Abstract
Shigella boydii is one of the four Shigella species that causes disease worldwide; however, there are few published studies that examine the genomic variation of this species. This study compares genomes of 72 total isolates; 28 S. boydii from Bangladesh and The Gambia that were recently isolated as part of the Global Enteric Multicenter Study (GEMS), 14 historical S. boydii genomes in the public domain and 30 Escherichia coli and Shigella reference genomes that represent the genomic diversity of these pathogens. This comparative analysis of these 72 genomes identified that the S. boydii isolates separate into three phylogenomic clades, each with specific gene content. Each of the clades contains S. boydii isolates from geographic and temporally distant sources, indicating that the S. boydii isolates from the GEMS are representative of S. boydii. This study describes the genome sequences of a collection of novel S. boydii isolates and provides insight into the diversity of this species in comparison to the E. coli and other Shigella species.
Keywords: Shigella boydii, microbial genomics, pathogenesis
This comparative genomic study identifies the diversity of Shigella boydii isolates when compared to reference isolates of closely related pathogens.
Shigella is a Gram-negative pathogen and the cause of shigellosis, a potentially deadly diarrheal disease whose symptoms range from mild intestinal discomfort to death depending on severity (Rasko et al. 2008; Sahl et al. 2015). Each year Shigella species cause 165 million cases of shigellosis with an estimated 1.1 million of those cases resulting in death (Kotloff et al. 2013). For 3 years, the Global Enteric Multicenter Study (GEMS) identified pathogens, such as Shigella and pathogenic Escherichia coli, believed to be a cause of moderate-to-severe diarrhea (MSD) in children aged 0–59 months in the endemic areas of sub-Saharan Africa and South Asia (Kotloff et al. 2013). GEMS was an age stratified, matched case-control study that demonstrated that Shigella were consistently in the top five of all cases of MSD in each of the age groups (Farag et al. 2013).
There are four species of Shigella: Shigella sonnei, S. flexneri, S. dysenteriae and Shigella boydii, each with their own global burdens and epidemiological profile (Livio et al. 2014). From the 1130 Shigella isolates collected during the 36 months of the GEMS, 5.4% (61/1130) were identified as S. boydii (Livio et al. 2014). While this is a proportionally small contribution to the overall observed cases of MSD compared to the other three Shigella species, S. boydii still makes up a significant component of the overall Shigella burden (Baker, Parkhill and Thomson 2015). By increasing the number and diversity of S. boydii genomes available to the scientific community, further functional studies can focused on this understudied and underreported pathogen.
Considering the burden of disease caused by Shigella, there are relatively few Shigella genomic studies (Wei et al. 2003; Yang et al. 2005); however, recent studies on S. sonnei (Holt et al. 2012, 2013) and S. flexneri (Connor et al. 2015) genomics have detailed the temporal and spatial virulence of these species. The 28 S. boydii isolates sequenced in this study represent all of the S. boydii isolates identified at the Bangladesh and The Gambia GEMS sites during the first 24 months. A total of 31 S. boydii isolates were identified at these two sites over the complete GEMS 36 month period (24 in Bangladesh and 7 in The Gambia; Livio et al. 2014), thus we have examined the majority of the isolates from these two sites. These 28 S. boydii genomes from GEMS were compared with 14 S. boydii isolates already in the public domain, labeled in this study as ‘historical isolates’ (Table 1). By presenting these genomes along with corresponding clinical and phylogenomic data, this study will begin to shed light on this pathogen and allow a deeper understanding of the role of this organism in the broader context of global Shigella infections.
Table 1.
