Abstract
Most phenome-wide association studies (PheWASs) to date have used a small to moderate number of SNPs for association with phenotypic data. We performed a large-scale single-cohort PheWAS, using electronic health record (EHR)-derived case-control status for 541 diagnoses using International Classification of Disease version 9 (ICD-9) codes and 25 median clinical laboratory measures. We calculated associations between these diagnoses and traits with ∼630,000 common frequency SNPs with minor allele frequency > 0.01 for 38,662 individuals. In this landscape PheWAS, we explored results within diseases and traits, comparing results to those previously reported in genome-wide association studies (GWASs), as well as previously published PheWASs. We further leveraged the context of functional impact from protein-coding to regulatory regions, providing a deeper interpretation of these associations. The comprehensive nature of this PheWAS allows for novel hypothesis generation, the identification of phenotypes for further study for future phenotypic algorithm development, and identification of cross-phenotype associations.
Keywords: bioinformatics, genomics, PheWAS, phenome-wide, genetic epidemiology, GWAS, EHR, biorepository
Introduction
The wealth of information within an electronic health record (EHR) can be leveraged to improve our understanding of the genetic architecture of human disease by characterizing a landscape of genetic associations across many different measures such as disease diagnosis codes and clinical laboratory tests. Phenome-wide association studies (PheWASs) have become a common tool for identifying comprehensive genetic associations between SNPs and a wide range of phenotypes, with successful implementation in phenotype data extracted/collected from EHRs, epidemiological studies, and clinical trials. Much of the past PheWAS literature has focused on smaller cohorts/datasets and often with small numbers of genetic variants.1, 2, 3, 4, 5, 6, 7, 8 Some studies obtained larger sample sizes by utilizing combined datasets from EHRs of different health care providers across the country.5, 9, 10, 11 These studies have shown the utility of PheWASs and identified new hypotheses for genetic associations and particularly cross-phenotype associations which can uncover pleiotropy. Integrating EHR data from different health care providers has been challenging for EHR-based PheWASs to date, due to the differences in coding practices and differing populations.
Currently, the Geisinger research program, the MyCode Community Health Initiative, consists of more than 160,000 consented individuals (Figure S1), tens of thousands of whom have genetic data linked to a single-source EHR, which presents an opportunity to perform a large-scale PheWAS. In our study, we utilized data from 50,726 individuals genotyped at the time of our study (April 2016) to execute a PheWAS using common frequency SNPs with greater than 1% minor allele frequency (MAF) and 541 International Code for Diseases version 9 (ICD-9) codes and 25 clinical laboratory measurements. Clinical laboratory measurements represent aspects of current health or disease state and are key decision-making elements for clinical diagnoses. In our previous work, we have shown that integrating both clinical lab measurements along with diagnosis codes can provide robustness to the interpretation of genetic association results.12
A PheWAS at this scale, where we computed a total of 343,819,025 associations for the diagnosis codes and 15,888,125 associations for the clinical lab measures, presented several big data challenges such as computational burden, high throughput result interpretation, and visualization of the results. In this analysis, we tested more than 300 million associations using SNPs and clinical phenotypes. The amount of storage and memory required exceeded the limits of most high-performance computing (HPC) clusters, even with software techniques to parallelize these association analyses. To execute a PheWAS at this scale, we used the distributed resources of a cloud computing platform through DNAnexus (see Web Resources) that uses Amazon Web Services (AWS). The scalability of cloud resources not only addresses the problem of dataset size but also allows for the computation of analyses at this scale in a reasonable amount of time. We ran more than 340 million models on 180 machines with 32 cores each using the “scatter-gather” approach. In contrast, if we had run our analyses using a single core machine, it would have taken 30 years to complete. The computational processing for all the model calculations took 3 days (more precisely, a total of 51 hours) for the entire study.
Further, this study addresses the issue of high-throughput interpretation of association results. In addition to the exhaustive associations evaluated in this study, we have expanded the resources and approaches for interpreting the resultant genetic associations. For replicating known associations, we used the EBI-GWAS13 catalog and GRASP14, 15 to compare our PheWAS associations to previously reported genetic associations. Also, with the number of PheWASs published in recent years, we compared our results to the results of eight recently published PheWASs.
The majority of associations reported in the GWAS catalog have focused on interpreting the results of genetic associations through identification of the nearest gene. However, the majority of highly significant GWAS associations are found outside of protein-coding genes.16 Exploring results beyond the nearest gene is imperative to improve the understanding of the relevance of a given genotype-phenotype association, even when it is not the most statistically significant finding. With the availability of public resources such as Roadmap Epigenomics,17 ENCODE,18 HaploReg,19 and others, we can explore the non-coding regions of the genome and annotate our results based on meaningful regulatory information including promoter, enhancer, and transcription sites, among others. In this paper, we present an application of a novel approach for prioritizing genetic association results using gene expression measures from RNA-seq data obtained from the Roadmap Epigenome. This method can be applied to other association studies to prioritize genetic associations. The annotation of our association results enabled us to explore beyond protein-coding regions of the genome and to improve our interpretation for understanding the potential effects of these genetic variations on the phenotypes.
For this large-scale single-cohort PheWAS, we have presented additional ways to expand the understanding of association findings from PheWASs including the use of existing expert knowledge. We investigated the landscape of associations independently across diseases and clinical lab measures as well as through an integrative approach to identify shared genetic patterns of associations between diseases and laboratory measures and to highlight potential pleiotropy. Further, epigenomics knowledge of non-coding regions of the genome helped us to refine the genetic associations, to illustrate the biological relevance to the associated disease. With these results, we provide a landscape of associations across diseases and quantitative traits, a series of potentially novel associations, and cross-phenotype associations, all within the context of protein-coding and regulatory impact of genetic variants.
Subjects and Methods
Geisinger, DiscovEHR, and MyCode Community Health Initiative
In this study, we used data from the DiscovEHR study, a collaboration between Geisinger and Regeneron Genetics Center as a part of DiscovEHR collaboration. Geisinger is one of the largest healthcare providers in Central Pennsylvania with more than 2.3 million individuals in the system (Figure S1). The Geisinger research biorepository, the MyCode Community Health Initiative, was launched in 2007 and is a collective resource of genomics data linked to the de-identified EHR data.20 Geisinger has been collecting EHR data since 1996 with digital information including diagnoses, laboratory, demographics, and medication use during the course of care at Geisinger. The Geisinger Institutional Review Board reviewed the study and declared it to be research with non-human subjects as all data were de-identified. All MyCode participants included in the study were consented.
Genotype Data and Quality Control Measures
At the time of this study, a total of 45,899 individuals were genotyped using the Illumina HumanOmniExpressExome bead chip by Regeneron Genetics Center. We applied standard genotype quality-control (QC) measures to eliminate any systematic errors that could lead to spurious associations. We filtered out variants with genotype and sample missingness (<99%) and we included only the common variants with minor allele frequency (MAF) > 1%. Further, to account for sample independence, one sample from each pair of related samples with kinship coefficient > 0.125 were dropped. The MyCode population was approximately 97% European American (EA) based on genetically informed ancestry, and since the sample size of other ancestral groups within our dataset was extremely limited, we did not include them in the study. We calculated principal components on EA samples using EIGENSOFT to further correct for confounding factors due to global ancestry within our genetic associations. After all the quality assessment, we had 635,525 SNPs in 38,622 unrelated samples available for genetic associations.
Phenotype Data
We collated 541 International Classification of Disease Version 9 (ICD-9) diagnosis codes as binary case/control disease outcomes and 25 median clinical laboratory measurements as continuous outcomes from the Geisinger EHR for all the 38,622 MyCode participants for this study. ICD-9 codes provide records of individuals’ disease diagnoses over the course of their clinic visits. We defined case and control groups based on the number of clinic visits for each ICD-9 code per individual. The individuals with at least three or more instances of a diagnosis code were considered case subjects and individuals with absence of that code were considered control subjects. Any individuals with an ICD-9 code that had between one and two visits were excluded from the association analysis for that ICD-9 code. Based on an independent PheWAS simulation study that we conducted,21 we considered only those ICD-9 codes with a sample size of greater or equal to 200 case or control subjects to reduce effect estimate inflation in regression models.
The decision-making and quality assessment for the 25 clinical laboratory measurements were previously published in our other studies using clinical lab measures for PheWAS.12, 22 Briefly, we used the median values of each lab value for each individual and we removed any values outside the range of 2.5 standard deviations. Standard statistical measures for data transformations were applied after inspecting the lab value distributions. We provide summary statistics for each lab measure used in the study in Table S1.
Association Testing
We used PLATO23 to perform association testing in this study. We performed two independent PheWAS analyses, a separate analysis for the binary outcomes using logistic regression and 541 ICD-9-based case/control diagnoses, and linear regression for the 25 median clinical laboratory measurements, with 632,574 genetic variants. We adjusted all regression models by sex, age, and first four principal components to account for any confounding bias due to these variables.
Calculating a total of more than 300 million genetic associations, even with parallelization of association testing, surpassed the capacity of standard computational resources such as high-performance clusters to run all of these associations in a reasonable amount of time. To address these challenges, we used DNAnexus, a genomic analysis platform built upon Amazon Web Services (AWS). We used the distributed cluster of computers on the cloud to reduce the computation time. We built a custom DNAnexus app for PLATO, using a scatter-process-gather implementation on the platform to compute regression models in a “perfectly parallel” manner. The scatter-gather approach invoked multiple AWS virtual machines to concurrently process the regression models. Once all calculations were completed, the application merged all results. We ran more than 340 million models on 180 machines with 32 cores each; it took 30 core years to complete the analysis (i.e., if we ran this analysis on a single core machine, it would have taken 30 years to complete). We finished all the model calculations in less than 3 days (51 hrs) for this entire study.
