but Flagellum Components Are Disorganized
(A and B) Light micrographs of semi-thin, EMbed812 sections of wild-type and Cfap69 KO testis stained with toluidine blue.
(A) Red arrows indicate spermatogonia, yellow arrows indicate spermatocytes, yellow arrowheads indicate round spermatids, red arrowheads indicate spermatozoa, and red asterisks indicate Sertoli cells, all of which are present in both wild-type and Cfap69 KO seminiferous epithelia.
(B) Seminiferous epithelia from both wild-type and Cfap69 KO mice of stages vii, ix, x, and xii are shown. Stages are determined by acrosome and nuclear morphology of germ cells, as well as by cell composition of the epithelium.
In (A) and (B), long flagella are conspicuously absent from the lumens of seminiferous tubules in Cfap69 KO sections.
(C–K) Transmission electron micrographs of testicular sperm from wild-type (C–F) and Cfap69 KO (G–K) mice. In (C)–(K), yellow arrows indicate mitochondria, yellow arrowheads indicate outer dense fibers, red arrows indicate fibrous sheath, red arrowheads indicates axoneme microtubules, and yellow “N” indicates the nucleus. In (H), yellow asterisk indicates split microtubule doublets.
Scale bars: (A) 10 μm; (B) 5 μm; (C) 1 μm; (D) 250 nm; (E and F) 100 nm; (G), 1 μm; (H–K) 100 nm.
To better understand the nature of the flagellum defects observed in Cfap69 knockout sperm, we examined their ultrastructure by TEM. Owing to an inability to obtain sufficient quantities of mutant sperm from epididymides for TEM, we instead analyzed sperm in testis sections. The components of the sperm flagellum and their native architecture, including the axoneme, the outer dense fibers (ODFs), the mitochondrial sheath of the midpiece, and the fibrous sheath of the principal piece, are readily observable in longitudinal and cross sections of wild-type, testicular sperm (Figures 6C–6F). However, sections of Cfap69 knockout sperm reveal flagellum components in a dramatic state of disarray within bodies of cytoplasm (Figures 6G–6K). Although some mitochondria can be observed to localize to regions of mutant sperm flagella that would ordinarily be the midpiece, many appear throughout the cytoplasm (Figure 6G), in contrast to what was observed in wild-type sperm. In addition, in Cfap69 knockout sperm, ODFs are irregular in number and organization (Figures 6G–6K). Notably, in longitudinal sections, the ODFs show coiled and tangled arrangements resembling the unusual flagellum morphology observed in whole Cfap69 knockout sperm (Figure 6G). Additionally, the fibrous sheath is disorganized and largely absent (Figure 6G). Finally, axoneme organization is severely disrupted. Doublet and singlet microtubules are still frequently found to associate with ODFs, but their numbers and their arrangement vary (Figures 6H–6K). Microtubule doublets that have been split are also observed (Figure 6I, asterisk). Large numbers of singlet microtubules, likely from the manchette, were also frequently observed (Figure 6K), indicating an abnormal confluence of the manchette and flagellum components. The abnormal organization of all sperm flagellum components indicates a failure of sperm flagellum development during spermiogenesis in Cfap69 knockout mice and likely accounts for the observed sperm morphology defects.
Discussion
We showed that, in humans and mice, the presence of biallelic truncating mutations in CFAP69 induces male infertility owing to MMAF, thus establishing CFAP69 as a MMAF-related gene along with DNAH1, CFAP43, and CFAP44. The two individuals in our cohort with CFAP69 mutations presented only with primary infertility without other clinical features, similar to the individuals with DNAH1, CFAP43, and CFAP44 mutations.7, 8, 9, 10, 11 Although CFAP69 regulates olfactory response kinetics, the individuals did not believe they had a deficient sense of smell and did not wish to participate in any olfaction-related studies.
In both humans and mice, dramatic sperm flagellum defects were observed (Figures 1 and 5). In the adult mouse testis, CFAP69 expression begins during meiosis and strongly increases in subsequent stages (Figure 4). Major processes of the spermatogenic cycle and some spermiogenesis processes appeared normal. TEM analysis of the testis showed dramatic disorganization of all sperm flagellum components (Figure 5), suggesting an essential role for CFAP69 in normal flagellum formation during spermiogenesis.