Shigella boydii isolates examined.
| Isolate | Shigella boydii | Number of | Total | GenBank | Short read | ||
|---|---|---|---|---|---|---|---|
| name | Origin | Year | phylogenetic clade | contigs | bp | accession | archive |
| 3083-94 | Arizona, USA | 1994 | 2 | 6 | 4874659 | NC_010658 | NDa |
| SB_3594-74 | Colorado, USA | 1974 | 3 | 96 | 4634068 | AFGC00000000 | SRA020641.2 |
| SB_965-58 | Minnesota, USA | 1958 | 1 | 96 | 5184598 | AKNA00000000 | SRA020850.2 |
| 248-1B | Chile | 1995 | 3 | 166 | 4788006 | AMKG00000000 | NDa |
| SB_08_0009 | British Columbia, Canada | 2008 | 3 | 165 | 4864228 | AMJZ00000000 | NDa |
| SB_08_0280 | Ontario, Canada | 2008 | 2 | 124 | 4835559 | AMKA00000000 | NDa |
| SB_08_2671 | Manitoba, Canada | 2008 | 3 | 185 | 4817878 | AMKB00000000 | NDa |
| SB_08_2675 | Alberta, Canada | 2008 | 2 | 335 | 4832830 | AMKC00000000 | NDa |
| SB_08_6341 | Ontario, Canada | 2008 | 2 | 138 | 4800746 | AMKD00000000 | NDa |
| SB_09_0344 | British Columbia, Canada | 2008 | 2 | 174 | 4821210 | AMKE00000000 | NDa |
| SB_4444-74 | Idaho, USA | 1974 | 3 | 314 | 4976495 | AKNB00000000 | SRS270182 |
| SB_5216-82 | Bulgaria | 1963 | 1 | 75 | 4882454 | AFGE00000000 | SRA020642.2 |
| SB_S6614 | Kenya | 2005 | 3 | 479 | 4610666 | AMJU00000000 | NDa |
| SB_S7334 | Kenya | 2007 | 3 | 249 | 4711626 | AMJX00000000 | NDa |
| 100705 | The Gambia | 2009 | 3 | 404 | 4361489 | LSCP00000000 | SRP072004 |
| 100706 | The Gambia | 2008 | 1 | 336 | 4475997 | LPSY00000000 | SRP072003 |
| 102252 | The Gambia | 2008 | 2 | 455 | 4380029 | LPSX00000000 | SRP072001 |
| 102265 | The Gambia | 2009 | 3 | 468 | 4547444 | LPSW00000000 | SRP072000 |
| 102309 | The Gambia | 2009 | 3 | 429 | 4384003 | LPSV00000000 | SRP072009 |
| 600080 | Bangladesh | 2008 | 1 | 358 | 4263951 | LSCB00000000 | SRP071936 |
| 600266 | Bangladesh | 2008 | 2 | 801 | 4158234 | LPTT00000000 | SRP071943 |
| 600375 | Bangladesh | 2008 | 2 | 473 | 4386200 | LPTS00000000 | SRP071984 |
| 600384 | Bangladesh | 2008 | 1 | 305 | 4367268 | LPTR00000000 | SRP071983 |
| 600657 | Bangladesh | 2008 | 2 | 467 | 4322758 | LSCA00000000 | SRP071982 |
| 600690 | Bangladesh | 2008 | 1 | 300 | 4742925 | LPTQ00000000 | SRP071994 |
| 600710 | Bangladesh | 2008 | 2 | 463 | 4333602 | LPTP00000000 | SRP071993 |
| 600746 | Bangladesh | 2009 | 1 | 386 | 4539801 | LPTO00000000 | SRP071992 |
| 601143 | Bangladesh | 2009 | 2 | 461 | 4291898 | LPTN00000000 | SRP071991 |
| 601276 | Bangladesh | 2009 | 2 | 472 | 4318093 | LPTM00000000 | SRP071985 |
| 601294 | Bangladesh | 2009 | 3 | 442 | 4353514 | LPTL00000000 | SRP071989 |
| 602068 | Bangladesh | 2009 | 3 | 432 | 4354983 | LPTK00000000 | SRP071988 |
| 602144 | Bangladesh | 2009 | 2 | 689 | 4246468 | LPTJ00000000 | SRP071986 |
| 602339_II | Bangladesh | 2009 | 3 | 413 | 4474771 | LPTI00000000 | SRP071999 |
| 602385 | Bangladesh | 2009 | 2 | 553 | 4292491 | LPTH00000000 | SRP071998 |
| 602404 | Bangladesh | 2009 | 2 | 471 | 4297951 | LPTG00000000 | SRP071997 |
| 602573 | Bangladesh | 2009 | 3 | 545 | 4500753 | LPTF00000000 | SRP071996 |
| 602682 | Bangladesh | 2009 | 3 | 414 | 4484949 | LPTE00000000 | SRP071995 |
| 602988 | Bangladesh | 2009 | 2 | 897 | 4165605 | LPTD00000000 | SRP072008 |
| 603122 | Bangladesh | 2009 | 3 | 925 | 4329955 | LPTC00000000 | SRP072007 |
| 603150 | Bangladesh | 2009 | 3 | 416 | 4516839 | LPTB00000000 | SRP072006 |
| 603210 | Bangladesh | 2009 | 3 | 542 | 4474419 | LPTA00000000 | SRP072005 |
| 603233 | Bangladesh | 2009 | 1 | 495 | 4758784 | LPSZ00000000 | SRP072002 |
Primary sequence was not deposited in the SRA.