Statistical Correction
We implemented a custom Bonferroni correction for multiple test correction to identify our most significant results for our phenome-wide correction p value. The Bonferroni correction can be overly conservative because of the assumption that all tests are independent and that all phenotypes are independent, thus not accounting for the correlation between the genetic variants due to linkage disequilibrium (LD) as well as the correlations between phenotypes. In this study, we used only genetically informed ancestry-based EA samples, and to account for correlation between SNPs, we identified the number of less correlated SNPs through LD pruning at an r2 threshold of 0.3, which is considered the optimal threshold for EA samples.24 Using the above threshold with PLINK (see Web Resources), we found a total of 172,690 independent SNPs. We used this to derive our phenome-wide significance threshold, α divided by the number of independent tests, i.e., 5.36 × 10−10 and 1.15 × 10−8 for ICD-9 codes and clinical lab measures, respectively.
The above LD pruning method accounts for the dependence between the genetic variants in a more accurate manner than the more conservative Bonferroni correction. However, it did not account for any underlying correlation patterns present in phenotype data such as correlation between cholesterol measurements (HDL, LDL, and total cholesterol) and the impact of power on associations due to sample sizes and case-control numbers. Thus, the phenome-wide correction threshold is still stringent. Hence, in addition to exploring results with a stringent Bonferroni correction, we further expanded our search space to look at a much wider landscape of associations by investigating at an exploratory threshold of 1 × 10−4 to study associations beyond only the most statistically significant results.
Genomic Annotations
Using various resources including the Ensembl Variant Effect Predictor (VEP)25 and Roadmap Epigenomics Project, we mapped genetic associations to these resources of functional genomics to obtain additional insights for the PheWAS results. In our first step, we used VEP to annotate the variants in our association results (p value < 1 × 10−4) with sequence ontology (SO) terms. Using VEP predictions, we systematically classified our variants by their biological consequences such as protein-coding (missense, synonymous, non-synonymous, splice site), non-coding, and regulatory (UTRs, transcription binding, intergenic variants, among others) region. We used these classifications to highlight the distribution of variants that are specifically mapping to protein coding, regulatory, and other non-coding regions of the genome.
In the second stage of mapping our results to functional information, we annotated variants in regulatory and non-coding regions defined by VEP to the Roadmap Epigenome. The Roadmap Epigenomics Project is a tissue-specific epigenomics dataset generated from 127 human tissues and 12 epigenetic marks such as histone modifications, H2A.z, and DNase. We used a model including 20 chromatin states predicted by a tool called the Integrative and Discriminative Epigenome Annotation System (IDEAS)26 from the 127 epigenomes dataset. IDEAS provides a predicted chromatin state across 200 base pair windows for each epigenome. To explore the overall representation of these variants in each chromatin state, we computed the most probable chromatin state across the 127 epigenomes. The most probable state for each 200 bp window was assigned to the state predicted most number of times across 127 tissues. We overlaid the SNP coordinates with the epigenome regions and annotated SNPs with the derived most probable chromatin state.
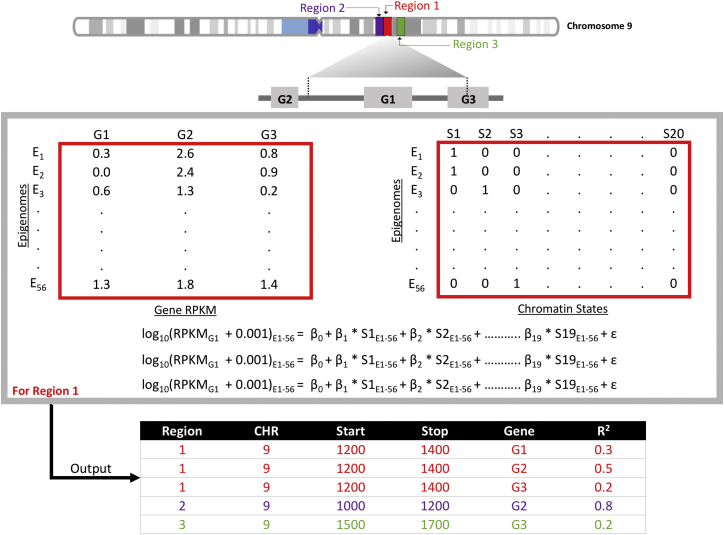
To understand the influence of the variants in this study on gene expression, we further extended our annotation approach. The Roadmap Epigenome Consortium also provides RNA-seq data on 56 epigenomes, and we used these data to evaluate the variance explained by each predicted chromatin state within gene expression data. The RNA-seq dataset consists of RPKM (reads per kilobase million) values for all the coding and noncoding genes including exons and introns. We used the same regions as in the chromatin state prediction by IDEAS, and for each region, we performed regression between the binary measures for each state and RPKM values for genes within ±100 kilobases of each 200 bp window across 56 epigenomes27 (Figure 1). We use adjusted r-squared (r2) value from the calculation to infer the correlation between the chromatin state and expression of nearby genes to obtain a measure of the contribution of predicted chromatin state to the expression of the gene. As there can be multiple genes near a region, and to simplify the annotations, we used only the gene with highest r2 value for a given genomic region. In this way, we could identify SNPs that were within regions that are most correlated with changes in gene expression of specific genes.
Figure 1.
Correlation between Chromatin State and Genes via RNA-Seq Data
For a given region such as region 1 shown on chromosome 9 in the figure, we calculated the correlation between the predicted chromatin state and gene expression using data from 56 tissues provided by the Roadmap Epigenome Consortium. The size of the regions was 200 bp in length, the same as used by IDEAS for chromatin state prediction. In this example, there are three genes in vicinity of “region 1” (±100 kb): G1, G2, and G3. Next, we generated a matrix of gene expression measures (RPKM values), represented in the matrix on the left in the middle of Figure 1. For each gene, we performed regression between the gene expression, log10(RPKM + 0.001), and the binary measures of a 20-state chromatin model from IDEAS (matrix on the right in the middle of figure). The output is the adjusted r2 between the “region 1” and the three genes. We used only the gene with highest r2 value for a given genomic region, which would be G2 in this example.
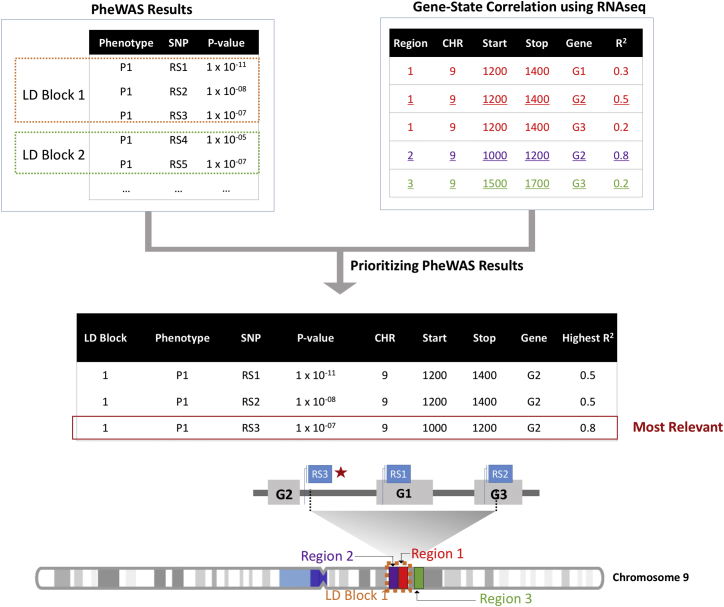
Fine Mapping PheWAS Haplotype Blocks
Due to the presence of linkage disequilibrium (LD) between the SNPs in a genome-wide scale study, many SNPs were in haplotype blocks associated with the same phenotype. We prioritized associations for SNPs in high LD using functional annotations from the above correlation between the chromatin state and gene expression (Figure 2). First, we generated haplotype blocks using pairwise LD calculations for all the SNPs with association p value < 1 × 10−4. For each haplotype block, we then mapped the SNP coordinates of correlated SNPs to the 200 bp regions of pre-computed chromatin state and gene-chromatin state correlations. Thus in a given haplotype block for a phenotype, we prioritized a single variant as the most biologically relevant SNP when it overlapped with a 200 bp region.
Figure 2.
Fine Mapping of PheWAS Results
We annotated our PheWAS associations with most probable chromatin state and the correlation of chromatin state with gene expression data. For each phenotype, we identified a haplotype block of variants with association p values < 1 × 10−4, then we annotated each variant within the haplotype block to identify the variant based on state with the highest value of r2 to the expression of a given gene. In this example, SNP RS3 is the variant overlapping region 2, the region that G2 is located within.
Results
Our results are from an EHR-based PheWAS from a single health care provider using 541 ICD-9 code-based diagnoses and 25 clinical laboratory measurements from 38,622 individuals, 58% female and 42% male with a mean age of 59 years. We executed two separate PheWASs, one using diagnosis codes and other with clinical laboratory measurements, and we investigated associations from each analysis independently as well as combined.
For the diagnosis code-focused PheWAS, we identified 1,118 associations passing our phenome-wide significance cutoff (p value < 1 × 10−11), with 902 SNPs (0.1% of total SNPs) and 27 diseases, listed in Table S1. We collapsed our case/control diagnoses for these results into major disease concept categories to group association results, and in Table 1 we present the most significant associations within each category. We observed that the majority of the top associations were with the type 1 diabetes diagnosis code “250.01.” The most significant association in our study was between SNP rs9273363 and “diabetes mellitus type 1” with p value 1.21 × 10−77, consistent with the previously reported associations of HLA-DQB1 variants with type 1 diabetes as well as other autoimmune disorders such as rheumatoid arthritis.28, 29, 30 Other significant associations included ICD-9 code 250.00 “type 2 diabetes” with SNP rs7903146, ICD-9 code 272.1 “pure hyperglyceridemia” with SNP rs964184, and ICD-9 code 702.0 “actinic keratosis” with SNP rs12203592. In the top associations, we identified two previously unreported associations between the SNP rs12207756 and ICD-9 code 696.1 “psoriasis” and between SNP rs2760985 and “rheumatoid arthritis” (RA) (Tables 1 and S2). The variant rs2760985 is near HLA-DRB1, and there are known GWAS findings with the variants mapped to the gene.31, 32 However, the variant identified in our study is not in linkage disequilibrium (LD) with any known genetic variant associated with RA.
