Abstract
The gut microbiota plays essential roles in human health and disease. In this review, we focus on the role of the intestinal microbiota in promoting resistance to infection by bacterial pathogens as well as how pathogens overcome this barrier. We discuss how the resident microbiota restricts growth and colonization of invading pathogens by limiting availability of nutrients and through generation of a hostile environment. Additionally, we examine how microbiota-derived signaling molecules interfere with bacterial virulence. In turn, we discuss how pathogens exploit non-competitive metabolites to replicate in vivo as well as to precisely control virulence and cause disease. This bacterial two step of creating and overcoming challenges important in preventing and establishing infection highlights the complexities of elucidating interactions between the commensal bacteria and pathogens. Better understanding of microbiota–pathogen interplay will have significant implications for developing novel therapeutics to treat infectious diseases.
Keywords: microbiota, pathogens, metabolites, signaling, virulence, competition
This review discusses how the microbiota functions as a barrier to invading pathogens as well as mechanism used by pathogens to avoid or exploit the microbiota to cause disease.
INTRODUCTION
The mammalian gastrointestinal (GI) tract is home to trillions of microbes collectively known as the microbiota that are essential for human health. The microbiota aids in nutrient uptake, vitamin production and in the development of the digestive and immune systems (Pédron and Sansonetti 2008; Round and Mazmanian 2009). Shortly after the emergence of antibiotics as treatment for bacterial infections, it became apparent that disturbances in the microbiota, or dysbiosis, resulted in susceptibility to bacterial infection. These findings suggested that the microbiota serves as a barrier against infection by pathogenic bacteria (Miller, Bohnhoff and Rifkind 1956). Indeed, the resident microbiota employs diverse mechanisms to promote resistance against bacterial pathogens; however, as in any arms race, pathogens evolved strategies to overcome these protective mechanisms and successfully establish infection. In this review, we discuss ways that the resident microbiota promotes resistance to and combats bacterial pathogens as well as how pathogens evade and exploit the microbiota.
Colonization resistance—more than a numbers game
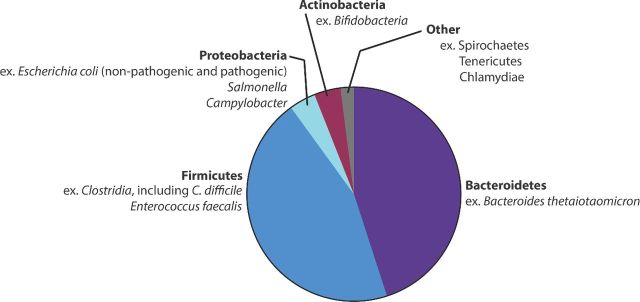
The microbiota directly provides protection against infection by invading pathogens by limiting access to nutrients (described below) as well as indirectly by bolstering host innate and adaptive immune responses (Macpherson and Uhr 2004; Duan et al. 2010; Lathrop et al. 2011; Chung et al. 2012; Hand et al. 2012; Olszak et al. 2012; Wingender et al. 2012; Diehl et al. 2013; Farache et al. 2013). This process has been termed colonization resistance (van der Waaij, Berghuis-de Vries and Lekkerkerk-van der Wees 1971). The microbiota is comprised of between 500 and 1000 different species of bacteria, the majority of which are localized to the large intestine (Savage 1977; Turnbaugh et al. 2010). Although the exact ratio of bacteria belonging to specific phyla varies among individuals, species belonging to the phyla Bacteroidetes and the Firmicutes are the most predominant with members of other phyla present in lower numbers (Eckburg et al. 2005; Andersson et al. 2008; Arumugam et al. 2011; Dominianni et al. 2015; Singh et al. 2015) (Fig. 1). This review focuses mainly on the roles of the Bacteroidetes, Firmicutes and Proteobacteria in enteric infections (for an in-depth review of the microbial diversity of the gut, we refer readers to Lozupone et al. 2012). Disturbances in the microbiota are associated with GI infections (Goldberg et al. 2014; Ling et al. 2014; Singh et al. 2015; Zhang et al. 2015; Gu et al. 2016; Kampmann et al. 2016). For example, mice colonized by a low-complexity microbiota (LCM) are more susceptible to Salmonella enterica serovar Typhimurium (Salmonella) infection compared to mice colonized by a normal microbiota. To confirm that susceptibility to infection was a result of the LCM, the authors reintroduced the normal microbiota to the LCM mice, which restored colonization resistance (Stecher et al. 2010). Additionally, the microbiota not only limits susceptibility to Salmonella, but also plays an additional role in mediating clearance of Salmonella and limiting infection (Endt et al. 2010). Moreover, the drug Metformin, used for treatment of diabetes, leads to increases in the proportion of Bacteroidetes and Firmicutes in patients, and is associated with a protective effect against the development of Clostridium difficile infections (CDI) (Eliakim-Raz et al. 2015). Overall, these studies suggest that a diverse population of microbes is necessary for protection against pathogens (Fig. 2).
Figure 1.
Relative proportions of the microbiota in the gastrointestinal tract. Bacteroidetes and Firmicutes are the most abundant phyla in the gut. Proteobacteria and Actinobacteria species are also frequently represented, but at lower numbers. Relative proportions are compiled as a summary of measurements, and do not represent exact numbers.
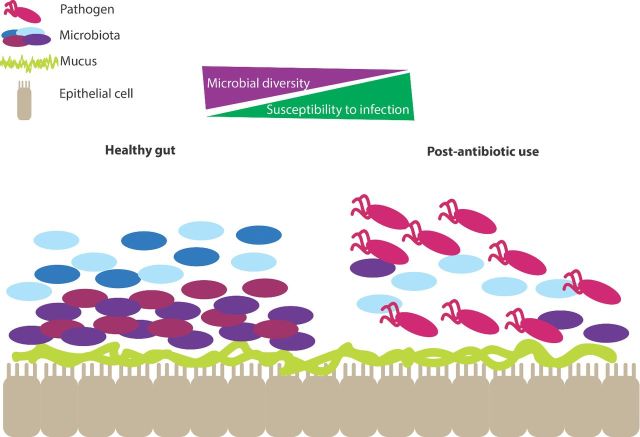
Figure 2.
Shifts in the composition of the microbiota allow for pathogen invasion. Diversity in the composition of the microbiota is protective against infection. A loss of diversity, such as that which occurs after antibiotic use, opens up a niche for pathogens to establish infection.
A key strategy to maintaining this diversity and consequent protective effects is based on the ability of each member of the microbiota to efficiently and specifically metabolize a limited repertoire of nutrients (Sperandio 2012). For example, anaerobic bacteria encode enzymes to break down polysaccharides present in the intestinal mucus and/or derived from the host. Commensal species of Bacteroides thetaiotaomicron or B. vulgatus, as well as commensal Escherichia coli, are better adapted to using these monosaccharides in the gut compared to intestinal pathogens such as enterohemorrhagic E. coli (EHEC), Salmonella or Shigella (Freter and Abrams 1972; Hudault, Guignot and Servin 2001; Miranda et al. 2004; Kamada et al. 2012). These invading pathogens are basically starved and unable to establish a foothold in the gut due to their poor efficiency in competing for these nutrients. Therefore, a diverse microbiota competing for a greater portion of the available nutrients restricts the ability of pathogens to replicate within a host.
