Abstract
Genome-wide association studies (GWASs) and functional genomics approaches implicate enhancer disruption in islet dysfunction and type 2 diabetes (T2D) risk. We applied genetic fine-mapping and functional (epi)genomic approaches to a T2D- and proinsulin-associated 15q22.2 locus to identify a most likely causal variant, determine its direction of effect, and elucidate plausible target genes. Fine-mapping and conditional analyses of proinsulin levels of 8,635 non-diabetic individuals from the METSIM study support a single association signal represented by a cluster of 16 strongly associated (p < 10−17) variants in high linkage disequilibrium (r2 > 0.8) with the GWAS index SNP rs7172432. These variants reside in an evolutionarily and functionally conserved islet and β cell stretch or super enhancer; the most strongly associated variant (rs7163757, p = 3 × 10−19) overlaps a conserved islet open chromatin site. DNA sequence containing the rs7163757 risk allele displayed 2-fold higher enhancer activity than the non-risk allele in reporter assays (p < 0.01) and was differentially bound by β cell nuclear extract proteins. Transcription factor NFAT specifically potentiated risk-allele enhancer activity and altered patterns of nuclear protein binding to the risk allele in vitro, suggesting that it could be a factor mediating risk-allele effects. Finally, the rs7163757 proinsulin-raising and T2D risk allele (C) was associated with increased expression of C2CD4B, and possibly C2CD4A, both of which were induced by inflammatory cytokines, in human islets. Together, these data suggest that rs7163757 contributes to genetic risk of islet dysfunction and T2D by increasing NFAT-mediated islet enhancer activity and modulating C2CD4B, and possibly C2CD4A, expression in (patho)physiologic states.
Keywords: islet, comparative genomics, stretch or super enhancer, NFAT, inflammation, C2CD4A, C2CD4B, proinsulin, GWAS, rs7163757
Introduction
Genome-wide association studies (GWASs) have identified >150 loci associated with type 2 diabetes (T2D [MIM: 125853]) and diabetes-related glycemic traits, such as fed and fasting glucose and insulin levels.1 The prevailing model that has emerged from collective analyses is that islet dysfunction plays a major role in T2D genetic risk.2 Protein-coding variants have been implicated as the most likely causal variants for only a handful of loci.3 Thus, the large majority of SNPs identified by T2D GWASs most likely reside in non-coding, regulatory regions of the genome. Identifying the causal variant(s), target gene(s), and direction-of-effect for each of these loci is critical for translating GWAS results into a mechanistic understanding of islet gene regulation and T2D pathogenesis and for identifying and developing new therapeutic targets and strategies.4
GWASs in different populations have reported multiple T2D index SNPs in C2 calcium-dependent domain-containing 4A (C2CD4A [MIM: 610343]), C2 calcium-dependent domain-containing 4B (C2CD4B [MIM: 610344]), and vacuolar protein sorting 13 homolog C (VPS13C [MIM: 608879]) on 15q22.2 (Table S1), each of which associated with a T2D odds ratio (OR) of ∼1.1 (range 1.06–1.14).5, 6, 7, 8 Physiologic studies in non-diabetic European individuals suggest that these variants do not affect insulin sensitivity but rather contribute to T2D genetic risk through islet dysfunction. Reported T2D index SNP risk alleles have been associated with increased fasting glucose9, 10, 11 and proinsulin levels11, 12 and with decreased 2-hr glucose,11, 13 HOMA-B,9 glucose-stimulated insulin secretion (GSIS) and glucose-stimulated insulin release (GSIR),10 the insulinogenic index,10, 11 and BIGTT (β cell function, insulin sensitivity, and glucose tolerance test)-based acute insulin release10 (Table S2). Conditional analysis of this locus in a Danish cohort indicated that rs7172432, or SNPs in high linkage disequilibrium (LD), exhibited stronger effects on fasting glucose and GSIR than several other SNPs reported.10 Similarly, the rs7172432 risk allele (A) was associated with increased proinsulin levels in 8,224 Metabolic Syndrome in Men (METSIM) study participants (β = 0.042 ± 0.005, p = 7.4 × 10−18).12 Most recently, this locus has been associated with altered first-phase insulin secretion.14 Increased proportions of proinsulin secretion and impaired first-phase insulin secretion are hallmarks of β cell dysfunction in T2D pathogenesis.15
Genome-wide functional genomic and epigenomic analyses revealed that T2D GWAS SNPs are specifically and significantly enriched in islet enhancers,16, 17, 18, 19 suggesting that these variants could perturb enhancer activity and transcriptional regulation in the islet to contribute genetic susceptibility to islet dysfunction and T2D.20 In particular, associated variants are enriched in islet stretch or super enhancers (SEs), which are large (≥3 kb), tissue-specific enhancer regions located in or nearby genes important for cell-type-specific functions, such as genes encoding insulin (INS [MIM: 176730]), pancreas/duodenum homeobox protein 1 (PDX1 [MIM: 600733]), and the regulatory and catalytic subunits of ATP-binding cassette sub-family C member 8 (ABCC8 [MIM: 600509]) and ATP-sensitive inward rectifier potassium channel 11 (KCNJ11 [MIM: 600937]) in islets.16 As such, SE chromatin-state signatures can be used for nominating most likely important regulatory regions in or nearby genes of unknown function.
In this study, we combined genetic and functional genomic approaches to investigate the GWAS association of T2D and T2D-related metabolic traits in chromosomal region 15q22.2. Together, our data identified rs7163757 as the most likely causal variant in this locus and implicate C2CD4B and C2CD4A induction in T2D genetic risk and diabetogenic islet stress responses. Genetic fine-mapping identified a single association signal consisting of 16 highly associated variants, which reside in an islet SE state between C2CD4A and C2CD4B. rs7163757 is the only variant overlapping an islet open chromatin site. The rs7163757 T2D risk allele (C) exhibited 2-fold higher enhancer activity than the non-risk allele (T), was differentially bound by β cell nuclear factors, and was associated with increased expression of C2CD4B, and potentially C2CD4A, in human pancreatic islets.
Material and Methods
Fine-Mapping Study Population and Phenotype
We report genetic association results for fasting proinsulin levels from up to 8,635 non-diabetic Finnish men from the population-based METSIM study.21 Study participants with previously diagnosed T1D or T2D (and on diabetes medication and with fasting glucose ≥ 7 mmol/L or 2-hr glucose ≥ 11.1 mmol/L) were excluded from the analysis. The mean age of the analyzed participants was 57.2 years (median = 57.0 years; range = 45–74 years), and the mean body mass index (BMI) was 26.8 kg/m2 (median = 26.3 kg/m2; range = 16.2–51.6 kg/m2). Blood samples were drawn after a 12-hr overnight fast, and fasting plasma-specific proinsulin (Human Proinsulin RIA Kit, Linco Research; no cross-reaction with insulin or C-peptide) and fasting insulin (ADVIA Centaur Insulin IRI, 02230141, Siemens Medical Solutions Diagnostics; minimal cross-reaction with proinsulin or C-peptide) were measured by immunoassay. The study was approved by the ethics committee of the University of Kuopio and Kuopio University Hospital, and informed consent was obtained from all study participants.
Genotyping and Genotype Imputation
METSIM samples were genotyped with the Illumina HumanOmniExpress BeadChip. We mapped the Illumina array probe sequences to the UCSC Genome Browser hg19 assembly with the Burrows-Wheeler Aligner (BWA).22 SNP quality-control steps prior to imputation included removing SNPs with ambiguously mapping probe sequences and SNPs with a call rate < 95% or a Hardy-Weinberg equilibrium test p value < 10−6. We followed a two-step genotype imputation strategy.23 First, we statistically estimated haplotypes by using SHAPEIT224 and then imputed genotypes into these estimated haplotypes by using minimac2.25 We used the haplotypes from 2,737 European individuals sequenced in the Genetics of Type 2 Diabetes project as the imputation reference panel.3 Participants were previously genotyped with the Illumina HumanExome BeadChip,12 which focuses on protein-altering variants selected from the exome sequences of >12,000 individuals. Exome chip variants were incorporated after imputation.
Fine-Mapping and Conditional Analysis
To account for relatedness between study participants and population structure, we tested associations between the phenotype and the estimated dosages (imputed variants) or additively coded genotypes (directly genotyped variants) by using a linear mixed model with empirical kinship matrix, as implemented in EMMAX.26 We adjusted log-transformed plasma proinsulin for age, BMI, and log-transformed insulin before association testing and analyzed rank-based inverse-normal-transformed residuals. We analyzed directly genotyped variants with a minor allele count (MAC) ≥ 5 and a Hardy-Weinberg equilibrium test p value ≥ 10−6 and imputed variants with an imputation quality score (R2) > 0.3 and a minor allele frequency (MAF) ≥ 0.5%.
To identify additional independent signals in the region, we carried out conditional analysis in which we included the allele count of the most strongly associated SNP (rs7172432) as a covariate in the model. Regional association results were visualized with LocusZoom.27 LD was estimated from the imputation reference panel.
For the Bayesian fine-mapping analysis, we followed Fuchsberger et al.3 In brief, we defined the candidate set of variants by identifying all analyzed variants with r2 ≥ 0.1 with the most associated variant and within a 5 Mb window centered on the most associated variant. We calculated an approximate Bayes’ factor (ABF) for each variant as follows:
where r = 0.04/(SEM2 + 0.04), z = β/SEM, and β and SEM are the estimated log OR and its associated standard error. We then calculated the posterior probability of being causal for each variant as ABF/T, where T is the sum of ABF values over all candidate variants. Next, we ranked variants in decreasing order by posterior probabilities and obtained the 99% credible set by including variants with the highest posterior probabilities until the cumulative posterior probability ≥ 99%.
Primary Islet and Cell Culture
Fresh human non-diabetic pancreatic islets were purchased from ProdoLabs (UNOS no. ABE1388) in accordance with the regulations of human subjects research. Upon arrival, the cells were transferred into PIM(S) media (ProdoLabs), supplemented with PIM(ABS) (ProdoLabs) and PIM(G) (ProdoLabs), and kept overnight in a T-150 non-treated tissue-culture flask (VWR) for recovery at 37°C and 5% CO2. ATAC-seq (assay for transposase accessible chromatin with sequencing) and RNA-seq (RNA sequencing) were performed the following day as described below.
Mouse MIN6 and rat INS-1(832/13) (a kind gift from C. Newgard) β cell lines were cultured as previously described.28, 29 In brief, MIN6 were grown in DMEM (4.5 g/L glucose; Life Technologies) supplemented with 1 mM sodium pyruvate (Life Technologies), 100 μM 2-mercaptoethanol (Sigma), and 10% fetal bovine serum (Seradigm). INS-1(832/13) cells were cultured in RPMI-1640 (11.1 mM d-glucose; Life Technologies) supplemented with 10% fetal bovine serum (Seradigm), 10 mM HEPES (Life Technologies), 2 mM l-glutamine, 1 mM sodium pyruvate, and 50 μM 2-mercaptoethanol (Sigma).
Chromatin Analyses: ChIP-Seq and ATAC-Seq
Crosslinking and chromatin immunoprecipitation sequencing (ChIP-seq) of human islets and MIN6 and INS-1(832/13) β cells were carried out as previously described.30 Human islet and MIN6 and INS-1(832/13) β cell line ATAC-seq libraries were prepared as previously described.31 Approximately 250 islet equivalents (∼250,000 islet cells), 250,000 INS-1(832/13) cells, and 50,000 MIN6 cells were transposed.