Table 1.
Top Associations with ICD-9 Disease Diagnoses Grouped by Disease Class
| ICD-9 Category | ICD-9 Code | ICD-9 Description | SNP | Gene | Case/Controls | Odds Ratio [CI] | p Value |
|---|---|---|---|---|---|---|---|
| Neoplasms | 238.2 | neoplasm of uncertain behavior of skin | rs12203592 | IRF4 | 2,237/26,680 | 1.39 [1.28,1.51] | 8.21 × 10−15 |
| Endocrine and metabolic | 244.9 | unspecified acquired hypothyroidism | rs965513 | PTCSC2 | 5,282/27,539 | 0.75 [0.71,0.78] | 7.30 × 10−34 |
| 250.00 | type II diabetes mellitus | rs7903146 | TCF7L2 | 9,117/24,057 | 1.29 [1.24,1.34] | 3.13 × 10−40 | |
| 250.01 | type I diabetes mellitus | rs9273363 | – | 756/33,165 | 2.79 [2.5,3.1] | 1.21 × 10−77 | |
| 268.9 | vitamin D deficiency | rs2282679 | GC | 4,423/25,600 | 1.24 [1.18,1.31] | 6.54 × 10−8 | |
| 272.1 | pure hyperglyceridemia | rs964184 | – | 615/33,195 | 1.76 [1.53,2.01] | 6.38 × 10−15 | |
| 272.4 | hyperlipidemia | rs7412 | APOE | 17,804/12,356 | 0.60 [0.56,0.65] | 1.81 × 10−46 | |
| 274.9 | gout | rs1014290 | SLC2A9 | 1,315/32,323 | 0.58 [0.53,0.65] | 1.43 × 10−25 | |
| 278.01 | morbid obesity | rs1421085 | FTO | 5,596/26,295 | 1.28 [1.23,1.34] | 5.31 × 10−32 | |
| Blood disorder | 289.81 | primary hypercoagulable state | rs6687813 | – | 273/34,102 | 6.26 [5.19,7.57] | 6.86 × 10−66 |
| Nervous system | 340 | multiple sclerosis | rs3129860 | – | 324/34,276 | 2.48 [2.09,2.95] | 5.56 × 10−22 |
| 362.5 | macular degeneration (senile) | rs572515 | CFH | 403/33,587 | 2.05 [1.77,2.37] | 6.18 × 10−23 | |
| 362.51 | nonexudative senile macular degeneration | rs395544 | CFH | 561/33,479 | 2.09 [1.84,2.37] | 4.02 × 10−32 | |
| 362.52 | exudative senile macular degeneration | rs1329424 | CFH | 287/34,239 | 2.20 [1.86,2.60] | 2.64 × 10−20 | |
| Circulatory | 427.31 | atrial fibrillation | rs2129982 | – | 3,130/30,481 | 1.43 [1.34,1.53] | 1.34 × 10−27 |
| Digestive | 571.8 | chronic nonalcoholic liver disease | exm1615904 | PNPLA3 | 609/31,328 | 1.98 [1.76,2.23] | 5.12 × 10−28 |
| Genitourinary | 585.3 | chronic kidney disease | rs12917707 | – | 4,076/29,131 | 0.72 [0.68,0.78] | 1.02 × 10−19 |
| 696.1 | psoriasis | rs12207756 | CDSN, PSORS1C1 | 6,60/33,277 | 2.07 [1.80,2.37] | 1.91 × 10−22 | |
| 702 | actinic keratosis | rs12203592 | IRF4 | 2,221/29,745 | 1.83 [1.68,1.99] | 5.17 × 10−43 | |
| Musculoskeletal | 714 | rheumatoid arthritis | rs2760985 | – | 970/33,273 | 1.86 [1.67,2.07] | 1.31 × 10−27 |
| Symptoms | 794.8 | abnormal results of function study of liver | exm1615904 | PNPLA3 | 554/32,681 | 1.68 [1.48,1.90] | 5.25 × 10−15 |
For the clinical lab-measurement PheWAS, our phenome-wide threshold was 1.15 × 10−8 (see Subjects and Methods for more details), and we observed 3,024 associations from 2,109 SNPs (0.3%) with 25 lab measures that were significant at that p value cut-off, listed in Table S2. In our top associations, we had 30 associations with a p value less than or equal to 3.29 × 10−83. In Table 2, we highlight the top statistically significant associations for each clinical laboratory measure investigated in this study. In our previous work, we studied these laboratory measures in a smaller study population and we found that the top results in this analysis are consistent with our previous findings.12 For example, we replicated the association between the UGT1A1 variant rs11568318 and bilirubin levels with p value = 3.29 × 10−83, and the intergenic variant rs7903146 associated with glucose levels with p value = 4.58 × 10−58. We also replicated many previous reported associations with mean corpuscular volume,33 white blood cell counts,33, 34, 35 hematocrit levels,36 and hemoglobin levels.37, 38, 39 Among these top associations, we also identified previously unreported genetic associations with the levels of carbon dioxide, chloride, serum protein, and serum sodium, as well as the measure of anion gap (Tables 2 and S2).
Table 2.
Top Associations for Each Clinical Laboratory from Clinical Lab PheWAS
| Clinical Lab | SNP | Gene | Beta | Sample Size | p Value |
|---|---|---|---|---|---|
| Anion gap | rs1260326 | GCKR | 0.156264 | 31,717 | 3.02 × 10−32 |
| Calcium | rs17251221 | CASR | 0.0678278 | 32,137 | 3.29 × 10−83 |
| Carbon dioxide | rs11465670 | IL18RAP | −0.184063 | 32,440 | 7.98 × 10−17 |
| Chloride | rs1808192 | – | 0.10478 | 32,261 | 6.56 × 10−10 |
| Hematocrit | rs7776054 | – | −0.202675 | 32,299 | 8.54 × 10−12 |
| Hemoglobin | rs855791 | TMPRSS6 | −0.100959 | 32,288 | 2.06 × 10−25 |
| Alanine amino-transferase | exm1615904 | – | 0.0535439 | 30,848 | 2.01 × 10−45 |
| Albumin | rs11671010 | HPN-AS1 | 0.00586762 | 30,879 | 2.38 × 10−12 |
| Alkaline phosphatase | rs635634 | – | −0.0699188 | 30,212 | 3.29 × 10−83 |
| Aspartate amino-transferase | rs35038329 | MRC1 | −0.0391737 | 30,649 | 1.38 × 10−70 |
| Bilirubin | rs11568318 | UGT1A10 | −0.173621 | 30,236 | 3.29 × 10−83 |
| Creatinine | rs12917707 | UMOD | −0.0200995 | 32,625 | 1.58 × 10−21 |
| RDW | rs855791 | TMPRSS6 | 0.00741513 | 31,880 | 6.74 × 10−53 |
| Glucose | rs7903146 | TCF7L2 | 0.0244817 | 32,241 | 4.58 × 10−57 |
| WBC counts | rs2227315 | CSF3 | 0.0301915 | 32,754 | 5.64 × 10−42 |
| Mean corpuscular hemoglobin concentration | rs855791 | TMPRSS6 | −0.103003 | 32,164 | 9.99 × 10−64 |
| Mean corpuscular hemoglobin | rs7775698 | – | 0.28882 | 32,186 | 3.29 × 10−83 |
| Mean corpuscular volume | rs9376092 | LOC105378010 | 0.686955 | 32,288 | 3.29 × 10−83 |
| Platelet counts | rs1354034 | ARHGEF3 | −5.81988 | 32,140 | 1.88 × 10−41 |
| Platelet mean volume | rs342293 | CTB-30L5.1 | 0.149491 | 32,439 | 2.42 × 10−64 |
| Potassium | rs4557401 | FAM13B | −0.0194743 | 32,322 | 1.99 × 10−13 |
| Serum protein | rs3132451 | AIF1 | −0.0294945 | 29,844 | 2.76 × 10−13 |
| RBC counts | rs7776054 | – | −0.0587365 | 32,230 | 1.56 × 10−61 |
| Serum sodium | rs10777939 | – | −0.0958484 | 32,277 | 9.42 × 10−13 |
| Urea nitrogen | rs2287921 | RASIP1 | −0.201762 | 32,104 | 1.24 × 10−10 |
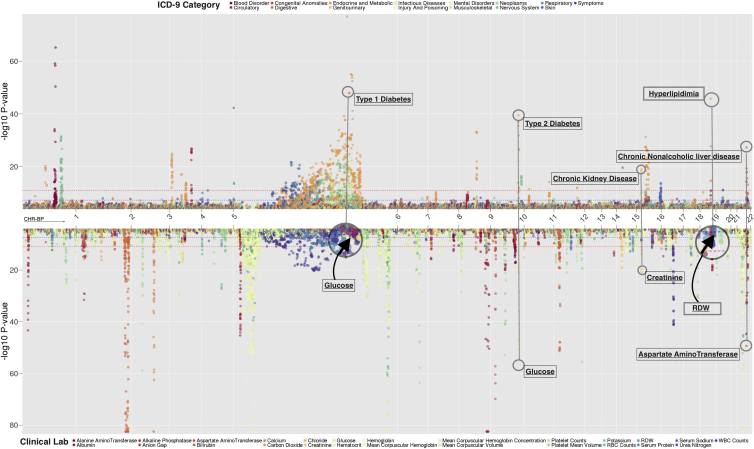
Landscape of PheWAS Associations
The high number of genetic associations of this study, coupled with the variations in power that occur within PheWASs due to varying sample sizes across phenotypes and differing effect sizes, can result in associations that do not pass a more stringent Bonferroni correction but that are still biologically relevant and plausible. Therefore, in this study we also studied all results passing a more exploratory p value of 1 × 10−4. In Figure 3, we showcase the landscape of our associations across the genome that include results confirming many previously reported associations as well as some previously unreported findings. For example, the diagnoses of “morbid obesity” (rs1421085 and p value = 5.31 × 10−32), “type 1 diabetes” (rs9273363 and p value = 1.21 × 10−77), and “primary hypercoagulable state” (rs6687813 and p value = 6.86 × 10−66) were the phenotypes of the most significant genetic associations and were supported by previously reported genetic associations with same or related phenotypes.28, 40, 41, 42 For the clinical laboratory measurements (Table 2), the median levels of bilirubin (rs11568318 and p value = 3.29 × 10−83), alkaline phosphatase (rs635634 and p value = 3.29 × 10−83), white blood cell count (rs2227315 and p value = 5.64 × 10−42), and mean corpuscular hemoglobin (rs855791 and p value = 9.99 × 10−64) replicated previously reported studies.43, 44
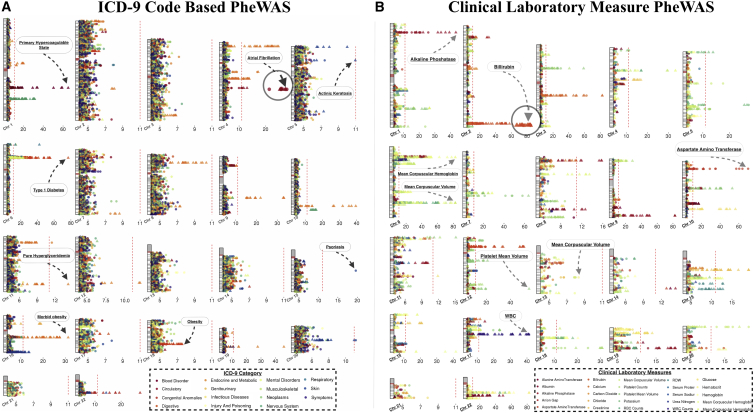
Figure 3.