Creating a hostile environment
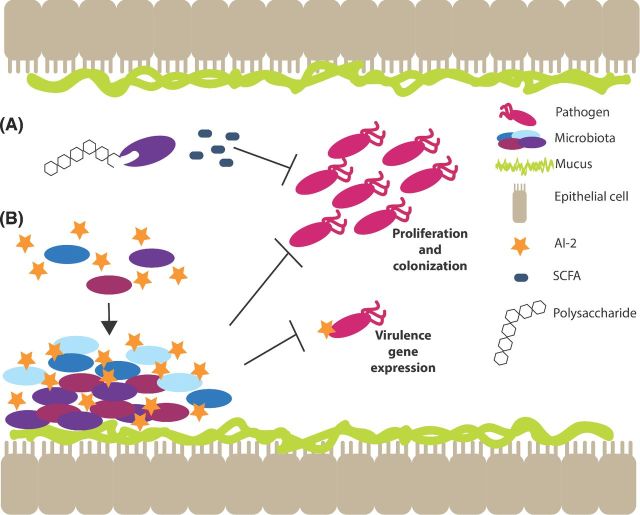
The microbiota can influence environmental conditions within the intestine, which consequently limits growth of invading pathogens. For example, microbiota-derived metabolites such as short chain fatty acids (SCFAs) provide a mechanism of resistance. The anaerobic members of the microbiota ferment polysaccharides, resulting in the production of SCFAs including butyrate, propionate and acetate (Tan et al. 2014). SFCAs have been shown to reduce disease severity associated with several enteric pathogens. Specifically, SCFAs inhibit the growth of EHEC, particularly at acidic pH and during anaerobic growth conditions (Shin, Suzuki and Morishita 2002). This is likely a result of the accumulation of SCFAs in the bacterial cytoplasm, which can be toxic especially at lower pH (Sun and O'Riordan 2013). Additionally, after infection with Shigella, rabbits given colonic infusions with a mixture of SFCAs (acetate, propionate, n-butyrate; 60:30:40 mM) displayed improved clinical symptoms and a correlating decrease of Shigella compared to the untreated control group (Rabbani et al. 1999), suggesting that SFCAs function to limit colonization by Shigella (Fig. 3A).
Figure 3.
Microbiota-derived molecules prevent colonization by pathogens. The microbiota produces molecules that contribute to colonization resistance. (A) Members of the microbiota consume complex polysaccharides and produce SCFAs, which prevent proliferation of and colonization by pathogens. (B) The QS molecule, autoinducer-2 (AI-2), helps the microbiota colonize the gut, which also prevents colonization by pathogens. Additionally, AI-2 negatively affects virulence gene regulation.
Secondary bile acids also contribute to colonization resistance. Bile acids are synthesized in the liver and are important for the metabolism of dietary lipids (Ridlon, Kang and Hylemon 2006). These primary bile acids may be resorbed in the small intestine or further metabolized by members of the microbiota to secondary bile acids (Ridlon, Kang and Hylemon 2006). Physiologically relevant concentrations of secondary bile acids inhibit C. difficile growth and spore germination during murine infection (Theriot, Bowman and Young 2016). Furthermore, members of the Firmicutes, specifically the Lachnospiraceae and Ruminococcaceae families as well as C. scindens, correlate positively with secondary bile acids generated within the intestine and resistance to C. difficile (Buffie et al. 2015; Theriot, Bowman and Young 2016). Studies that pinpoint beneficial attributes of particular members of the microbiota in promoting resistance to infection are necessary, as different strains of bacteria can provide varying degrees of resistance (Fukuda et al. 2011). These studies also indicate that bacterial-generated metabolites may have potential therapeutic applications for treatment of CDI and possibly other enteric infections.
In addition to creating an inhospitable environment for pathogens, the microbiota is armed to deploy direct assaults on invading pathogens. Commensal bacteria produce bacteriocins, which are small, ribosomally synthesized peptides that are active against other bacteria and against which the producer has a specific immunity mechanism (Cotter, Hill and Ross 2005). Bacteriocins were discovered nearly 100 years ago (Gratia 1925) and were presumed to function by eliminating competition during bacterial culture. Bacteriocins have been reported to inhibit important intestinal pathogens (Hechard and Sahl 2002; Snelling 2005; Kirkup 2006; Gillor, Etzion and Riley 2008). These data support the idea that bacteriocins influence the composition of the microbiota by providing a competitive advantage to bacteria that produce these molecules. The Firmicutes are the main producers of bacteriocins, and bacteria belonging to the Proteobacteria, Bacteroidetes and Actinobacteria also encode multiple bacteriocins (Drissi et al. 2015). Several bacteriocin-producing commensal bacteria are used as probiotics to bolster intestinal health (i.e. Bifidobacterium sp. and Lactobacillus sp.); however, for many of these strains and associated bacteriocins, it is still unclear whether the production of bacteriocins per se provides the probiotic properties of these bacteria (Martinez et al. 2013). Enterococcus sp. are common members of the microbiota; however, some strains are able to cause disease by translocating to deeper tissues and to the bloodstream (Buffie and Pamer 2013). Many enterococci carry conjugative plasmids that encode bacteriocins (Fujimoto et al. 1995). Recent evidence demonstrated Enterococcus faecalis carrying the conjugative plasmid pPD1, which expresses bacteriocin 21 (Fujimoto et al. 1995), can eliminate antibiotic resistant enterococci from the GI tract of mice (Kommineni et al. 2015). These data may have important implications for preventing severe, systemic infections associated with opportunistic enterococci. These findings also provide proof of principle that the production of bacteriocins, specifically, contributes to the probiotic properties of Enterococcus, and demonstrate that bacteriocins effectively inhibit growth of pathogens within the complex environment of the intestine.
A second mechanism that the microbiota has in its arsenal to combat pathogens is a type VI secretion system (T6SS). Gram-negative bacteria encode T6SSs, which enable bacteria to translocate effectors, including phospholipases, peptidoglycan hydrolases, nucleases and membrane pore-forming proteins, directly into the periplasm of a target bacterium (Russell, Peterson and Mougous 2014). Recent studies have shown that many members of the Bacteroidetes encode T6SSs (Russell et al. 2014; Coyne, Roelofs and Comstock 2016). Significantly, these genes are expressed during mammalian infection and inhibit growth of intestinal bacteria, suggesting that along with bacteriocins, T6SSs contribute to colonization resistance and stability of key members of the microbiota (Russell et al. 2014). Overall, these studies demonstrate that the microbiota plays indirect and direct roles to limit pathogen growth.
