Abstract
Incorporation of distant intronic sequences in mature mRNA is an underappreciated cause of genetic disease. Several disease-causing pseudoexons have been found to contain repetitive elements such as Alu elements. This study describes an original pathological mechanism by which a small intronic deletion leads to Alu exonization. We identified an intronic deletion, c.2113+461_2113+473del, in the F8 intron 13, in two individuals with mild hemophilia A. In vivo and in vitro transcript analysis found an aberrant transcript, with an insertion of a 122-bp intronic fragment (c.2113_2114ins2113+477_2113+598) at the exon 13–14 junction. This out-of-frame insertion is predicted to lead to truncated protein (p.Gly705Aspfs∗37). DNA sequencing analysis found that the pseudoexon corresponds to antisense AluY element and the deletion removed a part of the poly(T)-tail from the right arm of these AluY. The heterogenous nuclear riboprotein C1/C2 (hnRNP C) is an important antisense Alu-derived cryptic exon silencer and binds to poly(T)-tracts. Disruption of the hnRNP C binding site in AluY T-tract by mutagenesis or hnRNP C knockdown using siRNA in HeLa cells reproduced the effect of c.2113+461_2113+473del. The screening of 114 unrelated families with mild hemophilia A in whom no genetic event was previously identified found a deletion in the poly(T)-tail of AluY in intron 13 in 54% of case subjects (n = 61/114). In conclusion, this study describes a deletion leading to Alu exonization found in 6.1% of families with mild hemophila A in France.
Keywords: hemophilia A, Alu elements, hnRNP C, intronic deletion
Introduction
Hemophilia A (HA [MIM: 306700]) is an X-linked recessive bleeding disorder caused by deficiency of coagulation factor VIII (FVIII), encoded by F8 (MIM: 300841). The incidence of HA is 1 in 5,000 males worldwide.1 Based on residual FVIII activity (FVIII:C), the clinical phenotype is classified as severe (FVIII:C < 1 IU/dL), moderate (1 IU/dL ≤ FVIII:C < 5 IU/dL), or mild (5 IU/dL ≤ FVIII:C < 40 IU/dL) HA.2 HA is caused by a wide diversity of genetic events in F8 and more than 2,000 unique variants have been recorded in disease-specific databases (FVIII variant database; CHAMP Mutation List Database; accessed June 2017). In mildly affected case subjects, most of the variants are missense variations; however, 10% of mild HA is genetically unresolved.3, 4 In order to explore such instances, several studies have performed ectopic F8 messenger RNA (mRNA) analyses and these have reported the presence of intronic sequences in mature transcripts.5, 6
Incorporation of distant intronic sequences in mature mRNA is an underappreciated cause of genetic disease. Typically, this event results from intronic variation creating a de novo 3′ or 5′ splice site (ss) and activating a nearby pre-existing 3′ or 5′ ss, leading to cryptic exon (pseudoexon) activation.7, 8 Apart from creating de novo splice sites, intronic variation may activate cryptic exons by eliminating or creating splicing enhancers or silencers9 or by producing a more optimal branch point sequence.10 Among the disease-causing pseudoexons that have been characterized, several contain repetitive sequences such as Alu elements.11
Alu elements are the most abundant transposable elements in the human genome.12 A typical Alu is about 300 bp long, formed by two similar sequences (left and right arms) separated by a short A-rich linker and terminated by a long poly(A) tail of about 20–100 bases.13 Alu sequences are preferentially located within gene-rich regions and are particularly found within introns in both sense or antisense orientation relative to the mRNA.14 Vorechovsky et al.11 reported that more than 90% of Alu elements involved in disease-associated cryptic exons are in an antisense orientation. Exonization process can be facilitated by sequence motifs, including a pyrimidine-rich tract, generated by the reverse complement of the poly(A) tail and a CAG trinucleotide motif, in antisense Alu that are similar to a very strong acceptor splice site.13 Many Alu elements in an antisense orientation located in introns of genes have “perfect” 3′ and 5′ ss sequence requirements to undergo exonization.15, 16 However, there are molecular mechanisms for protecting the human transcriptome from the aberrant exonization of Alu elements. The heterogeneous nuclear riboprotein C1/C2 (hnRNP C) is one of the most important antisense Alu-derived cryptic exon silencers.17 hnRNP C particles bind on uninterrupted uridine (U) tracts of at least five nucleotides (such as the poly(T) tail of antisense Alu elements) on the pre-mRNA and competes with the core splicing factor U2AF65 in order to prevent the spliceosome binding.17, 18, 19, 20
In this study, we report a small intronic deletion in the poly(T) tail of AluY sequence in two people with mild HA in whom F8 mRNA analyses showed Alu exonization. These deletions might lead to decrease hnRNP C binding to pre-RNA causing alternative splicing and AluY exonization. Finally, we show that this genetic variation was found in 6.1% of our cohort consisting of families with mild HA in France.
