Abstract
Exome sequencing has markedly enhanced the discovery of genes implicated in Mendelian disorders, particularly for individuals in whom a known clinical entity could not be assigned. This has led to the recognition that phenotypic heterogeneity resulting from allelic mutations occurs more commonly than previously appreciated. Here, we report that missense variants in CDC42, a gene encoding a small GTPase functioning as an intracellular signaling node, underlie a clinically heterogeneous group of phenotypes characterized by variable growth dysregulation, facial dysmorphism, and neurodevelopmental, immunological, and hematological anomalies, including a phenotype resembling Noonan syndrome, a developmental disorder caused by dysregulated RAS signaling. In silico, in vitro, and in vivo analyses demonstrate that mutations variably perturb CDC42 function by altering the switch between the active and inactive states of the GTPase and/or affecting CDC42 interaction with effectors, and differentially disturb cellular and developmental processes. These findings reveal the remarkably variable impact that dominantly acting CDC42 mutations have on cell function and development, creating challenges in syndrome definition, and exemplify the importance of functional profiling for syndrome recognition and delineation.
Keywords: exome sequencing, mutation spectrum, functional profiling, developmental anomalies, phenotypic heterogeneity, genotype-phenotype correlations, thrombocytopenia, Noonan syndrome, microcephaly, cardiac defects
Main Text
The rate of identification of genes implicated in human disorders has dramatically increased with the use of second-generation sequencing technologies. In particular, exome sequencing has emerged as a feasible and efficient strategy to uncover the molecular basis of Mendelian disorders, particularly for individuals with a rare clinical presentation or for whom a unifying clinical diagnosis is not discerned.1 Mutations affecting the same gene but resulting in substantial phenotypic differences is a very well-known phenomenon, but the wide use of exome sequencing has led to the recognition that this event occurs much more commonly than previously appreciated.2, 3, 4 In the last few years, it has been recognized that the variable clinical manifestation of allelic mutations can often result from their differential impact on protein function, although the consequences of specific variants can be difficult to predict and may require substantial efforts to be fully understood.5, 6, 7 Here, we report that missense mutations in cell division cycle 42 (CDC42 [MIM: 116952]), a gene encoding a member of the RAS superfamily of low-molecular-weight GTP/GDP-binding proteins functioning as a major node in intracellular signaling, underlie a clinically heterogeneous group of developmental phenotypes. Our in silico, in vitro, and in vivo dissection of the structural and functional impact of disease-causing mutations documents that they variably perturb CDC42 biochemical behavior and differentially affect cellular and developmental processes, highlighting the variable impact of the functional dysregulation of this GTPase in cell physiology and development. Our findings also exemplify the importance of functional profiling for syndrome recognition and delineation.
A total of 15 subjects from 13 unrelated families were included in the study. Clinical data and DNA samples were collected from the participating families (after written informed consent was obtained) and stored and used under research projects approved by the Review Boards of the participating institutions. Investigators studying the affected individuals described here were connected via the MatchMaker Exchange (MME) network of web-based tools8 GeneMatcher and MyGene2.9, 10 Nine affected individuals (subjects 1 to 5 and 8 to 11), who exhibited a molecularly unexplained and clinically unrecognized multi-systemic disorder, were investigated by whole-exome sequencing (WES) using DNA samples obtained from either leukocytes or saliva specimens, and a child-parent trio-based strategy. Exome capture was carried out using the SureSelect Clinical Research Exome (Agilent) (subjects 1 and 8), SureSelect Human All Exon v.1, v.3, and v.5 (Agilent) (subjects 2, 10, and 5, respectively), Nextera Exome Enrichment Kit (Illumina) (subject 3), SeqCap EZ VCRome 2.0 (Roche) (subject 4), and SeqCap EZ MedExome v2 (Roche) (subjects 9 and 11) target enrichment kits, and sequencing was performed on a HiSeq 2000 platform (Illumina), using paired-end. WES data processing, sequence alignment to GRCh37, and variant filtering and prioritization by allele frequency, predicted functional impact, and inheritance models were performed as previously described.11, 12, 13 Mean coverage of target regions and average reads depth for individual samples are provided in Table S1. Subjects 12 (simplex case subject) and 13 to 15 (affected members of family 30153) (Figure S1) belonged to a cohort of 235 unrelated individuals with clinical features fitting Noonan syndrome (MIM: 163950) or overlapping with this disorder, followed at three participating genetic centers (Rome, Bologna, and Magdeburg),14, 15 who did not harbor mutations in previously identified genes implicated in RASopathies. Based on the hypothesis that mutations in CDC42 might be linked causally to Noonan syndrome (or a clinically related RASopathy), the entire CDC42 coding sequence was analyzed by targeted resequencing, using genomic DNA from blood, skin fibroblasts, hair bulbs, and/or epithelial cells from the oral mucosa. Target enrichment was performed using the Nextera Rapid Capture kit (Illumina), and sequencing was carried out on a NextSeq550 (Illumina) with a 2 × 150 bp paired-end read protocol. Alignment and variant calling were performed with the BWA Enrichment BaseSpace App (Illumina), and VCF output files were annotated using Variant Studio v.2.2 (Illumina). Finally, Sanger sequencing was used to screen the CDC42 coding exons in subjects 6 and 7, who showed clinical features suggestive for the condition associated with CDC42 group I mutations (see below).
Overall, nine different missense mutations distributed across the entire CDC42 coding sequence were identified (Table 1). Two amino acid substitutions affected the N-terminal α helix (residues Ile21 and Tyr23), three involved adjacent residues within the switch II motif (Tyr64, Arg66, and Arg68), two mapped to the fourth β strand (Cys81 and Ser83), and the remaining two were located close to the C terminus (Ala159 and Glu171) (Figure 1A). Four variants were recurrent, and all occurred as a de novo event in at least one family. Of note, the c.511G>A substitution (p.Glu171Lys) was shared by the four affected subjects with clinical features resembling Noonan syndrome, occurring de novo in subject 12, and co-segregating with the phenotype in family 30153 (subjects 13 to 15), consistent with dominant inheritance. None of these variants had been reported in ExAC/gnomAD, and all were predicted to be pathogenic and met the American College of Medical Genetics (ACMG) criteria to be considered disease causing (Table S2).19 One variant, c.191A>G (p.Tyr64Cys), had previously been reported in two subjects with syndromic thrombocytopenia (MIM: 616737).20, 21
Table 1.