In total, 28 S. boydii genomes were identified for sequencing and analysis from the 727 Shigella isolates obtained from Bangladesh and the Gambia during the GEMS (Kotloff et al. 2013; Livio et al. 2014). Multiple studies on the GEMS including the interpretation of the clinical, epidemiologic and microbial findings have been published (Farag et al. 2013; Lindsay et al. 2013; Baker, Parkhill and Thomson 2015; Sahl et al. 2015), and the genomic studies are now underway. In the current study, we have included 14 S. boydii genomes that had been published previously (Rasko et al. 2008; Sahl et al. 2015), including the first S. boydii genome that was in the public domain (strain BS512, (aka CDC 3083–94); serotype 18, Assembly Accession number NC_010658). For all GEMS isolates, genomic DNA was prepared as previously described with established methods (Sahl et al. 2011) taking precautions to minimize the passage number.
The genome sequence of each GEMS isolate was generated at the Institute for Genome Sciences, Genome Resource Center on the Illumina HiSeq2000 using paired-end libraries with 300 bp inserts. The draft genomes were assembled using Minimus (Sommer et al. 2007) to merge contigs generated using two different assemblers, Velvet assembly program (Zerbino and Birney 2008) (with kmer values determined using VelvetOptimiser v2.1.4 (http://bioinformatics.net.au/software.velvetoptimiser.shtml)), and the Edena v3 assembler (Hernandez et al. 2008). The metrics for the resulting assemblies corresponding GenBank accession and SRA numbers are presented in Table 1. For the S. boydi 3083–94, genomic DNA for sequencing was isolated from a stock culture and two Sanger sequencing libraries were constructed —a small insert library (4–5 kb) and a large insert library (10–12 kb) from which 20 656 and 47 348 reads were sequenced, respectively. The CDC 3083–94 genome was assembled as previously described (Rasko et al. 2008).
The 72 E. coli and Shigella genomes were aligned using Mugsy (Angiuoli and Salzberg 2011), and homologous blocks were concatenated using the bx-python toolkit (https://bitbucket.org/james_taylor/bx-python). The columns that contained one or more gaps were removed using Mothur (Schloss et al. 2009). These concatenated regions from each genome were used to construct a maximum-likelihood phylogeny with 100 bootstrap replicates using RAxML v7.2.8 (Stamatakis 2006) and visualized using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) (Fig. 1). Additionally, the level of similarity of protein-encoding genes was compared between the 42 S. boydii genomes in this study using a large-scale BLAST score ratio (LS-BSR) analysis as previously described (Hazen et al. 2013; Sahl et al. 2013, 2014). Protein-encoding genes were predicted for each genome sequence using Prodigal (Hyatt et al. 2010). The genes were then combined into gene clusters using uclust (Edgar 2010), and the gene clusters were assigned using a stringent nucleotide identity threshold of ≥90%. The protein-encoding genes that were considered present, with significant similarity had BSR values ≥0.8, while those with BSR values <0.8 but ≥0.4 were considered to be present but divergent and <0.4 were considered absent. The predicted protein function of each gene cluster was determined using an ergatis-based (Orvis et al. 2010) in-house annotation pipeline (Galens et al. 2011).