Material and Methods
Subjects
Two unrelated persons with mild HA referred to our laboratory for F8 mRNA analysis and in whom no detectable F8 sequence variation was found after direct sequencing of exons 1 to 26, including the intron/exon boundaries, the promotor region, and the 3′ untranslated region of the F8, were included in this study. Sanger sequencing was performed using Big Dye terminator chemistry (Applied Biosystems) and ABI 3130XL sequencer. For these two individuals, multiplex ligation-dependent probes amplification assay (MRC-Holland) was performed and found a normal allele copy number for F8 exons. Laboratory investigation of FVIII coagulant activity was measured using a 1-stage assay (FVIII:C; Instrumentation Laboratory) and chromogenic assay (FVIII:CR; Hyphen Biomed), von Willebrand factor antigen (VWF:Ag; Instrumentation Laboratory), von Willebrand factor ristocetin cofactor activity (VWF:RCo, Instrumentation Laboratory), von Willebrand Factor FVIII binding assay (VWF:FVIIIB; Asserachrom, Stago), and FV coagulant activity (FV:C, Instrumentation Laboratory) were performed.
134 individuals (119 mild HA males and 15 female carriers) from 114 unrelated families with history of mild HA and no detectable F8 genetic event were also included in this study. In most families mild HA is simplex case, but in four families several individuals with mild HA are known. All persons included in this study have French origin and were referred in our laboratory for HA genetic investigation by 16 different French Haemophilia centers.
Written informed consent for all subjects included in this study was obtained in accordance with protocols approved by the hospices Civils de Lyon.
F8 RNA Isolation and RT-PCR Assay
Blood was collected in a PAXgene blood RNA tube (PreAnalytix). Total cellular mRNA was extracted using the PAXgene blood RNA kit according to the manufacturer’s procedure (PreAnalytix). For the reverse transcription reaction (RT-PCR), F8 complementary DNA (cDNA) was divided into four different regions that cover all splice sites: region A (exon 1 to 7), region B (exon 7 to 14), region C (exon 14 to 22), and region D (exon 22 to 26). The isolated F8 mRNA was reverse transcribed to cDNA using four specific primers (one by region) and PrimeScript Reverse Transcriptase (Clonetech) according the manufacturer’s recommendations. An internal control from the human β-actin gene (ACTB) was also included to exclude any failure in the RT reaction.
We performed the amplification of F8 cDNA in two rounds of PCR (nested PCR approach) using PrimeSTAR GXL DNA polymerase (Clonetech). Each PCR were performed in the following cycling conditions: 30 cycles of 10 s at 98°C, 15 s at 55°C, and 2 min at 68°C. For the first PCR we used 4 μL (for a total of 50 μL PCR reaction) from the reverse transcription. For the second round of PCR, each of the above four regions (A–D) were divided into two or three segments (a total of nine overlapping regions) that were amplified using 4 μL (a total of 25 μL) from the first PCR as a template. The sequence of the primers and the product size of the RT-PCR are shown in Table S1. Some primers used were previously published by El-Maarri et al.21 The RT-PCR products were separated on 1% agarose gel. In case of multiple RT-PCR products detected on agarose gel, a gel-extraction was performed using NucleoSpin Gel and PCR Clean-up (Macherey-Nagel) and the different bands were sequenced using both the reverse and forward primers that were initially used for the nested PCR.
DNA Sequencing of the F8 Intron 13 Region of Interest
Genomic DNA was isolated from peripheral whole blood of the individuals and control subjects by the salting out method. The interest region of F8 intron 13 was amplified using specific primers (Table S2) and the PrimeSTAR GXL DNA polymerase (Clonetech). PCR were performed in the following cycling conditions: 30 cycles of 10 s at 98°C, 15 s at 55°C, and 2 min at 68°C. PCR products were sequenced using specific primers (Table S2) and Big Dye terminator chemistry (Applied Biosystems) on an ABI 3130XL sequencer. The resulting sequences were aligned and compared using Seqscape v.2.5 software (Applied Biosystems).
F8 Intron 13 Deletion Screening and Microsatellite Analysis
For haplotype analysis, six intragenic polymorphic microsatellite markers used routinely for HA linkage analysis were selected. These markers are located within intron 1 (STR1), intron 9 (STR9), intron 13 (STR13), intron 22 (STR22), and intron 25 (STR25A and STR25B).22, 23 Details of markers used are described in Table S3.
The same technical approach was used for haplotype analysis and F8 intron 13 deletion screening. PCR was performed using fluorescent end-labeled primers (FAM-labeled oligonucleotide, Table S4) and Optimase polymerase (Transgenomic). PCR products were analyzed by capillary electrophoresis using an ABI 3130XL Analyzer. For the F8 intron 13 deletion screening, fragment sizes of 272 bp and 260 bp corresponded to wild-type and c.2113+461_2113+473del, respectively. All deletion was confirmed by direct DNA sequencing.
Cell Culture
HeLa and HuH7 cells were grown in Dulbecco’s modified Eagle medium/nutriment mixture F-12 medium (DMEM-F12, Life Technologies) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin and cultured at 37°C in an humidified atmosphere containing 5% CO2.
In Vitro Minigene Splicing Reporter Assay
For F8 exon 13 minigene constructs, a 1,182-bp fragment including 202 bp of intron 12, the exon 13, and 770 bp of intron 13 was amplified from the two propositus and control DNA using specific primers (Table S2) and PrimeSTAR GXL DNA polymerase (Clonetech). Amplicons were inserted in the NdeI restriction site of the previously described pTB minigene vector24 using the In-fusion HD PCR cloning kit (Clontech). The correct sequences of wild-type (WT; pTB2ex13WT) and mutant plasmids (pTB2ex13DEL) were verified by DNA sequencing.