Landscape of Genome-wide PheWAS Results
We plotted the association results with p value < 1 × 10−4, using −log10(p value). Each association is represented in relation to the SNP location on each chromosome and the points are color-coded by ICD-9 code categories in (A) and clinical laboratory measures in (B). A triangle indicates that the association is previously reported and a circle represents a previously unreported association. The red line is at the phenome-wide significance p value threshold for each PheWAS. We indicated the phenotypes of a few of the most significant associations.
Comparing Results for ICD-9 Codes and Clinical Lab Measures
Clinical lab measures provide a representation of human health, and these measures play a critical role in disease diagnosis. In our previous work, we showed the robustness in the interpretation of disease associations by analyzing the ICD-9 code and median clinical lab measure association results in conjunction.12 In Figure 4, we present a position by position comparison between ICD-9 and clinical laboratory measure associations.
Figure 4.
Integrating ICD-9 and Clinical Lab PheWAS
We present a position-by-position comparison of genetic associations the two PheWASs, one with 541 ICD-9 diagnosis codes and the other with 25 clinical laboratory measures. The horizontal axis represents genomic locations by each chromosome and the vertical axis is the –log10(p value) of the associations. The red and blue dotted lines are the phenome-wide significance threshold for ICD-9 and clinical lab PheWAS, respectively. We annotated examples of associations between the same SNP and highly related phenotypes across the two PheWASs.
Blood glucose levels are common clinical tests to diagnose diabetes. We identified 220 associations where multiple loci are associated with median glucose levels as well as diabetes, including the intergenic variant rs9275495 downstream of MTCO3P1 that was phenome-wide significant for association with “type 1 diabetes” (p value = 7.83 × 10−56) and glucose levels (p value = 5.32 × 10−9, Table 3). MTCO3P1 is a pseudogene located in the MHC complex, and SNPs in multiple genes in the MHC complex have been known to have associations with type 1 diabetes (such as rs2647044 in HLA-DQB1).45 However, rs9275495 is 5.6 kb upstream of rs2647044, and there is weak LD detected between these two loci in our study population (r2 = 0.02), suggesting that this is a potential novel locus for type 1 diabetes.
Table 3.
Genetic Associations from Integrated Result Interpretation between ICD-9 Codes and Clinical Laboratory Measure PheWASs
| ICD-9 Code | ICD-9 Description | Clinical Lab | SNP | Gene | Case/Controls | Odds Ratio [CI] | p Value (ICD-9) | Lab Sample Size | Lab Beta | p Value (Lab) |
|---|---|---|---|---|---|---|---|---|---|---|
| 250.01 | type 1 diabetes | glucose | rs9275495 | MTCO3P1 (closest) | 756/33,131 | 2.90 [2.57,3.28] | 7.83 × 10−56 | 32,195 | 0.01 | 5.32 × 10−9 |
| 585.3 | chronic kidney disease | creatinine | rs12917707 | UMOD (closest) | 4,076/29,131 | 0.72 [0.68,0.78] | 1.02 × 10−19 | 32,625 | −0.02 | 1.5 × 10−21 |
| 285.9 | anemia | mean corpuscular hemoglobin levels | rs855791 | TMPRSS6 | 2,822/26,165 | 1.13 [1.07,1.20] | 1.1 × 10−05 | 32,149 | −0.26 | 3.29 × 10−83 |
| 272.2 | hyperlipidemia | erythrocytes distribution width (RDW) | rs7412 | APOE | 17,804/12,356 | 0.60 [0.56,0.65] | 1.8 × 10−46 | 31,766 | 0.005 | 8.74 × 10−9 |
For chronic kidney disease (ICD-9 585.3), the SNP rs12917707 (p value = 2.58 × 10−19, Table 3) upstream of UMOD was also associated with creatinine levels in serum plasma (p value = 1.58 × 10−21). For both creatinine and kidney-related diseases, we replicate previous findings.46, 47, 48
We also replicated the most significant associations with variants in the gene TMPRSS6 and the diagnosis of “anemia” as well as the levels of mean corpuscular hemoglobin concentration in red blood cells.37, 38, 49, 50 For example, in our study, we detected an association between missense variant rs855791 and the diagnosis of “anemia” (ICD-9 code 285.9, p value = 1.128 × 10−5, Table 3) as mean corpuscular hemoglobin levels (p value = 2.29 × 10−83, Table 3).
A previously reported association of APOE variant rs7412 with lipid traits was also replicated in our study with the diagnosis of “hyperlipidemia” (p value = 1.8 × 10−46).51, 52 In the clinical laboratory measurement analysis, we detected the same polymorphism associated with red blood cell distribution width (RDW) (p value = 8.74 × 10−9). The minor allele of the pathogenic missense variant rs7412 is commonly known as the E2 polymorphism of APOE, and many independent studies show association between this SNP and cardiovascular risks and early vascular diseases. Recent findings suggest that RDW has a high correlation with long-term cardiovascular events, supporting this association in our study population.53
Comparing Associations to Previous PheWASs
There have now been many published PheWASs, and we compared our ICD-9-based diagnosis results to previously reported PheWAS associations. PheWAS analyses have many challenges including the potential noise incorporated into analyses due to the fact that EHR were not designed for research purposes. Also, the differences in the sample size for each ICD-9 code can impact the statistical power to detect associations in PheWASs. As a result, the significance of associations for these PheWAS analyses may not reach the threshold for inclusion in genetic association catalogs such as the GWAS catalog and GRASP.
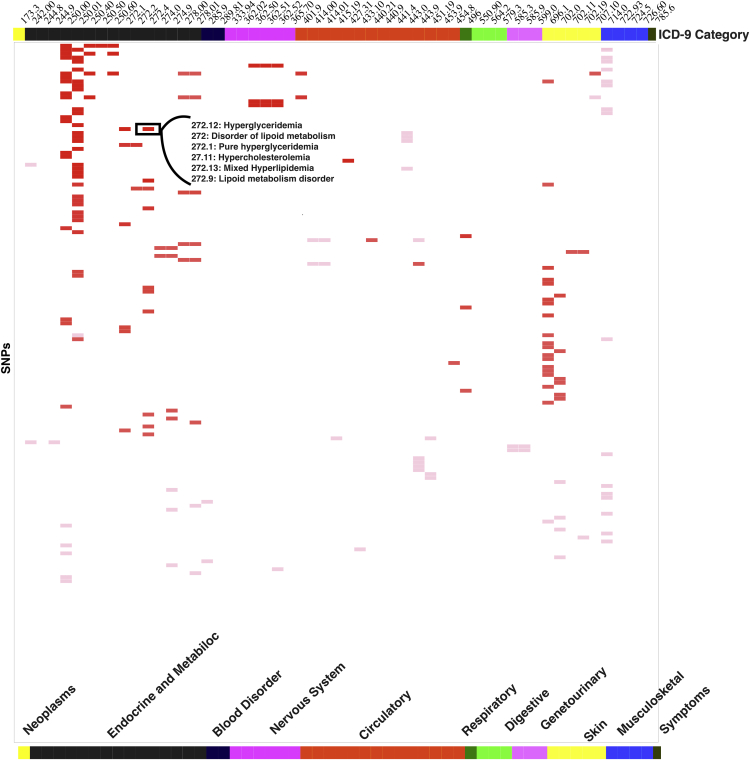
For this study, we further extended our investigation of results of this study by comparing results from our study to previously published PheWASs that used ICD-9-based case/control diagnoses, a total of eight other studies. We did this comparison using ICD-9 codes at the 3-digit level. This strategy has been found to be a robust way to compare PheWAS results across studies, as collapsing ICD-9 codes at the three-digit level identifies similar diagnoses but does not require the exact specificity of the five-digit level and accounts for the variability in ICD-9 use at different institutions.4 We replicated a total of 950 associations with previously published PheWASs. In Figure 5, we present associations replicated within each broader three-digit-based ICD-9 category. The majority (68%) of the replicating associations were with endocrine and metabolic disorders. For example, in Figure 5, the SNP rs964184 was associated with the diagnosis “pure hyperglyceridemia” (ICD-9 code 272.1), p value = 6.3 × 10−15 and we identified associations for this SNP with the 3-digit level of ICD-9 diagnosis code for “hyperlipidemia” (ICD-9 code: 272) in previous PheWASs.4, 10, 54, 55
Figure 5.
