Abstract
TLR2 heterodimers with TLR1 or TLR6 recognize distinct pathogen-associated molecules such as tri- and di-acylated lipopeptides. The activated TLR2 heterodimers recruit Toll-IL-1R domain- (TIR-) containing adapter proteins, TIRAP and MyD88, through the receptor TIR domains. Molecular recognition mechanisms responsible for agonist-driven, TIR domain-mediated receptor–adapter interactions as well as the structure of resultant signaling complexes remain unknown. We previously reported that the cell-permeable peptide derived from helix D of TLR2 TIR (2R9) specifically binds TIRAP in vitro and in cells and thereby inhibits TIRAP-dependent TLR signaling. This study demonstrates that cell-permeable peptides from D helix of TLR1 or TLR6, peptides 1R9 and 6R9 respectively, inhibit signaling mediated by cognate TLR2 co-receptors. Interestingly, 1R9 and 6R9 bind different TLR2 adapters, as they selectively bind MyD88 and TIRAP TIR, respectively. Both peptides block the agonist-induced co-immunoprecipitation (co-IP) of TLR2 with TIRAP or MyD88, but not TLR2 co-IP with co-receptors. Our data suggest that D helices of TLR1 and TLR6 TIR domains are adapter recruitment sites in both co-receptors; yet the sites recruit different adapters. The D helix in TLR1 is the MyD88 docking site, whereas in TLR6 this site recruits TIRAP.
Keywords: TLR1, TLR6, TLR2, TIRAP, MyD88, TIR domain
This research studies molecular mechanisms of initiation of intracellular TLR signaling and reports an important difference in adapter recruitment to TLR1 and TLR6.
ABBREVIATIONS
- TLR:
Toll-like receptor
- TIR:
Toll/IL-1 receptor homology domain
- TIRAP:
TIR domain-containing adapter protein
- Mal:
MyD88 adapter-like
- P3C:
S-(2,3-bis(palmitoyloxy)-(2R,2S)-propyl)-N-palmitoyl-(R)-Cys-Ser-Lys4-OH
- P2C:
S-(2,3-bis(palmitoyloxy)-(2R,2S)-propyl)-(R)-Cys-Ser-Lys4-OH
- LPS:
Lipopolysaccharide
- eCFP:
Enhanced cyan fluorescent protein
- Antp:
Antennapedia homeodomain translocation sequence
- IP:
Immunoprecipitation
INTRODUCTION
Toll-like receptors (TLRs) recognize microbial and danger molecules and activate host innate immune responses (Akira and Takeda 2004). TLRs are comprised of an ectodomain responsible for recognition of pathogen-associated molecules, a trans-membrane region and cytoplasmic Toll/interleukin-1 receptor (TIR) domain that initiates intracellular signaling. TLR agonists induce homo- or hetero-dimerization of cytoplasmic TIR domains of two receptor molecules leading to recruitment of adapters (Jin et al.2007; Jin and Lee 2008; Kang et al.2009). Recruitment of adapter proteins activates several signaling cascades and results in activation of proinflammatory cytokine production (Gay et al.2014). TIR domains of receptors and adapters interact via diverse structural regions (Pawson and Nash 2003; Toshchakov and Vogel 2007). Therapeutic targeting of TIR domains is an important strategy to control excessive TLR activation and can be useful in treatment of many diseases (O'Neill, Bryant and Doyle 2009; Kawai and Akira 2011; Brandes et al.2013).
TLR2 recognizes ligands specific for Gram-positive bacteria, mycobacteria and fungi (Means et al.1999; Takeuchi et al.1999; Underhill et al.1999; Werts et al.2001). TLR2 utilizes TLR1 or TLR6 as co-receptors. TLR2/1 and TLR2/6 heterodimers are activated by tri- and di-acylated lipopeptides, respectively. The dimers utilize two TIR domain-containing adapter proteins, MyD88 and TIRAP (also called Mal) and activate NF-κB and MAP kinase cascades leading to production of proinflammatory cytokines (Ozinsky et al.2000). MyD88 is an adapter common to all TIR-containing receptors, except TLR3. TIRAP was found dispensable for TLR3, TLR5, TLR7 and TLR9 signaling (Medzhitov et al.1998; Horng et al.2002; Yamamoto et al.2002; Kawai and Akira 2010). However, recent studies have indicated that TIRAP significantly enhances the endosomal TLR7 or TLR9 signaling in some cell lineages, such as immortalized bone marrow-derived macrophages, and in vivo (Bonham et al.2014; Piao et al.2015).
This study uses decoy peptide libraries derived from TIR domains of TLR1 and TLR6 to study TIR domain interactions and finds that the fourth helix of TIR is an adapter recruitment site for both TLR2 co-receptors. The sites, however, bind different adapter molecules. In TLR1, the site binds MyD88, whereas the site in TLR6 binds TIRAP. Presented data provide new insights into molecular mechanisms of differential adapter recruitment in TLR signaling.
MATERIALS AND METHODS
Animals, cells and cell culture
Preparation of peritoneal macrophages was described previously (Toshchakov et al.2005). Agonists of TLR2/1, S-(2, 3-bis (palmitoyloxy)-(2R, 2S)-propyl)-N-palmitoyl-(R)-Cys-Ser-Lys4-OH (P3C), TLR2/6, S-(2,3-bis(palmitoyloxy)-(2R,2S)-propyl)-(R)-Cys-Ser-Lys4-OH (P2C) and TLR9, ODN1668, were purchased from InvivoGen (San Diego, CA). Escherichia coli K235 LPS was phenol-purified (Hirschfeld et al.2000). Recombinant murine TNF-α was purchased from Biolegend, Inc. (San Diego, CA).