List of the Germline CDC42 Missense Mutations Identified in This Study
| Exon | Nucleotide Change | Amino Acid Change | Domain | Mutation Group | Subjects | Origin | MetaSVMa | CADD phreda | REVELa | ACMG |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | c.62T>C | p.Ile21Thr | α1 | III | 1 | de novo | 0.3729 | 27.1 | 0.901 | pathogenic |
| 1 | c.68A>G | p.Tyr23Cys | α1 | III | 2 | de novo | 0.7752 | 27.1 | 0.937 | pathogenic |
| 3 | c.191A>G | p.Tyr64Cys | switch II | I | 3 | de novo | 0.7976 | 23.4 | 0.834 | pathogenic |
| 3 | c.196A>G | p.Arg66Gly | switch II | I | 4, 5 | de novo | 0.5326 | 26.9 | 0.836 | pathogenic |
| 3 | c.203G>A | p.Arg68Gln | switch II | I | 6, 7 | de novo | 0.6586 | 26.3 | 0.827 | pathogenic |
| 3 | c.242G>T | p.Cys81Phe | β4 | II | 8 | de novo | 0.6280 | 30.0 | 0.840 | pathogenic |
| 3 | c.247T>C | p.Ser83Pro | β4 | II | 9, 10 | de novo | 0.8283 | 27.8 | 0.853 | pathogenic |
| 4 | c.476C>T | p.Ala159Val | NBP | II | 11 | de novo | 1.0179 | 34.0 | 0.916 | pathogenic |
| 5 | c.511G>Ab | p.Glu171Lys | CBR | III | 12, 13–15 | 1 de novo, 1 familial | 0.0158 | 24.7 | 0.768 | pathogenic |
Nucleotide numbering reflects cDNA numbering with 1 corresponding to the A of the ATG translation initiation codon in the CDC42 reference sequence (GenBank: NM_001791.3). No variants were reported in the public databases ExAC and GnomAD. All variants were predicted to be “deleterious” by Combined Annotation Dependent Depletion (CADD) v.1.3, Database for Nonsynonymous SNPs’ Functional Predictions (dbNSFP) Support Vector Machine (SVM) v.3.0, and REVEL algorithms.16, 17, 18 All changes satisfied the necessary criteria to be classified as pathogenic according to the American College of Medical Genetics guidelines.19 Abbreviations: NBP, nucleotide binding pocket; CBR, CRIB motif binding region.
Scores > 0 (MetaSVM), > 15 (CADDphred) or > 0.5 (REVEL) predict that the sequence change has a significant impact on protein structure and function.
This change affects transcript variant 1 (GenBank: NM_001791.3) and isoform 1 (GenBank: NP_001782.1), while it does not affect transcript variant 2 (GenBank: NM_044472.2) and isoform 2 (GenBank: NP_426359.1). The two isoforms have the same amino acid length but are characterized by a different C terminus.
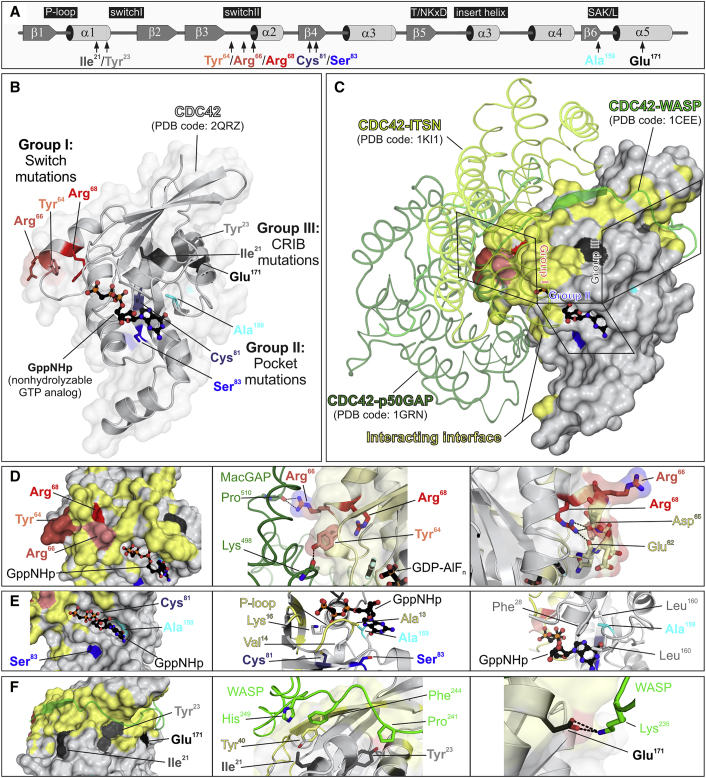
Figure 1.
Location of Disease-Causing CDC42 Mutations and Their Structural Impact
(A) Secondary structure elements (α helices and β strands), conserved motifs critical for tight guanine nucleotide binding and hydrolysis (G1–G5), and position of the identified disease-causing CDC42 mutations are illustrated.
(B) Variant residues are assigned to three groups according to their position in the context of CDC42 structure (PDB: 2QRZ): group I or switch mutations (Tyr64, Arg66, and Arg68) are part of the switch II loop; group II or pocket mutations (Cys81, Ser83, and Ala159) are located in the vicinity of nucleotide binding pocket; and group III or CRIB mutations (Ile21, Tyr23, and Glu171) are far outside of the major interaction sites of CDC42 with GTP/GDP and involve exposed residues located in or close to regions of the protein mediating binding to effectors containing a CRIB motif.
(C) The position of the mutant residues relative to CDC42 interactions is illustrated by overlaying three different crystal structures of CDC42 in complex with ARHGAP1 (p50GAP) (PDB: 1GRN), ITSN (PDB: 1KI1), and WAS (WASP) (PDB: 1CEE). Residues in reciprocal vicinity up to 4 Å were considered as part of binding interface. Residues of CDC42 mediating these interactions are shown in yellow.
(D) Group I mutations. Tyr64 and Arg66 are solvent-exposed residues and contribute to interactions with regulatory proteins and effectors (left). Interaction of both residues with ARHGAP18 (PDB: 5c2j) is shown as a representative for other interactions such as GEFs and effectors (middle). The disease-causing amino acid changes are predicted to affect this interaction. Arg68 participates in stabilizing the conformation of the switch II region via intramolecular interactions with Glu62 and Asp65 (right). The Arg-to-Gln change is predicted to destabilizes the switch II loop that is crucial for the interaction with signaling partners.
(E) Group II mutations. Cys81, Ser83, and Ala159 are in close vicinity of the phosphates (G1) and guanine base (G5) of bound GTP/GDP. Their substitutions are predicted to directly or indirectly affect the nucleotide binding affinity and to shift the balance between inactive and active CDC42 toward the latter.
(F) Group III mutations. Ile21, Tyr23, and Glu171 are part of a cavity on the CDC42 surface that accommodates the CRIB motif of bound effector proteins (e.g., WASP) (left). Ile21 and Tyr23 are critical for hydrophobic interactions (middle) with these type of proteins, while Glu171 contribute to binding mediating an electrostatic interaction (right).