We observed no significant differences between the semen parameters of the two individuals carrying mutations in CFAP69 compared to the parameters of individuals with mutations in other MMAF genes (DNAH1, CFAP43, and CFAP44). However, we found low sperm concentrations in the ejaculates (oligozoospermia) associated with a high rate of head malformations in both CFAP69-mutant individuals (Figure 1, Tables 1 and S1), which were also observed in the Cfap69 knockout mouse model (Figure 5). These observations suggest that mutations in CFAP69 lead to an atypical and more severe MMAF phenotype, and that CFAP69 could also be important for sperm head shaping during spermiogenesis. Whether CFAP69 is required for a process common to head and flagellum development or in distinct processes is not known.
Interestingly, immunostaining experiments with sperm from fertile control individuals showed that CFAP69 is located in the midpiece of the sperm flagellum (Figure 2). The midpiece-specific staining differs from the whole-flagellum staining observed for other axonemal components involved in the MMAF phenotype such as DNAH1, CFAP43, or CFAP447, 8, 10 (Figure S6). This localization suggests that CFAP69 is unlikely to belong to the core axoneme. The midpiece of the mammalian sperm flagellum is composed of the mitochondrial sheath surrounding nine outer dense fibers and the axoneme.29 However, CFAP69 is not necessarily, and may most likely not be, a mitochondrial or an ODF protein, as CFAP69 is found in other cilia in the mouse and in other organisms that contain neither outer dense fibers nor mitochondria.15, 30 For example, we recently showed that CFAP69 is present in the cilia of mouse olfactory sensory neurons, which do not contain mitochondria, and that it plays a role in regulating the odor-response kinetics of olfactory sensory neurons.15 Interestingly, in contrast to sperm flagella, which have severe morphology defects, the olfactory cilia lacking CFAP69 in mouse appeared morphologically normal,15 illustrating that the sperm flagellum is assembled and organized differently from other cilia to fulfill its unique functions.31 Additionally, proteomic analyses in Chlamydomonas rheinhardtii found CFAP69 enriched in the flagellum.32, 33 The role of CFAP69 in the function and assembly of these other cilia and its relationship to CFAP69 function in sperm flagellum assembly and function are under investigation.
The human and mouse sperm head and flagellum defects (Figures 1 and 5), as well as abnormally localized manchette microtubules (Figure 6), are reminiscent of defects arising from disruption of intramanchette transport (IMT) or intraflagellar (IFT) transport.23, 34, 35, 36, 37 Interestingly, IF staining of SPEF2, which is involved in IMT, sperm head shaping, and CPC assembly during spermiogenesis,22, 23, 38 is strongly reduced or totally absent from CFAP69-mutant individuals’ sperm cells (Figure 3B). Truncating mutations in Spef2 are also known to cause male infertility in the pig and mouse due to a short sperm tail phenotype associated with severe ultrastructural axonemal and peri-axonemal defects which closely resemble the MMAF phenotype observed in the Cfap69 knockout mouse model.22, 39 Additionally, reduced and atypical staining of the CPC protein SPAG6 in spermatozoa from our CFAP69-mutant individuals suggests that CFAP69 may be also involved in CPC organization (Figure 3A). We can therefore speculate that CFAP69 might partake in sperm tail biogenesis and CPC assembly through a role in flagellar protein transport (Figure S6). Although mass spectrometry analysis recently suggested that SPEF2 might interact with CFAP69 (Q8BH53),23 no protein interaction partners for CFAP69 have been confirmed. Bioinformatic analysis does suggest that CFAP69 contains an ARM-repeat domain and an AH/BAR domain (Figure 1D), indicating that CFAP69 function may indeed involve interaction with other proteins. The identity of these proteins and the processes they affect remain to be determined.