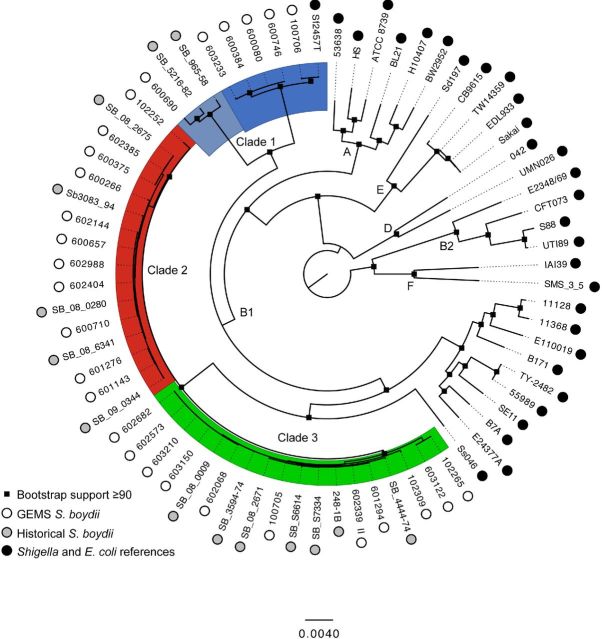
Figure 1.
Phylogenomic tree containing GEMS S. boydii isolates (white circles), S. boydii isolates that are currently in public databases (gray circles) and a collection of reference E. coli and Shigella species genomes (black circles). Three clades of S. boydii isolates are identified by color with clade 1 in blue (broken into two smaller subclades as denoted by the shades of blue), clade 2 in red and clade 3 in green. The tree was inferred with Figtree 1.4.2 with Bootstrap support values from 100 replicates are shown at the black square nodes. Distance for the number of nucleotide changes is shown to be 0.0040 with corresponding bar length.
The 42 S. boydii genomes from the 28 GEMS isolates and 14 historical isolates represent a collection of isolates that are geographically and temporally distributed (Table 1). The average genome size of the 28 GEMS S. boydii isolates examined is 4397 328 bp (range 4158 234–4758 784 bp). The average number of contigs in this collection is 493 (range 300–801 contigs), and the average GC percent is 50.75% (range 50.36%–51.19%). These values are typical of previously generated Shigella species genomes (Holt et al. 2012, 2013; Sangal et al. 2013; Connor et al. 2015; Sahl et al. 2015).
The phylogenomic analysis of the genomes was completed using the Mugsy algorithm (Angiuoli and Salzberg 2011). The alignment utilized in this reference-independent comparison contains ∼3.0 Mb which was a greater amount of genomic content than previous Shigella comparisons with this method (Sahl et al. 2015), suggesting a high level of conservation within the isolates of this species. The inferred phylogeny also identifies the S. boydii genomes as being separated from any of the E. coli reference genomes (Fig. 1). This E. coli and Shigella separation has been identified previously (Rasko et al. 2008; Tenaillon et al. 2010; Sahl et al. 2015). Additionally, phylogenomic analysis demonstrated that the S. boydii are separated into three clades (labeled clades 1, 2 and 3 in Fig. 1), with clade 1 potentially able to be further subdivided into two additional subgroups (Fig. 1). This data suggest that S. boydii clade 1 potentially diverged from clades 2 and 3 at an earlier point in the development of the S. boydii species (Fig. 1). This pattern is similarly observed in a global Shigella analyses recently published by our group (Sahl et al. 2015); however, the number of S. boydii isolates in that analyses were limited compared to our current study.
The comparisons included in this study are the greatest number of S. boydii compared in any one study, 42 isolates. Interestingly, the S. boydii isolates do not segregate by any specific geographic location or date of isolation, suggesting that this study has captured the genomic diversity of this species of Shigella. Both spatially and temporally, the new isolates from GEMS are distributed alongside the historic S. boydii isolates and are distributed between the three S. boydii clades (Table 1). This indicates that this GEMS collection of isolates has captured the diversity of the S. boydii species.