Microbiota-derived signals limit virulence
Bacteria rely on chemical and nutrient signaling to coordinate gene expression, which allows for successful adaptation of distinct host niches (Kendall and Sperandio 2016). To date, most research examining the impact of these signaling pathways on bacterial/host interactions has focused on their roles in bacterial pathogenesis (see below). However, increasing evidence suggests that chemical and nutrient signaling contribute to the establishment and maintenance of the resident microbiota as well as control of virulence of invading pathogens. During quorum sensing (QS), bacteria regulate gene expression in a manner that reflects population density (Nealson, Platt and Hastings 1970; Nealson and Hastings 1979). Briefly, a bacterial cell produces and secretes a signaling molecule, called an autoinducer. As the density of a particular bacterial population increases, the concentration of the autoinducer similarly increases. When the autoinducer concentration reaches a critical threshold, the autoinducer diffuses back into the cell and activates or represses certain target genes (Kendall and Sperandio 2009). An important QS system relies on the autoinducer AI-2, which is synthesized by the LuxS enzyme (Schauder et al. 2001). Several commensal bacteria, including Bifidobacterium sp. and Lactobacillus sp., encode a luxS homolog and synthesize AI-2 (DeKeersmaecker and Vanderleyden 2003; Kleerebezem et al. 2003; Sun et al. 2004; Altermann et al. 2005). Recently, AI-2 was detected in fecal contents of mice, confirming that the microbiota produces AI-2 in the intestine (Hsiao et al. 2014). Moreover, LuxS and AI-2 enhance biofilm formation in Bifidobacterium in vitro (Sun et al. 2014). Additionally, a luxS mutation resulted in a significant fitness defect during murine and nematode competition assays (Christiaen et al. 2014), suggesting that AI-2-dependent signaling enhances colonization by commensal bacteria (Fig. 3B).
Recent in vivo studies further support a protective role of AI-2 against pathogens by helping to restore the normal composition of the microbiota following antibiotic treatment. Thompson et al. colonized mice with recombinant E. coli strains that reduced or increased AI-2 concentration in the intestine. Significantly, an increase in AI-2 concentrations resulted in re-expansion of the Firmicutes, which had been depleted following streptomycin treatment. Bioinformatic analyses revealed that more than 80% of genomes classified in the Firmicutes contained putative luxS homologs, suggesting that AI-2 signaling may function in a feedback loop that restores colonization of AI-2 producing bacteria following dysbiosis (Thompson et al. 2015).
Another study extended these findings and showed that AI-2 produced by a member of the microbiota was not only associated with restoration of a healthy microbiota following acute infection by Vibrio cholerae, but also that AI-2 signaling dampened V. cholerae virulence (Hsiao et al. 2014). Specifically, Hsiao et al. (2014) demonstrated that recovery from V. cholerae infection correlated with an increase of bacterial taxa that is similar to the pattern of accumulation of the gut microbiota in healthy Bangladeshi children. One of the species consistently present in fecal samples following V. cholerae infection was Ruminococcus obeum (which is a member of the Firmicutes; Lawson and Finegold 2015). Because the relative abundance of R. obeum was consistently increased after V. cholerae infection, the authors focused on the impact of this bacterium on V. cholerae pathogenesis. Ruminococcus obeum restricted V. cholerae colonization and reduced expression of V. cholerae virulence factors in co-colonized mice. Additionally, RNAseq data revealed that expression of a R. obeum luxS homolog increased in the response to V. cholerae colonization. To confirm that AI-2-dependent signaling by R. obeum influenced V. cholerae virulence, the authors co-infected mice with V. cholerae and E. coli expressing R. obeum luxS, which led to the reduction of V. cholerae colonization and virulence gene expression (Hsiao et al. 2014).
The signaling molecule indole also regulates bacterial virulence. Species of commensal bacteria including E. coli, B. ovatus and C. bifermentans produce indole (Smith and Macfarlane 1996; Lee, Jayaraman and Wood 2007). In vitro studies demonstrated that indole represses EHEC chemotaxis, motility, adherence to epithelial cells and virulence gene expression (Bansal et al. 2007). Indole has been detected in human fecal samples (Karlin et al. 1985), suggesting that production of indole by the microbiota may limit intestinal colonization by pathogens. Finally, other microbiota-derived signals may also influence virulence gene expression, as an unidentified soluble factor secreted by B. thetaiotaomicron limits Shiga toxin expression in EHEC (deSablet et al. 2009). These studies reveal that signaling molecules produced by the gut microbiota play dual roles in enhancing resistance to pathogens: signaling molecules stabilize the microbiota and suppress bacterial virulence (Fig. 3B). Altogether, the microbiota is able to directly restrict pathogen colonization by shaping the intestinal environment to physically restrict pathogen growth, by attacking and killing pathogens, as well as by modulating regulatory circuits important for virulence.
Pathogens fight back—exploiting and promoting dysbiosis
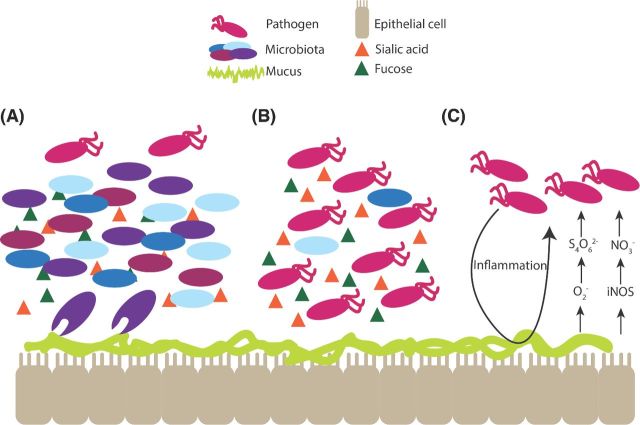
Despite the ability the microbiota to restrict pathogen invasion, pathogens have evolved mechanisms to overcome challenges posed by commensal bacteria. The microbiota remains relatively stable over time in healthy individuals, but changes in host environment, diet and use of antibiotics can cause reorganization of the community (Turnbaugh et al. 2009; David et al. 2014a,b), which in turn influences susceptibility to and severity of infections (Backhed et al. 2005; De Filippo et al. 2010; Lozupone et al. 2013) (Fig. 2). Several pathogens including C. difficile and Salmonella exploit dysbiosis to gain access to previously unavailable nutrients and expand in the perturbed intestine. For example, B. thetaiotaomicron cleaves host mucins, which produces monosaccharides such as fucose and sialic acid. In a healthy gut, commensal bacteria metabolize these sugars (Fig. 4A); however, after antibiotic treatment, C. difficile and Salmonella efficiently utilize these nutrients to establish infection (Ng et al. 2013) (Fig. 4B). B. thetaiotaomicron also produces succinate during the catabolism of dietary carbohydrates. Succinate is a fermentation intermediate that is metabolized to varying extents to SCFAs by cross-feeding species in the intestine (Bernalier, Dore and Durand 1999; Macfarlane and Macfarlane 2003). In general, succinate does not accumulate to significant levels in the gut; however, succinate levels increase during intestinal inflammation, such as during antibiotic-associated diarrhea and during CDI in humans and mice (Lawley et al. 2012). C. difficile takes advantage of the succinate accumulation during dysbiosis and couples the reduction of succinate to butyrate to expand in the intestine and cause disease (Ferreyra et al. 2014).
Figure 4.
Pathogens exploit microbiota-generated metabolites. Bacteroides thetaiotaomicron cleaves host mucin producing monosaccharides, including fucose and sialic acid. (A) Normally, the commensal bacteria consume these nutrients, preventing their consumption by pathogens. (B) If the microbiota is depleted or disturbed, such as after antibiotic use, pathogens are able to utilize these nutrients to establish infection. (C) Pathogens, such as Salmonella, take advantage of molecules generated by the host inflammatory response. Salmonella induces inflammation, which leads to the production of superoxide (O2−) and iNOS, which in turn lead to the formation of tetrathionate (S4O62−) and nitrate (NO3−), respectively. Salmonella utilizes tetrathionate and nitrate as electron acceptors during anaerobic respiration.