Sequence Alignment, Processing, and Analysis
Chromatin states were determined with ChromHMM as previously described.16 ATAC-seq reads were aligned to UCSC Genome Browser reference genomes hg19 (human islets), mm9 (MIN6), and rn5 (INS-1(832/13)) with the “mem” option in the BWA.22 Only reads uniquely mapping to their respective genomes were used in subsequent analysis. We removed reads mapping to the mitochondrial genome (chrM) and eliminated duplicate reads mapping to the nuclear genome to avoid potential artifacts of PCR amplification in ATAC-seq. Human islet, MIN6, and INS-1(832/13) ATAC-seq library and data statistics are shown in Table S3. ATAC-seq enriched regions (peaks) in each sample or merged replicates were identified with the MACS2 program32 with the following parameters: MACS/2.1.0.20151222/bin/macs2 callpeak -t <input tag file> -f BED -n <output peak file> -g ‘hs’ --nomodel --shift -100 --extsize 200 -B.
RNA-Seq
Human islet RNA-seq data and tracks have been previously described.16 Additionally, total RNA was extracted and purified from 32 human pancreatic islet samples of European descent from the Integrated Islet Distribution Program and NDRI, MIN6, and INS-1(832/13) β cell lines with the use of Trizol (Life Technologies) according to the manufacturer’s instructions and was sequenced with the Illumina TruSeq Kit (human islets and MIN6) or Kapa Biosystems KAPA Stranded mRNA-Seq Kit (INS-1(832/13)) according to the manufacturer’s instructions. MIN6 RNA-seq data were aligned to mm9 with tophat v.1.3.2.33 For INS-1(832/13) RNA-seq, the gene models for C2cd4a, C2cd4b, and Vps13c were absent in the current version of the UCSC Genome Browser rn5 gene annotations (Illumina iGenomes). We performed a reference-guided transcriptome assembly by using cufflinks (v.2.2.1) to construct the transcript models.33 The assembled transcripts were then visualized on the UCSC Genome Browser, and gene locations were identified by homology to mouse annotations. We collapsed transcript models for each locus to generate the corresponding gene model. These gene models were added to the existing rn5 gene annotations, and a reference index was created with RSEM (v.1.12.2).34 We realigned RNA-seq reads to this reference index by using RSEM to generate alignments and count matrices. INS-1(832/13) UCSC Genome Browser tracks were created with homer (v.4.6).35
Human Islet ATAC-Seq Footprint Analysis
Transcription factor (TF) binding in human islet ATAC-seq data was predicted as previously described.36 In brief, we generated matrices representing the Tn5 integration events ± 100 bp around PWM (position weight matrix) scan matches for a given motif. We used these matrices as input for CENTIPEDE37 to calculate the posterior probability that each motif instance was bound. Individual motifs were considered bound if the CENTIPEDE posterior was ≥0.99 and if the motif coordinates completely intersected an ATAC-seq peak in the sample of origin.
SE Constituent Sequence Cloning and Site-Directed Mutagenesis
Human SE constituent sequences from the region on chr15: 62,363,117–62,455,736 (GRCh37/hg19 coordinates), between C2CD4A and C2CD4B, were cloned. Table S4 contains sequence coordinates for the SE constituent open chromatin sites (based on ATAC-seq profiles in human islets) selected for cloning and the primers used to amplify them. SE constituent site sequences were PCR amplified from the islet genomic DNA (gDNA) of two individuals with different genotypes (haplotypes 1 and 2) at several of the SNPs in this region with Phusion High-Fidelity Polymerase (Thermo Scientific). Amplicons were cloned into pDONR201 and shuttled into modified pGL4.23 luciferase reporter vectors via Gateway cloning (Invitrogen) as previously described.30 HS3 rs7163757 alleles C and T were interconverted with the primer sequences indicated in Table S4 and the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer’s instructions.
Transfection and Dual Luciferase Reporter Assays
MIN6 or INS-1(832/13) cells were seeded at a density of 60,000 cells per well in 96-well plates 24 hr before transfection. Cells were co-transfected in triplicate with 200 ng pGL4.23 containing each human C2CD4A or C2CD4B SE sequence (Table S4) and 2 ng Renilla (pRL-TK) with Lipofectamine 2000 Transfection Reagent (Life Technologies) according to the manufacturer’s instructions. 4–16 clones were tested per C2CD4A or C2CD4B SE sequence and orientation. Each plasmid was transfected and measured in triplicate, and experiments were completed on at least three separate occasions. 38–40 hr after transfection, cells were lysed in 1× passive lysis buffer (PLB) with the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Luciferase was measured on a Synergy2 Microplate Reader (BioTek). We normalized firefly values to Renilla to control for differences in cell number or transfection efficiency. To determine glucose-stimulated activity of the reporters, we grew INS-1(832/13) cells in INS-1(832/13) media containing reduced glucose (3–5 mM) for 8–10 hr before transfection with the C2CD4A and C2CD4B HS3 reporter plasmids. Transfected cells were cultured in reduced glucose medium for an additional 16 hr, after which they were grown in high (15 mM) or low (3 mM) glucose-containing medium for an additional 24 hr before cell lysis in 1× PLB and luciferase reporter activity measurement as described above. At least three plasmid preparations were tested for each HS3 allele on three separate occasions.
Pharmacologic Inhibition of Calcineurin and NFAT
MIN6 cells were pre-treated with 10 ng/mL tacrolimus (FK506, Biotang) or ethanol vehicle control 30 min before transfection with 200 ng of the C2CD4A and C2CD4B SE firefly luciferase vectors and 2 ng Renilla (pRL-TK). Luciferase activity was tested as above for four or five clones on three separate occasions.
Molecular Manipulation of the Calcineurin-NFAT Pathway
200 ng of the C2CD4A and C2CD4B SE luciferase vectors was co-transfected in triplicate with 100 ng of plasmids expressing wild-type NFAT (EGFPC1-huNFATc1EE-WT;38 Addgene, plasmid no. 24219) or a mutant NFAT peptide sequence (EGFPN1-VIVIT;39 Addgene, plasmid no. 11106). We used pGL3-NFAT-luciferase40 (Addgene, plasmid no. 17870), which contains three canonical NFAT binding sequences, as a positive control to measure NFAT activity and transcriptional responses in these experiments. We documented GFP intensity (GFP-channel = 60% light intensity; transmitted = 53% light intensity) for all cells transfected with expression constructs EGFPC1-huNFATc1EE-WT, EGFPN1-VIVIT, and pEGFP-C1 (Clontech, catalog no. 6084-1) by using an EVOS FL Cell Imaging System (Life Technologies) to confirm GFP fusion protein levels for each co-transfected expression plasmid. For these experiments, four independent clones were selected and tested on four separate occasions.
Islet SNP Genotyping and Imputation
Islet DNA samples were genotyped at the Genetic Resources Core Facility of the Johns Hopkins Institute of Genetic Medicine on the HumanOmni2.5-4v1_H BeadChip array (Illumina). We applied the same quality-control criteria to SNPs for further analysis as described for the METSIM samples. In addition, we filtered out A/T or C/G SNPs with a MAF > 0.2 or SNPs that had an absolute alternate allele frequency difference > 0.2 with the 1000 Genomes EUR (European) population, yielding 2,057,703 SNPs for imputation. We performed SNP imputation and phasing by using the same strategy as described for METSIM samples. Haplotypes from 1000 Genomes phase 3 v.541 were used as the reference panel. To improve phasing quality given the small target sample set, we pre-phased our samples together with the 2,504 reference panel samples by using ShapeIT2.24
Islet eQTL Lookups and Conditional Analysis
cis-Expression quantitative trait locus (eQTL) data for each transcript (n = 10: C2CD4B, C2CD4A, VPS13C, NPM1P47, RP11-244F12.3, TPM1 [MIM: 191010], RP11-1069G10.1, RORA [MIM: 600825], TLN2 [MIM: 607349], and RP11-643M14.1) whose most upstream transcription start site (TSS) was within 1 Mb of rs7163757 were obtained from a parallel study of expression in 112 human islets36 (Table S10). To determine whether cis-eQTL associations between rs7163757 and nearby genes could be affected by other strong eQTLs in the region, we performed iterative conditional analysis on each of these genes. We used the following linear regression model, fit within the two islet studies (n = 31 and 81) from Varshney et al.33
Yij is the inverse-normalized and PEER-adjusted FPKM value for individual i and gene j, GiSNP is the imputed allele count of rs7163757, βjSNP is the regression coefficient of the imputed allele count for rs7163757, Gis is the set of all SNPs within 1 Mb of the most upstream TSS of the gene, and εij is an error term exhibiting normal distribution with mean 0 and variance σ2. We considered only SNPs present in both studies (MAC ≥ 1) and with a MAC ≥ 10 across all 112 samples. We combined the results from the two studies by meta-analysis weighted by sample size.33 If at least one SNP had a meta-analysis p value < 1.2 × 10−4 (corresponding to the p value threshold for cis-eQTLs with a false-discovery rate [FDR] < 5%), we retained the SNP with the most significant p value in the model and repeated the procedure until no added SNP had a p value < 1.2 × 10−4. This procedure corresponds to performing stepwise forward selection of SNPs within 1 Mb of the most upstream TSS on the basis of the results of the meta-analysis at each step (by using a stopping threshold p value of 1.2 × 10−4). The conditional p value for rs7163757 is the p value for βjSNP from the final model. We used the Benjamini-Hochberg method42 to adjust the conditional p values for multiple testing.
ASE QTL Analysis
Allele-specific expression (ASE) was quantified as previously described43 with RNA-seq data from FUSION36 and Groop44 human islet samples, except that stranded RNA-seq reads were considered together. We performed targeted analysis of ASE QTLs (aseQTLs) to identify transcripts affected by putative regulatory SNPs (rSNPs) in the C2CD4A-C2CD4B-VPS13C locus. The approach to statistically associate an rSNP genotype with the ASE of a transcribed SNP (tSNP) was performed as previously described45 with the following modifications: (1) coordinates for all TSSs for a given gene were merged and extended 100 kb upstream and downstream, (2) any SNPs in this window were tested as rSNPs, (3) any SNP that overlapped an exonic region of the gene and had at least 30× coverage was tested as a tSNP, and (4) rSNPs and tSNPs were tested in a pairwise, gene-by-gene fashion. For a tSNP-rSNP pair to be tested, we needed to observe with quantified ASE at least five samples each that were rSNP heterozygotes and homozygotes.
The fraction reference allele (fracRef) value measures ASE at tSNPs and is converted to the absolute allelic imbalance, a value representing the allelic skew from the expected fracRef, for aseQTL analysis. For example, a SNP with a fracRef of 0.8 (overexpressed reference allele) or 0.2 (overexpressed alternate allele) and an expected fracRef of 0.5 results in an absolute allelic imbalance of 0.3. Allelic imbalance is calculated from the absolute value of the difference between the observed fracRef and the sample-specific and allele-pair-specific expected fracRef. Absolute allelic-imbalance values range from 0 to 0.5; fracRef values range from 0 to 1. We compared the tSNP absolute allelic-imbalance values between homozygous and heterozygous rSNPs with a two-sided Wilcoxon rank-sum test (“wilcox.test” in R), producing a p value for every tested rSNP and tSNP pair. We performed Storey’s FDR to correct for multiple-testing burden on genes within the topologically associating domain (TAD)46 surrounding the C2CD4A-C2CD4B-VPS13C locus as opposed to genome-wide. q values were calculated for each rSNP-tSNP pair within the TAD coordinates chr15: 61,412,708–62,612,708 (hg19, as determined from hESC and IMR90 TADs46) with Bioconductor’s “qvalue” package.