Analysis of the gene content via LS-BSR (Sahl et al. 2014) identified total of 7355 gene clusters in the 28 GEMS Shigella genomes. Among those gene clusters, a core S. boydii genome of 2477 gene clusters that are present in all S. boydii genomes examined. Comparing the gene clusters of the 28 GEMS genomes to the 14 previously sequenced S. boydii genomes identifies a core genome of 2230 genes that were present with significant similarity in the 42 of the S. boydii genomes in this study.
When the phylogenomic data from Fig. 1 is combined with the LS-BSR data, protein-encoding genes that are clade specific were identified. Annotation of these specific regions also provided potential insight into the gene function for the unique genes in these three clades. Unique genes are present in all of the isolates in the clade of interest, but lacking in the isolates from the other clades. Shigella boydii clade 1 had 98 unique genes compared to S. boydii clades 2 and 3, which had only 4 and 12 unique genes, respectively (Table 2; Table S2, Supporting Information). The clade-specific S. boydii genes included inner membrane components for a transport system and zinc-binding proteins from clade 1, several phage component proteins from clade 2 and two integrase family proteins from clade 3 (Table S2, Supporting Information). There were also several hypothetical proteins that were identified as clade specific: 13 from clade 1, 4 from clade 2 and zero from clade 3. Why and how these unique genes arose solely in S. boydii clade one creates another reason S. boydii requires further functional analysis.
Table 2.
Gene prevalence in S. boydii clades.
| LS-BSR | |||
|---|---|---|---|
| ≥0.8a | |||
| Unique gene clusters | |||
| Clades | Total isolates | Uniqueb | Prevalentb |
| Clade 1 | 8 | 98 | 128 |
| Clade 2 | 18 | 12 | 38 |
| Clade 3 | 16 | 4 | 56 |
| Total | 42 | 114 | 209 |
LS-BSR cutoff for gene clusters was greater or equal to 0.8 in selected clade and less than or equal to 0.4 in the other three clades
Unique clusters have 100% similarity in one clade and 0% in the other two clades: Prevalent gene clusters are present in >90% of one clade and present in <20% of the other two clades.
If the criteria for clade specificity are broadened to identify prevalent genes (i.e. genes present in >90% of isolates of one clade but in <20% of the isolates of the other two clades), these numbers increases to 128 genes in S. boydii clade 1, 56 genes in S. boydii clade 2 and 38 genes in S. boydii clade 3 (Table 2; Table S2, Supporting Information). The relatively larger increase observed in S. boydii clades 2 and 3 suggests that there is more overlap within the gene content of these clades when compared to the content of clade 1. This further suggests a distinct separation of the members of S. boydii clade 1 from other S. boydii. This increased gene repertoire contains transmembrane proteins in clade 1, phage-associated proteins in clade 2 (tail subunits, head-tail connectors and phage portal proteins) and a collection of phage and metabolism proteins in clade 3 (Table S2, Supporting Information). Similar approaches to the ones used have been utilized in the past to identify common features that are in use as PCR-based diagnostics for Shigella (Sahl et al. 2015) and E. coli (Hazen et al. 2013; Sahl et al. 2013, 2014).
Shigella is a pervasive human pathogen that causes life-threatening disease. Genomics of understudied pathogens like S. boydii will allow continued development in the fields of pathogen identification, phylogenetic categorization and potential functional characterization of the identified clade- and species-specific genomic regions.
Supplementary Material
SUPPLEMENTARY DATA
FUNDING
This project was funded in part by federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract number HHSN272200900009C, grant number 1U19AI090873 and startup funds from the State of Maryland.
Conflict of interest. None declared.