CDC42 encodes a small GTPase of the RHO family modulating multiple signaling pathways controlling cell polarity and migration, endocytosis, and cell cycle progression, by cycling between an active (GTP-bound) and an inactive (GDP-bound) state.22, 23 It is characterized by five major highly conserved motifs, G1 to G5, which mediate GTP binding and hydrolysis (G4 and G5), phosphate binding (G1 and G3), and effector binding (G2) (Figure 1A).24, 25 Based on clinical heterogeneity (see below) and location of affected residues, we predicted that the mutations would have a variable functional impact. The structural consequences of the identified disease-causing mutations on CDC42 structure and function were assessed by Pymol molecular viewer (see Web Resources), using available PDB structures. This allowed us to inspect CDC42 interactions with ARHGAP1 (p50GAP/CDC42GAP; PDB: 1grn), ARHGAP18 (MacGAP; PDB: 5c2j), and ITSN1 (PDB: 1ki1) and WAS (WASP; PDB: 1cee), as representatives for CDC42’s GTPase activating proteins (GAPs), guanine nucleotide exchange factors (GEFs), and effectors, respectively, and to classify them structurally and functionally into three different groups. A first group of mutations affected the switch II region (p.Tyr64Cys, p.Arg66Gly, and p.Arg68Gln; group I), which mediates CDC42 binding to effectors and regulators (Figures 1B–1D).25 Tyr64 and Arg66 are located on the surface of CDC42 and directly participate in interactions with regulatory proteins and effectors. These changes were predicted to affect these interactions and, as a consequence, the catalytic activity of the GTPase and/or its capability to transduce signaling. Similarly, Arg68 is embedded in the protein interior and stabilizes the conformation of the switch II region via intramolecular interactions with multiple residues (Ala59, Gln61, and Glu100). The Arg-to-Gln change was assumed to strongly destabilize the switch II loop and the interaction with signaling partners. Group II included substitutions involving residues located within (Ala159) or close to (Cys81 and Ser83) the nucleotide-binding pocket (Figures 1B, 1C, and 1E). Ala159 faces the guanine base and replacement by valine was predicted to promote fast GDP/GTP cycling, favoring a GEF-independent active, GTP-bound state of the protein. A similar hyperactive behavior has been reported in RAS proteins.26, 27, 28 Similarly, Ser83 binds to Gln116, which interacts with the guanine base, predicting indirect perturbation of nucleotide binding properties of CDC42. Cys81 is an invariant residue among RHO GTPases located in proximity of the phosphate-binding loop, and its substitution to phenylalanine was expected to cause favorable hydrophobic interactions with this loop, dislocation of Gly12, and consequently defective GTP hydrolysis. Finally, group III (CRIB mutations) included variants at Ile21, Tyr23, and Glu171, which are exposed residues predicted to affect interactions with effectors containing a CDC42/RAC-interacting binding (CRIB) motif (Figures 1B, 1C, and 1F).29 Glu171 binds to Lys235 of WAS (WASP, hereafter) and plays a major role in the electrostatic binding network stabilizing the WASP-CDC42 association,30, 31 which was predicted to be disrupted by the Glu-to-Lys change. Tyr23 lies at the CDC42 surface implicated in PAK1 binding and stabilizes proper orientation of helix α5 mediating WASP binding.32, 33 Ile21 is located near the switch I region contributing to the hydrophobic pocket of helix α1 participating in WASP binding.30 The Ile-to-Thr substitution was predicted to perturb CDC42 binding to signaling partners.
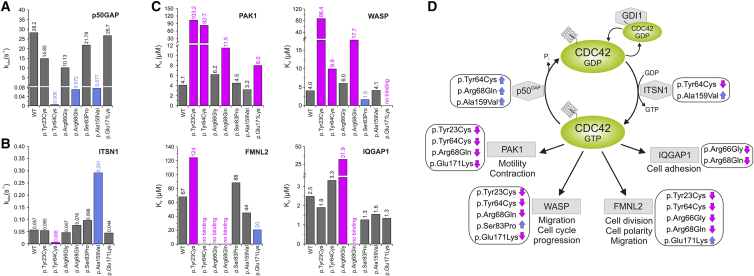
We assessed the effects of the disease-causing mutations on CDC42 GTPase activity, GDP/GTP exchange, and binding to effectors in vitro, using recombinant proteins. The p.Tyr23Cys, p.Tyr64Cys, p.Arg66Gly, p.Arg68Gln, p.Ser83Pro, p.Ala159Val, and p.Glu171Lys amino acid substitutions were selected as representative of the three mutation groups that were predicted to perturb differentially CDC42 function. pGEX vectors were used for bacterial overexpression of GST-tagged wild-type and mutant CDC42 proteins, and the GTPase-binding domains (GBD) of WASP (residues 154–321), PAK1 (residues 57–141), FMNL2 (residues 1–379), and IQGAP1 (residues 863–1657) in E. coli BL21 (DE3). Proteins were purified after cleavage of the GST tag (Superdex 75 or 200, GE Healthcare).31 Nucleotide-free and fluorescent nucleotide-bound CDC42 variants were prepared using alkaline phosphatase (Roche) and phosphodiesterase (Sigma Aldrich) at 4°C.31, 34 First, GTPase activity was measured basally and following ARHGAP1 (p50GAP, hereafter) stimulation by fluorescent experiments using tetramethylrhodamine (tamra-) GTP as substrate with a Hi-Tech Scientific (SF-61) stopped-flow instrument (Figures 2A and S2). The assays documented a variably increased basal GTP hydrolysis for CDC42Tyr64Cys, CDC42Arg68Gln, and CDC42Ala159Val. Each of these mutants, however, exhibited robust GAP insensitivity, showing respectively a 4,700-fold (CDC42Tyr64Cys), 392-fold (CDC42Arg68Gln), and 366-fold (CDC42Ala159Val) reduction in GAP-stimulated GTPase activity, compared to wild-type CDC42. A mildly decreased GAP-stimulated GTP hydrolysis was documented for CDC42Tyr23Cys and CDC42Arg66Gly. By using the same experimental approach, release of methylanthraniloyl (mant-) GDP was used to assess the basal and GEF-catalyzed nucleotide exchange reactions (Figures 2B and S3). The assays documented an increase of GDP release for CDC42Ala159Val and a slightly increased nucleotide exchange for CDC42Arg68Gln and CDC42Ser83Pro. By contrast, p.Tyr64Cys resulted in an almost completely abolished response to GEF. No substantial difference in GDP/GTP exchange behavior was observed for the other mutants. Then, fluorescence experiments were performed by using increasing amounts of the CDC42 interacting domains of WASP, PAK1, FMNL2, and IQGAP1 titrated to CDC42 proteins bound to mant-GppNHp, a non-hydrolyzable GTP analog, to assess the binding of mutants to four major CDC42 effectors and evaluate their ability to transduce signaling (Figures 2C and S4). Experiments were performed using a Fluoromax 4 fluorimeter in polarization mode, and the dissociation constants (Kd) were calculated by fitting the concentration-dependent binding curve using a quadratic ligand binding equation. Interaction with WASP was completely abolished in CDC42Glu171Lys and markedly decreased (21.6-fold) in CDC42Tyr23Cys. Decreased binding, albeit to a milder degree, was documented for CDC42Arg68Gln and CDC42Tyr64Cys, while CDC42Ser83Pro exhibited a slightly increased binding. Binding to PAK1 was impaired for CDC42Tyr23Cys and CDC42Tyr64Cys and reduced for CDC42Arg68Gln and CDC42Glu171Lys. Tyr64 and Arg66 contribute to CDC42 binding to FMNL2;35 consistently, CDC42Tyr64Cys and CDC42Arg66Gly had impaired FMNL2 binding, with the latter also having defective interaction with IQGAP1. Impaired binding to FMNL2 was also documented for CDC42Tyr23Cys and CDC42Arg68Gln, the latter also exhibiting defective IQGAP1 binding. By contrast, CDC42Glu171Lys showed enhanced FMNL2 binding. Overall, biochemical characterization of CDC42 mutants confirmed the heterogeneous clinical and structural impact of variants demonstrating a stabilized GTP-bound conformation but defective interaction with all tested partners for group I mutations, variable hyperactive behavior for group II mutations, and a diversified binding to effectors for group III mutations (Figure 2D).
Figure 2.
Assessment of the GTPase Activity, Nucleotide Exchange, and Binding to Effectors of Disease-Causing CDC42 Mutants
(A) Mean rate constants (kobs values) of p50GAP-stimulated GTP hydrolysis. Grey bars indicate non-significant differences compared to wild-type CDC42; blue bars indicate abolished/impaired GTP hydrolysis, which in turn results in an increased amount of active, GTP-bound CDC42 and thus enhanced signal flow. Data were obtained from >4 independent experiments.