Pathogens also exploit host inflammation to generate electron acceptors important in anaerobic respiration. Antibiotics as well as bacterial virulence factors can induce an inflammatory response (Barman et al. 2008; Spees et al. 2013). For example, following streptomycin treatment, bacteria belonging to the Enterobactericeae, including E. coli and Salmonella, are able to use inflammation-generated electron acceptors for robust growth in the murine GI tract, which enables these bacteria to outgrow the obligate anaerobes, such as the Bacteroidetes and Firmicutes (Stecher et al. 2007; Winter et al. 2010, 2013; Lopez et al. 2012; Spees et al. 2013). Notably, Salmonella uses two type III secretions systems (T3SSs) and effectors encoded within the Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2) to directly induce host inflammation (Stecher et al. 2007; Winter et al. 2010; Lopez et al. 2012). Salmonella-induced inflammation results in iNOS production that is converted to nitrate, an energetically favorable electron acceptor (Lopez et al. 2012) (Fig. 4C). Furthermore, superoxide production by infiltrating neutrophils generates tetrathionate, which is respired in conjunction with the non-competitive metabolite ethanolamine to outgrow the microbiota (Thiennimitr et al. 2011) (Fig. 4C).
Finally, accumulating data suggests that the toxin B, produced by C. difficile, alters the intestinal milieu and shapes the microbiota to create a beneficial environment for infection. C. difficile toxin B inhibits Rho GTPase in cells lines, resulting in internalization of the Na+/H+ exchanger isoform 3 (NHE3) (Hayashi et al. 2004). In mice, inhibition of NHE3 results in chronic diarrhea, elevated Na+ and alkaline luminal fluid, which is similar to conditions used to grow C. difficile in vitro (Engevik et al. 2015). Inhibition of NHE3 also results in an altered microbiota composition with decreased members of the Firmicutes and increased numbers of Bacteroidetes. Recently, Engevik et al. (2015) linked these findings and showed that NHE3 expression was decreased in biopsy specimens from patients experiencing CDI compared to healthy individuals. Additionally, CDI stool had elevated Na+ and alkaline pH, in conjunction with elevated Bacteroidetes and Proteobacteria and decreased Firmicutes (Engevik et al. 2015). These findings indicate a model in which C. difficile toxins create an alkaline environment in the gut, leading to the proliferation of Bacteroidetes that do not compete with C. difficile, as well as a lower number of Firmicutes, particularly those of the Clostridial species, which share similar nutrient preferences with C. difficile. These changes in bacterial composition during infection allow for enhanced expansion of C. difficile. Altogether, these findings highlight that pathogens actively and directly modulate the intestinal environment to enhance growth.
If you can't beat ‘em, avoid them—sidestepping competition
Besides eliminating competition or taking advantage of a disturbed microbiota, pathogens can metabolize non-competitive metabolites and/or colonize host niches devoid of microbial competition. For example, although commensal E. coli and EHEC will preferentially metabolize overlapping sugars for growth during murine infection, EHEC will also use a distinct repertoire of sugars (Fabich et al. 2008). Specifically, EHEC uses galactose, heuronates, mannose and ribose, whereas commensal E. coli uses gluconate and N-acetylneuraminic acid (Fabich et al. 2008). Furthermore, before the onset of inflammation, Salmonella uses microbiota-derived hydrogen as an electron donor coupled to fumarate reduction to establish infection in the gut (Maier et al. 2013). After the onset of inflammation, Salmonella uses ethanolamine as an electron donor coupled to tetrathionate reduction to outgrow the microbiota (Thiennimitr et al. 2011). Ethanolamine is derived from the breakdown of phosphatidylethanolamine, an abundant lipid in cell membranes, during normal turnover of bacteria and epithelial cells (Garsin 2010). In addition to Salmonella, ethanolamine metabolism provides a growth advantage to EHEC, E. faecalis and Listeria monocytogenes during infection (Maadani et al. 2007; Bertin et al. 2011; Mellin et al. 2014).
Another mechanism that pathogens use to establish infection is to colonize a distinct niche within the GI tract free of nutrient competition (Sperandio 2012). The epithelial cell surface is covered by mucus that physically excludes bacteria from contacting these cells (Sellers and Morton 2014). The composition of mucus is complex and includes glycosylated proteins (mucin), monosaccharides, enzymes as well as antimicrobial peptides (Becker and Lowe 2003; Robbe et al. 2004). The mucus layer chemically and physically excludes bacteria from contacting epithelial cells (Sellers and Morton 2014). However, some pathogens encode virulence factors that enable penetration of the mucus layer and adhesion to enterocytes, which is devoid of competing, commensal bacteria. For example, EHEC uses a T3SS and effectors encoded within the locus of enterocyte effacement (LEE) to intimately attach to enterocytes and cause attaching and effacing lesions (AE lesions) (Jerse et al. 1990; Jarvis et al. 1995; McDaniel et al. 1995; Kenny et al. 1997). Using Citrobacter rodentium, a murine pathogen that models EHEC mammalian infection, Kamada et al. (2012) demonstrated that the EHEC T3SS was only required during infection of conventional mice, but not during infection of germ-free mice. In conventional mice, Ci. rodentium T3SS deletion strains remained in the lumen and were outcompeted by other Proteobacteria, such as commensal E. coli; however, these deletion strains of Ci. rodentium were able to colonize and grow in germ-free mice. In contrast, Ci. rodentium expressing the T3SS were able to colonize both germ-free and conventional mice and localized to the epithelium (Kamada et al. 2012). Therefore, the ability of pathogens to bypass the robust barrier posed by the microbiota is an effective strategy to grow and replicate during infection.
Honing in on microbiota-derived signals to control pathogenesis
Bacterial pathogens must precisely control expression of virulence traits to conserve energy, avoid detection from the immune system and to coordinate expression of factors important for adhesion versus dissemination. Some E. faecalis strains produce abundant biofilms, which enhances intestinal colonization (Creti et al. 2006) and leads to severe biofilm-related infections, such as bacteremia, endocarditis and implant infections (Hufnagel et al. 2004; Tendolkar et al. 2004; Nallapareddy et al. 2006; Paganelli et al. 2016). Enterococcus faecalis uses AI-2 as a signal to induce virulence gene expression, including genes that belong to prophage 5. Phage expression induces dispersal of biofilm, suggesting that this is a mechanism to promote dissemination (Rossmann et al. 2015). Additionally, phage release can result in lysogeny of non-phage carrying probiotic strains of E. faecalis. Phage transduction resulted in augmented pathogenesis, as lysogenized E. faecalis were more virulent in mouse sepsis and rat endocarditis models of infection (Rossmann et al. 2015). Although E. faecalis produces AI-2, it is not clear whether E. faecalis also respond to AI-2 produced by commensal bacteria to modulate pathogenesis. Regardless, this is a mechanism in which E. faecalis can detect neighboring bacteria that are present in high numbers, and thus increase the likelihood of productive infection of these nearby bacteria with released phages (Rossmann et al. 2015).