Peptide design, synthesis and reconstitution
The peptide libraries of TLR1 and TLR6 were designed as previously described (Toshchakov et al.2011; Couture et al.2012; Piao, Vogel and Toshchakov 2013; Piao et al.2013, 2015). Each peptide contained a cell-penetrating segment of Antennapedia homeodomain (Antp) (RQIKIWFQNRRMKWKK) (Derossi et al.1994) placed at the N-terminus of the decoy sequence. Peptide sequences are provided in Table 1. The identical regions 1, 4, 6 and 7 in the TIR domains of TLR1 and TLR6 are designated as 16R1, 16R4, 16R6 and 16R7. Peptides of more than 95% purity were synthesized by AAPPTec (Louisville, KY) or GenScript (Piscataway, NJ), and were quantified by spectrophotometry (Pace et al.1995).
Table 1.
Sequences of TLR1 and TLR6 decoy peptides.
| Peptide name | Peptide sequence | Predominant structural region |
|---|---|---|
| 16R1 | HIPLEELQRNLQFH | Segment that precedes strand A |
| 1R2 | GHDSAWVKNELLPN | A helix |
| 6R2 | EHDSAWVKNELLPN | A helix |
| 1R3 | EKDDIQIC | AB loop |
| 6R3 | EKDDIRVC | AB loop |
| 16R4 | LHERNFVPGKSIVE | BB loop |
| 1R5 | NIINFIEKSYKS | B helix |
| 6R5 | NIINFIEKSYKA | B helix |
| 16R6 | PHFIQSEWCHYELY | C helix |
| 16R7 | FAHHNLFHEGSDNL | CD loop |
| 1R8 | LLAPIPQYSI | DD loop |
| 6R8 | LLEPILQNNI | DD loop |
| 1R9 | PTNYHKLKTLMSR | D helix |
| 6R9 | PSRYHKLRALMAQ | D helix |
| 1R10 | RTYLEWPTEKNKH | DE, EE loop and β-strand E |
| 1R1011 | NKHGLFWANLRAS | EE loop and E helix |
| 6R10 | RTYLEWPTEKGKR | DE, EE loop and β-strand E |
| 6R1011 | GKRGLFWANLRAS | EE loop and E helix |
| 16R10 | RTYLEWPTEK | DE loop and β-strand E |
| 1R11 | GLFWANLRASINV | E helix |
| 6R11 | GLFWANLRASFIM | E helix |
Expression vectors
TLR1 TIR-Cerulean (Cer) (Piao et al.2015), TLR6-Cer (Piao et al.2015), MyD88 TIR-Cer (Piao et al.2013) and TIRAP-Cer (Piao et al.2013) expression vectors were described previously. Full length TLR2 cDNA was amplified by PCR from mouse macrophage mRNA, and cloned into pCMV-Flag vector. Because of low expression level the full length TLR1-Cer vector was replaced with TLR1 TIR-Cer that does not encode the TLR1 ectodomain, but encodes the full transmembrane section and TIR domain of TLR1.
ELISA
A total of 1 × 106 peritoneal macrophages were plated in 12-well plates and treated with peptides for 30 min prior to stimulation. Mouse TNF-α or IL-6 levels were measured in supernatants using ELISA kits from Biolegend, Inc.
Immunoblotting and co-immunoprecipitation
HEK293T cells were transfected with indicated expression vectors using Superfect (Qiagen, CA). Cells were lysed 48 h after transfection in buffer containing 20 mM Hepes (pH 7.4), 150 mM NaCl, 10 mM NaF, 2 mM Na3VO4, 1 mM EDTA, 1 mM EGTA, 0.5% Triton X-100, 0.1 mM DTT, 1 mM PMSF and protease inhibitor cocktail (Roche, Indianapolis, IN). Antibodies against phospho-ERK, phospho-JNK, phospho-IKKα/β (Ser180/Ser181), MyD88, TIRAP and GAPDH were purchased from Cell Signaling Technology (Danvers, MA). Protein contents in cell extracts were quantified using protein quantification kit (Bio-Rad, Philadelphia, PA). Cell extracts were loaded into 10% acrylamide gels for SDS-PAGE. For co-immunoprecipitation (co-IP) assays, 500 μg total protein of cell extract was incubated with 1 μg of anti-mTLR2 antibody (T2.5) (Santa Cruz Biotechnology, Dallas, TX) overnight, followed by 4 h incubation with 25 μl protein G Agarose beads (Roche). The beads were then washed with lysis buffer and boiled in Laemmli sample buffer (Bio-Rad).
Peptide-protein co-IP assay
A total of 2 × 106 HEK293T cells were transfected with 10 μg of TLR2-Flag, TLR6-Cer construct or 1 μg of Cer-tagged TLR1 TIR, MyD88 TIR or TIRAP expression vectors. Cells were lysed 48 h after transfection. A total of 100 μg protein of cell lysates were incubated with or without peptides (20 μM) for 1 h at 4°C, followed by 3 h or overnight incubation with 0.5 μg of mouse anti-eCFP antibody (8A6) (Origene, Rockville, MD) or anti-Flag M2 antibody (Sigma-Aldrich, St. Louis, MO) and 4 h incubation with 25 μl protein G agarose beads. The washed beads were boiled in Laemmli sample buffer. Supernatants were spotted into PVDF membrane, followed by immunoblotting with rabbit anti-Antp Ab (Abcam, Cambridge, MA).