EMSA
Electrophoretic mobility shift assays (EMSAs) were carried out as previously described.47 In brief, 17-bp biotin-end-labeled complementary oligonucleotides were designed to surround the variant rs7163757 (5′ biotin-TGATTTTTC[C/T]ATTTTAAGC-3′, Integrated DNA Technologies), and double-stranded probes were generated for both alleles. Nuclear extract from mouse insulinoma MIN6 cells was prepared with the NE-PER Extraction Kit (Thermo Scientific). The LightShift Chemiluminescent EMSA Kit (Thermo Scientific) was used according to the manufacturer’s instructions. Binding reactions consisted of 1× binding buffer, 1 μg poly (dI-dC), 4 μg nuclear extract, and 200 fmol labeled probe. Reactions were incubated at room temperature for 25 min. For competition reactions, 64-fold excess of unlabeled probe for either allele was included and pre-incubated for 15 min. For supershift assays, 4 μg antibody (Nkx6.1, sc-15030X; PDX1, sc-14662X; YY1, sc-1703X; p300, sc-585x; FoxA2, sc-9187X; MafB, sc-22830X; NFATc2 (4G6-G5)x, sc-7296X [all Santa Cruz Biotechnology]; NFATc1(7A6), 556602 [BD Biosciences]) was added to the binding reaction and pre-incubated for 25 min. DNA-protein complexes were detected by chemiluminescence. EMSAs were repeated and yielded comparable results.
Inflammatory Cytokine Induction of C2CD4A, C2CD4B, and VPS13C in Human Islets, Rat Islets, and INS-1(832/13) β Cells
Processed RPKM (reads per kilobase of transcript per million mapped reads) values were obtained from published studies of five human islets treated with IL-1β (encoded by IL1B [MIM: 147720]) and IFN-γ (encoded by IFNG [MIM: 147570])48 and of rat islets treated with 0.1 or 20 U/mL IL-1β.49 INS-1(832/13) cells were incubated in reduced-serum (1% fetal bovine serum) INS-1(832/13) complete medium overnight and then treated for 2 hr with 0 or 2 U/mL recombinant rat IL-1β (BioLegend) before RNA harvest and processing for RNA-seq as described above.
Results
Fine-Mapping Identifies 16 Highly Linked Intergenic Variants Associated with Fasting Proinsulin and T2D on 15q22.2
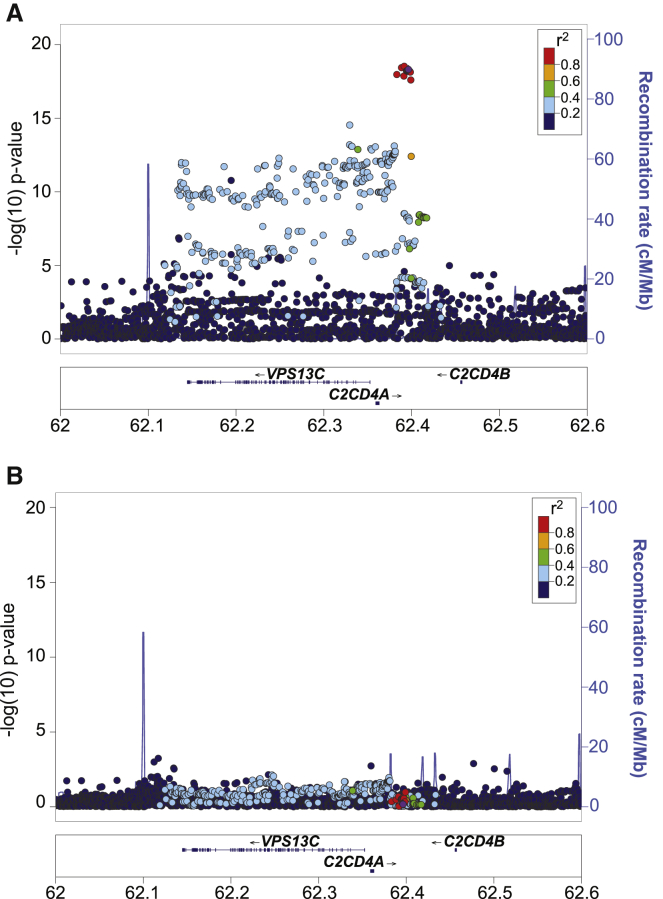
rs7172432 has been the most reported C2CD4A-C2CD4B-VPS13C index SNP associated with T2D (Table S1) and quantitative traits related to islet dysfunction (Table S2) in multiple populations.5, 6, 8, 10, 12 To identify a set of candidate causal variants for functional follow-up in this locus, we conducted genetic fine-mapping analysis of the fasting proinsulin association signal in the locus in 8,635 non-diabetic Finnish individuals from the METSIM study.11, 12, 21 Fine-mapping analysis identified a cluster of 16 strongly associated variants (p < 1 × 10−17; Figure 1A). These included the GWAS index SNP rs7172432 (purple dot) and 15 variants (red dots) in high LD (r2 > 0.8) as putative causal variants (see also Table 1 and Table S5). Of note, the set of 16 most strongly associated (p < 1 × 10−17) variants corresponded to the 99% “credible set” obtained from a more formal Bayesian fine-mapping analysis on the basis of approximate Bayes’ factors50, 51 (Figure S1 and Table S6). This 99% credible set contained, with ≥99% probability, the variant causal for the association signal, as long as this variant was included in the analysis. Importantly, these variants exhibited the strongest T2D association p values for this locus in the latest T2D meta-analysis52 (Table S7).
Figure 1.
Fine-Mapping Identifies a Single Signal Consisting of a Cluster of 16 Highly Linked Variants Associated with High Proinsulin Levels
(A) Locus Zoom plot showing the association between genetic variants and increased proinsulin levels in the VPS13C-C2CD4A-C2CD4B locus in the METSIM study population. The index SNP (rs7172432) is purple. The x axis displays the gene and variant position on chromosome 15 (chr15) in megabases (Mb). The y axis indicates association p values (left) and recombination rates (right) in centiMorgans (cM). Associated variants reside between two intergenic recombination hotspots (blue peaks) between C2CD4A and C2CD4B.
(B) Locus Zoom plot of proinsulin association in the locus after conditioning on rs7172432. The x and y axes are as described in (A).
Table 1.
Proinsulin-Associated C2CD4A and C2CD4B Intergenic Variants
| SNP ID | Position | p Value | Effect | Effect Allele | Other Allele | MAF | ATAC Positive |
|---|---|---|---|---|---|---|---|
| rs4502156 | chr15: 62,383,155 | 1.08 × 10−18 | −0.136 | C | T | 0.4355 | − |
| rs1881415 | chr15: 62,388,530 | 3.69 × 10−19 | −0.139 | C | T | 0.4369 | − |
| rs67818839 | chr15: 62,391,184 | 1.38 × 10−18 | −0.136 | AT | A | 0.44608 | − |
| rs7163757 | chr15: 62,391,608 | 2.98 × 10−19 | −0.138 | T | C | 0.44653 | + |
| rs8037894 | chr15: 62,394,264 | 6.13 × 10−19 | −0.137 | C | G | 0.45058 | − |
| rs8038040 | chr15: 62,394,339 | 6.01 × 10−19 | −0.137 | A | G | 0.45067 | − |
| rs6494307 | chr15: 62,394,690 | 6.31 × 10−19 | −0.137 | G | C | 0.45061 | − |
| rs7161785 | chr15: 62,395,224 | 6.21 × 10−19 | −0.137 | C | G | 0.45058 | − |
| rs7167878 | chr15: 62,396,189 | 6.24 × 10−19 | −0.137 | A | C | 0.45072 | − |
| rs7167932 | chr15: 62,396,278 | 6.30 × 10−19 | −0.137 | T | C | 0.45059 | − |
| rs7172432 | chr15: 62,396,389 | 4.88 × 10−19 | −0.138 | G | A | 0.44769 | − |
| rs7173964 | chr15: 62,396,942 | 6.86 × 10−19 | −0.137 | A | G | 0.45067 | − |
| rs4775466 | chr15: 62,397,118 | 6.65 × 10−19 | −0.137 | T | C | 0.4507 | − |
| rs4775467 | chr15: 62,397,398 | 6.64 × 10−19 | −0.137 | A | G | 0.45068 | − |
| rs10083587 | chr15: 62,398,533 | 7.39 × 10−19 | −0.137 | T | C | 0.45168 | − |
| rs11856307 | chr15: 62,399,093 | 2.54 × 10−18 | −0.135 | C | A | 0.45279 | − |
The following abbreviations are used: SNP, single-nucleotide polymorphism; MAF, minor allele frequency; and ATAC, assay for transposase accessible chromatin.
All 16 variants were located between two recombination hotspots in the intergenic region between C2CD4A and C2CD4B. Conditional analysis using rs7172432 as a covariate attenuated the strength of the proinsulin association in this locus (Figure 1B; p > 10−5 for all SNPs). Together, fine mapping and conditional analysis support a single proinsulin association signal in this locus and implicate 16 highly linked intergenic variants (r2 > 0.8) as the putative causal and/or functional variants underlying both increased proinsulin levels and T2D risk in this locus.
Putative Causal Variants Reside in an Evolutionarily Conserved Islet SE
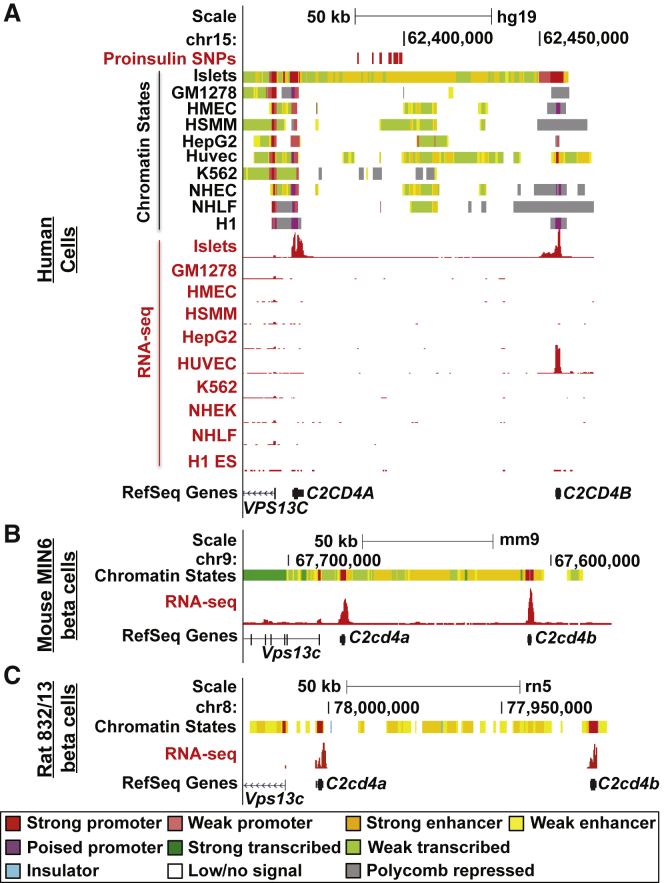
Because all 16 variants identified by fine-mapping were intergenic (Figure 1 and Table 1), we hypothesized that one or more of them could reside in islet regulatory elements. Analysis of ChromHMM-defined chromatin states16, 36 in this region revealed that these variants overlap islet-specific SEs between C2CD4A and C2CD4B (Figure 2A and Figure S2A, orange chromatin states). VPS13C was modestly expressed in multiple tissues (RPKM > 1), whereas C2CD4A and C2CD4B expression was restricted to human pancreatic islets and a few additional cell types or tissues (Figure 2A, RNA-seq red tracks; Figure S3). Interestingly, ChIP-seq and chromatin-state analyses of the orthologous region in mouse MIN6 (Figure 2B) and rat INS-1(832/13) (Figure 2C) β cell lines detected SE signatures in both species. Furthermore, both C2cd4a and C2cd4b were expressed in MIN6 and INS-1(832/13) β cells (Figures 2B and 2C, RNA-seq red tracks). The functional preservation of the chromatin structure and transcriptional output in this locus suggests that it plays an evolutionarily important role in islet and β cell function. Collectively, these results indicate that the putative causal variants overlap an evolutionarily conserved islet SE and support the hypothesis that one or more of them could contribute to β cell dysfunction by altering SE activity and target-gene expression.