REFERENCES
- Angiuoli SV, Salzberg SL. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics. 2011;27:334–42. doi: 10.1093/bioinformatics/btq665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KS, Parkhill J, Thomson NR. Draft genome sequence of 24570, the type strain of Shigella flexneri. Genome Announc. 2015;3 doi: 10.1128/genomeA.00393-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor TR, Barker CR, Baker KS, et al. Species-wide whole genome sequencing reveals historical global spread and recent local persistence in Shigella flexneri. eLife. 2015;4:e07335. doi: 10.7554/eLife.07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Farag TH, Faruque AS, Wu Y, et al. Housefly population density correlates with shigellosis among children in Mirzapur, Bangladesh: a time series analysis. PLoS Neglect Trop D. 2013;7:e2280. doi: 10.1371/journal.pntd.0002280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galens K, Orvis J, Daugherty S, et al. The IGS standard operating procedure for automated prokaryotic annotation. Stand Genomic Sci. 2011;4:244–51. doi: 10.4056/sigs.1223234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen TH, Sahl JW, Fraser CM, et al. Refining the pathovar paradigm via phylogenomics of the attaching and effacing Escherichia coli. P Natl Acad Sci USA. 2013;110:12810–5. doi: 10.1073/pnas.1306836110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez D, Francois P, Farinelli L, et al. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res. 2008;18:802–9. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt KE, Baker S, Weill FX, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet. 2012;44:1056–9. doi: 10.1038/ng.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt KE, Thieu Nga TV, Thanh DP, et al. Tracking the establishment of local endemic populations of an emergent enteric pathogen. P Natl Acad Sci USA. 2013;110:17522–7. doi: 10.1073/pnas.1308632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–22. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Lindsay B, Ochieng JB, Ikumapayi UN, et al. Quantitative PCR for detection of Shigella improves ascertainment of Shigella burden in children with moderate-to-severe diarrhea in low-income countries. J Clin Microbiol. 2013;51:1740–6. doi: 10.1128/JCM.02713-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livio S, Strockbine NA, Panchalingam S, et al. Shigella isolates from the global enteric multicenter study inform vaccine development. Clin Infect Dis. 2014;59:933–41. doi: 10.1093/cid/ciu468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvis J, Crabtree J, Galens K, et al. Ergatis: a web interface and scalable software system for bioinformatics workflows. Bioinformatics. 2010;26:1488–92. doi: 10.1093/bioinformatics/btq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Rosovitz MJ, Myers GS, et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 2008;190:6881–93. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl JW, Caporaso JG, Rasko DA, et al. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ. 2014;2:e332. doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl JW, Gillece JD, Schupp JM, et al. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS One. 2013;8:e54287. doi: 10.1371/journal.pone.0054287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl JW, Lloyd AL, Redman JC, et al. Genomic characterization of asymptomatic Escherichia coli isolated from the neobladder. Microbiology. 2011;157:1088–102. doi: 10.1099/mic.0.043018-0. [DOI] [PubMed] [Google Scholar]
- Sahl JW, Morris CR, Emberger J, et al. Defining the phylogenomics of Shigella species: a pathway to diagnostics. J Clin Microbiol. 2015;53:951–60. doi: 10.1128/JCM.03527-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangal V, Holt KE, Yuan J, et al. Global phylogeny of Shigella sonnei strains from limited single nucleotide polymorphisms (SNPs) and development of a rapid and cost-effective SNP-typing scheme for strain identification by high-resolution melting analysis. J Clin Microbiol. 2013;51:303–5. doi: 10.1128/JCM.02238-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microb. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer DD, Delcher AL, Salzberg SL, et al. Minimus: a fast, lightweight genome assembler. BMC Bioinformatics. 2007;8:64. doi: 10.1186/1471-2105-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tenaillon O, Skurnik D, Picard B, et al. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207–17. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- Wei J, Goldberg MB, Burland V, et al. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun. 2003;71:2775–86. doi: 10.1128/IAI.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XF, Zhou L, Zheng J, et al. Construction and characterization of a live attenuated Shigella flexneri 2a vaccine strain, sf301 Delta virG and dsbA33G. Wei Sheng Wu Xue Bao. 2005;45:748–52. [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.