Abstract
Chlamydiae exit via membrane-encased extrusion or through lysis of the host cell. Extrusions are novel, pathogen-containing structures that confer infectious advantages to Chlamydia, and are hypothesized to promote cell-to-cell spread, dissemination to distant tissues and facilitate immune evasion. The extrusion phenomenon has been characterized for several Chlamydia trachomatis serovars, but a thorough investigation of extrusion for additional clinically relevant C. trachomatis strains and Chlamydia species has yet to be performed. The key parameters investigated in this study were: (i) the conservation of extrusion across the Chlamydia genus, (ii) the functional requirement for candidate Chlamydia genes in extrusion formation i.e. IncA and CT228 and (iii) extrusion-mediated uptake, and consequent survival of Chlamydia inside macrophages. Inclusion morphology was characterized by live fluorescence microscopy, using an inverted GFP strategy, at early and mid-stages of infection. Enriched extrusions were used to infect bone marrow-derived macrophages, and bacterial viability was measured following macrophage engulfment. Our results demonstrate that extrusion is highly conserved across chlamydiae, including ocular, STD and LGV biovars and divergent Chlamydia species. Consequently, this exit mechanism for Chlamydia may fulfill common advantages important for pathogenesis.
Keywords: Chlamydia, extrusion, dissemination, exit, macrophage
The authors report that the extrusion mechanism of Chlamydia exit from host cells is broadly conserved across divergent Chlamydia species and clinical strains.
Chlamydiae are gram-negative obligate intracellular bacteria that inflict a tremendous public health burden globally. Chlamydia trachomatis is the causative agent of trachoma, the leading cause of infectious blindness (Burton and Mabey 2009), as well as the sexually transmitted disease (Gerbase, Rowley and Mertens 1998). Chlamydiae have evolved to infect a wide variety of host species including humans (Chlamydiae trachomatis, Chlamydiae pneumoniae), birds (Chlamydiae psittaci), rodents (Chlamydiae muridarum), guinea pigs (Chlamydiae caviae), horses and reptiles (C. pneumoniae) (Sachse et al.2015), and more. Despite this diverse host range, Chlamydia species are genetically closely related. Members of the family Chlamydiaceae share >91% similarity in 16S rRNA sequences, and >88% gene identity with C. trachomatis (Sachse et al.2014, 2015), with conservation of specific individual gene products limited in some cases. The extent to which virulence mechanisms of Chlamydia species are conserved is an active area of investigation.
The mechanisms by which Chlamydia spp. disseminate within a host, or transmit to new hosts, are not well understood. C. trachomatis replicates within an inclusion in host epithelial cells, then exits through two distinct mechanisms— extrusion and lysis (Hybiske and Stephens 2007). Extrusion occurs by pinching of the chlamydial inclusion and the plasma membrane, generating a detached, membrane-bound compartment containing infectious Chlamydia (Hybiske and Stephens 2007; Chin et al.2012). The benefits of extrusion for Chlamydia were recently explored, demonstrating that containment within an extrusion enhances extracellular survival of C. trachomatis (Zuck et al.2016). These novel pathogenic structures are also potentially involved in dissemination, immune evasion and transmission. Furthermore, extrusion is known to occur in C. trachomatis serovars L2 (CT L2) and D, C. caviae (Hybiske and Stephens 2007) and Chlamydia pecorum (Doughri, Storz and KP 1972) but had not been more widely explored. In this study, we investigated how broadly the extrusion exit mechanism is conserved among Chlamydia spp.
Inclusion morphology varies greatly between species of Chlamydia, ranging in phenotypes from: circular, amorphous and multivacuolar. We hypothesized that variation in inclusion number and morphology within host cells may affect whether these Chlamydia can extrude, or dictate extrusion size and downstream outcomes.