Replicating Published PheWASs
Here we plotted SNP phenotype associations replicating previously published PheWAS results from studies using ICD-9 code-based diagnoses. The top axis is all the ICD-9 codes from this study, and the rows represent SNPs. The gradient of the color in the matrix represents the number of associations replicating between our study and existing PheWAS results for each SNP-phenotype pair.
Coding and Non-coding Genomic Regions
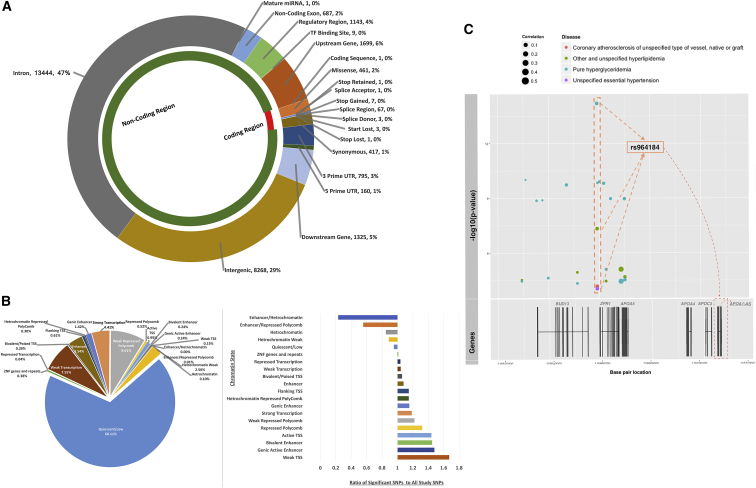
PheWAS provides a way to scan millions of associations between a wide range of phenotypes and genetic markers. However, an important aspect of PheWAS is leveraging the complex results to gain a greater understanding of the impact of the genetic architecture of an outcome beyond focusing on single SNP-phenotype associations and interpreting associations only through the impact of protein-coding genes. Approximately 90% of the genome-wide studies have identified statistically significant variants outside of protein-coding regions.16 We can use the amount of knowledge accumulated to date regarding activity and importance of the non-coding genome to better understand the results of this study, specifically for the variants outside of protein-coding regions. Using the Variant Effect Predictor (VEP), we characterized SNPs of the ICD-9 code PheWAS associations into coding and non-coding regions for results passing p < 1 × 10−4 (Figure 6A). We observed that almost 90% of the SNPs were represented in the non-coding or regulatory region of the genome. A total of 29% were intergenic variants, 11% were downstream or upstream of protein-coding regions, and the rest were located in untranslated, regulatory, non-coding exons, or splicing regions. In the protein-coding regions, we found 891 variants in exons, with the majority of these being missense variants (461 SNPs), synonymous variants (417 SNPs), and stop-gained variants (7 SNPs). When comparing to polymorphisms reported in the GWAS catalog, we found that the distribution of the location of the SNPs in the genome was similar to our results (Figure S2). We also compared our results to the overall genetic variants represented on the genotyping chip (Figure S2). Almost 80% of the variants found on the chip are located in the untranslated regions (49% 3′ UTR and 30% 5′ UTR).
Figure 6.
Functional Annotations (p Value < 1 × 10−4)
(A) We used Variant Effect Predictor (VEP) to identify functional consequences of the genetic variants in our study for the ICD-9-based PheWAS results. The plot shows the number of variants of each type of predicted consequence classifying SNPs across the coding and noncoding regions of the genome.
(B) The pie chart on the left is the representation of SNPs annotated to the most probable chromatin region across 127 epigenomes. The plot on the right shows the overall representation of each chromatin state for variants with significant results when compared to annotations of all the variants included in the study.
(C) In the plot, the scatterplot represents associations within a haplotype block on chromosome 11 where the horizontal axis is base pair location and –log10(p value) is shown on the vertical axis. The color of the circles represent phenotypes and size of the circle corresponds to pre-computed gene correlation measure (r2) for that region in roadmap epigenomes. The genes close by to that haplotype block are represented below the scatterplot. Based on haplotype block annotations, SNP rs964184 shows the highest correlation (highest r2) with the expression of APOA1.
Figure 6B shows genetic variation from non-protein coding regions mapped to 20 different chromatin states. By mapping genetic variation in non-coding regions to the tissue-specific data in the Roadmap Epigenomics, we identified that 68% of the associations in our study with a p value < 1 × 10−4 overlap with genomic locations in quiescent or low activity regions of the genome (Figure 6B). Of the rest of the SNPs, there were primarily four broad categories of region/activity that the SNPs mapped to: transcription start sites (2%), transcription (12%), enhancers (5%), and repressed polycomb (9%). As shown in Figure 6B, the pie chart represents the number of variants mapped to 20 different chromatin states based on chromosome base pair location. While the majority of variants map to the quiescent state, which is found consistent with previously reported mapping of variants to chromatin states in 127 epigenomes,56 there are important considerations that may explain how these SNPs have an impact. We averaged the predicted state across multiple tissues, and in some instances, these genetic variants may have a regulatory effect in a single or subset of tissues where a region is not in the quiescent state and that is not captured in our approach.
The number of significant variants mapping to chromatin states does not show how each chromatin state is represented in comparison to all variants used in analysis. Therefore, we calculated ratios of variants mapping to chromatin states for all genotype variants and also for variants at p value < 1 × 10−4 and then calculated the over- and under-representation of each state via the ratio. The bar plot in Figure 6B illustrates this representation for each chromatin state, and it highlights that even though the majority of the significant variants are in low activity state, such states are underrepresented when compared to chromatin state annotation of all the SNPs included in the study. We found that active chromatin states such as “weak transcription start site (TSS),” “genic active enhancer,” and “bivalent enhancer,” among others are the over-represented states.
PheWAS Associations (ICD-9) in Protein-Coding Regions
In a protein-coding region, we identified a stop-gained variant rs701884 in the gene HLA-DRB5, which was significantly associated with two autoimmune disorders: “type 1 diabetes” (p value = 2.53 × 10−28, Table S2) and multiple sclerosis (p value = 2.64 × 10−20, Table S2). There is a known SNP, rs26819262, in HLA-DRB5 in close LD with SNP rs701884 associated with multiple sclerosis, but not with type 1 diabetes. The gene HLA-DRB5 is one of the paralogs of HLA-DRB1, and many polymorphisms in HLA-DRB5 have previously reported the association with type 1 diabetes, suggesting that the finding in our study could be an indirect association with type 1 diabetes.28, 45, 57, 58, 59
A total of 48 associations were with missense variants, with the most significant association between the SNP rs7412 and “hyperlipidemia,” a well-reported finding in previous genome- and phenome-wide studies.10, 51, 52 For type 1 diabetes, we identified multiple missense variations in MHC complex significantly associated with this diagnosis including the SNPs rs1130399 (p value = 9.37 × 10−32, OR = 2.02 [1.80, 2.26]), rs1129740 (p value = 2.07 × 10−28, OR = 0.53 [0.48, 0.60]), rs1071630 (p value = 2.67 × 10−28, OR = 0.54 [0.48, 0.60]), and rs1049060 (p value = 3.56 × 10−28, OR = 0.53 [0.48, 0.60]) (Table S2). The locus of these variants lie within HLA-DRB1 and HLA-DRA1 and both of the genes have been previously reported with type 1 diabetes.28, 45, 59
For synonymous polymorphisms, we identified the SNP rs1061147 (CFH) to be significantly associated with “non-exudative senile macular degeneration” (p value = 1.04 × 10−30, OR = 2.05 [1.81, 2.33], Table S2). This SNP has been previously associated with age-related macular degeneration but with lower odds ratio (1.4 [1.32–1.48]).60 Additionally, other associations were found with synonymous polymorphisms including the SNP rs1475865 and “chronic kidney disease, stage III” (ICD-9 585.3, p value = 1.20 × 10−19, Table S2), between rs2076529 and “rheumatoid arthritis” (ICD-9 714.0, p value = 6.78 × 10−15, Table S2),29, 61 and between rs10939650 and “gout, unspecified” (ICD-9 274.9, p value = 3.24 × 10−24, Table S2).62
PheWAS Associations (ICD-9 Codes) with Non-Protein-Coding Regions
The SNP rs7850258 in a regulatory region had strongest association with ICD-9 code 244.9 “acquired hypothyroidism” (p value = 9.29 × 10−34, Table S2). Previous associations with hypothyroidism have been reported for FOXE1, which is downstream of the variant identified in our study.63 There were 21 associations within active transcription start sites (TSSs), with the most number of associations with the ICD-9 code 250.01 diagnosis “type 1 diabetes.” The SNP rs3749981 is a non-coding transcript variant and had the strongest association with type 1 diabetes (p value = 3.96 × 10−25, OR = 2.23 [1.95, 2.58], Table S2). The only known association for this active TSS variant is with autoimmune disorder rheumatoid arthritis.29 For ICD-9 code 696.1 “psoriasis,” there were three 5′ UTR SNPs near active TSSs, also in high LD with each other and representing a potential novel association for this disease diagnosis (rs2074510, rs1052693, rs9468842). The SNPs rs2074510 (p value = 9.4 × 10−12, OR = 1.56 [1.38, 1.77], Table S2) and rs1052693 (p value = 8.9 × 10−12, OR = 1.56 [1.38, 1.77], Table S2) map to GTF2H4 and the SNP rs9468842 (p value = 1.5 × 10−11, OR = 1.55 [1.37, 1.76]) is located within DDR1, and there are no known association of these genes with psoriasis.