Significantly, metabolites can function as cues that enable pathogens to sense niches within a host and modulate expression of virulence genes, independently of their roles in promoting bacterial growth (Luzader and Kendall 2015). EHEC and Salmonella sense end products of B. thetaiotaomicron metabolism, including fucose, succinate and SCFAs as cues to modulate expression of virulence genes important for host colonization. B. thetaiotaomicron cleaves fucose from the host mucin, and EHEC encodes the two-component system FusKR that senses fucose. In this two-component system, FusK is the histidine kinase that senses fucose and initiates a signaling cascade through the response regulator FusR. FusR in turn directs expression of virulence and metabolism genes, including repression of genes encoding the EHEC T3SS (Pacheco et al. 2012a). Fucose is abundant in the lumen, and presumably at this site, fucose acts as a signal that enables EHEC to repress virulence gene expression in the lumen where it would be energetically wasteful (Fig. 5). Succinate is a major by-product of fermentation by Bacteroides species (Macy, Ljungdahl and Gottschalk 1978), and EHEC senses succinate through the transcription factor Cra to gauge gluconeogenic versus glycolytic conditions within the intestine and modulate virulence gene expression (Njoroge et al. 2012; Curtis et al. 2014) (Fig. 5). The role of succinate in EHEC virulence was demonstrated in vivo using Ci. rodentium. Mice infected with Ci. rodentium displayed more severe clinical manifestations when reconstituted with B. thetaiotaomicron compared to mice in which the normal microbiota was depleted. Disease severity correlated with increased concentrations of succinate in mice with B. thetaiotaomicron compared to mice in which B. thetaiotaomicron was absent (Curtis et al. 2014).
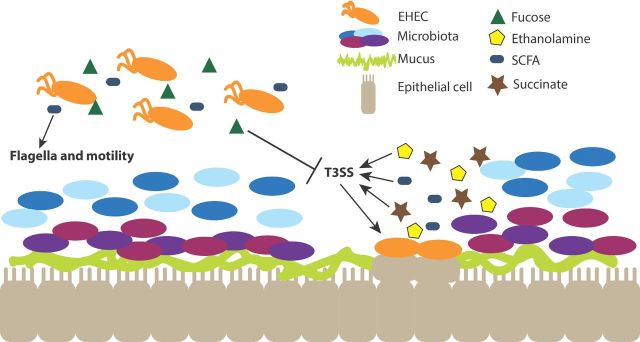
Figure 5.
Pathogens utilize host- and microbiota-derived molecules to regulate virulence gene expression. Pathogens sense different molecules within different niches of the GI tract (i.e. lumen and mucus layer), and either increase or decrease expression of virulence genes in response. For example, EHEC senses fucose in the lumen of the colon, which inhibits expression of the T3SS, preventing unnecessary expression of virulence genes in the wrong intestinal location. SCFAs in the lumen enhance expression of flagella and motility. In contrast, EHEC senses other metabolites near the epithelial layer, including succinate and the SCFA butyrate, which induce expression of the T3SS and the formation of AE lesions on host epithelial cells. Additionally, the metabolite ethanolamine, a component of host and bacterial cell membranes, induces expression of the T3SS.
SCFAs also provide information to bacterial pathogens concerning their location within the host. Concentrations of SCFAs vary throughout the GI tract, with the highest concentrations measured in the proximal colon (Tan et al. 2014). Pathogens exploit sensing SCFAs to differentially regulate gene expression. Salmonella senses acetate to promote expression of hilA and invF that encode regulators of SPI-1, a pathogenicity island required for invasion of epithelial cells (Durant, Corrier and Ricke 2000). Salmonella's response to acetate is due at least in part to the accumulation of acetate in the cytoplasm, likely as a result of increasing concentration of acetate in the distal ileum (Lawhon et al. 2002). Addition of propionate and butyrate to culture medium did not have significant effects on Salmonella virulence (Durant, Corrier and Ricke 2000; Lawhon et al. 2002). In EHEC, a mixture of SCFAs triggered expression of genes encoding flagella and motility (Tobe, Nakanishi and Sugimoto 2011); however, butyrate, specifically, enhanced LEE gene expression and adherence to epithelial cells (Nakanishi et al. 2009) (Fig. 5). The butyrate regulatory cascade is complex and involves several proteins. Upon sensing butyrate, the leucine-responsive regulatory (Lrp) protein initiates a signaling cascade that promotes expression of pchA (Nakanishi et al. 2009), which encodes a direct activator (PchA) of the LEE (Iyoda and Watanabe 2004; Abe et al. 2008). Additional studies demonstrated that Lrp directly regulates another transcription factor LeuO (Takao, Yen and Tobe 2014). Subsequently, LeuO binds the LEE promoter to activate gene expression and microcolony formation (Takao, Yen and Tobe 2014). LeuO activation of the LEE genes required PchA, and both PchA and Ler activated leuO expression. This positive feedback mechanism may prolong expression of the LEE (Takao, Yen and Tobe 2014), and thus enhance EHEC attachment to the host epithelium.
EHEC and Salmonella recognize ethanolamine as a signal to modulate virulence gene expression (Kendall et al. 2012; Luzader et al. 2013; Gonyar and Kendall 2014; Anderson et al. 2015) (Fig. 5). In EHEC, ethanolamine activates expression of genes important for colonization of the GI tract, including those encoding fimbrial adhesins, the T3SS encoded within the LEE pathogenicity island, and Shiga toxin (Kendall et al. 2012; Gonyar and Kendall 2014). In Salmonella, ethanolamine activates expression of the T3SS encoded within the Salmonella pathogenicity island 2, and thus augments Salmonella survival and replication within macrophages (Anderson et al. 2015). EHEC and Salmonella directly sense ethanolamine through the transcription factor EutR, which is encoded in the ethanolamine utilization (eut) operon (Roof and Roth 1992). Independently of its role in activating the eut operon, EutR directly activates transcription of virulence gene expression in EHEC and Salmonella (Luzader et al. 2013; Anderson et al. 2015). Interestingly, during murine infection, Salmonella EutR differentially regulates gene expression in response to distinct host environments. In the intestine, EutR promotes ethanolamine metabolism, enabling Salmonella to sidestep nutritional competition; however, in the spleen, EutR activates expression of SPI-2 and thus enhances systemic infection (Anderson et al. 2015). Ethanolamine is ubiquitous and constantly replenished in the host environment, suggesting that ethanolamine is a reliable indicator of the host environment. These studies highlight the complex roles of nutrients in bacterial pathogenesis, not only in promoting growth, but also serving as host recognition cues that enable proper spatiotemporal control of genes encoding colonization and virulence factors. Altogether, pathogens overcome the microbiota roadblock to infection by rapidly replicating during episodes of dysbiosis, scavenging metabolites and niches not readily consumed or inhabited by the microbiota, and exploit microbiota-derived signals, including metabolites, to drive virulence mechanisms.