Overall we identified two different pathogenic CFAP69 mutations in our cohort of 78 MMAF-affected individuals. Combined with other mutations identified in this cohort in the previously known MMAF-related genes DNAH1, CFAP43, and CFAP44,12 the diagnosis efficiency of MMAF phenotype by WES in our cohort is of 31%. These promising results support the implementation of WES as a routine practice in the genetic investigation of male infertility. This yield is comparable to that obtained in other genetic diseases with a high degree of genetic heterogeneity40, 41 and is consistent with the very large number of genes involved in the spermatogenesis.42 To improve this diagnosis rate and detect genes with lower mutation prevalence, genetic studies by WES should be performed in larger cohorts of individuals.
Acknowledgments
We thank all individuals for their participation. This work was mainly supported by the following grants: the “MAS-Flagella” project financed by French ANR and the DGOS for the program PRTS 2014 (P.F.R.), the “Whole genome sequencing of patients with Flagellar Growth Defects (FGD)” financed by the fondation maladies rares (FMR) for the program Séquençage à haut débit 2012 (P.F.R.), and the USA NIH grant DC007395 (H.Z.).
Published: March 29, 2018
Footnotes
Supplemental Data include Supplemental Material and Methods, six figures, and five tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.03.007.
Contributor Information
Haiqing Zhao, Email: hzhao@jhu.edu.
Charles Coutton, Email: ccoutton@chu-grenoble.fr.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
BioGPS, http://biogps.org/
ExAC Browser, http://exac.broadinstitute.org/
ExSQLibur, https://github.com/tkaraouzene/ExSQLibur
gnomAD Browser, http://gnomad.broadinstitute.org/
Human Splicing Finder, http://www.umd.be/HSF3/
International Mouse Phenotyping Consortium, http://www.mousephenotype.org/data/genes/
InterPro, https://www.ebi.ac.uk/interpro/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, http://genome.ucsc.edu
UniProt, http://www.uniprot.org/
Supplemental Data
References
- 1.Sharlip I.D., Jarow J.P., Belker A.M., Lipshultz L.I., Sigman M., Thomas A.J., Schlegel P.N., Howards S.S., Nehra A., Damewood M.D. Best practice policies for male infertility. Fertil. Steril. 2002;77:873–882. doi: 10.1016/s0015-0282(02)03105-9. [DOI] [PubMed] [Google Scholar]
- 2.Zorrilla M., Yatsenko A.N. The genetics of infertility: current status of the field. Curr. Genet. Med. Rep. 2013;1:1. doi: 10.1007/s40142-013-0027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ray P.F., Toure A., Metzler-Guillemain C., Mitchell M.J., Arnoult C., Coutton C. Genetic abnormalities leading to qualitative defects of sperm morphology or function. Clin. Genet. 2017;91:217–232. doi: 10.1111/cge.12905. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell M.J., Metzler-Guillemain C., Toure A., Coutton C., Arnoult C., Ray P.F. Single gene defects leading to sperm quantitative anomalies. Clin. Genet. 2017;91:208–216. doi: 10.1111/cge.12900. [DOI] [PubMed] [Google Scholar]
- 5.de Boer P., de Vries M., Ramos L. A mutation study of sperm head shape and motility in the mouse: lessons for the clinic. Andrology. 2015;3:174–202. doi: 10.1111/andr.300. [DOI] [PubMed] [Google Scholar]
- 6.Coutton C., Escoffier J., Martinez G., Arnoult C., Ray P.F. Teratozoospermia: spotlight on the main genetic actors in the human. Hum. Reprod. Update. 2015;21:455–485. doi: 10.1093/humupd/dmv020. [DOI] [PubMed] [Google Scholar]
- 7.Ben Khelifa M., Coutton C., Zouari R., Karaouzène T., Rendu J., Bidart M., Yassine S., Pierre V., Delaroche J., Hennebicq S. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amiri-Yekta A., Coutton C., Kherraf Z.-E., Karaouzène T., Le Tanno P., Sanati M.H., Sabbaghian M., Almadani N., Sadighi Gilani M.A., Hosseini S.H. Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum. Reprod. 2016;31:2872–2880. doi: 10.1093/humrep/dew262. [DOI] [PubMed] [Google Scholar]