In order to discontinue the T-tract of the AluY element, five substitutions (T>C) were generated in pTB2ex13WT through site-directed mutagenesis using the QuickChange XL reagent (Stratagene) according to the manufacturer’s instructions. The correct sequence of the pTB2ex13WT_5mut was verified by DNA sequencing.
HeLa and HuH7 cells were plated at a concentration of 105 cells/well in a 6-well cluster plate in 2 mL of growth medium. 16 hr after plating, the cells were transiently transfected with 1 μg of plasmid using JetPRIME (Polypus transfection) following the manufacturer’s recommendations. 48 hr after transfection, total RNA was extracted from the cells using RNAqueous-4PCR Total RNA Isolation Kit (Life Technologies) and then treated by DNase (DNA-free DNA Removal Kit, Life Technologies).
RT-PCR was performed with Transcriptor High Fidelity cDNA Synthesis Kit (Roche Molecular Diagnostics) using 250 ng of total RNA and random primers. cDNA amplification was performed using vector-specific primers surrounding the cloning site and Taq DNA polymerase (MP Biomedicals). The PCR products were resolved on a 2% agarose gel and sequenced to identify splicing events. All transfection experiments were performed in triplicate.
Knockdown of hnRNP C
For the RNA interference (siRNA) assays, the above protocol was modified as follows: the HeLa cells were seeded on day 0 and transfected on day 1 with hnRNP C siRNAs (FlexiTube GeneSolution GS3183 for HNRNPC, QIAGEN) or with AllStar negative control siRNA (QIAGEN) at a final concentration of 50 nM and transfected on day 2 with 1 μg of minigene plasmids. Then, RNA was extracted and studied on day 5 as described above.
The efficiency of the protein silencing and expression was evaluated by western blotting of cell lysates. For cell lysis, cells were washed twice with Dulbecco’s Phosphate-Buffered Saline (DPBS; Life Technologies), and lysis buffer (HEPES [pH 7.5] 50 mM, KCl 0.1 M, MgCl2 2 mM, Triton X-100 0.5%) containing protease inhibitors (cOmplete Mini Protease Inhibitor Cocktail, Roche) was added to each plate and incubated on ice 20 min. Then, the lysate was clarified with a centrifugation step and stored at −80°C. 20 μg of total protein was loaded onto 12% precasted MiniPROTEAN TGX gels (Bio-Rad). Proteins were then transferred on to a 0.45 μm polyvinylidene difluoride membrane. After blocking in 5% non-fat dried milk dissolved in TBS 1X, membranes were incubated overnight with a mouse monoclonal anti-human hnRNP C antibody (Anti-hnRNP C1 + C2 antibody [4F4], Abcam). After three wash steps with TBS, membranes were probed 1 hr at room temperature with horseradish peroxidase (HRP)-conjugated polyclonal anti-mouse antibody at 1:10,000 (GE Healthcare). Following three new wash steps with TBS, membranes were developed using the ECL reagent (GE Healthcare) and Fuji Medical X-Ray Film (FujiFilm). The GAPDH western blot signal (rabbit mAb anti GAPDH (D16H11) XP, Cell Signaling Technology) was used as expression and loading efficiency control.
Results
Characterization of the Two Index Case Subjects
The two index case subjects had a reduced FVIII:C, FVIII:CR, and FVIII:Ag. VWF:Ag, VWF:RCo, VWF:FVIIIB, and FV:C were normal, so combined FV and FVIII deficiency (MIM: 227300 and 613625) as well as type 1 (MIM:193400) and type 2N (MIM: 613554) von Willebrand disease were excluded. Neither proband had a history of inhibitor development. Proband 1 was completely responsive to 1-deamino-8-D-arginin vasopressin; 1 hr after infusion, FVIII:C increased from 26 to 166 IU/dL and these levels remained persistent 4 hr after treatment (Table 1).
Table 1.
Summary of Laboratory Measurements of the Two Index Case Subjects
| Subject | Age, years | FVIII:C, IU/dL | FVIII:CR, IU/dL | FVIII:Ag, IU/dL | VWF:RCo, IU/dL | VWF:Ag, IU/dL | VWF:FVIIIB | FV:C, IU/dL | Inhibitorsa | DDAVP Response |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 56 | 24 | 23 | 17 | 61 | 50 | normal | 81 | neg | yes |
| 2 | 11 | 40 | 41 | 37 | 62 | 88 | normal | 95 | neg | NA |
| Normal range | – | 59–167 | 50–140 | 63–199 | 48–240 | 42–176 | – | 73–188 | – | – |
Abbreviations: neg, <0.6 UB/mL; FVIII:C, FVIII activity measured using 1-stage assay; FVIII:CR, FVIII activity measured using chromogenic assay; FVIII:Ag, FVIII antigen; VWF:RCo, von Willebrand Factor ristocetin cofactor activity; VWF:Ag, von Willebrand Factor antigen; VWF:FVIIIB, von Willebrand Factor FVIII binding assay; FV:C, FV activity; DDAVP, 1-deamino-8-D-arginin vasopressin; neg, negative; NA, not available.