CONCLUSIONS AND OUTLOOK
In this review, we highlighted how the resident microbiota functions to antagonize growth and virulence of invading pathogens as well as ways that pathogens overcome and exploit the microbiota to successfully establish infection. Clearly, maintaining diversity of the microbiota is important to prevent and limit disease. Indeed, manipulation of the commensal community as a means to prevent and treat intestinal infections is an area of active investigation. Strategies for this include the use of prebiotics, foods that support the growth of the resident microbiota, as well as of probiotics, which are live bacteria. Recent evidence supports a role for the use of probiotics as a treatment against antibiotic-resistant enterococci (Kommineni et al. 2015). However, in most cases, the mechanism(s) and efficacy of orally ingested probiotics are poorly understood (Martinez et al. 2013). In the context of CDI, the standard treatment of oral vancomycin is approximately 30% effective (van Nood et al. 2013). By comparison, fecal transplant has been shown to cure 80%–90% of recurrent infections (Kelly et al. 2015), suggesting that direct repopulation of the microbiota is effective in treating CDI. Although the utility of fecal transplants has been most extensively studied in the context of treatment for CDI, growing evidence suggests that fecal transplants prevent infections by other pathogens. For example, a Microbial Ecosystem Therapeutic (MET-1) composed of 33 bacteria cultured from a healthy human volunteer not only cured recurrent CDI in humans (Petrof et al. 2013), but was also protective against Salmonella infection in a murine model of infection (Martz et al. 2015). Moreover, mice that are normally susceptible to Ci. rodentium infection become resistant upon receiving a fecal transplant from mice that are resistant to Ci. rodentium colonization (Willing et al. 2011). Overall, these studies highlight the feasibility of utilizing live bacteria as a preventative measure or treatment for infectious diseases as an alternative to conventional therapeutics. Additionally, utilizing microbiota-derived metabolites or small molecules that enhance the colonization resistance could be beneficial in limiting pathogen infection. These metabolites could be generated in vitro and administered orally to a patient, or single bacterial strains could be engineered to optimally produce pathogen-limiting metabolites and used to treat infection (Sonnenburg and Fischbach 2011). Longer term studies are necessary to fully understand the efficacy and safety of using live bacteria or bacterial-derived molecules as therapeutics (Hoffmann et al. 2013; Olle 2013; Choi and Cho 2016).
Although the enhancement and manipulation of the resident microbiota as a means to prevent and treat infectious diseases shows promise, the possibility exists that this type of treatment may have adverse outcomes. Invading pathogens exploit the microbiota to regulate expression of virulence traits, and even as reservoirs for harboring toxin-encoding phages (Gamage et al. 2006). Although it would be ideal to develop a ‘one size fits all’ approach to treating infectious diseases, the reality is that all hosts and their corresponding microbiota, as well as pathogenic mechanisms of invading pathogens, differ. Therefore, novel therapeutics developed to harness resistance mechanisms of the microbiota will need to consider not only a patient's genetic make-up and lifestyle but also incorporate the identification and knowledge of virulence mechanisms of the invading pathogen to effectively and safely treat infectious diseases.
Acknowledgments
We apologize to the numerous investigators whose work could not be cited due to space constraints.
FUNDING
Work in the Kendall lab is supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (AI118732); ESM also received NIH training grant support (5T32AI007046).
Conflict of interest. None declared.
REFERENCES
- Abe A, Miyahara A, Oshima T, et al. Global regulation by horizontally transferred regulators establishes the pathogenicity of Escherichia coli. DNA Res. 2008;15:25–38. doi: 10.1093/dnares/dsm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altermann E, Russell WM, Azcarate-Peril MA, et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. P Natl Acad Sci USA. 2005;102:3906–12. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CJ, Clark DE, Adli M, et al. Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLoS Pathog. 2015;11:e1005278. doi: 10.1371/journal.ppat.1005278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AF, Lindberg M, Jakobsson H, et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:1–8. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Ley RE, Sonnenburg JL, et al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- Bansal T, Englert D, Lee J, et al. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun. 2007;75:4597–607. doi: 10.1128/IAI.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman M, Unold D, Shifley K, et al. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–15. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker DJ, Lowe JB. Fucose: biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi: 10.1093/glycob/cwg054. [DOI] [PubMed] [Google Scholar]
- Bernalier A, Dore J, Durand M. Colonic microbiota, nutrition and health. In: Gibson G, Roberfroid MB, editors. Biochemistry of Fermentation. The Netherlands: Kluwer Academic Publishers; 1999. [Google Scholar]
- Bertin Y, Girardeau JP, Chaucheyras-Durand F, et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ Microbiol. 2011;13:365–77. doi: 10.1111/j.1462-2920.2010.02334.x. [DOI] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–8. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HH, Cho YS. Fecal Microbiota transplantation: current applications, effectiveness, and future perspectives. Clin Endosc. 2016;49:257–65. doi: 10.5946/ce.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen SE, O'Connell MM, Bottacini F, et al. Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PLoS One. 2014;9:e98111. doi: 10.1371/journal.pone.0098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–93. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3:777–88. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- Coyne MJ, Roelofs KG, Comstock LE. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics. 2016;17:58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creti R, Koch S, Fabretti F, et al. Enterococcal colonization of the gastro-intestinal tract: role of biofilm and environmental oligosaccharides. BMC Microbiol. 2006;6:60. doi: 10.1186/1471-2180-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MM, Hu Z, Klimko C, et al. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014;16:759–69. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Materna AC, Friedman J, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. P Natl Acad Sci USA. 2010;107:14691–6. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKeersmaecker SC, Vanderleyden J. Constraints on detection of autoinducer-2 (AI-2) signalling molecules using Vibrio harveyi as a reporter. Microbiology. 2003;149:1953–6. doi: 10.1099/mic.0.C0117-0. [DOI] [PubMed] [Google Scholar]
- deSablet T, Chassard C, Bernalier-Donadille A, et al. Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2009;77:783–90. doi: 10.1128/IAI.01048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl GE, Longman RS, Zhang JX, et al. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 2013;494:116–20. doi: 10.1038/nature11809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominianni C, Sinha R, Goedert JJ, et al. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One. 2015;10:1–14. doi: 10.1371/journal.pone.0124599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissi F, Buffet S, Raoult D, et al. Common occurrence of antibacterial agents in human intestinal microbiota. Front Microbiol. 2015;6:1–8. doi: 10.3389/fmicb.2015.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Chung H, Troy E, et al. Microbial colonization drives expansion of IL-1 receptor 1-expressing and IL-17-producing gamma/delta T cells. Cell Host Microbe. 2010;7:140–50. doi: 10.1016/j.chom.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant JA, Corrier DE, Ricke SC. Short-chain volatile fatty acids modulate the expression of the hilA and invF genes of Salmonella typhimurium. J Food Protect. 2000;63:573–8. doi: 10.4315/0362-028x-63.5.573. [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliakim-Raz N, Fishman G, Yahav D, et al. Predicting Clostridium difficile infection in diabetic patients and the effect of metformin therapy: a retrospective, case-control study. Eur J Clin Microbiol. 2015;34:1201–5. doi: 10.1007/s10096-015-2348-3. [DOI] [PubMed] [Google Scholar]