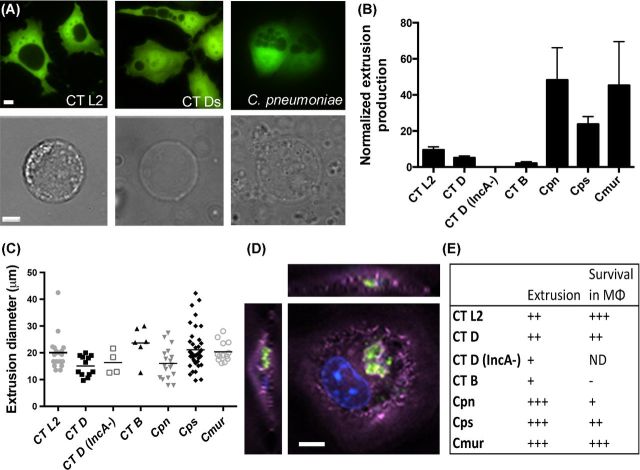
To compare inclusion morphological differences between the species in this study, green fluorescent protein (GFP)-expressing HeLa cells were infected with Chlamydia and imaged using an inverted GFP microscopy strategy, whereby fluorescent host cells reveal the nonfluorescent inclusion against the GFP labeled host cytoplasm (Zuck, Feng and Hybiske 2015). The following Chlamydia species were used in this study: C. trachomatis LGV L2/434, C. trachomatis serovar D, C. psittaci 6BC, C. muridarum Nigg and C. pneumoniae TW183. Two additional C. trachomatis strains were tested for functional requirement of candidate Chlamydia genes in extrusion; CT Ds/2923 (Suchland et al.2000) and CT B/Jali20/OT (validated IncA and CT228 truncation mutants, respectively, Seth-Smith et al.2009; Lutter et al.2013). CT B/Jali20/OT was included in this study based on published data involving CT228 in regulation of extrusion by interaction with host cell protein Myosin phosphatase target subunit 1 (MYPT1) (Lutter et al.2013). Inclusion morphology and numbers varied as expected; CT L2 displayed a single, round inclusion phenotype at 24 h post-infection (hpi), representative of the majority of strains tested (Fig. 1A, left). CT DincA− and C. pneumoniae displayed multiple inclusions per cell (Fig. 1A, middle, right, respectively).
Figure 1.
Conservation of extrusion production, size and survival in macrophages across Chlamydiae. (A) Inclusion morphology of Chlamydia spp.; HeLa cell (GFP), inclusion is shown by absence of GFP, at 60× magnification. Top row: CT L2 (left), CT DincA− (middle) and C. psittaci (right). Bottom row: single, isolated extrusions matching top row Chlamydia species shown in brightfield at 60× magnification. Scale bar = 5 μm. (B) Normalized extrusion production, calculated by counting number of extrusions relative to % of cells infected in original infection. Columns show mean ± SEM. n = 3 independent experiments. Statistics were performed by one-way ANOVA analysis using Tukey's multiple comparisons test, and no significance was observed between species or C. trachomatis strains. (C) Extrusion size graph displaying each extrusion as a single point on the graph. Extrusions counted by Volocity microscope software parameters to measure diameter (μm), n = 3 independent experiments. Statistics were performed using one-way ANOVA analysis using Tukey's multiple comparisons test. No significance was observed between species or C. trachomatis strain extrusion diameter or spread. (D) Extrusion uptake by bone marrow macrophages. CT L2 extrusion (GFP), host cell actin (purple) and nucleus (DAPI) at 60× magnification with z stacking. Scale bar = 5 μm. (E) Summary table displaying normalized extrusion production and survival within macrophages up to 6 h. +++ denotes >100 IFU/field, ++ denotes 50–100 IFU/field, + denotes < 50 IFU/field and – denotes no IFU seen. ND = no data.
To determine which Chlamydia species and C. trachomatis serovars utilize the extrusion exit pathway, all strains were grown in HeLa or Hep2 cells (for C. pneumoniae), and extrusions were collected over a time course from 0 to 48 hpi. For all strains, extrusion production was greatest at 48 hpi. (Fig. S1A-F, Supporting Information). Extrusions were collected using a centrifugation protocol (Zuck et al.2016), and counted via microscopy (Fig. 1A and B). Despite the presence of multiple inclusions or varied inclusion morphologies for some strains, the ability to extrude was conserved in all Chlamydia species and strains tested. Additionally, all extrusions had similar appearances via brightfield microscopy (Fig 1A).
Following confirmation that extrusion is a conserved exit mechanism among all Chlamydia species tested, we next sought to determine any differences in relative extrusion production. We collected extrusions from each strain at 48 hpi, enumerated extrusions by microscopy, and normalized extrusion quantity to the infection level of the monolayer that extrusions were collected from. Extrusion production differed among Chlamydia species but these differences were not statistically significant (Fig 1B; Fig. S1, Supporting Information). There was robust extrusion production in C. pneumoniae and C. muridarum but little extrusion production by the ocular strain CT B/Jali20 as previously reported (Fig. 1B; Lutter et al.2013). Interestingly, C. trachomatis D−incA did not extrude at 48 hpi; however, further experiments showed delayed extrusion production, peaking at 72 hpi. This delay could be due to attenuated growth, as previously shown for IncA mutants (Xia et al.2005). Alternatively, IncA could be required for normal extrusion kinetics, but other compensatory mechanisms are active after 48 hpi. The size of extrusions was also examined, and extrusion size of all Chlamydia species was conserved, at about ∼20 μm (Fig. 1C). We found no statistically significant difference in the mean or range of extrusion diameter among the strains tested, including the C. pneumoniae and CT D−incA species that harbor, on average, multiple and much smaller inclusions. Considering the substantial variation in developmental cycle among species, e.g. C. pneumoniae and C. muridarum, the absence of significant differences in extrusion sizes or numbers is intriguing. All Chlamydia species continually shed extrusions (Fig. S1, Supporting Information), possibly prior to inclusion fusion for C. pneumoniae, ultimately producing multiple extrusions from the same infected cell, though this has not been directly assessed.