(B) Mean rate constants (kobs values) of the GEF-catalyzed release of labeled GDP (mantGDP). Grey bars indicate non-significant differences compared to wild-type CDC42; blue and magenta bars indicate increased or abolished nucleotide exchange, respectively. The former is predicted to promote enhanced signaling, while the latter blocks CDC42 in its inactive state. Data were obtained from >4 independent experiments.
(C) CDC42 mutants variably affect binding to effectors. Dissociation constants (Kd) obtained for the interaction of CDC42 proteins with PAK1, WASP, IQGAP1, and FMNL2 determined by fluorescence polarization. Data were collected from titration of increasing concentrations of the respective effectors. They were obtained from >4 independent experiments and are illustrated as bar charts. Grey bars indicate non-significant differences compared to wild-type CDC42; blue and magenta bars indicate increased or decreased binding affinity, respectively.
(D) Scheme summarizing the functional dysregulation of disease-causing mutants on downstream signaling pathways and cellular processes. ITSN1 is a specific GEF for CDC42 promoting the active state of the GTPase by catalyzing GDP release. p50GAP negatively controls CDC42 function by stimulating the GTP hydrolysis reaction. CDC42 interaction with PAK1, WASP, FMNL2, and IQGAP1 activates signaling pathways controlling different cellular processes. For each specific function, the blue and magenta arrows indicate the hyperactive or defective behavior, respectively.
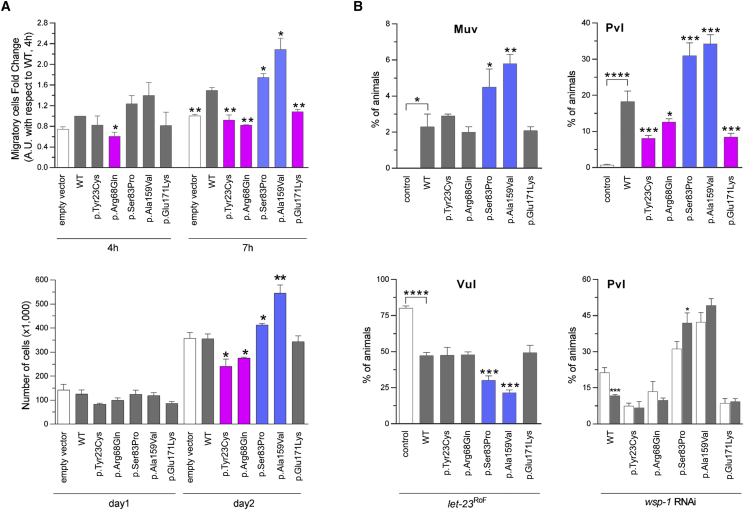
CDC42 is a master regulator of cell polarization and controls cell migration and growth.36, 37 The impact of CDC42 mutations on polarized migration was assessed by an in vitro wound-healing assay on fibronectin-coated wells (Sigma-Aldrich) (Figures 3A and S5). Monolayers of NIH 3T3 cells (American Type Culture Collection) cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS, 2 mM L-glutamine, and 10 U/mL penicillin/streptomycin (Sigma-Aldrich) were transiently transfected using Fugene 6 (Roche) to express wild-type FLAG-tagged CDC42 isoform 1 (GenBank: NP_001782.1) or each of the p.Tyr23Cys, p.Arg68Gln, p.Ser83Pro, p.Ala159Val, and p.Glu171Lys mutants. 24 hr after transfection, cells were scratched and incubated in low serum medium in the presence of thymidine (Sigma-Aldrich) to inhibit cell proliferation. Cells that had migrated in the wounded area were counted 4 and 7 hr after scratch (four fields per well). Comparable transfection efficiency was verified by western blot analysis of the protein lysates. Only cells expressing group II mutants exhibited enhanced wound closure ability compared to cells expressing wild-type CDC42. Other tested mutants failed to increase migration, suggesting loss of function of this CDC42-mediated process, in line with defective binding of these mutants to WASP, a mediator of polarized migration.38 Mutants also affected cell proliferation differentially (Figure 3A). CDC42Ala159Val and CDC42Ser83Pro variably enhanced cell growth, while CDC42Tyr23Cys and CDC42Arg68Gln significantly impaired proliferation, indicating a dominant-negative effect.
Figure 3.
In Vitro and In Vivo Functional Characterization of CDC42 Mutations
(A) CDC42 mutations differentially impact polarized migration and cell proliferation. Wound-healing assays (above) and proliferation assays (below) were performed using NIH 3T3 cells transiently transfected to express wild-type CDC42 or each of the indicated mutants. Mean ± SD densitometry values of three independent experiments are shown. The wound was generated 24 hr after transfection, and migration in the wounded area was evaluated after 4 and 7 hr. Cells expressing exogenous wild-type CDC42 migrate more rapidly into the scratched area than cells transfected with the empty vector (EV). Mutants differentially perturb polarized migration, with CDC42Ser83Pro and CDC42Ala159Val overexpression variably enhancing the wound closure ability of transfected cells compared to the wild-type protein, whereas CDC42Tyr23Cys, CDC42Arg68Gln, and CDC42Glu171Lys fail to do that, supporting a gain-of-function and a loss-of-function effect of these mutants, respectively. Cell proliferation was evaluated in transfected cells at the indicated time points and quantified by manual counting using a Neubauer hemocytometer. The trypan blue dye exclusion test was used to consider viable cells only. While the CDC42Ala159Val and CDC42Ser83Pro mutants variably enhance proliferation compared to cells expressing wild-type CDC42, no effect on proliferation (CDC42Glu171Lys) and reduced proliferation (CDC42Tyr23Cys and CDC42Arg68Gln) is documented for the other mutants, indicating a loss-of-function and a dominant-negative effect, respectively. Asterisks indicate significant differences compared with wild-type CDC42 (∗p < 0.05; ∗∗p < 0.01; Student’s t test).
(B) Consequences of CDC-42 expression on vulval development in C. elegans. Ectopic expression of wild-type CDC-42 at the L2/L3 stage elicits a multivulva (Muv) phenotype (left, upper panel), and CDC-42 overexpression in a LET-23/EGFR hypomorphic background reduces the penetrance of the vulvaless (Vul) phenotype (left, lower panel). Compared to animals expressing wild-type CDC-42, those expressing CDC-42Ser83Pro and CDC-42Ala159Val show higher prevalence of the Muv phenotype and lower prevalence of the Vul phenotype, indicating a gain-of-function role on LET-60/RAS signaling. Animals expressing the other tested CDC-42 mutants do not significantly differ from those expressing wild-type CDC-42. Ectopic expression of wild-type CDC-42 at the early L3 stage elicits a protruding vulva (Pvl) phenotype (right, upper panel). Animals expressing CDC-42Ser83Pro and CDC-42Ala159Val show a higher prevalence of the phenotype compared to worms expressing wild-type CDC-42, while a less penetrant phenotype was scored for animals expressing CDC-42Tyr23Cys, CDC-42Arg68Gln, or CDC-42Glu171Lys mutants. RNA interference (RNAi) experiments show that the Pvl phenotype associated with overexpression of wild-type CDC-42 is mediated, in part, by WSP-1/WASP (right, lower panel). White and gray bars indicate the penetrance of Pvl in non-interfered and interfered animals, respectively. Error bars indicate SEM of four independent experiments, and asterisks specify significance differences between animals expressing CDC-42 mutants and those expressing wild-type CDC-42 or between interfered and non-interfered nematodes (∗p < 0.05; ∗∗p < 0.001; ∗∗∗p < 0.0001; ∗∗∗∗p < 0.00005; two-tailed Fisher’s exact test). Comparisons between worms expressing wild-type CDC-42 and control animals are also shown. RNAi was performed by feeding using HT115 E. coli bacteria expressing double stranded wsp-1 RNA (Ahringer’s C. elegans RNAi feeding library) and optimized to overcome lethality. As a control of the efficiency of the modified RNAi protocol, let-60 RNAi experiments were performed on animals carrying the let-60 gain-of-function allele n1046 (p.Gly13Glu), and the prevalence of the Muv phenotype was scored at a dissecting microscope (Table S4).