- Endt K, Stecher B, Chaffron S, et al. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 2010;6:e1001097. doi: 10.1371/journal.ppat.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engevik MA, Engevik KA, Yacyshyn MB, et al. Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am J Physiol-Gastr L. 2015;308:G497–509. doi: 10.1152/ajpgi.00090.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabich AJ, Jones SA, Chowdhury FZ, et al. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008;76:1143–52. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farache J, Koren I, Milo I, et al. Luminal bacteria recruit CD103+ dendritic cells into the intestinal epithelium to sample bacterial antigens for presentation. Immunity. 2013;38:581–95. doi: 10.1016/j.immuni.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreyra JA, Wu KJ, Hryckowian AJ, et al. Gut microbiota-produced succinate promotes C. difficile Infection after antibiotic treatment or motility disturbance. Cell Host Microbe. 2014;16:770–7. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter R, Abrams GD. Function of various intestinal bacteria in converting germfree mice to the normal state. Infect Immun. 1972;6:119–26. doi: 10.1128/iai.6.2.119-126.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S, Tomita H, Wakamatsu E, et al. Physical mapping of the conjugative bacteriocin plasmid pPD1 of Enterococcus faecalis and identification of the determinant related to the pheromone response. J Bacteriol. 1995;177:5574–81. doi: 10.1128/jb.177.19.5574-5581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Toh H, Hase K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–7. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- Gamage SD, Patton AK, Strasser JE, et al. Commensal bacteria influence Escherichia coli O157:H7 persistence and Shiga toxin production in the mouse intestine. Infect Immun. 2006;74:1977–83. doi: 10.1128/IAI.74.3.1977-1983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nature Rev Microbiol. 2010;8:290–5. doi: 10.1038/nrmicro2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillor O, Etzion A, Riley MA. The dual role of bacteriocins as anti- and probiotics. Appl Microbiol Biot. 2008;81:591–606. doi: 10.1007/s00253-008-1726-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg E, Amir I, Zafran M, et al. The correlation between Clostridium-difficile infection and human gut concentrations of Bacteroidetes phylum and clostridial species. Eur J Clin Microbiol. 2014;33:377–83. doi: 10.1007/s10096-013-1966-x. [DOI] [PubMed] [Google Scholar]
- Gonyar LA, Kendall MM. Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2014;82:193–201. doi: 10.1128/IAI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratia A. Sur un remarquable exemple d'antagonisme entre deux souches de colibacille. C R Soc Biol. 1925;93:1040–1. [Google Scholar]
- Gu S, Chen Y, Zhang X, et al. Identification of key taxa that favor intestinal colonization of Clostridium difficile in an adult Chinese population. Microbes Infect. 2016;18:30–8. doi: 10.1016/j.micinf.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Hand TW, Dos Santos LM, Bouladoux N, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–536. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Szaszi K, Coady-Osberg N, et al. Inhibition and redistribution of NHE3, the apical Na+/H+ exchanger, by Clostridium difficile toxin B. J Gen Physiol. 2004;123:491–504. doi: 10.1085/jgp.200308979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hechard Y, Sahl HG. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie. 2002;84:545–7. doi: 10.1016/s0300-9084(02)01417-7. [DOI] [PubMed] [Google Scholar]
- Hoffmann DE, Fraser CM, Palumbo FB, et al. Science and regulation. Probiotics: finding the right regulatory balance. Science. 2013;342:314–5. doi: 10.1126/science.1244656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao A, Ahmed AM, Subramanian S, et al. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–6. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudault S, Guignot J, Servin AL. Escherichia coli strains colonising the gastrointestinal tract protect germfree mice against Salmonella typhimurium infection. Gut. 2001;49:47–55. doi: 10.1136/gut.49.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel M, Koch S, Creti R, et al. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. J Infect Dis. 2004;189:420–30. doi: 10.1086/381150. [DOI] [PubMed] [Google Scholar]
- Iyoda S, Watanabe H. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology. 2004;150:2357–571. doi: 10.1099/mic.0.27100-0. [DOI] [PubMed] [Google Scholar]
- Jarvis KG, Giron JA, Jerse AE, et al. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. P Natl Acad Sci USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse AE, Yu J, Tall BD, et al. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. P Natl Acad Sci USA. 1990;87:7839–43. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Kim Y, Sham HP, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science. 2012;336:1325–9. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann C, Dicksved J, Engstrand L, et al. Composition of human faecal microbiota in resistance to Campylobacter infection. Clin Microbiol Infect. 2016;22:e1–8. doi: 10.1016/j.cmi.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Karlin DA, Mastromarino AJ, Jones RD, et al. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin. 1985;109:135–41. doi: 10.1007/BF00391888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CR, Kahn S, Kashyap P, et al. Update on fecal microbiota transplantation 2015: indications, methodologies, mechanisms, and outlook. Gastroenterology. 2015;149:223–37. doi: 10.1053/j.gastro.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall MM, Gruber CC, Parker CT, et al. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio. 2012;3:e00050–12. doi: 10.1128/mBio.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall MM, Sperandio V. Cell-to-cell signaling in Escherichia coli and Salmonella. In: Böck A, Curtiss R, III, Kaper JB, et al., editors. EcoSal-Escherichia coli and Salmonella: cellular and molecular biology. Washington, DC: ASM Press; 2009. [Google Scholar]
- Kendall MM, Sperandio V. What a dinner party! Mechanisms and functions of interkingdom signaling in host-pathogen associations. mBio. 2016;7:e01748–15. doi: 10.1128/mBio.01748-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny B, DeVinney R, Stein M, et al. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;14:511–20. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- Kirkup BCJ. Bacteriocins as oral and gastrointestinal antibiotics: theoretical considerations, applied research, and practical applications. Curr Med Chem. 2006;13:3335–50. doi: 10.2174/092986706778773068. [DOI] [PubMed] [Google Scholar]
- Kleerebezem M, Boekhorst J, van Kranenburg R, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. P Natl Acad Sci USA. 2003;100:1990–5. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommineni S, Bretl DJ, Lam V, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–22. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop SK, Bloom SM, Rao SM, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–4. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawhon SD, Maurer R, Suyemoto M, et al. Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol. 2002;46:1451–64. doi: 10.1046/j.1365-2958.2002.03268.x. [DOI] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson PA, Finegold SM. Reclassification of Ruminococcus obeum as Blautia obeum comb. nov. Int J Syst Evol Micr. 2015;65:789–93. doi: 10.1099/ijs.0.000015. [DOI] [PubMed] [Google Scholar]
- Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Z, Liu X, Jia X, et al. Impacts of infection with different toxigenic Clostridium difficile strains on faecal microbiota in children. Sci Rep. 2014;4:1–10. doi: 10.1038/srep07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CA, Winter SE, Rivera-Chávez F, et al. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. mBio. 2012;3:e00143–12. doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh J, Gonzalez A, et al. Meta-analyses of studies of the human microbiota. Genome Res. 2013;23:1704–14. doi: 10.1101/gr.151803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzader DH, Clark DE, Gonyar LA, et al. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol. 2013;195:4947–53. doi: 10.1128/JB.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzader DH, Kendall MM. Commensal ‘trail of bread crumbs’ provide pathogens with a map to the intestinal landscape. Curr Opin Microbiol. 2015;29:68–73. doi: 10.1016/j.mib.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maadani A, Fox KA, Mylonakis E, et al. Enterococcus faecalis mutations affecting virulence in Caenorhabditis elegans model host. Infect Immun. 2007;75:2634–7. doi: 10.1128/IAI.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel TK, Jarvis KG, Donnenberg MS, et al. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci. 1995;92:1664–8. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67–72. doi: 10.1079/PNS2002207. [DOI] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–5. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- Macy JM, Ljungdahl LG, Gottschalk G. Pathway of succinate and propionate formation in Bacteroides fragilis. J Bacteriol. 1978;134:84–91. doi: 10.1128/jb.134.1.84-91.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L, Vyas R, Cordova CD, et al. Microbiota-derived hydrogen fuels Salmonella typhimurium invasion of the gut ecosystem. Cell Host Microbe. 2013;14:641–51. doi: 10.1016/j.chom.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Martinez FAC, Balciunas EM, Converti A, et al. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv. 2013;31:482–8. doi: 10.1016/j.biotechadv.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Martz SL, McDonald JA, Sun J, et al. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci Rep. 2015;5:1–14. doi: 10.1038/srep16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellin JR, Koutero M, Dar D, et al. Riboswitches. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science. 2014;345:940–3. doi: 10.1126/science.1255083. [DOI] [PubMed] [Google Scholar]