- 9.Sha Y., Yang X., Mei L., Ji Z., Wang X., Ding L., Li P., Yang S. DNAH1 gene mutations and their potential association with dysplasia of the sperm fibrous sheath and infertility in the Han Chinese population. Fertil. Steril. 2017;107:1312–1318.e2. doi: 10.1016/j.fertnstert.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Tang S., Wang X., Li W., Yang X., Li Z., Liu W., Li C., Zhu Z., Wang L., Wang J. Biallelic mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2017;100:854–864. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X., Jin H., Han F., Cui Y., Chen J., Yang C., Zhu P., Wang W., Jiao G., Wang W. Homozygous DNAH1 frameshift mutation causes multiple morphological anomalies of the sperm flagella in Chinese. Clin. Genet. 2016;91:313–321. doi: 10.1111/cge.12857. [DOI] [PubMed] [Google Scholar]
- 12.Coutton C., Vargas A.S., Amiri-Yekta A., Kherraf Z.-E., Ben Mustapha S.F., Le Tanno P., Wambergue-Legrand C., Karaouzène T., Martinez G., Crouzy S. Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in Trypanosoma and human. Nat. Commun. 2018;9:686. doi: 10.1038/s41467-017-02792-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Yang J., Jia Y., Xiong C., Meng T., Guan H., Xia W., Ding M., Yuchi M. Variability in the morphologic assessment of human sperm: use of the strict criteria recommended by the World Health Organization in 2010. Fertil. Steril. 2014;101:945–949. doi: 10.1016/j.fertnstert.2013.12.047. [DOI] [PubMed] [Google Scholar]
- 14.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Talaga A.K., Dong F.N., Reisert J., Zhao H. Cilia- and flagella-associated protein 69 regulates olfactory transduction kinetics in mice. J. Neurosci. 2017;37:5699–5710. doi: 10.1523/JNEUROSCI.0392-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitsky K.L., Toledo-Aral J.J., López-Barneo J., Villadiego J. Direct confocal acquisition of fluorescence from X-gal staining on thick tissue sections. Sci. Rep. 2013;3:2937. doi: 10.1038/srep02937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivliev A.E., ’t Hoen P.A.C., van Roon-Mom W.M.C., Peters D.J.M., Sergeeva M.G. Exploring the transcriptome of ciliated cells using in silico dissection of human tissues. PLoS ONE. 2012;7:e35618. doi: 10.1371/journal.pone.0035618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnoni R., Palmfeldt J., Hansen J., Christensen J.H., Corydon T.J., Bross P. The Hsp60 folding machinery is crucial for manganese superoxide dismutase folding and function. Free Radic. Res. 2014;48:168–179. doi: 10.3109/10715762.2013.858147. [DOI] [PubMed] [Google Scholar]
- 21.Sapiro R., Tarantino L.M., Velazquez F., Kiriakidou M., Hecht N.B., Bucan M., Strauss J.F., 3rd Sperm antigen 6 is the murine homologue of the Chlamydomonas reinhardtii central apparatus protein encoded by the PF16 locus. Biol. Reprod. 2000;62:511–518. doi: 10.1095/biolreprod62.3.511. [DOI] [PubMed] [Google Scholar]
- 22.Sironen A., Kotaja N., Mulhern H., Wyatt T.A., Sisson J.H., Pavlik J.A., Miiluniemi M., Fleming M.D., Lee L. Loss of SPEF2 function in mice results in spermatogenesis defects and primary ciliary dyskinesia. Biol. Reprod. 2011;85:690–701. doi: 10.1095/biolreprod.111.091132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehti M.S., Zhang F.-P., Kotaja N., Sironen A. SPEF2 functions in microtubule-mediated transport in elongating spermatids to ensure proper male germ cell differentiation. Development. 2017;144:2683–2693. doi: 10.1242/dev.152108. [DOI] [PubMed] [Google Scholar]
- 24.Wu C., Macleod I., Su A.I. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41:D561–D565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Margolin G., Khil P.P., Kim J., Bellani M.A., Camerini-Otero R.D. Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics. 2014;15:39. doi: 10.1186/1471-2164-15-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.da Cruz I., Rodríguez-Casuriaga R., Santiñaque F.F., Farías J., Curti G., Capoano C.A., Folle G.A., Benavente R., Sotelo-Silveira J.R., Geisinger A. Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genomics. 