Quantitative real-time PCR
cDNA was synthesized from 1 μg of total RNA isolated with Nucleospin RNA II kits (Macherey-Nagel, Inc., Bethlehem, PA) using Goscript transcriptase (Promega, Madison, WI), and subjected to real-time PCR (RT-PCR) with gene-specific primers for HPRT, TNF-α or IL-6 (Couture et al.2012) using Fast SYBR®Green master mix (Applied Biosystems, Foster City, CA).
MTT viability assay
A total of 5 × 104 mouse peritoneal macrophages were plated into 96-well tissue culture plates, incubated overnight, and treated with peptides with or without TLR2 agonist for 2 or 5 h, followed by 3 h incubation with 0.5 mg ml−1 MTT (3-(4, 5-Dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide) (Sigma-Aldrich). A total of 50 μl DMSO was added to cells before reading OD at 540 nm.
Data representation
Numerical data are presented as means ±SEM. Significance of differences was evaluated using one-way ANOVA and Prizm 5 software. Asterisks mark data statistically different from corresponding controls with probability more than 99%.
RESULTS
Identification of TLR inhibitory peptides
Inhibition of TLR2 by peptides derived from TLR1 and TLR6 TIR domains was first evaluated in primary peritoneal macrophages based on peptide effects on cytokine transcription and secretion following stimulation of cells by P3C, a TLR2/1 agonist, or P2C, a TLR2/6 agonist. TNF-α and IL6 mRNA were measured 1 and 4 h, respectively, after stimulation because kinetics of activation of these genes by TLR agonists is different (DeForge and Remick 1991; Shcheblyakov et al.2011; Piao et al.2013). IL6 mRNA expression was low and barely detectable in macrophage cultures 1 h after TLR stimulation (not shown); whereas TNF-α is an ‘immediately activated’ gene, expression of which is strong and clearly detectable as early as 1 h after TLR stimulation. We included both time points in evaluation of cytokine expression to verify that both early and late TLR-induced cytokine expression is suppressed by peptides. 1R9, peptide derived from the D helix of TLR1 TIR, and 1R10, peptide from the region that includes DE loop, β-strand E and EE loop, inhibited the TLR2/1-activated transcription of TNF-α and IL-6 mRNA (Fig. 1A and B). Accordingly, both peptides inhibited TLR2/1-activated cytokine production when used at 40 μM, although the effect of peptides at 20 μM did not reach the level of statistical significance (Fig. 1D and E). Screening of the TLR6 TIR library produced similar results in that 6R9 and 6R10, which are structurally homologous to 1R9 and 1R10, inhibited TLR2/6-driven cytokine activation, whereas other peptides did not inhibit (Fig. 1F, G, I and J). Peptides derived from TLR1 or TLR6 were less potent inhibitors compared to 2R9, a previously reported TLR2-inhibitory peptide derived from the D helix of TLR2 TIR (Piao et al.2015) (Fig. 1). Ten N-terminal amino acids of 1R10 and 6R10 are identical. Interestingly, peptide 16R10 that represents the common sequence of 1R10 and 6R10 did not inhibit TLR2-mediated cytokine activation (Fig. 1C and H). This finding indicates that three N-terminal residues that are dissimilar in TIRs of TLR1 (763NKH) and TLR6 (768GKR) (these residues are shown underlined in peptide sequences of Table 1) are critical for inhibitory effect of corresponding peptides. We tested two additional peptides which include three residues that are dissimilar in 1R10 and 6R10 together with 10 subsequent C terminal residues of TLR1 and TLR6, respectively. These additional peptides (1R1011 and 6R1011) exhibited inhibitory activity comparable to that of peptides from regions 9 and 10 (Fig. 1C and H). TLR2 inhibitory peptides from TLR1 and TLR6 did not affect cell viability evaluated by MTT test (Fig. S1, Supporting Information).
Figure 1.
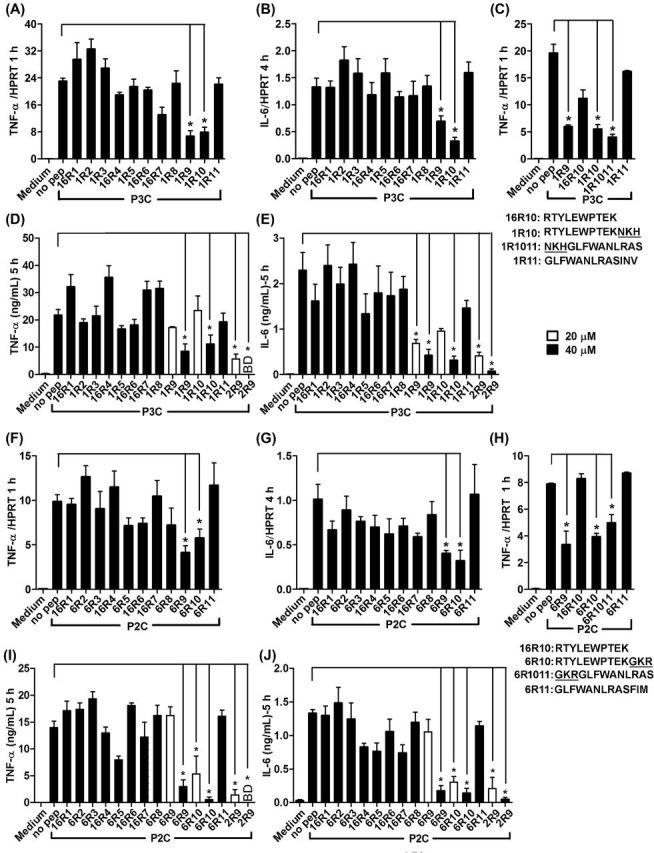
Peptides derived from D helix and region that includes DE loop, E strand and EE loop of TLR1 or TLR6 TIR inhibit TLR2-mediated macrophage activation. (A–J) Mouse peritoneal macrophages were treated with 20 μM or 40 μM of indicated peptides for 30 min prior to P3C (500 ng ml−1) (A–E) or P2C (50 ng ml−1) (F–J) stimulation. Cytokine gene transcription was measured by RT-PCR 1 h (TNF-α) (A, C, F and H) and 4 h (IL-6) (B and G) after stimulation. Secretion of TNF-α (D and I) and IL-6 (E and J) was measured by ELISA in supernatants collected 5 h after stimulation. Means ±SEM of more than three independent experiments are shown. * P < 0.01.