Based on Bethesda assay
AluY-Derived Pseudoexon Insertion between Exon 13 and Exon 14 in Both Mild HA Probands
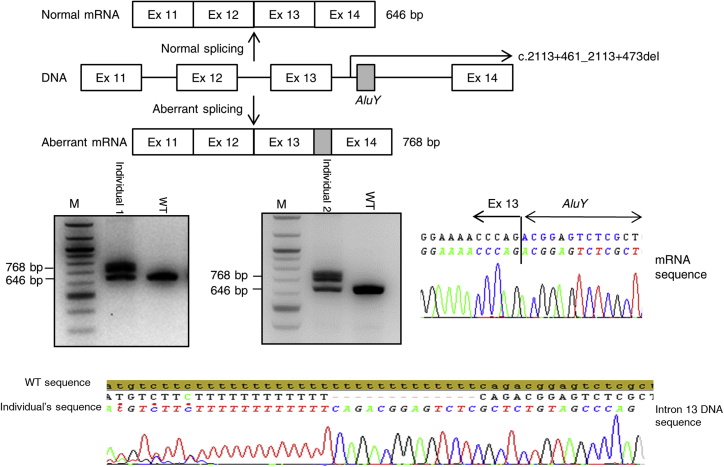
F8 (GenBank: NM_000132.3) mRNA analyses were performed in the two index case subjects. There was no difference in either the number or the size of the cDNA of either probands compared with wild-type except for the region spanning exon 11–14. In this region, two cDNA fragments were observed, one of 646 bp that corresponded to a wild-type sequence and the other of 768 bp (Figure 1). Sequencing of the 768 bp fragment found the insertion of an F8 intron 13 fragment of 122 bp (c.2113_2114ins2113+477_2113+598) at the exon 13-14 junction. This out-of-frame insertion is predicted to lead to truncated protein (p.Gly705Aspfs∗37). F8 intron 13 DNA sequencing in both probands found that the pseudoexon was flanked by canonical AG/GT splice sites and found a short deletion (c.2113+461_2113+473del) in a poly(T) tract located 13 nucleotides upstream of the pseudoexon. Two genome databases, the Bravo database (search terms: chrX: 154,947,205–154,947,265; hg38) and the GnomAD database (search terms chrX:154,175,480–154,175,540; hg19), were analyzed for c.2113+461_2113+473del. However, the region of interest was not adequately covered in either database, precluding interpretation of the data. Analysis of the sequence determined that the pseudoexon corresponded to an antisense AluY element and the deletion removed a part of the poly(T)-tail from the right arm of antisense AluY.
Figure 1.
Identification of a c.2113+461_2113+473del Variation in the Two Index Case Subjects
Two mRNA fragments were observed when amplifying fragments spanning exons 11–14 (wild-type 646 bp; mutant 768 bp). Sequencing of the 768 bp fragment found the insertion of intron 13 fragment of 122 bp (c.2113_2114ins2113+477_2113+598). Sequencing of the intron 13 found a c.2113+461_2113+473del short deletion in the poly(T)-tail of the right arm of antisense AluY. Abbreviations: M, marker size; WT, wild-type; Ex, exon, bp; base pair.
Reporter Minigene Assay Confirmed the Role of c.2113+461_2113+473del in AluY Exonization
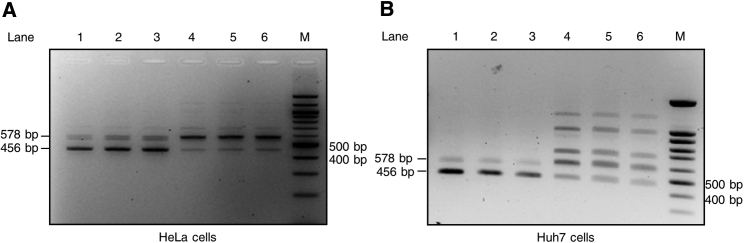
To confirm the role of the c.2113+461_2113+473del in AluY exonization, we prepared WT (pTB2ex13WT) and mutated (pTB2ex13DEL) splicing reporter constructs and analyzed their splicing pattern 48 hr following transient transfection of HeLa cells (Figure 2A). In addition to normal transcripts (major product, 456 bp), the pTB2ex13WT construct generated traces of RNA product with AluY inclusion (578 bp). In contrast, the reporter construct with the deletion (pTB2ex13DEL) produced mostly a larger product with the pseudoexon and traces of normal products (Figure 2A). To determine whether these observations were cell-line specific, the same experiments were repeated in Huh7 cells and similar results were obtained. In addition to traces of normal product, several aberrant transcripts were observed in Huh7 cells transfected with pTB2ex13DEL including the 578 bp transcript corresponding to AluY inclusion (Figure 2B).
Figure 2.
Molecular Characterization of the Splicing Effect of c.2113+461_2113+473del
HeLa (A) and Huh7 (B) cells were transiently transfected with wild-type (pTB2ex13WT) and mutated (pTB2ex13DEL) splicing reporter constructs. 48 hr after transfection, RNA products were analyzed by RT-PCR (normal exon 13 splicing 456 bp; AluY inclusion 578 bp). Lanes 1 to 3: pTB2ex13WT; lanes 4 to 6: pTB2ex13DEL. Abbreviations: M, marker size; bp, base pair.