Within a strong transcription region, an intronic variant rs4823173 in PNPLA3 is the strongest association with ICD-9 code 571.8 “other chronic non-alcoholic liver disease” (p value = 4.67 × 10−20, OR = 1.87 [1.65, 2.13], Table S2). There are known associations between variants in PNPLA3 and non-alcoholic liver diseases and the genetic variant found in our analysis is in strong LD with previously identified variants.64
Identifying Genetic Associations Most Correlated with Gene Expression
Using PLINK, we first identified haplotype blocks from LD correlations between the SNPs in the genotype data from 38,622 European American populations in our study. We found a total of 14,764 haplotype blocks with block size ranging from 2 bp to 900 kb. Then we identified the genetic variants from the PheWAS results most correlated with gene expression changes in specific genes by using the correlations between chromatin state and gene expression measures from 56 epigenomes available through Roadmap Epigenomics Project (see Subjects and Methods for more details). For example, in a haplotype block of 88.6 kb size on chromosome 6 with 37 SNPs in LD, we identified that DDR1 variant rs9501032 was associated with ICD-9 code 696.1 “psoriasis” (p value = 1.44 × 10−11), and it is located in an enhancer region with correlation (r2) of 0.45 with DDR1 expression. There is no previously reported association for this finding.
For another example, the intergenic variant rs2038024 in strong LD with two SNPs is associated with the primary hypercoagulable state (ICD-9 289.81), and it is located in an active TSS and has a low correlation with the expression of the pseudogene RP1-206D15.5 (r2 = 0.174). It is a pseudogene, so potentially the association effect might be a result of links with variants in other paralogous genes.
Within a 52 kb haplotype block on chromosome 11, an intergenic variant rs964184 had a strong association with hyperlipidemia (ICD-9 272.1, 272.4) as well as coronary atherosclerosis (ICD-9 414.00) and essential hypertension (ICD-9 401.9) at an exploratory significance cutoff (Table S3). This variant is 359 base pairs upstream of ZPR1, and it is predicted to be in strong transcription region (Figure 6C). We also identified that the 200 bp region of strong transcription has a slight LD correlation with a long non-coding RNA (lncRNA) APOA1-AS (r2 = 0.22). Although recent studies suggest that lncRNA are involved in the function of protein-coding genes, there are only few that have been found involved in disease mechanism. APOA1-AS has been found to influence APOA1 and APOC3 genes by initiating the transcriptional upregulation.65
Discussion
We present one of the most comprehensive PheWAS analyses to date using EHR from a single health care provider linked to a genetic biobank through the Geisinger biorepository, the MyCode Community Health Initiative. Many findings of our study replicated previously reported associations in non-EHR-based genetic studies with the same or similar phenotypes, supporting the results of our EHR-based genome-wide PheWASs. We observed that the most significant associations were identified in the MHC complex on chromosome 6, which reflects the importance of this region on human health and impact on disease pathogenesis. Most of the associations in the MHC gene region were found associated with autoimmune diseases such as type 1 diabetes, rheumatoid arthritis, and multiple sclerosis.
Additionally, using the genome-wide landscape of this comprehensive PheWAS, we detected multiple previously unreported genetic associations with essential hypertension, occlusion and stenosis of carotid, psoriasis, and depressive disorders. For example, a polymorphism on chromosome 7p21.1 in HDAC9 was suggestively significantly associated with ICD-9 433.10 “occlusion and stenosis of carotid” (the narrowing of arteries due to plaque deposits) (p value = 2.26 × 10−8). Variations in HDCA9 have previous associations with coronary artery disease (CAD) such as ischemic stroke,66 but there are no previously reported genetic associations between rs2074633 and our specific phenotype or other CAD phenotypes. The expression of HDAC9 is increased in carotid atherosclerotic plaques, in greater quantities than even femoral plaques. There is also a potential involvement of this gene in a mechanistic pathway, where HDAC9 has been shown to inhibit FOXP3 expression and the function of regulatory T cells, which protect against atherosclerosis.67
Another previously unreported association, between SNP rs1275980 located within intronic region of KCNK3, was associated with ICD-9 code 401.9 “essential hypertension” (p value = 9.27 × 10−8). The diagnosis code 401.9 has the highest case number within our PheWAS (17,975), making it is one of the most well-powered associations of this study. The gene KCNK3 (potassium two-pore domain channel subfamily K member 3) encodes a protein of potassium channel family, and the gene product contributes to regulation of blood pH levels. Variants in this gene have been found linked to aldosterone hormone production, systolic blood pressure,68 body mass index, and mean arterial pressure.69, 70 Aldosterone affects the body’s ability to regulate blood pressure and is one of the causative factors in hypertension. Based on the regulatory annotations, we found that the SNP rs1275980 is located in the repressed polycomb chromatin state, and correlation (r2) of 0.25 with KCNK3 suggests a minor relationship to the gene expression of KCNK3. Also, the SNP rs1275980 was significantly associated with blood CO2 levels in the PheWAS with clinical laboratory measures. A previous study suggests that CO2 levels in blood plasma reflect different cerebrovascular responses, and more importantly there is a positive correlation between increased blood pressure and CO2 levels.71
CRHR-2 is one of the receptors of corticotropin-releasing hormone (CRH) and is found in various regions of the brain that play an important role in responding to stress, anxiety, fear, and arousal,72 and the receptor is synthesized in the brain in response to stress. An intronic variant rs255112 within CRHR-2 was associated with the diagnosis ICD-9 296.90 “episodic mood disorder” (p value = 1.28 × 10−6) in our study. The variant is located in the second intron of CRHR2 and this gene has previous known associations with lower physiological responses to anxiety and stress and psychiatric disorders including depression, PTSD, and bipolar disorder.72 The variant is also enriched in various tissues including brain, and it maps to the non-coding region with a predicted weak-repressed-polycomb chromatic state.
Lastly, we identified a previously unreported association with the USP8 SNP on chromosome 15 (rs148783236) associated with the diagnosis of psoriasis (ICD-9 code 696.1, p value = 2.54 × 10−20). This finding may present new insights into underlying mechanism of the disease. Ubiquitin-specific-processing protease 8 (USP8) is involved in cell proliferation and plays an important regulatory role in epidermal growth factor receptor (EFGR) degradation.73 EFGR binds with epidermal growth factor (EGF) and regulates cell growth, proliferation, and differentiation of cells in the epidermis. The increase in EGF and decrease in EGFR are reported in individuals with psoriasis, suggesting a critical role in the pathogenesis of the disease.74 Of note, there are many pseudogenes across the genome processed within the chromosome 15q21 region including the pseudogene USP8P1, upstream of the HLA-C in major histocompatibility complex (MHC). HLA-C is also known as psoriasis susceptibility 1 (PSORS1) and is a part of the cluster of genes in MHC region on chromosome 6 with susceptibility to psoriasis and systemic sclerosis. However, there is no sequence similarity between the two regions of USP8 and USP8P1, suggesting that this observation was not due to cross-hybridization.75, 76 It is also worth noting that rs148783236 is located within a strong transcription region, which supports that it could provide regulation for USP8.
There were some limitations of this study, which can be addressed through future analyses with these data. Genome-wide array technology identifies tag SNPs, so many of the SNPs of this study may be highly correlated with the actual causative genetic variation, and thus our functional annotation is not necessarily for the SNP impacting phenotypic variability. Our future directions include further analysis of the novel results of this study using imputed data, as well as exploring rare genetic variations within genes using whole-exome sequencing data. We incorporated chromatin state predictions for the non-coding region by averaging the probability of chromatin state across multiple tissues. Thus, there were many variants located in the low activity regions of the genome, also referred to as the quiescent state. However, there are likely many scenarios where genetic regions are not quiescent in one or more tissues, but their specificity was lost when activity levels are averaged across all tissues. A future direction is to further delve into the impact of variants on individual tissues and link that back to the diagnosis and trait associations that we identified in this study.
In conclusion, we provide a landscape of associations across diseases and quantitative traits through a comprehensive PheWAS using EHR data from a single health care provider. For this study, we also presented additional ways to expand the understanding of association findings through the use of existing expert knowledge. We addressed the computational challenges of such studies at such large scale by utilizing resources through cloud computing. With ever-increasing genomic data and study participation, we believe that use of the cloud computing will become more common. The findings of our study serve as an excellent resource for hypothesis generation for targeted future studies.
Acknowledgments
This project is funded, in part, under a grant with the Pennsylvania Department of Health (#SAP 4100070267). The Department specifically disclaims responsibility for any analyses, interpretations, or conclusions.
Published: March 29, 2018
Footnotes
Supplemental Data include two figures and three tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.02.017.
Accession Numbers
Additional information for reproducing the results described in the article is available upon reasonable request and subject to a data use agreement.