In Caenorhabditis elegans, CDC-42 controls early and late developmental programs (see WormBook in Web Resources), including vulval development,39, 40, 41 a process that is regulated by LET-60/RAS-dependent and -independent signals.42 To explore the impact of the disease-causing CDC42 mutations in vivo, transgenic lines were generated to conditionally express wild-type CDC-42 or a selected subset of mutations for each mutation group (p.Tyr23Cys, p.Arg68Gln, p.Ser83Pro, p.Ala159Val, and p.Glu171Lys) affecting residues conserved in the nematode ortholog (Figure S6).43 The wild-type cdc-42 cDNA (ORF clone R07G3.1; ThermoScientific) was subcloned into the pPD49.83 heat shock-inducible vector (gift of A. Fire, Stanford University School of Medicine), and the generated constructs were injected at 100 ng/μL. The pJM67 plasmid (pelt-2::NLS::GFP) (gift from J.D. McGhee, University of Calgary), which drives green fluorescent protein (GFP) expression in intestinal cell nuclei, was used as co-injection marker (30 ng/μL). To analyze vulval induction and morphogenesis, synchronized animals from at least three independent lines for each construct were grown at 20°C and heat-shocked (90 min at 33°C followed by 30 min at 30°C) at late L2/early L3 larval stages and scored for vulval induction and morphogenesis from late L3 to mid L4 stages. The presence of a protruding vulva (Pvl phenotype), multiple ectopic pseudovulvae (multivulva [Muv] phenotype), and lack of a vulva (vulvaless [Vul] phenotype) was analyzed at the adult stage. Lines were scored in triplicate experiments using a Nikon Eclipse 80i instrument equipped with Nomarski differential interference contrast optics and used for further analyses and crosses. After each cross, the genotype of individual alleles was confirmed by direct sequencing of the appropriate genomic region. Isogenic animals that had lost the transgene were cloned separately and used as controls in each experiment. Overexpression of wild-type CDC-42 at the L2/L3 stage elicited a low-penetrant Muv phenotype, exacerbated the Muv phenotype associated with a let-60 gain-of-function allele, let-60(n1046) IV, and partially rescued the Vul phenotype of animals carrying a hypomorphic let-23/EGFR allele, let-23(sy1) II, indicating LET-60/RAS signaling hyperactivation (Figures 3B and S7, Table S3). Compared to wild-type CDC-42, group II mutations induced a more severe Muv phenotype and more efficiently rescued the Vul phenotype of let-23(sy1) animals, indicative of enhanced signal flow through LET-60. Overexpression of wild-type CDC-42 also engendered aberrant vulva morphogenesis, generating a Pvl phenotype that was mediated, in part, by WSP-1/WASP (Figures 3B and S7, Tables S3 and S4). The same phenotype had previously been reported in C. elegans lines expressing the RASopathy-causing SHOC2S2G and RRASG39dup mutants.44, 45 Like those animals, a variable proportion of CDC-42 hermaphrodites exhibiting Pvl displayed egg-laying defects (Egl phenotype) and accumulation of larvae inside the mother (Bag-of-worms phenotype) (data not shown). Of note, this phenotype was markedly promoted by group II mutations in a WASP-independent manner, indicating a gain-of-function effect of these changes. By contrast, CDC-42Tyr23Cys, CDC-42Arg68Gln, and CDC-42Glu171Lys significantly reduced the Pvl phenotype, supporting a selective hypomorphic behavior. Pvl was not modulated by wsp-1 RNAi in animals expressing CDC-42Tyr23Cys and CDC-42Glu171Lys and was only slightly reduced in those expressing CDC-42Arg68Gln, consistent with the biochemical data indicating an abolished or strongly reduced binding to WASP of those mutants, respectively. Nomarski observations of L3 and L4 control larvae showed that only P6.p descendants detached from the cuticle generating a single, symmetric invagination; by contrast, a variable proportion of larvae expressing wild-type and mutant CDC-42 displayed asymmetric and/or multiple invaginations (Figure S7), which represent the earliest signs of the Pvl and Muv phenotypes, respectively. Overall, the data indicate that group II mutations upregulate multiple signaling pathways, including LET-60/RAS, while the other variants behave as hypomorphic mutations on WASP-dependent signaling.
The cohort of individuals carrying heterozygous mutations in CDC42 had an unusually broad spectrum of anomalies. Core clinical features included defective growth, intellectual disability (ID), facial dysmorphism, hearing/vision problems, cardiac malformations, immune, hematologic, and lymphatic abnormalities, and brain malformations (Figures 4 and S8, Tables S5–S7, Supplemental Note). Correlating the functional impact of mutations on clinical phenotypes observed in affected individuals permitted a preliminary analysis of genotype-phenotype relationships (Table 2). Individuals with group I mutations manifested with ID, muscle tone abnormalities, and variable other less common features, including cardiac defects. All individuals within this group had thrombocytopenia, similar to two previously reported individuals.20, 21 Individuals with group II mutations manifested with strikingly dysmorphic facial features: subject 11 (p.Ala159Val) had marked hypertelorism, prominent forehead, bitemporal narrowing, and downslanting palpebral fissures with coarse thick hair, resembling a RASopathy (Figures 4K and 4L). Features within the Noonan syndrome phenotypic spectrum were observed in all affected individuals from two unrelated families carrying the c.511G>A (p.Glu171Lys) change (subjects 12 to 15), with a particularly striking gestalt of this disorder occurring in subject 12 (Figure 4Q). Notably, brain malformations occurred in all groups, and four individuals manifested with cerebellar-posterior fossa abnormalities. Subjects 8 and 11 (with group II mutations) had a large cerebellum with evidence of posterior fossa crowding and cerebellar tonsillar ectopia, features commonly reported in RASopathies.46 It should be noted that notwithstanding the occurrence of a clinical overlap within each mutation group, intra-group phenotypic variability was observed, which would suggest a specific impact of individual mutations on developmental processes.
Figure 4.
Facial Features of Individuals with Heterozygous CDC42 Mutations
(A–C) Subject 3 (p.Tyr64Cys) at age 2 years and 6 months (A) and 15 years (B and C) showing upslanted palpebral fissures, smooth philtrum, flaring alae nasi, thin upper vermilion, and wide mouth with widely spaced teeth.
(D) Subject 4 (p.Arg66Gly) at 15 years showing broad forehead and broad nasal bridge with bulbous nasal tip.
(E and F) Subject 6 (p.Arg68Gln) at 24 months (E) and 4 years (F) showing a prominent broad forehead, hypertelorism, long philtrum, and thin upper vermilion.
(G and H) Subject 9 (p.Ser83Pro) at age 2 (G) and 6 (H) years showing prominent forehead, hypertelorism, wide mouth with cupid’s bow, thin upper vermilion, and widely spaced teeth.