Figure 2.
Fine-Mapped Variants Reside in an Evolutionarily Conserved Islet Stretch Enhancer
UCSC Genome Browser views of chromatin states (upper panels) and RNA-seq (lower panels, red) in (A) human pancreatic islets and nine ENCODE cell types (GM12878, lymphoblastoid cells; HMECs, human mammary epithelial cells; HSMMs, human smooth muscle myoblasts; HepG2, hepatocellular carcinoma cells; HUVECs, human umbilical vein endothelial cells; K562, erythroleukemia cells; NHEKs, normal human epidermal keratinocytes; NHLFs, normal human lung fibroblasts; and H1 ES, embryonic stem cells), (B) mouse MIN6 β cells, and (C) rat INS-1(832/13) beta cells. RefSeq Genes indicate the location of C2CD4A, C2CD4B, and VPS13C in the region. Chromatin-state colors are as indicated. Increased low or no-signal states in 832/13 are due to low mappability. 16 proinsulin-associated variants are indicated in (A) (upper panel in red). UCSC Genome Browser coordinates correspond to hg19, mm9, or rn5.
rs7163757 Resides in an Islet SE Constituent Open Chromatin Site
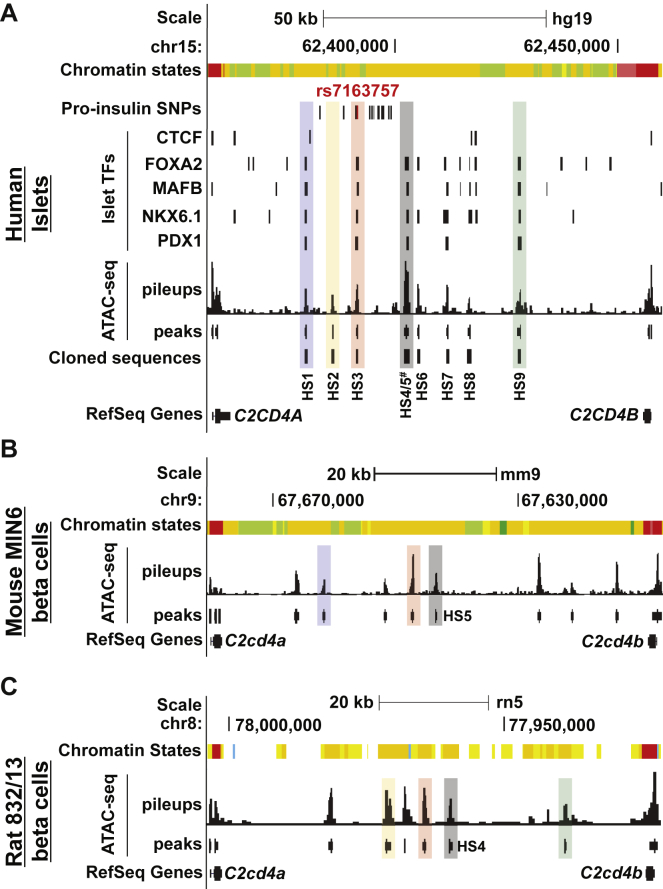
SEs can encompass multiple, discrete open chromatin sites.16, 17, 53 We profiled chromatin accessibility in human islets by using ATAC-seq31 to determine the constituent open chromatin sites within the C2CD4A-C2CD4B-VPS13C islet SE and to identify any overlaps with the putative causal and/or functional variant(s) among the 16 variants in this locus. As shown in Figure 3A, we detected nine intergenic open chromatin sites (HS1–HS9) between C2CD4A and C2CD4B in human islets (sequence pileups and MACS2 peak calls). All sites overlapped islet active enhancer states (Figure 3A and Figure S2A, orange chromatin state), and some of them also exhibited evidence of chromatin accessibility or enhancer chromatin states in other tissues (Figure S2A). By analyzing reported genome-wide binding sites for islet TFs FOXA2, MAFB, NKX6.1, and PDX1,18 we found that about half (4/9) of these sites were bound by all four islet TFs (Figure 3A). Only one of the 16 fine-mapped variants, rs7163757, overlapped an islet open chromatin site (Figure 3A, HS3), suggesting this is the functional variant among the multiple linked variants.
Figure 3.
rs7163757 Resides in an Evolutionarily Conserved Islet (β Cell) Open Chromatin Site
UCSC Genome Browser views of C2CD4A-C2CD4B-VPS13C SE constituent open chromatin sites determined by ATAC-seq in (A) human islets, (B) mouse MIN6 β cells, and (C) rat INS-1(832/13) β cells. Open chromatin sites were detected in this region and are indicated by the ATAC-seq pileups and the peaks (MACS peak calls) in each species between C2CD4A and C2CD4B (RefSeq Genes). Chromatin states are indicated and color coded as in Figure 2. One of the 16 proinsulin variants (rs7163757, red) overlaps an open chromatin site (HS3) bound by islet TFs NKX6.1, FOXA2, PDX1, and MAFB, but not CTCF, according to published islet ChIP-seq data.18 Black tick marks in (A) indicate the locations of the additional 15 variants associated with higher proinsulin (pro-insulin SNPs). Human genomic DNA sequences overlapping open chromatin sites (HS1–HS9) were selected and cloned as indicated. Human open chromatin DNA sequences mapping to mouse and/or rat open chromatin sites are indicated with colored shading. The HS3 DNA sequence maps to open chromatin sites in both MIN6 and INS-1(832/13) (red shading). HS1 and HS5 map only to MIN6 open chromatin sites (blue and gray shading, respectively); HS2, HS4 and HS9 map only to INS-1(832/13) open chromatin sites (orange-yellow, gray, and green shading, respectively). HS4/5# stands for the individually cloned sites HS4 and HS5 and the combined site HS4+HS5. UCSC Genome Browser coordinates correspond to hg19, mm9, or rn5.
Sequences overlapping seven of the islet SE constituent sites exhibited evidence of sequence constraint through the vertebrate lineage (Figure S2A). We sought to determine which of the islet SE constituent sites in this locus were functionally preserved in rodents. ATAC-seq profiling identified eight open chromatin sites in mouse MIN6 cell lines (Figure 3B) and six sites in rat INS-1(832/13) β cell lines (Figure 3C). We used the UCSC Genome Browser liftOver and bnmapper54 comparative sequence analysis tools to identify mouse and rat sequences orthologous to the human islet HS1-9 open chromatin sites (Table S8) and to determine which of them overlapped β cell SE constituent sites in MIN6 (Figure 3B) and INS-1(832/13) (Figure 3C) β cell lines. Two adjacent open chromatin site sequences, HS4 and HS5 (Figure 3, gray shading) mapped to an open chromatin site in INS-1(832/13) or MIN6, respectively (Figure S2B). One open chromatin site sequence, HS1, mapped only to an open chromatin site in MIN6 (Figures 3A and 3B, blue shading). HS2 and HS9 mapped only to open chromatin sites in INS-1(832/13) (Figures 3A and 3C, orange-yellow and green shading, respectively). Only HS3 sequence mapped to open chromatin sites in the β cell lines from both species (Figure 3, red shading; Table S8). Thus, these comparative analyses highlight HS3, which contains the proinsulin-associated variant rs7163757, as the only sequence preserved as a functional regulatory site in both mouse and rat β cells.
The rs7163757 Risk Allele (C) Exhibits Higher Enhancer Activity Than the Non-risk Allele (T)
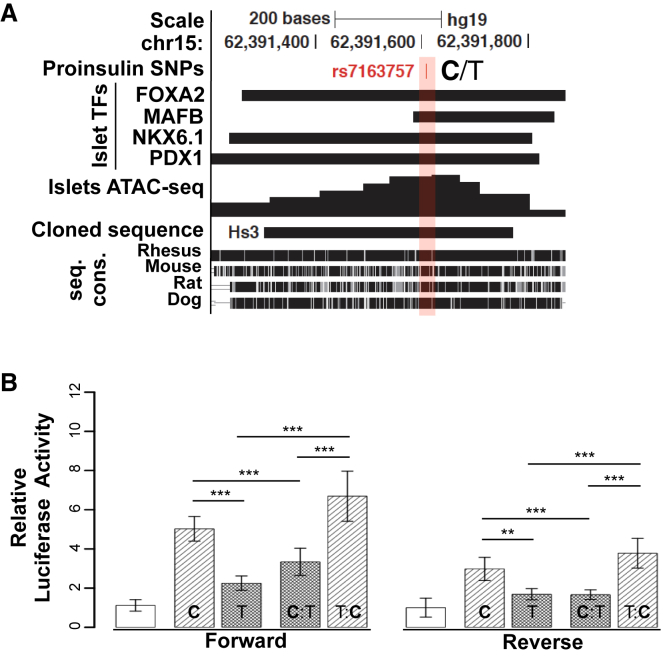
On the basis of chromatin profiling and cross-species analyses, we hypothesized that rs7163757 alleles alter HS3 regulatory element activity. To test this, we cloned the HS3 sequence (Figure 4A) from human islet gDNA containing either the risk (C) or non-risk (T) allele and measured each sequence’s enhancer activity in MIN6 and INS-1(832/13) β cells by using luciferase reporter assays. Compared with the empty vector, both HS3 alleles exhibited enhancer activity (Figures 4B and S4A). However, the rs7163757 risk allele (C) conferred 2-fold more enhancer activity than the non-risk allele (T) (Figure 4B). We identified an additional sequence variant (rs28578604) in the HS3 amplicon (Table S9). To rule out a role for rs28578604 in the observed differences in HS3 allelic enhancer activity, we used site-directed mutagenesis to experimentally convert the rs7163757 genotypes (T:C and C:T) while leaving rs28578604 unchanged. The mutagenized rs7163757 T:C and C:T constructs exhibited enhancer activity comparable to that of the original risk and non-risk haplotypes, respectively (Figure 4B and Table S4).
Figure 4.
The rs7163757 Risk Allele (C) Exhibits 2-fold Higher Enhancer Activity Than the Non-risk Allele (T)
(A) UCSC Genome Browser view of islet SE constituent HS3. Islet TF ChIP-seq,18 ATAC-seq pileup, and cross-species sequence conservation (seq. cons.) are indicated. Proinsulin- and T2D-associated rs7163757 (C/T) is indicated by red shading and font. The DNA sequence cloned and tested for enhancer activity is indicated. UCSC Genome Browser coordinates correspond to hg19.