Web Resources
DNAnexus, https://www.dnanexus.com/
Supplemental Data
References
- 1.Hall M.A., Verma A., Brown-Gentry K.D., Goodloe R., Boston J., Wilson S., McClellan B., Sutcliffe C., Dilks H.H., Gillani N.B. Detection of pleiotropy through a phenome-wide association study (PheWAS) of epidemiologic data as part of the Environmental Architecture for Genes Linked to Environment (EAGLE) study. PLoS Genet. 2014;10:e1004678. doi: 10.1371/journal.pgen.1004678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M., Crawford D.C. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pendergrass S.A., Brown-Gentry K., Dudek S., Frase A., Torstenson E.S., Goodloe R., Ambite J.L., Avery C.L., Buyske S., Bůžková P. Phenome-wide association study (PheWAS) for detection of pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network. PLoS Genet. 2013;9:e1003087. doi: 10.1371/journal.pgen.1003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verma A., Basile A.O., Bradford Y., Kuivaniemi H., Tromp G., Carey D., Gerhard G.S., Crowe J.E., Jr., Ritchie M.D., Pendergrass S.A. Phenome-wide association study to explore relationships between immune system related genetic loci and complex traits and diseases. PLoS ONE. 2016;11:e0160573. doi: 10.1371/journal.pone.0160573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Namjou B., Marsolo K., Caroll R.J., Denny J.C., Ritchie M.D., Verma S.S., Lingren T., Porollo A., Cobb B.L., Perry C. Phenome-wide association study (PheWAS) in EMR-linked pediatric cohorts, genetically links PLCL1 to speech language development and IL5-IL13 to eosinophilic esophagitis. Front. Genet. 2014;5:401. doi: 10.3389/fgene.2014.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doss J., Mo H., Carroll R.J., Crofford L.J., Denny J.C. Phenome-wide association study of rheumatoid arthritis subgroups identifies association between seronegative disease and fibromyalgia. Arthritis Rheumatol. 2017;69:291–300. doi: 10.1002/art.39851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebbring S.J., Schrodi S.J., Ye Z., Zhou Z., Page D., Brilliant M.H. A PheWAS approach in studying HLA-DRB1∗1501. Genes Immun. 2013;14:187–191. doi: 10.1038/gene.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hebbring S.J., Rastegar-Mojarad M., Ye Z., Mayer J., Jacobson C., Lin S. Application of clinical text data for phenome-wide association studies (PheWASs) Bioinformatics. 2015;31:1981–1987. doi: 10.1093/bioinformatics/btv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarty C.A., Chisholm R.L., Chute C.G., Kullo I.J., Jarvik G.P., Larson E.B., Li R., Masys D.R., Ritchie M.D., Roden D.M., eMERGE Team The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma A., Verma S.S., Pendergrass S.A., Crawford D.C., Crosslin D.R., Kuivaniemi H., Bush W.S., Bradford Y., Kullo I., Bielinski S.J. eMERGE Phenome-Wide Association Study (PheWAS) identifies clinical associations and pleiotropy for stop-gain variants. BMC Med. Genomics. 2016;9(Suppl 1):32. doi: 10.1186/s12920-016-0191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Canela-Xandri, O., Rawlik, K., and Tenesa, A. (2017). An atlas of genetic associations in UK Biobank. bioRxiv. https://doi.org/10.1101/176834. [DOI] [PMC free article] [PubMed]
- 12.Verma A., Leader J.B., Verma S.S., Frase A., Wallace J., Dudek S., Van Hout C.V., Dewey F.E., Penn J., Lopez A. Integrating clinical laboratory measures and ICD-9 code diagnoses in phenome-wide association studies. Pac. Symp. Biocomput. 2016;21:168–179. [PMC free article] [PubMed] [Google Scholar]
- 13.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eicher J.D., Landowski C., Stackhouse B., Sloan A., Chen W., Jensen N., Lien J.-P., Leslie R., Johnson A.D. GRASP v2.0: an update on the genome-wide repository of associations between SNPs and phenotypes. Nucleic Acids Res. 2015;43:D799–D804. doi: 10.1093/nar/gku1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leslie R., O’Donnell C.J., Johnson A.D. GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics. 2014;30:i185–i194. doi: 10.1093/bioinformatics/btu273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward L.D., Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carey D.J., Fetterolf S.N., Davis F.D., Faucett W.A., Kirchner H.L., Mirshahi U., Murray M.F., Smelser D.T., Gerhard G.S., Ledbetter D.H. The Geisinger MyCode community health initiative: an electronic health record-linked biobank for precision medicine research. Genet. Med. 2016;18:906–913. doi: 10.1038/gim.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verma, A., Bradford, Y., Dudek, S., Verma, S.S., Pendergrass, S.A., and Ritchie, M.D. (2017). A simulation study investigating power estimates in phenome-wide association studies. bioRxiv. https://doi.org/10.1101/115550. [DOI] [PMC free article] [PubMed]
- 22.Verma S.S., Lucas A.M., Lavage D.R., Leader J.B., Metpally R., Krishnamurthy S., Dewey F., Borecki I., Lopez A., Overton J. Identifying genetic associations with variability in metabolic health and blood count laboratory values: diving into the quantitative traits by leveraging longitudinal data from an EHR. Pac. Symp. Biocomput. 2017;22:533–544. doi: 10.1142/9789813207813_0049. [DOI] [PubMed] [Google Scholar]
- 23.Hall M.A., Wallace J., Lucas A., Kim D., Basile A.O., Verma S.S., McCarty C.A., Brilliant M.H., Peissig P.L., Kitchner T.E. PLATO software provides analytic framework for investigating complexity beyond genome-wide association studies. Nat. Commun. 2017;8:1167. doi: 10.1038/s41467-017-00802-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobota R.S., Shriner D., Kodaman N., Goodloe R., Zheng W., Gao Y.-T., Edwards T.L., Amos C.I., Williams S.M. Addressing population-specific multiple testing burdens in genetic association studies. Ann. Hum. Genet. 2015;79:136–147. doi: 10.1111/ahg.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y., An L., Yue F., Hardison R.C. Jointly characterizing epigenetic dynamics across multiple human cell types. Nucleic Acids Res. 2016;44:6721–6731. doi: 10.1093/nar/gkw278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Hardison R.C. Accurate and reproducible functional maps in 127 human cell types via 2D genome segmentation. Nucleic Acids Res. 2017;45:9823–9836. doi: 10.1093/nar/gkx659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stahl E.A., Raychaudhuri S., Remmers E.F., Xie G., Eyre S., Thomson B.P., Li Y., Kurreeman F.A.S., Zhernakova A., Hinks A., BIRAC Consortium. YEAR Consortium Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Rooy D.P.C., Zhernakova A., Tsonaka R., Willemze A., Kurreeman B.A., Trynka G., van Toorn L., Toes R.E.M., Huizinga T.W.J., Houwing-Duistermaat J.J. A genetic variant in the region of MMP-9 is associated with serum levels and progression of joint damage in rheumatoid arthritis. Ann. Rheum. Dis. 2014;73:1163–1169. doi: 10.1136/annrheumdis-2013-203375. [DOI] [PubMed] [Google Scholar]
- 31.Scott R.A., Scott L.J., Mägi R., Marullo L., Gaulton K.J., Kaakinen M., Pervjakova N., Pers T.H., Johnson A.D., Eicher J.D., DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nischwitz S., Cepok S., Kroner A., Wolf C., Knop M., Müller-Sarnowski F., Pfister H., Roeske D., Rieckmann P., Hemmer B. Evidence for VAV2 and ZNF433 as susceptibility genes for multiple sclerosis. J. Neuroimmunol. 2010;227:162–166. doi: 10.1016/j.jneuroim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Kamatani Y., Matsuda K., Okada Y., Kubo M., Hosono N., Daigo Y., Nakamura Y., Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 34.Nalls M.A., Couper D.J., Tanaka T., van Rooij F.J.A., Chen M.-H., Smith A.V., Toniolo D., Zakai N.A., Yang Q., Greinacher A. Multiple loci are associated with white blood cell phenotypes. PLoS Genet. 2011;7:e1002113. doi: 10.1371/journal.pgen.1002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okada Y., Kamatani Y., Takahashi A., Matsuda K., Hosono N., Ohmiya H., Daigo Y., Yamamoto K., Kubo M., Nakamura Y., Kamatani N. Common variations in PSMD3-CSF3 and PLCB4 are associated with neutrophil count. Hum. Mol. Genet. 2010;19:2079–2085. doi: 10.1093/hmg/ddq080. [DOI] [PubMed] [Google Scholar]
- 36.van Rooij F.J.A., Qayyum R., Smith A.V., Zhou Y., Trompet S., Tanaka T., Keller M.F., Chang L.-C., Schmidt H., Yang M.-L., BioBank Japan Project Genome-wide trans-ethnic meta-analysis identifies seven genetic loci influencing erythrocyte traits and a role for RBPMS in erythropoiesis. Am. J. Hum. Genet. 2017;100:51–63. doi: 10.1016/j.ajhg.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganesh S.K., Zakai N.A., van Rooij F.J.A., Soranzo N., Smith A.V., Nalls M.A., Chen M.-H., Kottgen A., Glazer N.L., Dehghan A. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat. Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chambers J.C., Zhang W., Li Y., Sehmi J., Wass M.N., Zabaneh D., Hoggart C., Bayele H., McCarthy M.I., Peltonen L. Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. Nat. Genet. 2009;41:1170–1172. doi: 10.1038/ng.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodonsky C.J., Jain D., Schick U.M., Morrison J.V., Brown L., McHugh C.P., Schurmann C., Chen D.D., Liu Y.M., Auer P.L. Genome-wide association study of red blood cell traits in Hispanics/Latinos: The Hispanic Community Health Study/Study of Latinos. PLoS Genet. 2017;13:e1006760. doi: 10.1371/journal.pgen.1006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Germain M., Saut N., Greliche N., Dina C., Lambert J.-C., Perret C., Cohen W., Oudot-Mellakh T., Antoni G., Alessi M.-C. Genetics of venous thrombosis: insights from a new genome wide association study. PLoS ONE. 2011;6:e25581. doi: 10.1371/journal.pone.0025581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler E., Huang N., Bochukova E.G., Keogh J.M., Lindsay S., Garg S., Henning E., Blackburn H., Loos R.J.F., Wareham N.J. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat. Genet. 