(I and J) Subject 10 (p.Ser83Pro) at 13 (I) and 32 (J) years showing prominent forehead, wide nasal bridge, ptosis, flared nostrils, and wide mouth with widely spaced teeth.
(K and L) Subject 11 (p.Ala159Val) at 2 years (K) and at 3 years and 7 months (L) showing very broad and prominent forehead, bulbous nasal tip, flared nostrils, cupid’s bow, and downturned corners of the mouth.
(M–O) Subject 1 (p.Ile21Thr) at age 3 months (M), 2 years (N), and 10 years (O) showing synophrys, wide palpebral fissures, high and narrow nasal bridge, bulbous nasal tip, wide mouth with downturned corners, and mildly laterally prominent ears.
(P) Subject 2 (p.Tyr23Cys) at 14 years showing wide palpebral fissures, high nasal bridge with elevated nasal tip, short philtrum, and long neck.
(Q) Subject 12 (p.Glu171Lys) at 12 years showing typical facial features of Noonan syndrome, including broad forehead, hypertelorism, low-set ears, bulbous nasal tip, and flared nostrils.
(R) Subject 13 (p.Glu171Lys) showing ptosis, broad neck, and pectus deformity.
Note that individuals fitting the different mutation groups share some facial characteristics, and that intragroup variability is also observed.
Table 2.
Summary of the Clinical Features of CDC42 Mutation-Positive Subjects
| Mutation group | Group I | Group II | Group III |
|---|---|---|---|
| Number of individuals | 5 | 4 | 6 |
| Amino acid substitutions | p.Tyr64Cys, p.Arg66Gly, p.Arg68Gln | p.Cys81Phe, p.Ser83Pro, p.Ala159Val | p.Ile21Thr, p.Tyr23Cys, p.Glu171Lys |
| Growth | |||
| Prenatal – weight at birth ≤ 2 SD | 1/4 | 2/4 | 1/4 |
| Prenatal – OFC at birth ≤ 2 SD | 1/3 | 1/2 | 1/2 |
| Postnatal – weight ≤ 2 SD | 4/5 | 2/3 | 1/4 |
| Postnatal – OFC ≤ 2 SD | 3/5 | 2/4 | 3/4 |
| Postnatal – growth deficiency | 3/5 | 4/4 | 4/6 |
| Intellectual disability | 5/5 | 4/4 | 2/6 |
| Seizures | 1/4 | 2/4 | 1/6 |
| MRI brain anomaliesa | 4/4 | 4/4 | 1/2 |
| Tone anomalies | 3/4 | 2/4 | 2/6 |
| Optic atrophy | 1/4 | 0/4 | 2/6 |
| Endocrine anomalies | 2/4 | 1/4 | 1/5 |
| Facial dysmorphismb | 4/5 | 4/4 | 6/6 |
| Pectus deformity | 1/5 | 0/4 | 4/5 |
| Scoliosis/vertebral anomalies | 2/5 | 1/4 | 2/6 |
| Camptodacyly | 1/5 | 2/4 | 1/5 |
| Cardiac anomalies | 3/5 | 2/4 | 2/5 |
| GU anomalies | 2/5 | 2/4 | 1/6 |
| Lymphatic anomalies | 1/5 | 1/4 | 0/6 |
| Recurrent infections | 4/5 | 3/4 | 1/6 |
| Platelet anomalies (thrombocytopenia, macrothrombocytes) | 4/4 | 1/3 | 0/5 |
Abbreviations: OFC, occipito-frontal circumference; SD, standard deviation; GU, genitourinary. Detailed phenotypic description of subjects is reported in the Supplemental Note and Table S5.
For details regarding the facial features, see Table S6.
While traditionally CDC42 has been functionally linked to remodeling of the actin cytoskeletal architecture,47 its role in controlling intracellular signaling has recently been broadened.48 Such complex modulatory function is accomplished by interactions with a wide array of signaling partners functioning in distinct signal cascades. Cdc42 loss of function is embryonic lethal, and its targeted deletion has been shown to disrupt cell fate decision, differentiation, and function of multiple cell lineages as well as tissue homeostasis.49 Here, we report that dominantly acting mutations differentially perturb CDC42 function and cause clinically heterogeneous phenotypes affecting development and growth. Group I mutations associated with impaired binding to regulators and effectors cause a syndromic form of thrombocytopenia, while the variably hyperactive group II mutations are associated with a variable developmental disorder characterized by striking dysmorphic features, and one specific amino acid change among the group III mutations, which affects only one of the two CDC42 isoforms and specifically impairs binding to WASP, results in an overall milder clinical phenotype that phenocopies Noonan syndrome.14
Noonan syndrome, the most common and clinically variable among the RASopathies, is caused by dysregulated signaling through RAS and the MAPK cascade. This disorder and its clinically related phenotypes result from heterozygous germline mutations affecting RAS genes or genes coding proteins functioning as RAS effectors, regulators of RAS function, or more generally as modulators of RAS-MAPK signaling.15 More recently, the family of genes implicated in RASopathies has been extended to include LZTR1 (MIM: 600574), RIT1 (MIM: 609591), and RRAS (MIM: 165090), which encode for signal transducers whose direct link to the RAS and the MAPK cascade had not previously been appreciated.45, 50, 51 While it is possible that functional dysregulation of these proteins may impact RAS signaling directly or indirectly, these findings raise also the possibility that other pathways may contribute to disease pathogenesis. The present in vivo data provide evidence for enhanced signaling through RAS for group II mutations, indicating that upregulated CDC42 function is able to perturb signal flow through RAS; however, no effect on RAS-mediated signaling was inferred for the p.Glu171Lys change, here identified to be associated with a phenotype resembling Noonan syndrome. While it is possible that the used in vivo model failed in providing informative data for the specific effect of CDC42Glu171Lys on RAS signaling, our finding suggests that other processes, including aberrant cytoskeletal rearrangement, may represent a previously unappreciated aspect contributing to disease pathogenesis in Noonan syndrome. Consistent with this possibility, SOS1, SHOC2, and RRAS function has been linked to cell migration and other cellular processes strictly dependent on cytoskeletal rearrangement.52, 53, 54 Further studies are thus required to specifically address the impact of dysregulated CDC42 function on RAS signaling as well as on the cellular and developmental processes that are altered in Noonan syndrome.
Overall, the present work links different classes of dominantly acting mutations of CDC42, a master regulator of actin cytoskeleton and major node in intracellular signaling, to a heterogeneous set of developmental and multi-system phenotypes, demonstrating the critical requirement of proper CDC42 function in a large array of developmental processes. This study also exemplifies current challenges in syndrome delineation in the post-WES era and emphasizes the relevance of functional profiling in syndrome recognition and delineation.
Acknowledgments
We thank the families and referring physicians for their participation in this study. We thank Dr. David Wilson (Washington University, St. Louis) for providing clinical expertise and Dr. Serenella Venanzi (Istituto Superiore di Sanità, Rome) for technical assistance. This project was supported by the National Institute of Neurological Disorders and Stroke (NINDS) (K08NS092898 to G.M.M.), the Associazione Italiana per la Ricerca sul Cancro (AIRC) (IG17583 to M.T.), Fondazione Bambino Gesù (Vite Coraggiose to M.T.), Italian Ministry of Health (RF-2011-02349938 and Ricerca Corrente 2017 to M.T.), E-Rare (NSEuroNet to M.Z., M.R.A., and M.T.), International Research Training Group 1902 Intra- and Interorgan Communication of the Cardiovascular System (IRTG 1902 to E.A., M.B., and M.R.A.), and Medical Faculty of the Heinrich-Heine University Duesseldorf (9772617 to K.N., O.H.F.K., R.K., and M.R.A.). Exome sequencing was performed at the University of Washington Center for Mendelian Genomics (UW-CMG) and was funded by the National Human Genome Research Institute and the National Heart, Lung, and Blood Institute grant HG006493 (to D.A.N. and M.J.B.). This work was also supported by grants U01HL131003, UM1HL098147, UM1HL098123, UM1HL128761, UM1HL128711, and UM1HL098162 in support of the Pediatric Cardiac Genomics Consortium from the National Heart, Lung, and Blood Institute and the Eunice Kennedy Shriver National Institute of Child Health and Human Development. C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). We also thank WormBase, the contributors to MyGene2, and the University of Washington Center for Mendelian Genomics for use of data.