(B) Luciferase reporter assay of the HS3 sequences (cloned in forward and reverse orientations) containing the rs7163757 risk allele (C) or non-risk allele (T) (haplotype 1 or 2, respectively), the C allele mutated to the T allele (C:T), and vice versa (T:C). All luciferase activity was normalized to that of the empty vector. HS3 enhancer activity was compared with the normalized activity of the pGL4.23 vector containing Gateway sequence in the forward and reverse orientations. All cloned HS3 enhancer sequences were significantly more active than the control vectors (p values not indicated). p values are indicated for the HS3 C allele versus T allele, the T allele versus T:C, the C allele versus C:T, and T:C versus C:T in the forward and reverse orientations. There was no significant difference between mutated alleles T:C and C and between C:T and versus T (p values not indicated). Each construct was measured in triplicate; a total of 16 biological replicates were measured per haplotype over seven independent experiments. Data represent the mean ± SEM; ∗∗p < 0.01, ∗∗∗p < 0.001 (two-sided unpaired t test).
We tested the additional C2CD4A and C2CD4B SE constituent sites (Figure 3A, HS1-9; Table S4) and found that HS1, HS7, and HS9 also enhanced luciferase reporter activity in both orientations (Figure S4B). HS1, HS2, and HS6 also contained sequence variants. However, we did not detect significant haplotype-specific differences in enhancer activity of these islet SE constituent sequences (Figure S4C). Thus, multiple islet SE constituent sites exhibited enhancer activity. However, only HS3, which contains the putative causal variant rs7163757, exhibited allelic differences in activity. Allelic analyses of ATAC-seq data from rs7163757 C/T heterozygous islet samples36 did not reveal consistent or significant over- or underrepresentation of the risk allele in accessible or open chromatin, and we found both alleles present in published islet active enhancer histone modifications and TF ChIP-seq datasets (data not shown).18, 36 Together, these data suggest that the rs7163757 alleles do not create or destroy the transcriptional enhancer. We conclude from these experiments that the rs7163757 risk allele (C) potentiates HS3 enhancer activity, potentially by creating a binding site for a stimulus- or stress-responsive TF.
Increased C2CD4B and C2CD4A Expression Is Linked to the Fine-Mapped Putative Causal Variant(s) and Inflammatory Cytokine Response in Islets
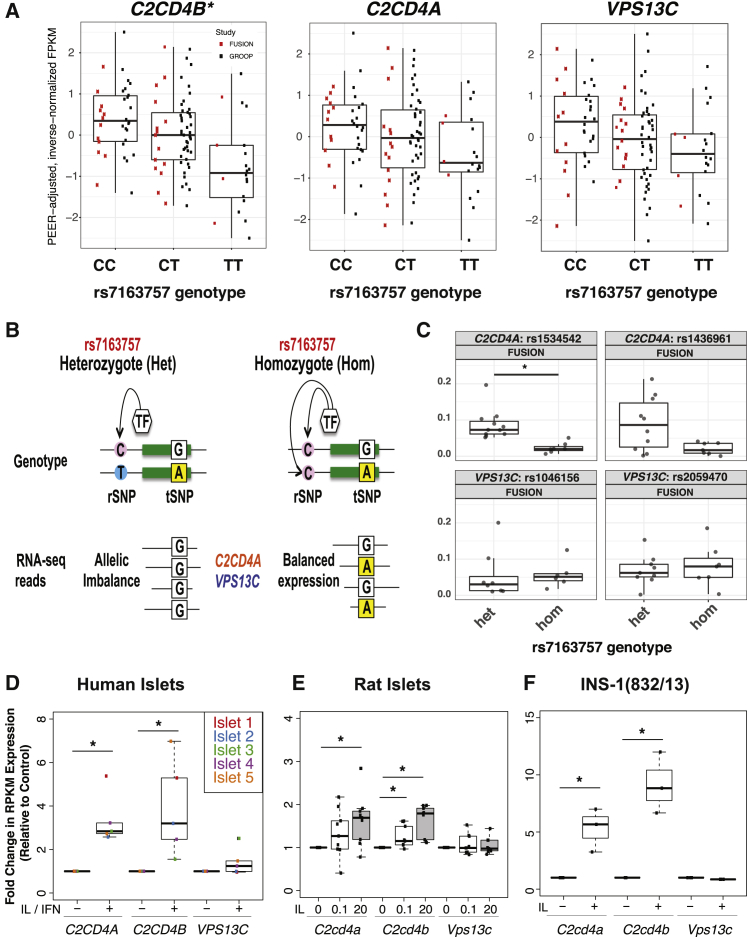
To identify the putative target gene(s) of the fine-mapped variants (Table 1 and Tables S5–S7), we examined associations between the rs7163757 genotype and the expression of genes with TSSs residing within 1 Mb of this putative functional variant (Material and Methods) in our recent study of 112 islet samples from individuals of European descent36 and in multiple tissues examined by the Genotype-Tissue Expression (GTEx) Consortium.55, 56, 57 C2CD4A and C2CD4B were robustly expressed in islets and only selectively among other GTEx tissues, contrasting with the wide expression of VPS13C (Figure S3). The rs7163757 risk allele (C) was significantly associated with increased C2CD4B expression in both islet cohorts (Figure 5A, Figure S3, and Table S10). Conditional analysis (Material and Methods) indicated that rs7163757 was associated with C2CD4B expression independently of other variants in the region (Table S10). Although it didn’t reach statistical significance after genome-wide testing correction, C2CD4A also exhibited a trend of increased expression with increasing risk allele dosage in islets.
Figure 5.
Islet C2CD4B and C2CD4A Expression Is Increased by the rs7163757 T2D Risk Allele (C) and Induced by Diabetogenic Inflammatory Cytokines
(A) Boxplots of C2CD4B, C2CD4A, and VPS13C expression (PEER-adjusted, inverse-normalized FPKM) in 112 human islets grouped by rs7163757 genotype. Red and black dots indicate FPKM values from FUSION and Groop islet cohorts, respectively; the asterisk indicates a genome-wide significant association (see also Table S10).
(B) Schematic depicting the principles of AEI. The heterozygous rSNP genotype leads to an expression imbalance of the tSNP in RNA-seq data for genes regulated by the rSNP (left). The homozygous rSNP genotype manifests as equivalent transcription of both tSNP alleles.
(C) AEI between rSNP rs7163757 and tSNPs in C2CD4A and VPS13C in FUSION human pancreatic islet RNA-seq data. Dots indicate the value of each islet sample, which have been grouped into heterozygous (Het) and homozygous (Hom) rs7163757 genotypes on the x axis. The absolute value of allelic imbalance for each tSNP is indicated on the y axis; ∗q < 0.05 (two-sided Wilcoxon rank-sum test adjusted for multiple testing) (see Material and Methods and Table S11). C2CD4A, not VPS13C, exhibits a rs7163757 genotype-dependent allelic imbalance, as depicted schematically in (B).
(D–F) C2CD4A, C2CD4B, and VPS13C expression in (D) human islets before and after IL-1β (IL) and IFN-γ (IFN) exposure,48 (E) rat islets exposed to 0, 0.1, or 20 ng/mL IL-1β,49 or (F) INS-1(832/13) cells exposed to 2 U/mL IL-1β for 2 hr. Boxplots represent fold changes in expression (RPKM) in relation to that of untreated controls; ∗p < 0.05 (Wilcoxon rank-sum test).
To assess further the potential links between the rs7163757 genotype and C2CD4A and/or VPS13C expression, we completed allele-specific eQTL (aseQTL) analyses45 of tSNPs (Figure 5B) to determine the relative abundance of C2CD4A and VPS13C mRNA transcripts produced from the maternal and paternal chromosomes within each islet sample. Specifically, we sought to determine whether RNA-seq reads from deeply sequenced FUSION islet samples heterozygous for the putative rs7163757 rSNP (Figure 5B) exhibited a higher allelic expression imbalance (AEI) for C2CD4A or VPS13C than rs7163757 homozygous islet samples. rs7163757 heterozygous FUSION islet samples exhibited significantly higher AEI for the C2CD4A tSNP rs1534542 (Storey’s FDR < 0.01) and a trend of increased AEI for C2CD4A tSNP rs1436961 (Figure 5C, Figure S5, and Table S11). These tSNPs did not exhibit significant AEI differences in Groop islet samples (Figure S5 and Table S11), which could reflect differences in islet processing and culture times or the substantially lower RNA-seq coverage of tSNPs in these samples than in FUSION samples (Figure S5B). In both islet cohorts, no significant AEI differences between rs7163757 heterozygote and homozygote islet samples were observed for any VPS13C tSNPs (Figure 5C, Figure S5, and Table S11).
Both C2CD4A and C2CD4B are induced by inflammatory cytokines in endothelial cells.58 To determine whether C2CD4A, C2CD4B, and/or VPS13C are induced by these (patho)physiologic signals48, 49, 59, 60, 61 in islets and β cells, we analyzed RNA-seq data from human48 and rat49 islets exposed to inflammatory cytokines. As shown in Figures 5D and 5E, both C2CD4A and C2CD4B, but not VPS13C, were induced by inflammatory cytokines in human and rat islets. Similarly, we found that IL-1β induced both C2cd4a and C2cd4b in INS-1(832/13), whereasVps13c was unaffected (Figure 5F). Together, these data link the rs7163757 proinsulin-raising and T2D risk allele (C) to increased expression of C2CD4B, and possibly C2CD4A, and implicate induction of C2CD4A and C2CD4B, but not VPS13C, in islet and β cell responses to (patho)physiologic signals.
NFAT Potentiates Enhancer Activity of Risk Alleles
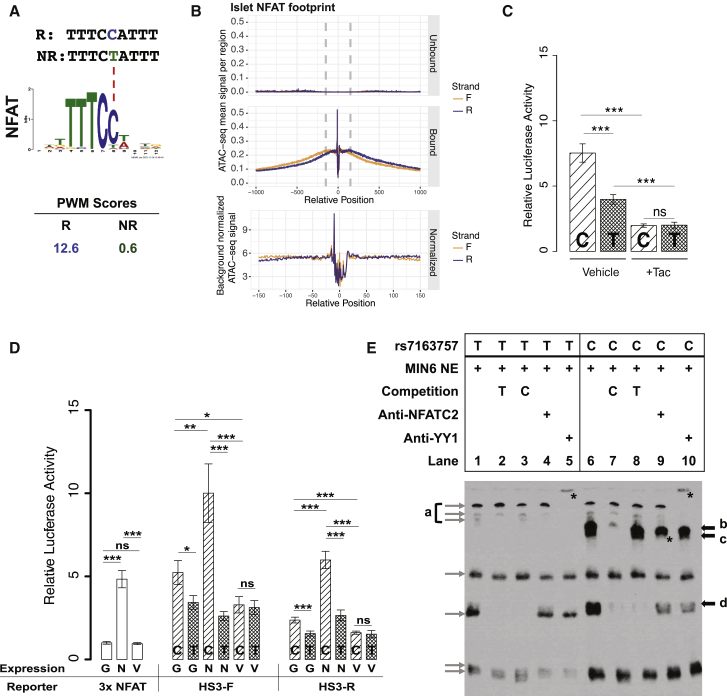
We next sought to identify plausible trans-factors that could mediate the increased rs7163757 risk allele (C) enhancer activity observed in Figure 4B. HaploReg v.4.162 predicts that the rs7163757 risk allele (C), which exhibited increased enhancer activity (Figure 4B), creates high-affinity DNA binding motifs for the TFs NFAT and zinc finger protein Helios (Figure 6A and Table S12). Genes encoding zinc finger protein Helios (IKZF2 [MIM: 606234]) and multiple members of the NFAT TF family (NFATC1, NFATC2, and NFATC3 [MIM: 602698] and NFATC4 [MIM: 602699]) are expressed in human and mouse islet cells, MIN6, and INS-1(832/13) (data not shown). We hypothesized that the NFAT family mediates the effects of the rs7163757 risk allele because (1) the calcineurin-NFAT pathway has well-described roles in islet physiology63, 64, 65, 66 and pathophysiology,67, 68, 69 including a recent report implicating NFAT isoforms as regulators of several putative T2D genes in mouse and human islets;70 (2) NFAT modulates transcriptional inflammatory responses,68 and the putative rs7163757 target genes, C2CD4B and C2CD4A, are induced by inflammatory cytokines in islets48, 49 and β cells (Figures 5D–5F); (3) the rs7163757 risk and non-risk alleles alter a site of high information content in the NFAT PWM (Figure 6A); and (4) we detected NFAT footprints (Figure 6B) in the ATAC-seq open chromatin site overlapping rs7163757 in both human islets (Figure 3 and Figure S2A), which were both C/T heterozygotes.