2013;45:513–517. doi: 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berndt S.I., Gustafsson S., Mägi R., Ganna A., Wheeler E., Feitosa M.F., Justice A.E., Monda K.L., Croteau-Chonka D.C., Day F.R. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson A.D., Kavousi M., Smith A.V., Chen M.-H., Dehghan A., Aspelund T., Lin J.-P., van Duijn C.M., Harris T.B., Cupples L.A. Genome-wide association meta-analysis for total serum bilirubin levels. Hum. Mol. Genet. 2009;18:2700–2710. doi: 10.1093/hmg/ddp202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Harst P., Zhang W., Mateo Leach I., Rendon A., Verweij N., Sehmi J., Paul D.S., Elling U., Allayee H., Li X. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hakonarson H., Grant S.F.A., Bradfield J.P., Marchand L., Kim C.E., Glessner J.T., Grabs R., Casalunovo T., Taback S.P., Frackelton E.C. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 46.Pattaro C., Teumer A., Gorski M., Chu A.Y., Li M., Mijatovic V., Garnaas M., Tin A., Sorice R., Li Y., ICBP Consortium. AGEN Consortium. CARDIOGRAM. CHARGe-Heart Failure Group. ECHOGen Consortium Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat. Commun. 2016;7:10023. doi: 10.1038/ncomms10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorski M., Tin A., Garnaas M., McMahon G.M., Chu A.Y., Tayo B.O., Pattaro C., Teumer A., Chasman D.I., Chalmers J. Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int. 2015;87:1017–1029. doi: 10.1038/ki.2014.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Köttgen A., Pattaro C., Böger C.A., Fuchsberger C., Olden M., Glazer N.L., Parsa A., Gao X., Yang Q., Smith A.V. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J., Glessner J.T., Zhang H., Hou C., Wei Z., Bradfield J.P., Mentch F.D., Guo Y., Kim C., Xia Q. GWAS of blood cell traits identifies novel associated loci and epistatic interactions in Caucasian and African-American children. Hum. Mol. Genet. 2013;22:1457–1464. doi: 10.1093/hmg/dds534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oexle K., Ried J.S., Hicks A.A., Tanaka T., Hayward C., Bruegel M., Gögele M., Lichtner P., Müller-Myhsok B., Döring A. Novel association to the proprotein convertase PCSK7 gene locus revealed by analysing soluble transferrin receptor (sTfR) levels. Hum. Mol. Genet. 2011;20:1042–1047. doi: 10.1093/hmg/ddq538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kettunen J., Demirkan A., Würtz P., Draisma H.H.M., Haller T., Rawal R., Vaarhorst A., Kangas A.J., Lyytikäinen L.-P., Pirinen M. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun. 2016;7:11122. doi: 10.1038/ncomms11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Y., Marvelle A.F., Li J., Croteau-Chonka D.C., Feranil A.B., Kuzawa C.W., Li Y., Adair L.S., Mohlke K.L. Genetic association with lipids in Filipinos: waist circumference modifies an APOA5 effect on triglyceride levels. J. Lipid Res. 2013;54:3198–3205. doi: 10.1194/jlr.P042077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danese E., Lippi G., Montagnana M. Red blood cell distribution width and cardiovascular diseases. J. Thorac. Dis. 2015;7:E402–E411. doi: 10.3978/j.issn.2072-1439.2015.10.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J., Ye Z., Mayer J.G., Hoch B.A., Green C., Rolak L., Cold C., Khor S.-S., Zheng X., Miyagawa T. Phenome-wide association study maps new diseases to the human major histocompatibility complex region. J. Med. Genet. 2016;53:681–689. doi: 10.1136/jmedgenet-2016-103867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye Z., Mayer J., Ivacic L., Zhou Z., He M., Schrodi S.J., Page D., Brilliant M.H., Hebbring S.J. Phenome-wide association studies (PheWASs) for functional variants. Eur. J. Hum. Genet. 2015;23:523–529. doi: 10.1038/ejhg.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mero I.-L., Gustavsen M.W., Sæther H.S., Flåm S.T., Berg-Hansen P., Søndergaard H.B., Jensen P.E.H., Berge T., Bjølgerud A., Muggerud A., International Multiple Sclerosis Genetics Consortium Oligoclonal band status in Scandinavian multiple sclerosis patients is associated with specific genetic risk alleles. PLoS ONE. 2013;8:e58352. doi: 10.1371/journal.pone.0058352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat. Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 59.Cooper J.D., Smyth D.J., Smiles A.M., Plagnol V., Walker N.M., Allen J.E., Downes K., Barrett J.C., Healy B.C., Mychaleckyj J.C. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holliday E.G., Smith A.V., Cornes B.K., Buitendijk G.H.S., Jensen R.A., Sim X., Aspelund T., Aung T., Baird P.N., Boerwinkle E., Wellcome Trust Case Control Consortium 2 Insights into the genetic architecture of early stage age-related macular degeneration: a genome-wide association study meta-analysis. PLoS ONE. 2013;8:e53830. doi: 10.1371/journal.pone.0053830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steer S., Abkevich V., Gutin A., Cordell H.J., Gendall K.L., Merriman M.E., Rodger R.A., Rowley K.A., Chapman P., Gow P. Genomic DNA pooling for whole-genome association scans in complex disease: empirical demonstration of efficacy in rheumatoid arthritis. Genes Immun. 2007;8:57–68. doi: 10.1038/sj.gene.6364359. [DOI] [PubMed] [Google Scholar]
- 62.Köttgen A., Albrecht E., Teumer A., Vitart V., Krumsiek J., Hundertmark C., Pistis G., Ruggiero D., O’Seaghdha C.M., Haller T., LifeLines Cohort Study. CARDIoGRAM Consortium. DIAGRAM Consortium. ICBP Consortium. MAGIC Consortium Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat. Genet. 2013;45:145–154. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Denny J.C., Crawford D.C., Ritchie M.D., Bielinski S.J., Basford M.A., Bradford Y., Chai H.S., Bastarache L., Zuvich R., Peissig P. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am. J. Hum. Genet. 2011;89:529–542. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Speliotes E.K., Yerges-Armstrong L.M., Wu J., Hernaez R., Kim L.J., Palmer C.D., Gudnason V., Eiriksdottir G., Garcia M.E., Launer L.J., NASH CRN. GIANT Consortium. MAGIC Investigators. GOLD Consortium Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Halley P., Kadakkuzha B.M., Faghihi M.A., Magistri M., Zeier Z., Khorkova O., Coito C., Hsiao J., Lawrence M., Wahlestedt C. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6:222–230. doi: 10.1016/j.celrep.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Markus H.S., Mäkelä K.-M., Bevan S., Raitoharju E., Oksala N., Bis J.C., O’Donnell C., Hainsworth A., Lehtimäki T. Evidence HDAC9 genetic variant associated with ischemic stroke increases risk via promoting carotid atherosclerosis. Stroke. 2013;44:1220–1225. doi: 10.1161/STROKEAHA.111.000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Azghandi S., Prell C., van der Laan S.W., Schneider M., Malik R., Berer K., Gerdes N., Pasterkamp G., Weber C., Haffner C., Dichgans M. Deficiency of the stroke relevant HDAC9 gene attenuates atherosclerosis in accord with allele-specific effects at 7p21.1. Stroke. 2015;46:197–202. doi: 10.1161/STROKEAHA.114.007213. [DOI] [PubMed] [Google Scholar]
- 68.Manichaikul A., Rich S.S., Allison M.A., Guagliardo N.A., Bayliss D.A., Carey R.M., Barrett P.Q. KCNK3 variants are associated with hyperaldosteronism and hypertension. Hypertension. 2016;68:356–364. doi: 10.1161/HYPERTENSIONAHA.116.07564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kato N., Loh M., Takeuchi F., Verweij N., Wang X., Zhang W., Kelly T.N., Saleheen D., Lehne B., Leach I.M., BIOS-consortium. CARDIo GRAMplusCD. LifeLines Cohort Study. InterAct Consortium Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat. Genet. 2015;47:1282–1293. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R., Powell C., Vedantam S., Buchkovich M.L., Yang J., LifeLines Cohort Study. ADIPOGen Consortium. AGEN-BMI Working Group. CARDIOGRAMplusC4D Consortium. CKDGen Consortium. GLGC. ICBP. MAGIC Investigators. MuTHER Consortium. MIGen Consortium. PAGE Consortium. ReproGen Consortium. GENIE Consortium. International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Battisti-Charbonney A., Fisher J., Duffin J. The cerebrovascular response to carbon dioxide in humans. J. Physiol. 2011;589:3039–3048. doi: 10.1113/jphysiol.2011.206052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wolf E.J., Mitchell K.S., Logue M.W., Baldwin C.T., Reardon A.F., Humphries D.E., Miller M.W. Corticotropin releasing hormone receptor 2 (CRHR-2) gene is associated with decreased risk and severity of posttraumatic stress disorder in women. Depress. Anxiety. 2013;30:1161–1169. doi: 10.1002/da.22176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Row P.E., Liu H., Hayes S., Welchman R., Charalabous P., Hofmann K., Clague M.J., Sanderson C.M., Urbé S. The MIT domain of UBPY constitutes a CHMP binding and endosomal localization signal required for efficient epidermal growth factor receptor degradation. J. Biol. Chem. 2007;282:30929–30937. doi: 10.1074/jbc.M704009200. [DOI] [PubMed] [Google Scholar]
- 74.Flisiak I., Szterling-Jaworowska M., Baran A., Rogalska-Taranta M. Effect of psoriasis activity on epidermal growth factor (EGF) and the concentration of soluble EGF receptor in serum and plaque scales. Clin. Exp. Dermatol. 2014;39:461–467. doi: 10.1111/ced.12356. [DOI] [PubMed] [Google Scholar]
- 75.Veal C.D., Capon F., Allen M.H., Heath E.K., Evans J.C., Jones A., Patel S., Burden D., Tillman D., Barker J.N.W.N., Trembath R.C. Family-based analysis using a dense single-nucleotide polymorphism-based map defines genetic variation at PSORS1, the major psoriasis-susceptibility locus. Am. J. Hum. Genet. 2002;71:554–564. doi: 10.1086/342289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu Y., Helms C., Liao W., Zaba L.C., Duan S., Gardner J., Wise C., Miner A., Malloy M.J., Pullinger C.R. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4:e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.