hnRNP C Was Involved in F8 Intron 13 AluY Repression, and Discontinuation of AluY U-Tract and hnRNP C Knockdown Reproduced the Effect of c.2113+461_2113+473del
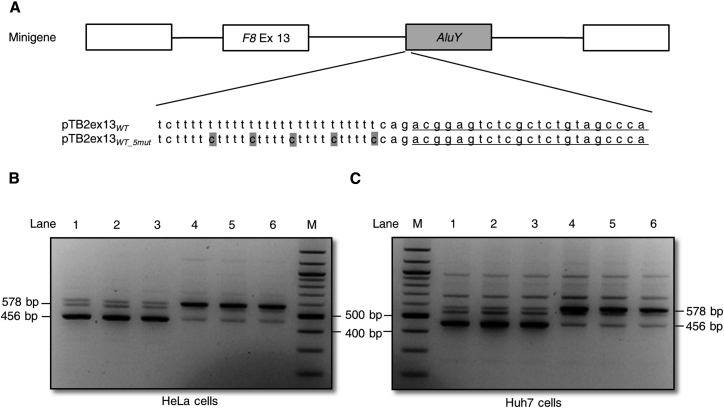
A hnRNP C is the most important antisense Alu-derived cryptic exon silencer. hnRNP C binding site consisted of uninterrupted T-tracts of at least five nucleotides. As c.2113+461_113+473del removed a part of the poly(T)-tail of the AluY, we hypothesized that this deletion might decrease hnRNP C binding, leading to AluY exonization. To investigate the role of hnRNP C in AluY repression, we first disrupted the hnRNP C binding sites in the pTB2ex13WT inserting 5 T>C substitutions by site-directed mutagenesis in the poly(T)-tail of the AluY. Then, the pTB2ex13WT_5mut plasmid was transfected in HeLa cells. We found that these substitutions promoted AluY inclusion (Figures 3A and 3B). To determine whether the observations were cell line specific, the same experiments were repeated in Huh7 cells and no difference was observed (Figure 3C).
Figure 3.
Substitutions in Poly(T) Tail Promote Inclusion of the AluY Exon
(A) Schematic representation of the minigene including AluY exon (gray square), exon 13 (white square), intronic region (black lines). The wild-type sequence (pTB2ex13WT) as well as the mutated sequence (pTB2ex13WT_5mut) are depicted below. Point variations are highlighted in gray. The AluY exonized sequence is underlined.
(B and C) RT-PCR results obtained after transfection of HeLa (B) or Huh7 (C) cells with pTB2ex13WT (lines 1 to 3) and pTB2ex13WT_5mut (lines 4 to 6). Abbreviations: Ex, exon; M, marker size; bp, base pair.
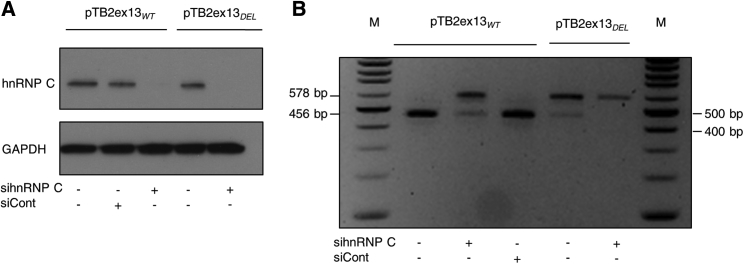
We next confirmed the effects of the hnRNP C on repression of F8 intron 13 AluY by siRNA-mediated downregulation in HeLa cells transfected with pTB2ex13WT. We first confirmed efficient downregulation of hnRNP C (Figure 4A). Downregulation of hnRNP C resulted in a significant AluY inclusion in HeLa cells (Figure 4B).
Figure 4.
hnRNP C Repression Promotes Inclusion of the AluY Exon
(A) Western blotting using the indicated antibodies after hnRNP C knockdown with siRNA (sihnRNP C) against control (siCont) in HeLa cells.
(B) RT-PCR results obtained after transfection with pTB2ex13WT or pTB2ex13DEL minigenes in HeLa cells treated with the indicated siRNA. Abbreviations: M; marker size; bp, base pair.
These experiments showed that the alteration of the hnRNP C function, either by decreasing the DNA binding or by decreasing the protein biosynthesis, led to the same impact on Alu inclusion as c.2113+461_2113+473del. These observations gave some evidence that c.2113+461_2113+473del might lead to decreased hnRNP C binding to pre-RNA, causing alternative splicing and AluY exonization.
c.2113+461_2113+473del Was a Recurrent Genetic Variation in Persons with Mild Hemophilia A in France
Among the 1,970 families registered in our laboratory for HA genetic investigations, 992 have a history of mild HA. We screened for c.2113+461_2113+473del in 119 individuals from 114 different families with genetically unresolved mild HA. Short deletions, ranging from 11 to 13 nucleotides, in the T-tail of the AluY element within F8 intron 13 were found for 66 individuals from 61 unrelated families (61/114). c.2113+461_2113+473del (deletion of 13T) was found in 57 families, c.2113+462_2113+473del (deletion of 12T) was in 1 family, and c.2113+463_2113+473del (deletion of 11T) was in 3 families. Maternal DNA was available for 15 samples and c.2113+463_2113+473del was found in heterozygous state. Familial segregation could be assessed in 4 families with c.2113+461_2113+473del allele where the deletion appropriately segregated with mild HA.
Therefore, short deletion in the T-tail of AluY element in F8 intron 13 is a recurrent variant with a prevalence of 6.1% (61/992) in families with mild HA in France. HA-affected individuals carrying such deletion exhibited a FVIII:C close to 30 IU/dL (mean ± standard deviation, 30 ± 6 IU/dL; range: 15–47 IU/dL).