- Miller CP, Bohnhoff M, Rifkind D. The effect of an antibiotic on the susceptibility of the mouse's intestinal tract to Salmonella infection. T Am Clin Climatol Assoc. 1956;68:51–5. [PMC free article] [PubMed] [Google Scholar]
- Miranda RL, Conway T, Leatham MP, et al. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect Immun. 2004;72:1666–76. doi: 10.1128/IAI.72.3.1666-1676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Tashiro K, Kuhara S, et al. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology. 2009;155:521–30. doi: 10.1099/mic.0.023499-0. [DOI] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Sillanpaa J, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116:2799–807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson KH, Hastings JW. Bacterial bioluminescence: its control and ecological significance. Microbiol Rev. 1979;43:496–518. doi: 10.1128/mr.43.4.496-518.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminsescent system. J Bacteriol. 1970;104:313–22. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–101. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njoroge JW, Nguyen Y, Curtis MM, et al. Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. mBio. 2012;3:e00280–12. doi: 10.1128/mBio.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olle B. Medicines from microbiota. Nat Biotechnol. 2013;31:309–15. doi: 10.1038/nbt.2548. [DOI] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco AR, Curtis MM, Ritchie JM, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–7. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganelli FL, Huebner J, Singh KV, et al. Genome-wide screening identifies PTS permease BepA to be involved in Enterococcus faecium endocarditis and biofilm formation. J Infect Dis. 2016 doi: 10.1093/infdis/jiw108. [DOI] [PubMed] [Google Scholar]
- Pédron T, Sansonetti P. Commensals, bacterial pathogens and intestinal inflammation: an intriguing ménage à trois. Cell Host Microbe. 2008;3:344–7. doi: 10.1016/j.chom.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Petrof EO, Gloor GB, Vanner SJ, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1:3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani GH, Albert MJ, Hamidur Rahman ASM, et al. Short-chain fatty acids improve clinical, pathologic, and microbiologic features of experimental shigellosis. J Infect Dis. 1999;179:390–7. doi: 10.1086/314584. [DOI] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–59. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- Robbe C, Capon C, Coddeville B, et al. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochemical J. 2004;384:307–16. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DM, Roth JR. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J Bacteriol. 1992;174:6634–43. doi: 10.1128/jb.174.20.6634-6643.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann FS, Racek T, Wobser D, et al. Phage-mediated dispersal of biofilm and distribution of bacterial virulence genes is induced by quorum sensing. PLoS Pathog. 2015;11:e1004653. doi: 10.1371/journal.ppat.1004653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–23. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12:137–48. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AB, Wexler AG, Harding BN, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16:227–36. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–33. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Schauder S, Shokat K, Surette MG, et al. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–76. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Sellers RS, Morton D. The colon: from banal to brillant. Toxicol Pathol. 2014;42:67–81. doi: 10.1177/0192623313505930. [DOI] [PubMed] [Google Scholar]
- Shin R, Suzuki M, Morishita Y. Influence of intestinal anaerobes and organic acids on the growth of enterohaemorrhagic Escherichia coli O157:H7. J Med Microbiol. 2002;51:201–6. doi: 10.1099/0022-1317-51-3-201. [DOI] [PubMed] [Google Scholar]
- Singh P, Teal TK, Marsh TL, et al. Intestinal microbial communities associated with acute enteric infections and disease recovery. Microbiome. 2015;3:1–12. doi: 10.1186/s40168-015-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EA, Macfarlane GT. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol. 1996;81:288–302. doi: 10.1111/j.1365-2672.1996.tb04331.x. [DOI] [PubMed] [Google Scholar]
- Snelling AM. Effects of probiotics on the gastrointestinal tract. Curr Opin Infect Dis. 2005;18:420–6. doi: 10.1097/01.qco.0000182103.32504.e3. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Fischbach MA. Community health care: therapeutic opportunities in the human microbiome. Sci Transl Med. 2011;3:1–6. doi: 10.1126/scitranslmed.3001626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spees AM, Wangdi T, Lopez CA, et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio. 2013;4:e00430–13. doi: 10.1128/mBio.00430-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V. Virulence or competition? Science. 2012;336:1238–9. doi: 10.1126/science.1223303. [DOI] [PubMed] [Google Scholar]
- Stecher B, Chaffron S, Käppeli R, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–89. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Daniel R, Wagner-Döbler I, et al. Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol Biol. 2004;4:36. doi: 10.1186/1471-2148-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, O'Riordan MXD. Chapter three - regulation of bacterial pathogenesis by intestinal short-chain fatty acids. In: Sariaslani S, Gadd GM, editors. Advances in Applied Microbiology. Vol. 85. San Diego, CA: Academic Press; 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Brancaccio VF, Yuan J, et al. Bifidobacteria exhibit LuxS-dependent autoinducer 2 activity and biofilm formation. PLoS One. 2014;9:e88260. doi: 10.1371/journal.pone.0088260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Yen H, Tobe T. LeuO enhances butyrate-induced virulence expression through a positive regulatory loop in enterohaemorrhagic Escherichia coli. Mol Microbiol. 2014;93:1302–13. doi: 10.1111/mmi.12737. [DOI] [PubMed] [Google Scholar]
- Tan J, McKenzie C, Potamitis M, et al. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9. [DOI] [PubMed] [Google Scholar]
- Tendolkar PM, Baghdayan AS, Gilmore MS, et al. Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect Immun. 2004;72:6032–9. doi: 10.1128/IAI.72.10.6032-6039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot CM, Bowman AA, Young VB. Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere. 2016;1:e00045–15. doi: 10.1128/mSphere.00045-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. P Natl Acad Sci USA. 2011;108:17480–5. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JA, Oliveira RA, Djukovic A, et al. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 2015;10:1861–71. doi: 10.1016/j.celrep.2015.02.049. [DOI] [PubMed] [Google Scholar]
- Tobe T, Nakanishi N, Sugimoto N. Activation of motility by sensing short-chain fatty acids via two steps in a flagellar gene regulatory cascade in enterohemorrhagic Escherichia coli. Infect Immun. 2011;79:1016–24. doi: 10.1128/IAI.00927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Quince C, Faith JJ, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. P Natl Acad Sci USA. 2010;107:7503–8. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Waaij D, Berghuis-de Vries JM, Lekkerkerk-van der Wees JEC. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J Hyg. 1971;69:405–11. doi: 10.1017/s0022172400021653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–15. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Willing BP, Vacharaksa A, Croxen M, et al. Altering host resistance to infections through microbial transplantation. PloS One. 2011;6:e26988. doi: 10.1371/journal.pone.0026988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender G, Stepniak D, Krebs P, et al. Intestinal microbes affect phenotypes and functions of invariant natural killer T cells in mice. Gastroenterology. 2012;143:418–28. doi: 10.1053/j.gastro.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–9. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–11. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Dong D, Jiang C, et al. Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization. Anaerobe. 2015;34:1–7. doi: 10.1016/j.anaerobe.2015.03.008. [DOI] [PubMed] [Google Scholar]