The data for extrusion's survival benefits to Chlamydia, coupled with the conservation of extrusion in a diverse set of Chlamydia species, suggest that this mechanism plays an important part in pathogenesis. In a previous study (Zuck et al.2016), we demonstrated that extrusions are engulfed by primary bone marrow-derived macrophages from C57Bl/6 mice, and subsequently survive within macrophages much longer than free Chlamydia elementary bodies (EB). To determine if this is a conserved survival strategy, we examined uptake of extrusions within macrophages. Extrusions were collected and co-cultured with macrophages for 1 h at 37°C, then thoroughly rinsed and analyzed via immunofluorescence. Fluorescence microscopy revealed that all C. trachomatis serovars and Chlamydia species tested were capable of being engulfed by macrophages. Figure 1D shows a CT L2 extrusion within a macrophage at 1 hpi. The majority of extrusions were intact upon engulfment, but some engulfed extrusions displayed GFP fluorescence localized into several distinct foci or scattered throughout the cytoplasm of the host macrophage. This range in morphology was characteristic of all strains observed (Fig. S2, Supporting Information).
Lastly, we characterized the extrusion-mediated enhancement of Chlamydia survival within macrophages. We co-cultured primary macrophages with extrusions, rinsed, then incubated macrophages at 37°C for 6 h to measure short-term survival of extrusion-delivered Chlamydia. Notably, all extrusions were engulfed and facilitated short-term survival of their bacteria within macrophages (Fig. 1D and E). Survival of CT B/Jali20 within macrophages was not detected, but could be a result of our inability to harvest high numbers of extrusions for this strain. Survival of C. pneumoniae in macrophages was surprisingly low, considering previously published data demonstrating the ability of free EB to infect macrophages (Gaydos et al.1996; Redecke et al.1998; Marangoni et al.2014). These data demonstrate the conservation of the extrusion exit mechanism across a diverse set of C. trachomatis serovars and Chlamydia species.
Our results reveal that extrusion is a broadly conserved exit mechanism within Chlamydiae. Among the species and serovars tested, there was no significant difference in the ability to extrude or the size of extrusions between Chlamydiae. The absence of functional proteins CT228 and IncA hindered extrusion production, though this effect was not statistically significant. These genes may be involved in extrusion formation, but are not absolute requirements for this exit mechanism. Given the natural history of Chlamydiae as obligate intracellular bacteria, the evolution of promoting exit in an extracellular compartment makes sense as a pathogenesis strategy. It was recently shown that there are tangible benefits for Chlamydia that exit within extrusions (Zuck et al.2016), including enhanced survival in the extracellular environment compared to free EB. The extrusion compartment may function as an ‘intracellular-like’ environment, retaining vital nutrients and protecting against extracellular damage. The broad conservation of extrusion suggests that this exit strategy is universally beneficial to Chlamydia species in a common stage of pathogenesis.
Additionally, our data indicate that uptake of extrusions into primary macrophages is conserved among Chlamydiae. Spread of Chlamydia within the host from the original infection site is a pathogenesis trait for several species including C. trachomatis and C. pneumoniae (Moazed et al.1998; Darville and Hiltke 2010). However, as a non-motile bacterium, the mechanism by which Chlamydia spreads in its host is not well understood. Migration of other non-motile bacteria can be assisted by hitching a ride in a host phagocyte, as shown for Mycobacterium marinum, Neisseria gonorrhoeae and Staphylococcus aureus (Davis and Ramakrishnan 2009; Thwaites and Gant 2011; Criss and Seifert 2012). Extrusion, and the conservation of this mechanism throughout the Chlamydia species tested, is a strategy to allow survival within phagocytic cells, perhaps to sustain Chlamydia until they disseminate to new mucosal tissues or distant sites.
Supplementary Material
Acknowledgments
We would like to acknowledge Ted Hackstadt for use of the C. trachomatis B/Jali20/OT strain for these studies.
SUPPLEMENTARY DATA
FUNDING
This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health [R01 AI095603 to K.H. and T32 AI007140 to A.S.].
Conflict of interest. None declared.