To evaluate the possibility of a founder effect, a haplotype analysis was performed in individuals with c.2113+461_2113+473del (n = 29), c.2113+463_2113+473del (n = 3), and c.2113+462_2113+473del (n = 1) (Table 2). A single haplotype, characterized by the alleles 248 (STR1), 201 (STR9), 149 (STR13), 178 (STR22), 134 (STR25A), and 255 (STR25B), was identified in all individuals carrying c.2113+461_2113+473del, suggesting a founder effect for c.2113+461_2113+473del. However, four different haplotypes were found in the four individuals carrying c.2113+461_2113+471del or c.2113+461_2113+472del, suggesting that these alleles arose independently.
Table 2.
Haplotypes Flanking the Short Deletion in the T-Tail of the AluY Element within F8 Intron 13
| Deletion | STR 1 | STR 9 | STR 13 | STR 22 | STR 25A | STR 25B | Frequency, n (%) |
|---|---|---|---|---|---|---|---|
| c.2113+461_2113+473del | 248 | 201 | 149 | 178 | 134 | 255 | 29a (100) |
| c.2113+461_2113+471del | 246 | 201 | 147 | 180 | 134 | 257 | 1 (33) |
| 250 | 201 | 153 | 172 | 134 | 253 | 1 (33) | |
| 246 | 201 | 145 | 180 | 134 | 257 | 1 (33) | |
| c.2113+461_2113+472del | 246 | 201 | 145 | 180 | 134 | 257 | 1 (100) |
Haplotype analysis was performed on 29 of the 57 families in which c.2113+461_2113+473del was found
Discussion
In the present study, we identified the same intronic insertion in F8 in two unrelated probands with mild HA (c.2113_2114ins2113+477_2113+598). Interestingly, cDNA analysis also found the presence of a normal F8 transcript in addition of the aberrant transcript, explaining the mild phenotype of these probands. Additionally, sequencing the F8 intron 13 identified a short deletion (c.2113+461_2113+473del) located 13 nucleotides upstream of the pseudoexon in both probands. Previous work has described several pseudoexon inclusions in HA-affected individuals but they were associated with intronic point variants. These substitutions either created a de novo 3′ or 5′ ss (c.143+1567A>G, c.5587−93C>T, and c.1537+325A>G) or increased the strength of a pre-existing 3′ or 5′ ss located close to the substitution (c.5999−277G>A, c.5998+941G>A, and 2113+1152delA).5, 6, 25, 26
Sequence analysis of the pseudoexon found that it corresponded to antisense AluY elements. Alu elements can integrate within protein-coding regions by two mechanisms. The first is through de novo retrotransposition into an exon. This phenomenon has been reported in two case subjects with severe HA.27, 28 The second mechanism involves splicing of an intronic sequence into the coding region of mRNA. As in the case in this report, the antisense AluY identified in F8 intron 13 was spliced to a pre-existing 3′ ss at Alu position 279 that corresponded to 3′ ss used by 24% of exonized Alu and a pre-existing 5′ ss at Alu position 157 that corresponded to 5′ ss used by 65% of exonized Alu.15, 16, 29
The c.2113+461_2113+473del deletion removed a part of the poly(T)-tail from the right arm of antisense AluY. Recently, it has been shown that the poly(T)-tail of antisense Alu element is the binding site of the hnRNP C, the most important antisense Alu-derived cryptic exon silencers.17, 18 We hypothesized that c.2113+461_2113+473del may be responsible for the decreased hnRNP C binding leading AluY exonization. Our minigene experiments found that both the disruption the U-tract of AluY by site-directed mutagenesis (loss of the hnRNP C binding site) and downregulation of hnRNP C using specific siRNA led to partial AluY exonization identical to that observed in the persons carrying the c.2113+461_2113+473del deletion. These results suggested that c.2113+461_2113+473del might lead to decrease hnRNP C binding to pre-RNA causing alternative splicing and AluY exonization. A similar molecular mechanism has been previously reported in a proband with hyperphenylalaninemia (MIM: 261640), in whom a deletion removing the entire poly(T)-tail from the right arm of antisense AluSq in intron 2 of the PTS gene leads to strong AluSq exon inclusion in PTS (MIM: 212719) transcripts.17, 30 In the two index case subjects herein, it was interesting to note that only partial deletion of the poly(T)-tail was sufficient to lead to significant pathological consequences.
The main limitation of our study was the fact that both the in vivo and the in vitro transcript analysis were performed from tissues that are not constitutively producing FVIII. FVIII is produced by endothelial hepatic cells. No cell line from this source is available and due to the relative inaccessibility of this tissue in HA-affected persons, F8 mRNA studies are usually performed in leukocytes as they contain an amount of ectopic mRNA transcripts.21, 31 Due to the aberrant RNA expression, it cannot be excluded that our results might not reflect what happens in vivo. However, Alu exonization was observed in each experimental model used in this study.