2016;17:294. doi: 10.1186/s12864-016-2618-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Björkgren I., Saastamoinen L., Krutskikh A., Huhtaniemi I., Poutanen M., Sipilä P. Dicer1 ablation in the mouse epididymis causes dedifferentiation of the epithelium and imbalance in sex steroid signaling. PLoS ONE. 2012;7:e38457. doi: 10.1371/journal.pone.0038457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merkwitz C., Blaschuk O., Schulz A., Ricken A.M. Comments on methods to suppress endogenous β-galactosidase activity in mouse tissues expressing the LacZ reporter gene. J. Histochem. Cytochem. 2016;64:579–586. doi: 10.1369/0022155416665337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fawcett D.W. The mammalian spermatozoon. Dev. Biol. 1975;44:394–436. doi: 10.1016/0012-1606(75)90411-x. [DOI] [PubMed] [Google Scholar]
- 30.Tewari R., Bailes E., Bunting K.A., Coates J.C. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 2010;20:470–481. doi: 10.1016/j.tcb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Lee L. Mechanisms of mammalian ciliary motility: Insights from primary ciliary dyskinesia genetics. Gene. 2011;473:57–66. doi: 10.1016/j.gene.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 32.Pazour G.J., Agrin N., Leszyk J., Witman G.B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang P., Diener D.R., Yang C., Kohno T., Pazour G.J., Dienes J.M., Agrin N.S., King S.M., Sale W.S., Kamiya R. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehti M.S., Sironen A. Formation and function of the manchette and flagellum during spermatogenesis. Reproduction. 2016;151:R43–R54. doi: 10.1530/REP-15-0310. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Z., Li W., Zhang Y., Zhang L., Teves M.E., Liu H., Strauss J.F., 3rd, Pazour G.J., Foster J.A., Hess R.A., Zhang Z. Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. Mol. Biol. Cell. 2016 doi: 10.1091/mbc.E16-05-0318. mbc.E16-05-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.San Agustin J.T., Pazour G.J., Witman G.B. Intraflagellar transport is essential for mammalian spermiogenesis but is absent in mature sperm. Mol. Biol. Cell. 2015;26:4358–4372. doi: 10.1091/mbc.E15-08-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H., Li W., Zhang Y., Zhang Z., Shang X., Zhang L., Zhang S., Li Y., Somoza A.V., Delpi B. IFT25, an intraflagellar transporter protein dispensable for ciliogenesis in somatic cells, is essential for sperm flagella formation. Biol. Reprod. 2017;96:993–1006. doi: 10.1093/biolre/iox029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sironen A., Hansen J., Thomsen B., Andersson M., Vilkki J., Toppari J., Kotaja N. Expression of SPEF2 during mouse spermatogenesis and identification of IFT20 as an interacting protein. Biol. Reprod. 2010;82:580–590. doi: 10.1095/biolreprod.108.074971. [DOI] [PubMed] [Google Scholar]
- 39.Sironen A., Thomsen B., Andersson M., Ahola V., Vilkki J. An intronic insertion in KPL2 results in aberrant splicing and causes the immotile short-tail sperm defect in the pig. Proc. Natl. Acad. Sci. USA. 2006;103:5006–5011. doi: 10.1073/pnas.0506318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farwell K.D., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Tippin Davis B., Baxter R.M., Zeng W., Mroske C., Parra M.C. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015;17:578–586. doi: 10.1038/gim.2014.154. [DOI] [PubMed] [Google Scholar]
- 41.Fogel B.L., Lee H., Strom S.P., Deignan J.L., Nelson S.F. Clinical exome sequencing in neurogenetic and neuropsychiatric disorders. Ann. N Y Acad. Sci. 2016;1366:49–60. doi: 10.1111/nyas.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matzuk M.M., Lamb D.J. The biology of infertility: research advances and clinical challenges. Nat. Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper T.G., Noonan E., von Eckardstein S., Auger J., Baker H.W.G., Behre H.M., Haugen T.B., Kruger T., Wang C., Mbizvo M.T., Vogelsong K.M. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 44.Auger J., Jouannet P., Eustache F. Another look at human sperm morphology. Hum. Reprod. 2016;31:10–23. doi: 10.1093/humrep/dev251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.