Specificity of signaling inhibition by TLR1 and TLR6-derived peptides
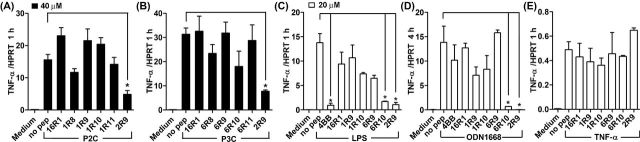
We next tested specificity of signaling inhibition by TLR1 and TLR6 peptides. TLR1 peptides were tested for inhibition of P2C-induced signaling and TLR6 peptides for P3C signaling inhibition. Experiments demonstrated that TLR1 inhibitory peptides do not inhibit TLR2/6-mediated cytokine activation even at the high dose of 40 μM (Fig. 2A). Analogously, TLR6 inhibitory peptides did not inhibit TLR2/1-mediated signaling (Fig. 2B).
Figure 2.
Specificity of signaling inhibition by TLR1- and TLR6-derived peptides. (A–E) Experimental conditions are same as in Fig. 1. Mouse peritoneal macrophages were treated with 40 μM (A, B) or 20 μM (C–E) of indicated peptides for 30 min prior to P2C (50 ng ml−1) (A), P3C (500 ng ml−1) (B), LPS (100 ng ml−1) (C), ODN1668 (3 μM) (D) or TNF-α (5 ng ml−1) (E) stimulation. TNF-α mRNA expression was measured 1 h after cell stimulation. Peptides 4BB (derived from TLR4 BB loop) and 2R9 (derived from TLR2 D helix) are included as additional specificity controls. Means ±SEM of more than three independent experiments are shown. * P < 0.01.
We also tested if TLR2-inhibitory peptides inhibit TLR4, TLR9 and TNF-α signaling. Only 6R10 and 2R9, but not other peptides, inhibited TLR4- or TLR9-mediated cytokine activation in macrophages at 20 μM (Fig. 2C and D). The tested peptides did not affect TNF-α expression induced by TNF-α (Fig. 2E).
Peptides derived from D helix of TLR1 or TLR6 TIRs bind TLR adapters, and prevent adapter recruitment to TLR2 receptor complex
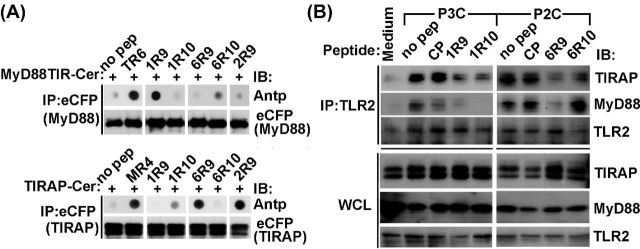
We used peptide-protein co-IP dot blot assay to identify the binding targets of inhibitory peptides. In this approach, tagged TIR domains are expressed in cells and then immunoprecipitated from cell lysates supplemented with decoy peptides (Piao et al.2013, 2015). Peptide contents in precipitates were measured by dot blot assay using antibody to Antennapedia translocating sequence (Antp). Obtained data suggested that 1R9 binds MyD88, whereas 6R9 binds TIRAP (Fig. 3A). The binding of 1R9 to MyD88 TIR domain is comparable to that of TR6, peptide from TIRAP C helix that binds MyD88 TIR (Couture et al.2012; Piao et al.2013). 6R9 binding to TIRAP was only slightly weaker than binding of TIRAP to 2R9, peptide from the D helix of TLR2 TIR. TLR2 inhibitory peptides diminished agonist-induced co-IP of TIRAP or MyD88 to TLR2 receptor complex in primary macrophages (Fig. 3B).
Figure 3.
Peptides representing the D helix and subsequent region that includes DE loop, E strand and EE loop of TLR1 or TLR6 selectively binds TLR adapters and prevent ligand-induced adapter recruitment to TLR2 receptor complex. (A) Peptide-protein co-IP. Lysates of HEK293T cells that express indicated Cerulean- (Cer-) fused proteins were incubated with 20 μM of indicated peptides for 1 h and immunoprecipitated with anti-eCFP Ab. Peptide contents in the precipitated immune complex were detected by dot blotting using anti-Antp antibody. Peptides TR6, derived from C helix of TIRAP TIR, and MR4, derived from BB loop of MyD88 TIR, were used as a positive binding control for MyD88 and TIRAP, respectively. 2R9 is TLR2 D helix peptide. CP is a scrambled control peptide. (B) Mouse peritoneal macrophages were pretreated with 20 μM of indicated peptides for 30 min and stimulated with P3C (500 ng ml−1) or P2C (50 ng ml−1) for 20 min. Cell lysates were immunoprecipitated with anti-TLR2 Ab and immune complexes assessed using anti-TIRAP or anti-MyD88 Ab. One representative of three independent experiments is shown in each panel.