REFERENCES
- Burton MJ, Mabey DCW. The global burden of trachoma: a review. PLoS Neglect Trop D. 2009;3:e460. doi: 10.1371/journal.pntd.0000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin E, Kirker K, Zuck M, et al. Actin Recruitment to the Chlamydia inclusion is spatiotemporally regulated by a mechanism that requires host and bacterial factors. PLoS One. 2012;7:1–12. doi: 10.1371/journal.pone.0046949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criss AK, Seifert HS. A bacterial siren song: intimate interactions between Neisseria and neutrophils. Nat Rev Microbiol. 2012;10:178–90. doi: 10.1038/nrmicro2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darville T, Hiltke TJ. Pathogenesis of genital tract disease due to Chlamydia trachomatis. J Infect Dis. 2010;201(Suppl.):S114–25. doi: 10.1086/652397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JM, Ramakrishnan L. The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell. 2009;136:37–49. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doughri A, Storz J, KP A. Mode of entry and release of chlamydiae in infections of intestinal epithelial cells. J Infect Dis. 1972;126:652–7. doi: 10.1093/infdis/126.6.652. [DOI] [PubMed] [Google Scholar]
- Gaydos CA, Summersgill JT, Sahney NN, et al. Replication of Chlamydiapneumoniae in vitro in human macrophages, endothelial cells, and aortic artery smooth muscle cells. Infect Immun. 1996;64:1614–20. doi: 10.1128/iai.64.5.1614-1620.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbase aC, Rowley JT, Mertens TE. Global epidemiology of sexually transmitted diseases. Lancet. 1998;351(Suppl.):2–4. doi: 10.1016/s0140-6736(98)90001-0. [DOI] [PubMed] [Google Scholar]
- Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. P Natl Acad Sci USA. 2007;104:11430–5. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter EI, Barger AC, Nair V, et al. Chlamydia trachomatis inclusion membrane protein CT228 recruits elements of the myosin phosphatase pathway to regulate release mechanisms. Cell Rep. 2013;3:1921–31. doi: 10.1016/j.celrep.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangoni A, Bergamini C, Fato R, et al. Infection of human monocytes by Chlamydiapneumoniae and Chlamydia trachomatis: an in vitro comparative study. BMC Res Notes. 2014;7:230. doi: 10.1186/1756-0500-7-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed TC, Kuo CC, Grayston JT, et al. Evidence of systemic dissemination of Chlamydiapneumoniae via macrophages in the mouse. J Infect Dis. 1998;177:1322–5. doi: 10.1086/515280. [DOI] [PubMed] [Google Scholar]
- Redecke V, Dalhoff K, Bohnet S, et al. Interaction of Chlamydiapneumoniae and human alveolar macrophages: infection and inflammatory response. Am J Resp Cell Mol. 1998;19:721–7. doi: 10.1165/ajrcmb.19.5.3072. [DOI] [PubMed] [Google Scholar]
- Sachse K, Bavoil PM, Kaltenboeck B, et al. Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognized species. Syst Appl Microbiol. 2015;38:99–103. doi: 10.1016/j.syapm.2014.12.004. [DOI] [PubMed] [Google Scholar]
- Sachse K, Laroucau K, Riege K, et al. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydiaavium sp. nov. and Chlamydiagallinacea sp. nov. Syst Appl Microbiol. 2014;37:79–88. doi: 10.1016/j.syapm.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Seth-Smith HMB, Harris SR, Persson K, et al. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics. 2009;10:239. doi: 10.1186/1471-2164-10-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchland RJ, Rockey DD, Bannantine JP, et al. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect Immun. 2000;68:360–7. doi: 10.1128/iai.68.1.360-367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thwaites GE, Gant V. Are bloodstream leukocytes Trojan Horses for the metastasis of Staphylococcus aureus? Nat Rev Microbiol. 2011;9:215–22. doi: 10.1038/nrmicro2508. [DOI] [PubMed] [Google Scholar]
- Xia M, Suchland RJ, Bumgarner RE, et al. Chlamydia trachomatis variant with nonfusing inclusions: growth dynamic and host-cell transcriptional response. J Infect Dis. 2005;192:1229–36. doi: 10.1086/444394. [DOI] [PubMed] [Google Scholar]
- Zuck M, Ellis TC, Venida A, et al. Extrusions promote engulfment and Chlamydia survival within macrophages. bioRxiv. 2016 doi: 10.1101/041079. [DOI] [PubMed] [Google Scholar]
- Zuck M, Feng C, Hybiske K. Using fluorescent proteins to visualize and quantitate Chlamydia vacuole growth dynamics in living cells. J Vis Exp. 2015:1–9. doi: 10.3791/51131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.