Figure 6.
NFAT Modulates Enhancer Activity of the rs7163757 Risk Allele (C)
(A) PWM scores (HaploReg) identify NFAT as the most plausible TF binding to the risk allele (C), but not to the non-risk allele (T), of rs7163757. Sequence is shown for the risk allele (R, blue) and the non-risk allele (NR, green). A dashed red line points to the affected position in the NFAT binding motif.
(B) Islet NFAT footprints. Density plots indicate normalized sequence coverage of ATAC-seq at the NFAT motifs predicted as bound (including the rs7163757 site) or unbound in two human islet samples.
(C) Luciferase reporter assay of HS3 (cloned in the forward orientation) containing the rs7163757 risk allele (C) or non-risk allele (T) in MIN6 cells treated with 10 ng/mL tacrolimus (Tac) or ethanol control (vehicle). Four to five biological replicates were measured in three independent experiments. Luciferase activity of HS3 constructs were normalized to that of the corresponding vehicle-control- and tacrolimus-treated pGL4.23 empty vector. Error bars represent mean luciferase activity ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (unpaired two-sided t test).
(D) Luciferase reporter assay of HS3 (cloned in forward and reverse orientations) containing the rs7163757 risk allele (C) or non-risk allele (T) in the presence of EGFPC1-huNFATc1EE-WT or the NFAT inhibitor construct EGFPN1-VIVIT. pGL3-NFAT-Luciferase (NFAT reporter), containing three canonical NFAT binding sites upstream of a luciferase reporter, plus EGFP plasmid served as a control for NFAT activity of the expression constructs. For all cloned HS3 enhancer sequences co-transfected with EGFP, EGFPC1-huNFATc1EE-WT, or EGFPN1-VIVIT, eight biological replicates were measured in four independent experiments. Luciferase activity of NFAT expression constructs was normalized to the NFAT reporter plus EGFP. Luciferase activity of HS3 expression constructs with EGFP or NFAT expression constructs was normalized to that of the pGL4.23 empty vector with EGFP or NFAT expression constructs, respectively. HS3-F and HS3-R denote the HS3 sequence (C or T allele) cloned into pGL4.23 in the forward and reverse orientations, respectively; G, EGFP; N, EGFPC1-huNFATc1EE-WT; and V, EGFPN1-VIVIT. Error bars represent mean luciferase activity ± SEM; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (unpaired two-sided t test).
(E) Allele-specific protein binding of sequences containing different rs7163757 alleles. EMSA with biotin-labeled probes containing the rs7163757 T or C allele incubated with MIN6 nuclear extract (NE). Black arrows indicate allelic differences in protein binding. Gray arrows show non-specific binding or no clear differences between alleles. Bands labeled b, c, and d indicate proteins that bind specifically to the C allele. The asterisk in lane 9 indicates the disruption of band c by NFATc2 antibody. Asterisks in lanes 5 and 10 indicate supershift of non-allele specific band a by YY1 antibody.
To test this hypothesis, we first examined how pharmacologic inhibition of the calcineurin-NFAT pathway by tacrolimus (FK506) would affect enhancer activity of HS3 risk and non-risk alleles. Calcineurin-mediated dephosphorylation is essential for NFAT translocation from the cytosol to the nucleus,71 and tacrolimus inhibits this process. We tested the effect of tacrolimus treatment on enhancer activity in MIN6 and found that it abrogated the higher enhancer activity of the rs7163757 risk allele (C) to that of the rs7163757 non-risk allele (T) (Figure 6C; compare the activity of the C and T alleles between “Tac” and “Veh” controls). These data thus provide pharmacologic evidence that the calcineurin-NFAT pathway mediates the effects of the rs7163757 risk allele.
Next, we tested how molecular manipulation of NFAT affects HS3 enhancer activity. We co-transfected MIN6 cells with the HS3 luciferase vector and plasmids expressing GFP-tagged NFATc1. Furthermore, given that a peptide sequence (“VIVIT”) has been shown to selectively inhibit calcineurin-NFAT interactions and NFAT activity,39, 71 we also tested HS3 activity when HS3 was co-transfected with GFP-tagged VIVIT to inhibit NFAT activity in the cells. As a positive control, we co-transfected each expression vector with a luciferase reporter plasmid containing three canonical NFAT binding sites (NFAT reporter). Expression of GFP alone (G), NFATc1 alone (N), or VIVIT alone (V) did not alter transcriptional activity of the empty luciferase vector (pGL4.23, not shown). As expected, NFATc1 expression in MIN6 cells enhanced luciferase activity of the NFAT reporter by 5-fold (Figure 6D). Importantly, NFATc1 expression specifically potentiated enhancer activity of the HS3 sequence containing the rs7163757 risk allele (C) rather than that containing the non-risk allele (T) in both forward and reverse orientations (Figure 6D; compare alleles in the N groups). Expression of the inhibitory GFP-tagged VIVIT peptide reduced the enhancer activity of the HS3 risk allele to that of the non-risk allele in both orientations (Figure 6D; compare alleles in the V groups). Microscopic analysis of the transfected cells confirmed comparable GFP-VIVIT and EGFP levels, both of which were more robust than the levels of the GFP-NFATc1 fusion (Figure S6). This strongly suggests that NFAT-potentiated enhancer activity of the HS3 risk allele was not merely the consequence of more robust NFAT expression. Together, these results support the hypothesis that NFAT mediates increased enhancer activity of the rs7163757 risk allele.
Finally, to determine whether the risk and non-risk alleles are bound by different β cell nuclear proteins and complexes, we incubated DNA probes containing the rs7163757 risk allele (C) or the non-risk allele (T) with MIN6 β cell line nuclear extracts and looked for electrophoretic mobility differences of the resulting protein-DNA complexes. As shown in Figure 6E, EMSAs revealed that the risk allele was bound by β cell nuclear factors not bound to the non-risk allele (bands b, c, and d; compare lanes 1 and 6). Incubation of each labeled probe with excess unlabeled probe containing the risk or non-risk allele allowed us to determine the allele specificity of these complexes, revealing that complexes b and c were specifically disrupted by the risk allele (Figure 6E; compare lanes 7 and 8). Finally, we tested for the presence of candidate TFs and cofactors in the complexes binding this transcriptional enhancer sequence. We observed supershift and disruption of complex a when antibodies YY1 (Figure 6E, asterisks in lanes 5 and 10) and p300 (Figure S7A, asterisks in lanes 4 and 9) were co-incubated with the extract and probes, respectively, providing in vitro support that this sequence is bound by enhancer-associated cofactors. The addition of anti-FOXA2, -MAFB, -NKX6.1, and -PDX1 antibodies disrupted several of the complexes formed with risk and non-risk alleles (Figure S7B, asterisks in lanes 4, 5, 9, and 10), providing in vitro support of the in vivo binding of this site, as determined by ChIP-seq in human islets.18 Importantly, NFATc2 antibody completely disrupted the risk allele (C)-specific complex c (Figure 6E, asterisk in lane 9) and reduced complex d binding (Figure 6E, lane 9). These data indicate that rs7163757 risk and non-risk alleles are both bound by enhancer-associated cofactors YY1 and p300, consistent with in vivo chromatin accessibility and published islet TF binding, but that the risk allele (C) is bound by additional nuclear factors, including NFAT. These data do not eliminate the possibility that other factors, such as zinc finger protein Helios, might bind and modulate risk-allele enhancer activity. However, the luciferase reporter and EMSA results together provide pharmacologic, molecular, and biochemical evidence supporting one or more NFAT isoforms as factors that bind to and potentiate enhancer activity of the rs7163757 risk allele.
Discussion
GWASs have identified over 150 loci contributing genetic susceptibility to islet dysfunction and T2D. The vast majority of GWAS SNPs associated with T2D and related molecular traits (e.g., fasting glucose, fasting insulin, 2-hr glucose, and GSIS) reside in non-coding regions of the genome. Identifying (1) the causal variant(s), (2) their target gene(s), and (3) their direction(s) of effect for T2D risk (i.e., gain or loss of function) at each associated locus is important for better understanding the molecular genetic pathology underlying islet dysfunction and T2D and determining therapeutic suitability. In this study, we applied genetic fine-mapping and functional genomic techniques to refine the proinsulin and T2D association signal on 15q22.2 and address these three important questions. After identifying 16 intergenic non-coding variants, we applied functional (epi)genomics and experimental analyses to implicate rs7163757 as the most likely functional variant, to identify C2CD4B, and possibly C2CD4A, as a putative target gene of this human pancreatic islet enhancer SNP, and to link C2CD4A and C2CD4B induction in islets to inflammatory cytokines. The T2D risk and proinsulin-raising allele (C) exhibited increased enhancer activity and was differentially bound by β cell nuclear factors in vitro. Moreover, pharmacologic and molecular manipulation of the calcineurin-NFAT pathway in combination with in vitro evidence suggests that one or more of the NFAT TFs can bind to the risk allele and potentiate its enhancer activity. Together, these results implicate gain-of-function effects of the rs7163757 risk allele (C) on an evolutionarily conserved islet SE and increased expression of C2CD4B (and most likely C2CD4A) as a molecular mechanism underlying the 15q22.2 genetic association with increased proinsulin levels and T2D.