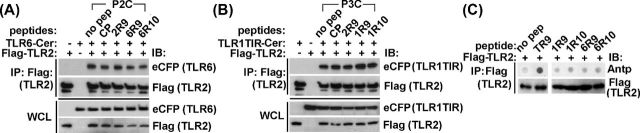
TLR1- and TLR6-derived TLR2 inhibitory peptides do not affect receptor heterodimerization
Inhibitory peptides were further examined for possible disruption of receptor dimerization. HEK293T cells were transfected with Flag-tagged TLR2 together with Cerulean-tagged whole length TLR6 or TLR1 TIR. Cells were treated with peptides 48 h after transfection for 30 min and then challenged with corresponding TLR2 agonist for 20 min. The receptor complex was immunoprecipitated with anti-Flag Ab, and TLR1 or TLR6 contents in precipitates were measured using anti-eCFP Ab. TLR2 associates with TLR1 or TLR6 in agonist-independent manner (Ozinsky et al.2000; Takeuchi et al.2002). TLR2 interaction with co-receptors was not affected by inhibitory peptides (Fig. 4A and B). In accordance with this finding, the tested peptides did not bind TLR2 in the dot blot assay (Fig. 4C).
Figure 4.
Peptides derived from the D helix or subsequent region that includes DE loop, E strand and EE loop of TIR of TLR1 or TLR6 do not block TLR2 co-IP with co-receptors. (A, B) HEK293T cells were transfected with Flag-tagged mouse TLR2 and Cer-tagged mouse TLR1 TIR or TLR6 proteins. Cells were pretreated for 30 min with 20 μM of indicated peptides prior to stimulation with P3C (500 ng ml−1) or P2C (50 ng ml−1) for 20 min. The cell lysates were immunoprecipitated with anti-Flag antibody and the immune complexes assessed using anti-eCFP Ab. CP is a scrambled control peptide. (C) Peptide-protein co-IP assay was performed as described in Fig. 3A. Cell lysates of HEK293T cells that express Flag-tagged mouse TLR2 were immunoprecipitated with anti-Flag Ab. TR9 is a peptide derived from D helix of TIRAP TIR (Couture et al.2012). One representative of three (panel A and B) or two (panel C) individual experiments is shown.
Thus, obtained data suggest that 1R9 and 6R9 directly bind MyD88 and TIRAP, respectively, and prevent adapter recruitment to activated receptors. 1R10 and 6R10, however, do not bind TIRAP, MyD88 or TLR2 TIR domains, so their targets remain unknown. In conclusion, D helices of TLR1 and TLR6 are adapter recruitment sites in both TLRs; the sites, however, recruit different TLR adapters.
DISCUSSION
TLR2 functions as a heterodimer with TLR1 or TLR6 (Ozinsky et al.2000; Takeuchi et al.2002). TLR2 activation by bacterial lipopeptides changes the conformation of the receptor heterodimers so that intracellular TIR domains of two receptors may contact directly. The intracellular TIR domains of TLR1 and TLR6 have 90% of sequence identity. Consequently, TLR2/TLR1 and TLR2/TLR6 heterodimers activate similar signaling cascades, leading to similar gene activation profiles in general (Farhat et al.2008); however, some differences in signaling and sensitivity to inhibitors have also been reported for different TLR2 heterodimers (Reschner et al.2003; Couture et al.2012; Liu et al.2012; Ma et al.2013; Mistry et al.2015). TLR2/1, but not TLR2/6 heterodimer induces CXCL10 secretion and dendritic cell maturation (Reschner et al.2003), while TLR2/6 induces a distinct set of chemokines (Reschner et al.2003) and macrophage membrane ruffling (Santos-Sierra et al.2009). Moreover, TLR2/1 and TLR2/6 agonists activate NF-κB, MAP kinases and cytokine transcription with different kinetics (Piao et al.2015) (Fig. S2, Supporting Information).
Ability of TLR2 to dimerize with different co-receptors not only broadens the spectrum of pathogens sensed by TLRs, but also diversifies downstream signaling. It is still unclear why TLR2 TIR homodimer does not induce signaling, whereas TLR4 homodimer does (Ozinsky et al.2000). Little is known about molecular details of functional interactions of TLR2 co-receptors with downstream signaling molecules. This study through screening of TLR1 and TLR6 TIR peptide libraries has revealed that two peptides in each library inhibit signaling mediated by corresponding receptors. Interestingly, inhibitory peptides represent structurally homologous regions of TLR1 and TLR6 (Fig. 1). 1R9 and 6R9, peptides derived from the fourth helix of TLR1 and TLR6 TIR respectively, suppressed adapter recruitment to activated TLR2 receptor complex. Interestingly, despite structural homology and some sequence similarity (1R9 and 6R9 have 7 identical amino acids of 13 total) these D helix peptides bind different adapters. 1R9 binds MyD88, whereas 6R9 targets TIRAP (Fig. 3). This finding suggests that the D helix of TLR1 TIR is the MyD88 binding site, whereas homologous helix of TLR6 is the site for TIRAP. Interestingly, 2R9, a TLR2-derived peptide, which potently inhibits several TLRs, is also derived from the D helix (Piao et al.2015). This peptide, similarly to 6R9, binds TIRAP and prevents TIRAP and MyD88 recruitment to TLR2 receptor complex (Piao et al.2015). The TIRAP binding affinity of 6R9 is apparently lower than that of 2R9 (Figs 1, 2 and 3A).