Published: January 25, 2018
Footnotes
Supplemental Data include a supplemental note (clinical data), eight figures, and seven tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2017.12.015.
Contributor Information
Marco Tartaglia, Email: marco.tartaglia@opbg.net.
Ghayda M. Mirzaa, Email: gmirzaa@u.washington.edu.
Accession Numbers
ClinVar accession ID for data provided herein are SCV000572034.2 (c.62T>C), SCV000244190.3 (c.68A>G), SCV000577577.2 (c.191A>G), SCV000244118.3 (c.196A>G), SCV000589746.1 (c.242G>T), SCV000678254 (c.203G>A), SCV000678255 (c.247T>C), SCV000678256 (c.476C>T), and SCV000678257 (c.511G>A).
Web Resources
ExAC Browser, http://exac.broadinstitute.org/
gnomAD Browser, http://gnomad.broadinstitute.org/
NCBI Gene, https://www.ncbi.nlm.nih.gov/gene
OMIM, http://www.omim.org/
PyMOL, https://pymol.org/2
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
WormBase, http://www.wormbase.org/
WormBook, http://www.wormbook.org
Supplemental Data
References
- 1.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 2.Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D., Harrell T.M., McMillin M.J., Wiszniewski W., Gambin T., Centers for Mendelian Genomics The genetic basis of Mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 2015;97:199–215. doi: 10.1016/j.ajhg.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Menke L.A., van Belzen M.J., Alders M., Cristofoli F., Ehmke N., Fergelot P., Foster A., Gerkes E.H., Hoffer M.J., Horn D., DDD Study CREBBP mutations in individuals without Rubinstein-Taybi syndrome phenotype. Am. J. Med. Genet. A. 2016;170:2681–2693. doi: 10.1002/ajmg.a.37800. [DOI] [PubMed] [Google Scholar]
- 4.Lee C.S., Fu H., Baratang N., Rousseau J., Kumra H., Sutton V.R., Niceta M., Ciolfi A., Yamamoto G., Bertola D., Baylor-Hopkins Center for Mendelian Genomics Mutations in fibronectin cause a subtype of spondylometaphyseal dysplasia with “corner fractures”. Am. J. Hum. Genet. 2017;101:815–823. doi: 10.1016/j.ajhg.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niceta M., Stellacci E., Gripp K.W., Zampino G., Kousi M., Anselmi M., Traversa A., Ciolfi A., Stabley D., Bruselles A. Mutations impairing GSK3-mediated MAF phosphorylation cause cataract, deafness, intellectual disability, seizures, and a Down syndrome-like facies. Am. J. Hum. Genet. 2015;96:816–825. doi: 10.1016/j.ajhg.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reijnders M.R.F., Ansor N.M., Kousi M., Yue W.W., Tan P.L., Clarkson K., Clayton-Smith J., Corning K., Jones J.R., Lam W.W.K., Deciphering Developmental Disorders Study RAC1 missense mutations in developmental disorders with diverse phenotypes. Am. J. Hum. Genet. 2017;101:466–477. doi: 10.1016/j.ajhg.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinelli S., Torreri P., Tinti M., Stella L., Bocchinfuso G., Flex E., Grottesi A., Ceccarini M., Palleschi A., Cesareni G. Diverse driving forces underlie the invariant occurrence of the T42A, E139D, I282V and T468M SHP2 amino acid substitutions causing Noonan and LEOPARD syndromes. Hum. Mol. Genet. 2008;17:2018–2029. doi: 10.1093/hmg/ddn099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Philippakis A.A., Azzariti D.R., Beltran S., Brookes A.J., Brownstein C.A., Brudno M., Brunner H.G., Buske O.J., Carey K., Doll C. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum. Mutat. 2015;36:915–921. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong J.X., Yu J.H., Lorentzen P., Park K.M., Jamal S.M., Tabor H.K., Rauch A., Saenz M.S., Boltshauser E., Patterson K.E. Gene discovery for Mendelian conditions via social networking: de novo variants in KDM1A cause developmental delay and distinctive facial features. Genet. Med. 2016;18:788–795. doi: 10.1038/gim.2015.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homsy J., Zaidi S., Shen Y., Ware J.S., Samocha K.E., Karczewski K.J., DePalma S.R., McKean D., Wakimoto H., Gorham J. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka A.J., Cho M.T., Millan F., Juusola J., Retterer K., Joshi C., Niyazov D., Garnica A., Gratz E., Deardorff M. Mutations in SPATA5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am. J. Hum. Genet. 2015;97:457–464. doi: 10.1016/j.ajhg.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farwell Hagman K.D., Shinde D.N., Mroske C., Smith E., Radtke K., Shahmirzadi L., El-Khechen D., Powis Z., Chao E.C., Alcaraz W.A. Candidate-gene criteria for clinical reporting: diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genet. Med. 2017;19:224–235. doi: 10.1038/gim.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts A.E., Allanson J.E., Tartaglia M., Gelb B.D. Noonan syndrome. Lancet. 2013;381:333–342. doi: 10.1016/S0140-6736(12)61023-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tartaglia M., Gelb B.D. Disorders of dysregulated signal traffic through the RAS-MAPK pathway: phenotypic spectrum and molecular mechanisms. Ann. N Y Acad. Sci. 2010;1214:99–121. doi: 10.1111/j.1749-6632.2010.05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takenouchi T., Kosaki R., Niizuma T., Hata K., Kosaki K. Macrothrombocytopenia and developmental delay with a de novo CDC42 mutation: Yet another locus for thrombocytopenia and developmental delay. Am. J. Med. Genet. A. 2015;167A:2822–2825. doi: 10.1002/ajmg.a.37275. [DOI] [PubMed] [Google Scholar]
- 21.Takenouchi T., Okamoto N., Ida S., Uehara T., Kosaki K. Further evidence of a mutation in CDC42 as a cause of a recognizable syndromic form of thrombocytopenia. Am. J. Med. Genet. A. 2016;170A:852–855. doi: 10.1002/ajmg.a.37526. [DOI] [PubMed] [Google Scholar]
- 22.Etienne-Manneville S. Cdc42--the centre of polarity. J. Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- 23.Heasman S.J., Ridley A.J. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 24.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci. STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dvorsky R., Ahmadian M.R. Always look on the bright site of Rho: structural implications for a conserved intermolecular interface. EMBO Rep. 2004;5:1130–1136. doi: 10.1038/sj.embor.7400293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janakiraman M., Vakiani E., Zeng Z., Pratilas C.A., Taylor B.S., Chitale D., Halilovic E., Wilson M., Huberman K., Ricarte Filho J.C. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 2010;70:5901–5911. doi: 10.1158/0008-5472.CAN-10-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gremer L., Merbitz-Zahradnik T., Dvorsky R., Cirstea I.C., Kratz C.P., Zenker M., Wittinghofer A., Ahmadian M.R. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum. Mutat. 2011;32:33–43. doi: 10.1002/humu.21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang M.T., Asthana S., Gao S.P., Lee B.H., Chapman J.S., Kandoth C., Gao J., Socci N.D., Solit D.B., Olshen A.B. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 2016;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirone D.M., Carter D.E., Burbelo P.D. Evolutionary expansion of CRIB-containing Cdc42 effector proteins. Trends Genet. 2001;17:370–373. doi: 10.1016/s0168-9525(01)02311-3. [DOI] [PubMed] [Google Scholar]