Human pancreatic islet eQTL and aseQTL analyses link increased expression of C2CD4B, and potentially C2CD4A, to the T2D risk and proinsulin-increasing rs7163757 allele (C). Whereas rs7163757 has been detected as a lung eQTL for C2CD4A by the GTEx Consortium,55, 56, 57 the link between the rs7163757 genotype and C2CD4B expression appears to be unique to islets. C2CD4A and C2CD4B, but not VPS13C, are induced by proinflammatory cytokines in islets and β cells. Moreover, our in vitro and in vivo data strongly suggest that the risk allele increases enhancer activity and C2CD4B and C2CD4A expression. Notably, the trend of increased expression was consistent between RNA-seq profiles obtained from two of three independent islet cohorts,36, 44, 72 supporting the robust and reproducible nature of these observations. These results contrast with those of a previous report linking the rs7163757 risk allele to decreased enhancer activity and female-specific decreases in VPS13C and C2CD4A expression among 40 female islet samples.73 Carefully controlled meta-analyses of expression data from hundreds of islets and targeted functional studies will provide important insights to resolve this apparent discrepancy; these data, however, challenge the existing model suggesting that female-specific decreases in VPS13C and C2CD4A expression underlie the association between the rs7163757 risk allele and islet dysfunction and T2D. Together, these data clearly call for future studies to determine (1) rs7163757 variant effects on C2CD4A and C2CD4B expression in (patho)physiologic responses and (2) the role(s) that C2CD4B and/or C2CD4A might play in pathogenic or compensatory islet stress responses and to determine how these could be exploited for preventing and/or treating T2D.
SEs16, 17, 53 are important transcriptional regulatory regions that govern cell-type-specific functions. In human islets, genes encoding proteins involved in glucose sensing (e.g., GCK), insulin secretion (e.g., INS, ABCC8, and KCNJ11), and islet cell identity (e.g., PDX1, MAFA [MIM: 610303], and NKX6.1) overlap or are nearby SE chromatin signatures. Evolutionary conservation of the C2CD4A-C2CD4B-VPS13C SE signature reported here suggests that this locus is also an important region for islet function. rs7163757 resides in this conserved islet SE, and the surrounding sequence, HS3, is an open chromatin site and an active enhancer independent of the rs7163757 genotype in human islets, as indicated by in vivo islet ATAC-seq and TF ChIP-seq data and in vitro luciferase reporter activity. Consistent with this finding, the rs7163757 risk (C) and non-risk (T) alleles are both empirically bound by multiple islet TFs according to islet ChIP-seq data.18 This implies that the risk allele (C) does not create or destroy this open chromatin site but that the observed increase in enhancer activity of the risk allele (C) (gain-of-function effect) is facilitated by the recruitment of an additional TF, in this case NFAT, to a canonical binding site created by the risk allele. Such gain-of-function effects have been identified for other T2D-associated loci,74 and “enhancer hijacking” is emerging as a tumorigenic mechanism in cancer.75, 76
Together, the pharmacologic, molecular, and in vitro experiments in this study strongly suggest that NFAT is a TF family that mediates these gain-of-function risk-allele effects. The NFAT TF family is linked to both physiologic and pathophysiologic transcriptional responses in islets and β cells. Physiologically, it directly regulates Ins transcription in pancreatic β cells in response to increased Ca2+ levels and calcineurin activation,66 and pharmacologic calcineurin inhibition decreases human β cell survival and murine β cell proliferation.65 β-cell-specific deletion of the murine calcineurin regulatory subunit (Cnb1, also known as Ppp3r1 [human homolog PPP3R1 [MIM:601302]) results in age-dependent diabetes characterized by decreased β cell proliferation and mass and by reduced pancreatic insulin content; conditional expression of active Nfatc1 in Cnb1−/− mice rescues these defects and prevents diabetes.64 Conversely, islet expression of constitutively active calcineurin in mice results in glucose intolerance and loss of β cell mass as a result of decreased proliferation and increased apoptosis.69 Additionally, the NFAT TF family has been implicated in the pancreatic islet and β cell inflammation response, wherein it mediates TNF (MIM: 191160) expression after exposure to the pro-inflammatory cytokine IL-1β.67 Recent data suggest that binding partners, such as ERK and JNK, might recruit NFAT to distinct cis-regulatory elements to mediate physiologic and pathophysiologic or inflammatory gene expression responses.68 Interestingly, C2CD4A and C2CD4B were both identified as inflammation-responsive genes in endothelial cells, suggesting that they could indeed be co-regulated: C2CD4A and C2CD4B expression increased 30- and 18-fold, respectively, after 2 hr of treatment with IL-1β in endothelial cells.58 C2CD4A and C2CD4B expression in human islets was 3- to 5-fold and 2- to 7-fold higher, respectively, after exposure to pro-inflammatory cytokines IL-1β and IFN-γ than before exposure;48 palmitate treatment induced C2CD4A expression 3- to 4-fold more than no treatment.77 We detected 3-fold higher expression of C2CD4A and C2CD4B in human pancreatic islets after IL-1β treatment than before treatment (data not shown). Similarly, we observed 2- and 3-fold higher expression of C2cd4a and C2cd4b, respectively, in INS-1(832/13) β cells treated with 2 U/mL IL-1β for 2 hr than in untreated controls. Finally, C2cd4a expression was 3-fold higher in diabetes-sensitive New Zealand Obese (NZO) mouse islets than in those of diabetes-resistant B6-ob/ob mice after a carbohydrate challenge.78
We propose a working model wherein the rs7163757 T2D risk allele creates NFAT-mediated or -dependent enhancer gain of function and inappropriate, enhanced, or extended expression of C2CD4B (and most likely C2CD4A) in response to islet and/or β cell (patho)physiologic signals. Multiple NFAT TF paralogs (NFATC1, NFATC2, and NFATC3) are expressed in islets. Thus, it will be important in future studies to elucidate the specific NFAT family member(s) mediating the effects of the rs7163757 risk allele and clarify the condition(s) that elicit NFAT binding to this regulatory site in vivo. Moreover, follow-up studies to define the molecular functions of C2CD4B and/or C2CD4A in human islets and to determine the effect of their overexpression and inactivation on islet and β cell (patho)physiology will be critical to understanding their roles in T2D pathogenesis and their utility as therapeutic targets for preventing and/or treating T2D.
Acknowledgments
We thank members of our laboratories and colleagues at the Jackson Laboratory for Genomic Medicine, University of Michigan, and National Human Genome Research Institute and members of the FUSION Consortium for helpful comments and suggestions throughout this study, as well as the anonymous reviewers whose comments and suggestions improved the quality and clarity of this manuscript. We are especially grateful to the pancreatic organ donors and their families, without whom this study of the genetics of human islet dysfunction and type 2 diabetes would not have been possible. Finally, we extend our appreciation to Drs. van de Bunt, McCarthy, and Gloyn for providing summary islet eQTL data from their PLoS Genetics study for analysis. This study was made possible by generous financial support from the National Institute of Diabetes and Digestive and Kidney Diseases under award numbers DK092251 (to M.L.S.), DK099240 (to S.C.J.P.), DK072193 and DK105561 (to K.L.M.), and DK062370 (to M.B.), the NIH Intramural Program (ZIAHG000024 to F.S.C.), and the American Diabetes Association Pathway to Stop Diabetes (grants 1-14-INI-07 to S.C.J.P. and 1-18-ACE-15 to M.L.S.). Opinions, interpretations, conclusions, and recommendations are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or American Diabetes Association.
Published: April 5, 2018
Footnotes
Supplemental Data include 7 figures and 12 tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.02.020.
Accession Numbers
The accession numbers for the mouse MIN6 and rat INS-1(832/13) ATAC-seq, ChIP-seq, and RNA-seq data reported in this paper are NCBI BioProject: PRJNA398550 and NCBI Sequence Read Archive: SRP116556.
Web Resources
Genotype-Tissue Expression (GTEx) Portal, https://www.gtexportal.org/home/
Islet eQTL Explorer, http://theparkerlab.org/tools/isleteqtl/
OMIM, http://www.omim.org
UCSC Genome Browser liftOver tool, https://genome.ucsc.edu/cgi-bin/hgLiftOver
Supplemental Data
References
- 1.Mohlke K.L., Boehnke M. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum. Mol. Genet. 2015;24(R1):R85–R92. doi: 10.1093/hmg/ddv264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashcroft F.M., Rorsman P. Diabetes mellitus and the β cell: the last ten years. Cell. 2012;148:1160–1171. doi: 10.1016/j.cell.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchsberger C., Flannick J., Teslovich T.M., Mahajan A., Agarwala V., Gaulton K.J., Ma C., Fontanillas P., Moutsianas L., McCarthy D.J. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawlor N., Khetan S., Ucar D., Stitzel M.L. Genomics of Islet (Dys)function and Type 2 Diabetes. Trends Genet. 2017;33:244–255. doi: 10.1016/j.tig.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamauchi T., Hara K., Maeda S., Yasuda K., Takahashi A., Horikoshi M., Nakamura M., Fujita H., Grarup N., Cauchi S. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat. Genet. 2010;42:864–868. doi: 10.1038/ng.660. [DOI] [PubMed] [Google Scholar]
- 6.Cui B., Zhu X., Xu M., Guo T., Zhu D., Chen G., Li X., Xu L., Bi Y., Chen Y. A genome-wide association study confirms previously reported loci for type 2 diabetes in Han Chinese. PLoS ONE. 2011;6:e22353. doi: 10.1371/journal.pone.0022353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shu X.O., Long J., Cai Q., Qi L., Xiang Y.-B., Cho Y.S., Tai E.S., Li X., Lin X., Chow W.-H. Identification of new genetic risk variants for type 2 diabetes. PLoS Genet. 2010;6:e1001127. doi: 10.1371/journal.pgen.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahajan A., Go M.J., Zhang W., Below J.E., Gaulton K.J., Ferreira T., Horikoshi M., Johnson A.D., Ng M.C., Prokopenko I., DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium. Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium. South Asian Type 2 Diabetes (SAT2D) Consortium. Mexican American Type 2 Diabetes (MAT2D) Consortium. Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupuis J., Langenberg C., Prokopenko I., Saxena R., Soranzo N., Jackson A.U., Wheeler E., Glazer N.L., Bouatia-Naji N., Gloyn A.L., DIAGRAM Consortium. GIANT Consortium. Global BPgen Consortium. Anders Hamsten on behalf of Procardis Consortium. MAGIC investigators New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat. Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grarup N., Overvad M., Sparsø T., Witte D.R., Pisinger C., Jørgensen T., Yamauchi T., Hara K., Maeda S., Kadowaki T. The diabetogenic VPS13C/C2CD4A/C2CD4B rs7172432 variant impairs glucose-stimulated insulin response in 5,722 non-diabetic Danish individuals. Diabetologia. 2011;54:789–794. doi: 10.1007/s00125-010-2031-2. [DOI] [PubMed] [Google Scholar]