We analyzed available structural models of TLR1, TLR2 and TLR6 TIR domains (PDB ID 1FYV, 1FYW and 4OM7, respectively) to determine residues that form the side of the helix which is adjacent to the central β-sheet and thus is unlikely to play a role in adapter recognition and recruitment. The TLR1 and TLR6 helix D residues, which are buried and have surface exposure of 10% or less, i.e. tyrosine in the fourth position, leucines in positions 7 and 10, and methionine at position 11 (Fig. 5C), are fully conserved not only in human TLR1 and TLR6, but also in the mouse homologs of the proteins (not shown). This structural conservancy suggests that region 9 residues, which are responsible for adapter recognition in TLR2, include three positively charged basic residues (R748, K751 and K754); whereas TLR6 does not have the third charged residue of the sequence (R747, K750 and A753) and TLR1 TIR D helix is most dissimilar to 2R9 and additionally misses the conserved N-terminal arginine (N742, K745 and T748) (Fig. 5C). These data illustrate structural differences that account for differences in affinity and specificity of TIR–TIR and TIR-peptide binding.
Figure 5.
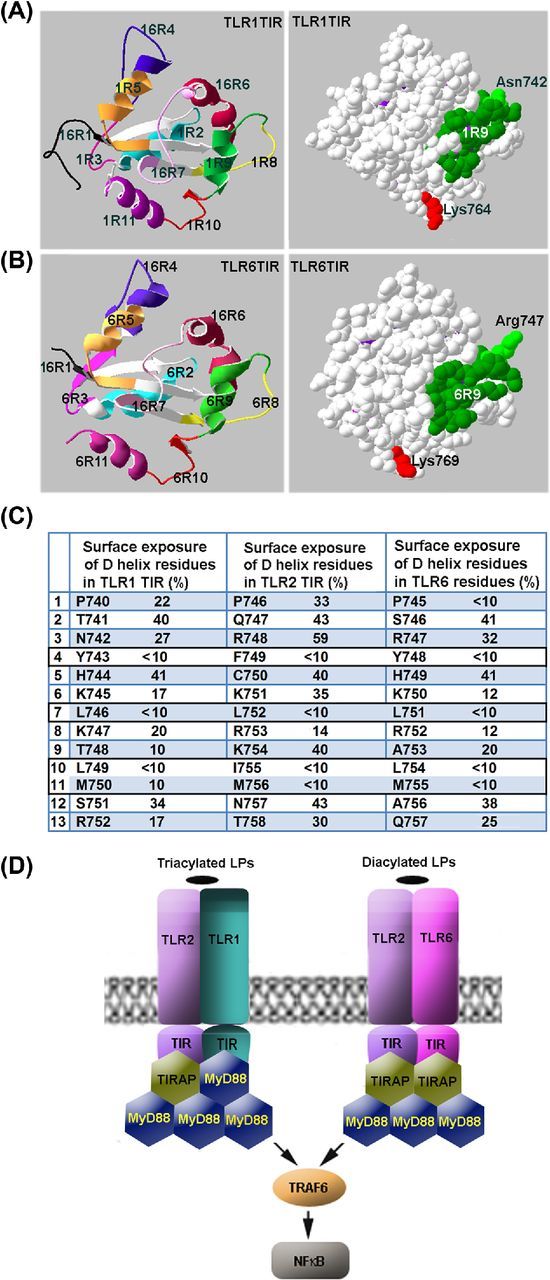
Mechanisms of adapter recruitment in TLR2 signaling. (A, B) TLR1 and TLR6 TIR domain regions that correspond to decoy peptides. (C) Surface exposure of region 9 residues in TIR domains of TLR1, TLR2 and TLR6. Surface exposure of the residues was calculated based on the available crystal structures of TIR domains of human TLRs. It should be noted that the D helix residues that have surface exposure <10% are 100% conserved not only in human TLR1 and TLR6, but also in mouse homologs of the proteins. (D) Schematic models of adapter recruitment to TLR2/1 and TLR2/6 heterodimers. The TLR2/1 heterodimer directly binds TIRAP and MyD88. The TLR2/6 heterodimers bind two TIRAP TIR domains through direct interactions mediated by D helices of TIR domains of both receptors. Additional MyD88 molecules are recruited to the primary complex through TIRAP-MyD88 and MyD88-MyD88 interactions. Disruption of any TIR–TIR interaction involved in the assembly of TLR signaling complexes abolishes signaling. Differences in composition of TLR2/1 and TLR2/6 complexes may account for kinetical differences in activation of NFκB and MAPKs by TLR2/1 and TLR2/6 agonists.
Presented data also suggest that TLR2 heterodimers directly recruit at least two adapter molecules (Fig. 5D). In case of TLR2/1 heterodimer, the two adapters are different. The first directly recruited adapter is TIRAP, which is recruited to TLR2 TIR (Piao et al.2015). The second adapter is MyD88, recruited through D helix of TLR1 TIR (Fig. 5D). The TLR2/6 heterodimer apparently recruits two TIRAP molecules, which are recruited through D helices of TLR2 and TLR6 TIR domains. Direct recruitment of two TIRAP molecules to TLR2/6 heterodimer is suggested by similarity of the surface-exposed region 9 residues of TLR2 and TLR6 (particularly, one common feature of 2R9 and 6R9 is arginines in positions 3 and 8; Fig. 5C). This similarity implies that both TIRs bind same site on TIRAP surface and thus suggest recruitment of two separate TIRAP molecules. Direct TIRAP-TLR6 binding is in accordance with recent observation of Jang, Narayanan and Park (2016) that recombinant TLR6 TIR domain, unlike TLR5 TIR domain, does not bind MyD88 TIR.