The importance of these findings was then evaluated by the screening of 114 unrelated families with mild HA in whom no genetic events were previously detected, and the deletion in the poly(T)-tail of AluY was found in just over half of them. Furthermore, among all variants leading to mild HA phenotype characterized in our laboratory, this deletion is the most frequent with a prevalence of 6.1%. Such variations that are found at a relatively high frequency are either due to a founder effect or due to recurrent molecular mechanism (such as intron 22 and intron 1 inversions). Founder effects have been described for several variants in HA-affected individuals and occurred in isolated populations.23, 32, 33 Haplotype analysis performed in persons carrying c.2113+461_2113+473del found that all individuals presented the same haplotype. This observation may suggest a founder effect for c.2113+461_2113+473del. However, the fact that other deletions in the poly(T)-tail of inverted AluY within F8 intron 13 with variable size (c.2113+462_2113+473del and c.2113+463_2113+473del) were found in persons carrying different haplotypes may indicate the existence of a recurring molecular mechanism. Thus, it would be interesting to investigate these short intronic deletions in other HA populations.
In conclusion, this study found an original molecular mechanism by which a mild HA-causing short intronic deletion in poly(T)-tail of inverted AluY leads to AluY exonization in F8 transcripts. Due to its high prevalence, we recommend that this deletion be specifically investigated in affected individuals with mild HA in whom no genetic abnormality has been detected by standard genetic analysis.
Acknowledgments
The authors would like to thank Dr. M. Raponi and Dr. D. Baralle for providing the pTB minigene. We also thank D. Pellecchia and H. Lazare for their technical assistance and P. Robinson for English reviewing.
Published: January 11, 2018
Footnotes
Supplemental Data include four tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.12.010.
Web Resources
Factor VIII Variant Database, http://www.factorviii-db.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
OMIM, http://www.omim.org/
Supplemental Data
References
- 1.Oldenburg J., Ananyeva N.M., Saenko E.L. Molecular basis of haemophilia A. Haemophilia. 2004;10(Suppl 4):133–139. doi: 10.1111/j.1365-2516.2004.01005.x. [DOI] [PubMed] [Google Scholar]
- 2.White G.C., 2nd, Rosendaal F., Aledort L.M., Lusher J.M., Rothschild C., Ingerslev J., Factor VIII and Factor IX Subcommittee Definitions in hemophilia. Recommendation of the scientific subcommittee on factor VIII and factor IX of the scientific and standardization committee of the International Society on Thrombosis and Haemostasis. Thromb. Haemost. 2001;85:560. [PubMed] [Google Scholar]
- 3.Bogdanova N., Markoff A., Eisert R., Wermes C., Pollmann H., Todorova A., Chlystun M., Nowak-Göttl U., Horst J. Spectrum of molecular defects and mutation detection rate in patients with mild and moderate hemophilia A. Hum. Mutat. 2007;28:54–60. doi: 10.1002/humu.20403. [DOI] [PubMed] [Google Scholar]
- 4.Castaman G., Mancuso M.E., Giacomelli S.H., Tosetto A., Santagostino E., Mannucci P.M., Rodeghiero F. Molecular and phenotypic determinants of the response to desmopressin in adult patients with mild hemophilia A. J. Thromb. Haemost. 2009;7:1824–1831. doi: 10.1111/j.1538-7836.2009.03595.x. [DOI] [PubMed] [Google Scholar]
- 5.Inaba H., Koyama T., Shinozawa K., Amano K., Fukutake K. Identification and characterization of an adenine to guanine transition within intron 10 of the factor VIII gene as a causative mutation in a patient with mild haemophilia A. Haemophilia. 2013;19:100–105. doi: 10.1111/j.1365-2516.2012.02906.x. [DOI] [PubMed] [Google Scholar]
- 6.Castaman G., Giacomelli S.H., Mancuso M.E., D’Andrea G., Santacroce R., Sanna S., Santagostino E., Mannucci P.M., Goodeve A., Rodeghiero F. Deep intronic variations may cause mild hemophilia A. J. Thromb. Haemost. 2011;9:1541–1548. doi: 10.1111/j.1538-7836.2011.04408.x. [DOI] [PubMed] [Google Scholar]
- 7.Buratti E., Chivers M., Královicová J., Romano M., Baralle M., Krainer A.R., Vorechovsky I. Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007;35:4250–4263. doi: 10.1093/nar/gkm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vorechovský I. Aberrant 3′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2006;34:4630–4641. doi: 10.1093/nar/gkl535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishii S., Nakao S., Minamikawa-Tachino R., Desnick R.J., Fan J.-Q. Alternative splicing in the alpha-galactosidase A gene: increased exon inclusion results in the Fabry cardiac phenotype. Am. J. Hum. Genet. 2002;70:994–1002. doi: 10.1086/339431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Klein A., Riegman P.H., Bijlsma E.K., Heldoorn A., Muijtjens M., den Bakker M.A., Avezaat C.J., Zwarthoff E.C. A G-->A transition creates a branch point sequence and activation of a cryptic exon, resulting in the hereditary disorder neurofibromatosis 2. Hum. Mol. Genet. 1998;7:393–398. doi: 10.1093/hmg/7.3.393. [DOI] [PubMed] [Google Scholar]