- 11.Strawbridge R.J., Dupuis J., Prokopenko I., Barker A., Ahlqvist E., Rybin D., Petrie J.R., Travers M.E., Bouatia-Naji N., Dimas A.S., DIAGRAM Consortium. GIANT Consortium. MuTHER Consortium. CARDIoGRAM Consortium. C4D Consortium Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huyghe J.R., Jackson A.U., Fogarty M.P., Buchkovich M.L., Stančáková A., Stringham H.M., Sim X., Yang L., Fuchsberger C., Cederberg H. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat. Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saxena R., Hivert M.-F., Langenberg C., Tanaka T., Pankow J.S., Vollenweider P., Lyssenko V., Bouatia-Naji N., Dupuis J., Jackson A.U., GIANT consortium. MAGIC investigators Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat. Genet. 2010;42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wood A.R., Jonsson A., Jackson A.U., Wang N., van Leewen N., Palmer N.D., Kobes S., Deelen J., Boquete-Vilarino L., Paananen J., Diabetes Research on Patient Stratification (DIRECT) A Genome-Wide Association Study of IVGTT-Based Measures of First-Phase Insulin Secretion Refines the Underlying Physiology of Type 2 Diabetes Variants. Diabetes. 2017;66:2296–2309. doi: 10.2337/db16-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boland B.B., Rhodes C.J., Grimsby J.S. The dynamic plasticity of insulin production in β-cells. Mol. Metab. 2017;6:958–973. doi: 10.1016/j.molmet.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker S.C.J., Stitzel M.L., Taylor D.L., Orozco J.M., Erdos M.R., Akiyama J.A., van Bueren K.L., Chines P.S., Narisu N., Black B.L., NISC Comparative Sequencing Program Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. USA. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasquali L., Gaulton K.J., Rodríguez-Seguí S.A., Mularoni L., Miguel-Escalada I., Akerman İ., Tena J.J., Morán I., Gómez-Marín C., van de Bunt M. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 2014;46:136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stitzel M.L., Kycia I., Kursawe R., Ucar D. Transcriptional Regulation of the Pancreatic Islet: Implications for Islet Function. Curr. Diab. Rep. 2015;15:66. doi: 10.1007/s11892-015-0635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stancáková A., Javorský M., Kuulasmaa T., Haffner S.M., Kuusisto J., Laakso M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009;58:1212–1221. doi: 10.2337/db08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaneau O., Marchini J., Zagury J.-F. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 25.Fuchsberger C., Abecasis G.R., Hinds D.A. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.-Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyazaki J., Araki K., Yamato E., Ikegami H., Asano T., Shibasaki Y., Oka Y., Yamamura K. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127:126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 29.Hohmeier H.E., Mulder H., Chen G., Henkel-Rieger R., Prentki M., Newgard C.B. Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes. 2000;49:424–430. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
- 30.Stitzel M.L., Sethupathy P., Pearson D.S., Chines P.S., Song L., Erdos M.R., Welch R., Parker S.C.J., Boyle A.P., Scott L.J., NISC Comparative Sequencing Program Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varshney A., Scott L.J., Welch R.P., Erdos M.R., Chines P.S., Narisu N., Albanus R.D., Orchard P., Wolford B.N., Kursawe R., NISC Comparative Sequencing Program Genetic regulatory signatures underlying islet gene expression and type 2 diabetes. Proc. Natl. Acad. Sci. USA. 2017;114:2301–2306. doi: 10.1073/pnas.1621192114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pique-Regi R., Degner J.F., Pai A.A., Gaffney D.J., Gilad Y., Pritchard J.K. Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res. 2011;21:447–455. doi: 10.1101/gr.112623.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beals C.R., Clipstone N.A., Ho S.N., Crabtree G.R. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- 39.Aramburu J., Yaffe M.B., López-Rodríguez C., Cantley L.C., Hogan P.G., Rao A. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- 40.Clipstone N.A., Crabtree G.R. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 41.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 43.Scott L.J., Erdos M.R., Huyghe J.R., Welch R.P., Beck A.T., Wolford B.N., Chines P.S., Didion J.P., Narisu N., Stringham H.M. The genetic regulatory signature of type 2 diabetes in human skeletal muscle. Nat. Commun. 2016;7:11764. doi: 10.1038/ncomms11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fadista J., Vikman P., Laakso E.O., Mollet I.G., Esguerra J.L., Taneera J., Storm P., Osmark P., Ladenvall C., Prasad R.B. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc. Natl. Acad. Sci. USA. 2014;111:13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Battle A., Mostafavi S., Zhu X., Potash J.B., Weissman M.M., McCormick C., Haudenschild C.D., Beckman K.B., Shi J., Mei R. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fogarty M.P., Cannon M.E., Vadlamudi S., Gaulton K.J., Mohlke K.L. Identification of a regulatory variant that binds FOXA1 and FOXA2 at the CDC123/CAMK1D type 2 diabetes GWAS locus. PLoS Genet. 2014;10:e1004633. doi: 10.1371/journal.pgen.1004633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eizirik D.L., Sammeth M., Bouckenooghe T., Bottu G., Sisino G., Igoillo-Esteve M., Ortis F., Santin I., Colli M.L., Barthson J. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012;8:e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arous C., Ferreira P.G., Dermitzakis E.T., Halban P.A. Short term exposure of beta cells to low concentrations of interleukin-1β improves insulin secretion through focal adhesion and actin remodeling and regulation of gene expression. J. Biol. Chem. 2015;290:6653–6669. doi: 10.1074/jbc.M114.611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maller J.B., McVean G., Byrnes J., Vukcevic D., Palin K., Su Z., Howson J.M., Auton A., Myers S., Morris A., Wellcome Trust Case Control Consortium Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am. J. Hum. Genet. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott R.A., Scott L.J., Mägi R., Marullo L., Gaulton K.J., Kaakinen M., Pervjakova N., Pers T.H., Johnson A.D., Eicher J.D., DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Denas O., Sandstrom R., Cheng Y., Beal K., Herrero J., Hardison R.C., Taylor J. Genome-wide comparative analysis reveals human-mouse regulatory landscape and evolution. BMC Genomics. 2015;16:87. doi: 10.1186/s12864-015-1245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melé M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J., GTEx Consortium Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.GTEx Consortium Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warton K., Foster N.C., Gold W.A., Stanley K.K. A novel gene family induced by acute inflammation in endothelial cells. Gene. 2004;342:85–95. doi: 10.1016/j.gene.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 59.Halban P.A., Polonsky K.S., Bowden D.W., Hawkins M.A., Ling C., Mather K.J., Powers A.C., Rhodes C.J., Sussel L., Weir G.C. β-cell failure in type 2 diabetes: postulated mechanisms and prospects for prevention and treatment. Diabetes Care. 2014;37:1751–1758. doi: 10.2337/dc14-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hajmrle C., Smith N., Spigelman A.F., Dai X., Senior L., Bautista A., Ferdaoussi M., MacDonald P.E. Interleukin-1 signaling contributes to acute islet compensation. JCI Insight. 2016;1:e86055. doi: 10.1172/jci.insight.86055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hajmrle C., Ferdaoussi M., Plummer G., Spigelman A.F., Lai K., Manning Fox J.E., MacDonald P.E. SUMOylation protects against IL-1β-induced apoptosis in INS-1 832/13 cells and human islets. Am. J. Physiol. Endocrinol. Metab. 2014;307:E664–E673. doi: 10.1152/ajpendo.00168.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward L.D., Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goodyer W.R., Gu X., Liu Y., Bottino R., Crabtree G.R., Kim S.K. Neonatal β cell development in mice and humans is regulated by calcineurin/NFAT. Dev. Cell. 2012;23:21–34. doi: 10.1016/j.devcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heit J.J., Apelqvist A.A., Gu X., Winslow M.M., Neilson J.R., Crabtree G.R., Kim S.K. Calcineurin/NFAT signalling regulates pancreatic beta-cell growth and function. Nature. 2006;443:345–349. doi: 10.1038/nature05097. [DOI] [PubMed] [Google Scholar]
- 65.Soleimanpour S.A., Crutchlow M.F., Ferrari A.M., Raum J.C., Groff D.N., Rankin M.M., Liu C., De León D.D., Naji A., Kushner J.A., Stoffers D.A. Calcineurin signaling regulates human islet beta-cell survival. J. Biol. Chem. 2010;285:40050–40059. doi: 10.1074/jbc.M110.154955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lawrence M.C., Bhatt H.S., Easom R.A. NFAT regulates insulin gene promoter activity in response to synergistic pathways induced by glucose and glucagon-like peptide-1. Diabetes. 2002;51:691–698. doi: 10.2337/diabetes.51.3.691. [DOI] [PubMed] [Google Scholar]
- 67.Lawrence M.C., Naziruddin B., Levy M.F., Jackson A., McGlynn K. Calcineurin/nuclear factor of activated T cells and MAPK signaling induce TNF-alpha gene expression in pancreatic islet endocrine cells. J. Biol. Chem. 2011;286:1025–1036. doi: 10.1074/jbc.M110.158675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lawrence M.C., Borenstein-Auerbach N., McGlynn K., Kunnathodi F., Shahbazov R., Syed I., Kanak M., Takita M., Levy M.F., Naziruddin B. NFAT targets signaling molecules to gene promoters in pancreatic β-cells. Mol. Endocrinol. 2015;29:274–288. doi: 10.1210/me.2014-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bernal-Mizrachi E., Cras-Méneur C., Ye B.R., Johnson J.D., Permutt M.A. Transgenic overexpression of active calcineurin in beta-cells results in decreased beta-cell mass and hyperglycemia. PLoS ONE. 2010;5:e11969. doi: 10.1371/journal.pone.0011969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keller M.P., Paul P.K., Rabaglia M.E., Stapleton D.S., Schueler K.L., Broman A.T., Ye S.I., Leng N., Brandon C.J., Neto E.C. The Transcription Factor Nfatc2 Regulates β-Cell Proliferation and Genes Associated with Type 2 Diabetes in Mouse and Human Islets. PLoS Genet. 2016;12:e1006466. doi: 10.1371/journal.pgen.1006466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aramburu J., Garcia-Cózar F., Raghavan A., Okamura H., Rao A., Hogan P.G. Selective inhibition of NFAT activation by a peptide spanning the calcineurin targeting site of NFAT. Mol. Cell. 1998;1:627–637. doi: 10.1016/s1097-2765(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 72.van de Bunt M., Manning Fox J.E., Dai X., Barrett A., Grey C., Li L., Bennett A.J., Johnson P.R., Rajotte R.V., Gaulton K.J. Transcript Expression Data from Human Islets Links Regulatory Signals from Genome-Wide Association Studies for Type 2 Diabetes and Glycemic Traits to Their Downstream Effectors. PLoS Genet. 2015;11:e1005694. doi: 10.1371/journal.pgen.1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mehta Z.B., Fine N., Pullen T.J., Cane M.C., Hu M., Chabosseau P., Meur G., Velayos-Baeza A., Monaco A.P., Marselli L. Changes in the expression of the type 2 diabetes-associated gene VPS13C in the β-cell are associated with glucose intolerance in humans and mice. Am. J. Physiol. Endocrinol. Metab. 2016;311:E488–E507. doi: 10.1152/ajpendo.00074.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gaulton K.J., Ferreira T., Lee Y., Raimondo A., Mägi R., Reschen M.E., Mahajan A., Locke A., Rayner N.W., Robertson N., DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat. Genet. 2015;47:1415–1425. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Northcott P.A., Lee C., Zichner T., Stütz A.M., Erkek S., Kawauchi D., Shih D.J.H., Hovestadt V., Zapatka M., Sturm D. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weischenfeldt J., Dubash T., Drainas A.P., Mardin B.R., Chen Y., Stütz A.M., Waszak S.M., Bosco G., Halvorsen A.R., Raeder B. Pan-cancer analysis of somatic copy-number alterations implicates IRS4 and IGF2 in enhancer hijacking. Nat. Genet. 2017;49:65–74. doi: 10.1038/ng.3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cnop M., Abdulkarim B., Bottu G., Cunha D.A., Igoillo-Esteve M., Masini M., Turatsinze J.-V., Griebel T., Villate O., Santin I. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014;63:1978–1993. doi: 10.2337/db13-1383. [DOI] [PubMed] [Google Scholar]
- 78.Kluth O., Matzke D., Schulze G., Schwenk R.W., Joost H.-G., Schürmann A. Differential transcriptome analysis of diabetes-resistant and -sensitive mouse islets reveals significant overlap with human diabetes susceptibility genes. Diabetes. 2014;63:4230–4238. doi: 10.2337/db14-0425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.