The primary receptor–adapter complexes are subsequently enlarged by recruitment of additional MyD88 molecules through homotypic and heterotypic TIRAP-MyD88 TIR interactions. We previously demonstrated that peptides that represent AB loop (TR3, EGSQASLRCF) and helix C of TIRAP (TR6, PGFLRDPWCKYQML) inhibit TLR2/1, but not TLR2/6 signaling (Couture et al.2012). It was found later that TR6 binds MyD88 (Piao et al.2015). Functional importance of AB loop and C helices of TIRAP was confirmed by systematic mutagenesis of surface-exposed residues of TIRAP (Lin et al.2012). Lin et al. (2012) found that mutations in TIRAP regions represented by TR3 (E108A, R115A and F117A) and TR6 (W156A, Y159A and L162A) impaired TIRAP functions (Valkov et al.2011). Given that TIRAP can bind MyD88 directly and both adapters can homodimerize, it is difficult to predict the higher order structure of the complex assembled at the primary platforms at this time.
In accordance with the notion that inhibitory peptides 1R9 and 6R9 represent adapter recruitment sites and block recruitment of adapters to receptors, these peptides do not interfere with TLR2 association with co-receptors and do not bind TLR2 (Fig. 4). Notably, the screening of peptide libraries derived from TLR1 and TLR6 TIR domains, unlike the screening of TLR4 TIR library (Toshchakov et al.2011), has failed to identify an inhibitory peptide that would bind the TIR of co-receptor, i.e. TLR2. This finding indicates that interaction of TIR domains in TLR2/1 and TLR2/6 receptor pairs is either weak or cannot be represented sufficiently fully by a peptide fragment.
Studies of selectivity of TLR inhibition by 1R9 and 6R9 have found that the peptides block only TLR2-mediated signaling, but not TIRAP-dependent signaling by TLR4 or TLR9. This pattern of inhibition is dissimilar to that demonstrated by 2R9, peptide from D helix of TLR2 TIR, which is a potent inhibitor of multiple TLR, including TLR2, TLR4, TLR7 and TLR9 (Piao et al.2015). This finding might suggest that TIRAP is recruited to TLR6 through a docking site which is different from the site used for binding to TLR2, TLR4 or TLR9. An example of a unique mode of adapter recruitment to TLR2/6 heterodimer has been published. Thus, study of Jiang et al. (2006) has found that macrophages from mice that carry homozygous Pococurante mutation, which is located in the first helix of MyD88 TIR (I179N), lose all MyD88-dependent TLR signaling, except TLR2/6-mediated signaling. These findings of Jiang et al. (2006) together with our findings presented here suggest that MyD88 and TIRAP are recruited to activated TLR2/6 heterodimer in a mode that differs from that in TLR4, TLR2/1 or TLR9 signaling.
In conclusion, our data suggest that, similarly to TLR2, TLR1 and TLR6 recruit adapters primarily through the fourth helix of TIR domain. These sites, however, bind different adapters; the D helix of TLR6 and TLR 2 TIR recruit TIRAP, whereas the fourth helix of TLR1 TIR recruits MyD88. Together, these findings shed new light on molecular mechanisms of adapter recruitment in TLR2 signaling.
Supplementary Material
SUPPLEMENTARY DATA
FUNDING
This work was supported by NIH grants AI-082299 (VYT).
Conflict of interest. None declared.
REFERENCES
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Bonham KS, Orzalli MH, Hayashi K, et al. A promiscuous lipid-binding protein diversifies the subcellular sites of toll-like receptor signal transduction. Cell. 2014;156:705–16. doi: 10.1016/j.cell.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes M, Klauschen F, Kuchen S, et al. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154:197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture LA, Piao W, Ru LW, et al. Targeting Toll-like receptor (TLR) signaling by Toll/interleukin-1 receptor (TIR) domain-containing adapter protein/MyD88 adapter-like (TIRAP/Mal)-derived decoy peptides. J Biol Chem. 2012;287:24641–8. doi: 10.1074/jbc.M112.360925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForge LE, Remick DG. Kinetics of TNF, IL-6, and IL-8 gene expression in LPS-stimulated human whole blood. Biochem Bioph Res Co. 1991;174:18–24. doi: 10.1016/0006-291x(91)90478-p. [DOI] [PubMed] [Google Scholar]
- Derossi D, Joliot AH, Chassaing G, et al. The third helix of the Antennapedia homeodomain translocates through biological membranes. J Biol Chem. 1994;269:10444–50. [PubMed] [Google Scholar]
- Farhat K, Riekenberg S, Heine H, et al. Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J Leukocyte Biol. 2008;83:692–701. doi: 10.1189/jlb.0807586. [DOI] [PubMed] [Google Scholar]
- Gay NJ, Symmons MF, Gangloff M, et al. Assembly and localization of Toll-like receptor signaling complexes. Nat Immunol. 2014;2014:546–58. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- Hirschfeld M, Ma Y, Weis JH, et al. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J Immunol. 2000;165:618–22. doi: 10.4049/jimmunol.165.2.618. [DOI] [PubMed] [Google Scholar]