- 30.Abdul-Manan N., Aghazadeh B., Liu G.A., Majumdar A., Ouerfelli O., Siminovitch K.A., Rosen M.K. Structure of Cdc42 in complex with the GTPase-binding domain of the ‘Wiskott-Aldrich syndrome’ protein. Nature. 1999;399:379–383. doi: 10.1038/20726. [DOI] [PubMed] [Google Scholar]
- 31.Hemsath L., Dvorsky R., Fiegen D., Carlier M.F., Ahmadian M.R. An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol. Cell. 2005;20:313–324. doi: 10.1016/j.molcel.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 32.Morreale A., Venkatesan M., Mott H.R., Owen D., Nietlispach D., Lowe P.N., Laue E.D. Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat. Struct. Biol. 2000;7:384–388. doi: 10.1038/75158. [DOI] [PubMed] [Google Scholar]
- 33.Gizachew D., Guo W., Chohan K.K., Sutcliffe M.J., Oswald R.E. Structure of the complex of Cdc42Hs with a peptide derived from P-21 activated kinase. Biochemistry. 2000;39:3963–3971. doi: 10.1021/bi992646d. [DOI] [PubMed] [Google Scholar]
- 34.Hemsath L., Ahmadian M.R. Fluorescence approaches for monitoring interactions of Rho GTPases with nucleotides, regulators, and effectors. Methods. 2005;37:173–182. doi: 10.1016/j.ymeth.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 35.Kühn S., Erdmann C., Kage F., Block J., Schwenkmezger L., Steffen A., Rottner K., Geyer M. The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation. Nat. Commun. 2015;6:7088. doi: 10.1038/ncomms8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melendez J., Liu M., Sampson L., Akunuru S., Han X., Vallance J., Witte D., Shroyer N., Zheng Y. Cdc42 coordinates proliferation, polarity, migration, and differentiation of small intestinal epithelial cells in mice. Gastroenterology. 2013;145:808–819. doi: 10.1053/j.gastro.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zegers M.M., Friedl P. Rho GTPases in collective cell migration. Small GTPases. 2014;5:e28997. doi: 10.4161/sgtp.28997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beaty B.T., Condeelis J. Digging a little deeper: the stages of invadopodium formation and maturation. Eur. J. Cell Biol. 2014;93:438–444. doi: 10.1016/j.ejcb.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welchman D.P., Mathies L.D., Ahringer J. Similar requirements for CDC-42 and the PAR-3/PAR-6/PKC-3 complex in diverse cell types. Dev. Biol. 2007;305:347–357. doi: 10.1016/j.ydbio.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohmer L.L., Clay M.R., Naegeli K.M., Chi Q., Ziel J.W., Hagedorn E.J., Park J.E., Jayadev R., Sherwood D.R. A sensitized screen for genes promoting invadopodia function in vivo: CDC-42 and Rab GDI-1 direct distinct aspects of invadopodia formation. PLoS Genet. 2016;12:e1005786. doi: 10.1371/journal.pgen.1005786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi M.S., Yoo A.S., Greenwald I. sel-11 and cdc-42, two negative modulators of LIN-12/Notch activity in C. elegans. PLoS ONE. 2010;5:e11885. doi: 10.1371/journal.pone.0011885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid T., Hajnal A. Signal transduction during C. elegans vulval development: a NeverEnding story. Curr. Opin. Genet. Dev. 2015;32:1–9. doi: 10.1016/j.gde.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cordeddu V., Di Schiavi E., Pennacchio L.A., Ma’ayan A., Sarkozy A., Fodale V., Cecchetti S., Cardinale A., Martin J., Schackwitz W. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat. Genet. 2009;41:1022–1026. doi: 10.1038/ng.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flex E., Jaiswal M., Pantaleoni F., Martinelli S., Strullu M., Fansa E.K., Caye A., De Luca A., Lepri F., Dvorsky R. Activating mutations in RRAS underlie a phenotype within the RASopathy spectrum and contribute to leukaemogenesis. Hum. Mol. Genet. 2014;23:4315–4327. doi: 10.1093/hmg/ddu148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gripp K.W., Hopkins E., Doyle D., Dobyns W.B. High incidence of progressive postnatal cerebellar enlargement in Costello syndrome: brain overgrowth associated with HRAS mutations as the likely cause of structural brain and spinal cord abnormalities. Am. J. Med. Genet. A. 2010;152A:1161–1168. doi: 10.1002/ajmg.a.33391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nobes C.D., Hall A. Rho, rac and cdc42 GTPases: regulators of actin structures, cell adhesion and motility. Biochem. Soc. Trans. 1995;23:456–459. doi: 10.1042/bst0230456. [DOI] [PubMed] [Google Scholar]
- 48.Arias-Romero L.E., Chernoff J. Targeting Cdc42 in cancer. Expert Opin. Ther. Targets. 2013;17:1263–1273. doi: 10.1517/14728222.2013.828037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pedersen E., Brakebusch C. Rho GTPase function in development: how in vivo models change our view. Exp. Cell Res. 2012;318:1779–1787. doi: 10.1016/j.yexcr.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Aoki Y., Niihori T., Banjo T., Okamoto N., Mizuno S., Kurosawa K., Ogata T., Takada F., Yano M., Ando T. Gain-of-function mutations in RIT1 cause Noonan syndrome, a RAS/MAPK pathway syndrome. Am. J. Hum. Genet. 2013;93:173–180. doi: 10.1016/j.ajhg.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto G.L., Aguena M., Gos M., Hung C., Pilch J., Fahiminiya S., Abramowicz A., Cristian I., Buscarilli M., Naslavsky M.S. Rare variants in SOS2 and LZTR1 are associated with Noonan syndrome. J. Med. Genet. 2015;52:413–421. doi: 10.1136/jmedgenet-2015-103018. [DOI] [PubMed] [Google Scholar]
- 52.Sini P., Cannas A., Koleske A.J., Di Fiore P.P., Scita G. Abl-dependent tyrosine phosphorylation of Sos-1 mediates growth-factor-induced Rac activation. Nat. Cell Biol. 2004;6:268–274. doi: 10.1038/ncb1096. [DOI] [PubMed] [Google Scholar]
- 53.Holly S.P., Larson M.K., Parise L.V. The unique N-terminus of R-ras is required for Rac activation and precise regulation of cell migration. Mol. Biol. Cell. 2005;16:2458–2469. doi: 10.1091/mbc.E03-12-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaduwal S., Jeong W.J., Park J.C., Lee K.H., Lee Y.M., Jeon S.H., Lim Y.B., Min S., Choi K.Y. Sur8/Shoc2 promotes cell motility and metastasis through activation of Ras-PI3K signaling. Oncotarget. 2015;6:33091–33105. doi: 10.18632/oncotarget.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.