- 11.Vorechovsky I. Transposable elements in disease-associated cryptic exons. Hum. Genet. 2010;127:135–154. doi: 10.1007/s00439-009-0752-4. [DOI] [PubMed] [Google Scholar]
- 12.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 13.Hancks D.C., Kazazian H.H., Jr. Roles for retrotransposon insertions in human disease. Mob. DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C., Gentles A.J., Jurka J., Karlin S. Genes, pseudogenes, and Alu sequence organization across human chromosomes 21 and 22. Proc. Natl. Acad. Sci. USA. 2002;99:2930–2935. doi: 10.1073/pnas.052692099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sorek R., Lev-Maor G., Reznik M., Dagan T., Belinky F., Graur D., Ast G. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- 16.Lev-Maor G., Sorek R., Shomron N., Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 17.Zarnack K., König J., Tajnik M., Martincorena I., Eustermann S., Stévant I., Reyes A., Anders S., Luscombe N.M., Ule J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cieniková Z., Damberger F.F., Hall J., Allain F.H.-T., Maris C. Structural and mechanistic insights into poly(uridine) tract recognition by the hnRNP C RNA recognition motif. J. Am. Chem. Soc. 2014;136:14536–14544. doi: 10.1021/ja507690d. [DOI] [PubMed] [Google Scholar]
- 19.König J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan L., Kim J.K., Pollard V.W., Dreyfuss G. Mutational definition of RNA-binding and protein-protein interaction domains of heterogeneous nuclear RNP C1. J. Biol. Chem. 2001;276:7681–7688. doi: 10.1074/jbc.M010207200. [DOI] [PubMed] [Google Scholar]
- 21.El-Maarri O., Herbiniaux U., Graw J., Schröder J., Terzic A., Watzka M., Brackmann H.H., Schramm W., Hanfland P., Schwaab R. Analysis of mRNA in hemophilia A patients with undetectable mutations reveals normal splicing in the factor VIII gene. J. Thromb. Haemost. 2005;3:332–339. doi: 10.1111/j.1538-7836.2005.01140.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim J.-W., Park S.-Y., Kim Y.-M., Kim J.-M., Kim D.-J., Ryu H.-M. Identification of new dinucleotide-repeat polymorphisms in factor VIII gene using fluorescent PCR. Haemophilia. 2005;11:38–42. doi: 10.1111/j.1365-2516.2005.01064.x. [DOI] [PubMed] [Google Scholar]
- 23.Garagiola I., Seregni S., Mortarino M., Mancuso M.E., Fasulo M.R., Notarangelo L.D., Peyvandi F. A recurrent F8 mutation (c.6046C>T) causing hemophilia A in 8% of northern Italian patients: evidence for a founder effect. Mol. Genet. Genomic Med. 2015;4:152–159. doi: 10.1002/mgg3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baralle D., Baralle M. Splicing in action: assessing disease causing sequence changes. J. Med. Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pezeshkpoor B., Zimmer N., Marquardt N., Nanda I., Haaf T., Budde U., Oldenburg J., El-Maarri O. Deep intronic ‘mutations’ cause hemophilia A: application of next generation sequencing in patients without detectable mutation in F8 cDNA. J. Thromb. Haemost. 2013;11:1679–1687. doi: 10.1111/jth.12339. [DOI] [PubMed] [Google Scholar]
- 26.Bagnall R.D., Waseem N.H., Green P.M., Colvin B., Lee C., Giannelli F. Creation of a novel donor splice site in intron 1 of the factor VIII gene leads to activation of a 191 bp cryptic exon in two haemophilia A patients. Br. J. Haematol. 1999;107:766–771. doi: 10.1046/j.1365-2141.1999.01767.x. [DOI] [PubMed] [Google Scholar]
- 27.Sukarova E., Dimovski A.J., Tchacarova P., Petkov G.H., Efremov G.D. An Alu insert as the cause of a severe form of hemophilia A. Acta Haematol. 2001;106:126–129. doi: 10.1159/000046602. [DOI] [PubMed] [Google Scholar]
- 28.Ganguly A., Dunbar T., Chen P., Godmilow L., Ganguly T. Exon skipping caused by an intronic insertion of a young Alu Yb9 element leads to severe hemophilia A. Hum. Genet. 2003;113:348–352. doi: 10.1007/s00439-003-0986-5. [DOI] [PubMed] [Google Scholar]
- 29.Sela N., Mersch B., Gal-Mark N., Lev-Maor G., Hotz-Wagenblatt A., Ast G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu’s unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meili D., Kralovicova J., Zagalak J., Bonafé L., Fiori L., Blau N., Thöny B., Vorechovsky I. Disease-causing mutations improving the branch site and polypyrimidine tract: pseudoexon activation of LINE-2 and antisense Alu lacking the poly(T)-tail. Hum. Mutat. 2009;30:823–831. doi: 10.1002/humu.20969. [DOI] [PubMed] [Google Scholar]
- 31.Martorell L., Corrales I., Ramirez L., Parra R., Raya A., Barquinero J., Vidal F. Molecular characterization of ten F8 splicing mutations in RNA isolated from patient’s leucocytes: assessment of in silico prediction tools accuracy. Haemophilia. 2015;21:249–257. doi: 10.1111/hae.12562. [DOI] [PubMed] [Google Scholar]
- 32.Winter P.C., Egan H., McNulty O., Jones F.G.C., O’Donnell J., Jenkins P.V. A recurrent F8 mutation in Irish haemophilia A patients: evidence for a founder effect. Haemophilia. 2008;14:394–395. doi: 10.1111/j.1365-2516.2007.01639.x. [DOI] [PubMed] [Google Scholar]
- 33.Xie Y.G., Zheng H., Leggo J., Scully M.F., Lillicrap D. A founder factor VIII mutation, valine 2016 to alanine, in a population with an extraordinarily high prevalence of mild hemophilia A. Thromb. Haemost. 2002;87:178–179. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.