- Horng T, Barton GM, Flavell RA, et al. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 2002;420:329–33. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- Jang TH, Narayanan KB, Park HH. In vitro reconstitution of the toll/interleukin-1 receptor (TIR) domain complex between TLR5/6 and Myd88. Protein Peptide Lett. 2016;23:55–62. doi: 10.2174/0929866523666151106123613. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Georgel P, Li C, et al. Details of Toll-like receptor:adapter interaction revealed by germ-line mutagenesis. Proc Natl Acad Sci U S A. 2006;103:10961–6. doi: 10.1073/pnas.0603804103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin MS, Kim SE, Heo JY, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–82. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Jin MS, Lee JO. Structures of the toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–91. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Kang JY, Nan X, Jin MS, et al. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31:873–84. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–84. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–50. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Lin Z, Lu J, Zhou W, et al. Structural insights into TIR domain specificity of the bridging adaptor Mal in TLR4 signaling. PLoS One. 2012;7:e34202. doi: 10.1371/journal.pone.0034202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Liu Y, Hao W, et al. TLR2 is a primary receptor for Alzheimer's amyloid beta peptide to trigger neuroinflammatory activation. J Immunol. 2012;188:1098–107. doi: 10.4049/jimmunol.1101121. [DOI] [PubMed] [Google Scholar]
- Ma D, Jin S, Li E, et al. The neurotoxic effect of astrocytes activated with toll-like receptor ligands. J Neuroimmunol. 2013;254:10–8. doi: 10.1016/j.jneuroim.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Means TK, Lien E, Yoshimura A, et al. The CD14 ligands lipoarabinomannan and lipopolysaccharide differ in their requirement for Toll-like receptors. J Immunol. 1999;163:6748–55. [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Kopp E, et al. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell. 1998;2:253–8. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- Mistry P, Laird MH, Schwarz RS, et al. Inhibition of TLR2 signaling by small molecule inhibitors targeting a pocket within the TLR2 TIR domain. Proc Natl Acad Sci U S A. 2015;112:5455–60. doi: 10.1073/pnas.1422576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA, Bryant CE, Doyle SL. Therapeutic targeting of Toll-like receptors for infectious and inflammatory diseases and cancer. Pharmacol Rev. 2009;61:177–97. doi: 10.1124/pr.109.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozinsky A, Underhill DM, Fontenot JD, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A. 2000;97:13766–71. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CN, Vajdos F, Fee L, et al. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–23. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–52. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Piao W, Ru LW, Piepenbrink KH, et al. Recruitment of TLR adapter TRIF to TLR4 signaling complex is mediated by the second helical region of TRIF TIR domain. Proc Natl Acad Sci U S A. 2013;110:19036–41. doi: 10.1073/pnas.1313575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao W, Shirey KA, Ru LW, et al. A decoy peptide that disrupts TIRAP recruitment to TLRs is protective in a murine model of Influenza. Cell Rep. 2015;11:1941–52. doi: 10.1016/j.celrep.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao W, Vogel SN, Toshchakov VY. Inhibition of TLR4 signaling by TRAM-derived decoy peptides in vitro and in vivo. J Immunol. 2013;190:2263–72. doi: 10.4049/jimmunol.1202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschner A, Moretta A, Landmann R, et al. The ester-bonded palmitoyl side chains of Pam3CysSerLys4 lipopeptide account for its powerful adjuvanticity to HLA class I-restricted CD8+ T lymphocytes. Eur J Immunol. 2003;33:2044–52. doi: 10.1002/eji.200323776. [DOI] [PubMed] [Google Scholar]
- Santos-Sierra S, Deshmukh SD, Kalnitski J, et al. Mal connects TLR2 to PI3Kinase activation and phagocyte polarization. EMBO J. 2009;28:2018–27. doi: 10.1038/emboj.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcheblyakov DV, Logunov DY, Rakovskaya IV, et al. Triggering of toll-like Receptor-2 in mouse myelomonocytic leukaemia cells WEHI-3B leads to the suppression of apoptosis and promotes tumor progression in vivo. Acta Naturae. 2011;3:83–93. [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–51. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Sato S, Horiuchi T, et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–4. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- Toshchakov VY, Basu S, Fenton MJ, et al. Differential involvement of BB loops of toll-IL-1 resistance (TIR) domain-containing adapter proteins in TLR4- versus TLR2-mediated signal transduction. J Immunol. 2005;175:494–500. doi: 10.4049/jimmunol.175.1.494. [DOI] [PubMed] [Google Scholar]
- Toshchakov VY, Szmacinski H, Couture LA, et al. Targeting TLR4 signaling by TLR4 Toll/IL-1 receptor domain-derived decoy peptides: identification of the TLR4 Toll/IL-1 receptor domain dimerization interface. J Immunol. 2011;186:4819–27. doi: 10.4049/jimmunol.1002424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshchakov VY, Vogel SN. Cell-penetrating TIR BB loop decoy peptides a novel class of TLR signaling inhibitors and a tool to study topology of TIR-TIR interactions. Expert Opin Biol Th. 2007;7:1035–50. doi: 10.1517/14712598.7.7.1035. [DOI] [PubMed] [Google Scholar]
- Underhill DM, Ozinsky A, Hajjar AM, et al. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–5. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- Valkov E, Stamp A, Dimaio F, et al. Crystal structure of Toll-like receptor adaptor MAL/TIRAP reveals the molecular basis for signal transduction and disease protection. Proc Natl Acad Sci U S A. 2011;108:14879–84. doi: 10.1073/pnas.1104780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werts C, Tapping RI, Mathison JC, et al. Leptospiral lipopolysaccharide activates cells through a TLR2-dependent mechanism. Nat Immunol. 2001;2:346–52. doi: 10.1038/86354. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Hemmi H, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420:324–9. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.