Abstract
Increasing morbidity and mortality from Clostridium difficile infection (CDI) present an enormous challenge to healthcare systems. Clostridium difficile express type IV pili (T4P), but their function remains unclear. Many chronic and recurrent bacterial infections result from biofilms, surface-associated bacterial communities embedded in an extracellular matrix. CDI may be biofilm mediated; T4P are important for biofilm formation in a number of organisms. We evaluate the role of T4P in C. difficile biofilm formation using RNA sequencing, mutagenesis and complementation of the gene encoding the major pilin pilA1, and microscopy. RNA sequencing demonstrates that, in comparison to other growth phenotypes, C. difficile growing in a biofilm has a distinct RNA expression profile, with significant differences in T4P gene expression. Microscopy of T4P-expressing and T4P-deficient strains suggests that T4P play an important role in early biofilm formation. A non-piliated pilA1 mutant forms an initial biofilm of significantly reduced mass and thickness in comparison to the wild type. Complementation of the pilA1 mutant strain leads to formation of a biofilm which resembles the wild-type biofilm. These findings suggest that T4P play an important role in early biofilm formation. Novel strategies for confronting biofilm infections are emerging; our data suggest that similar strategies should be investigated in CDI.
Keywords: Clostridium difficile, type IV pili, biofilm
We demonstrate that type IV pili play an important role in the initial stages of Clostridium difficile biofilm formation, which has direct implications for future research into novel CDI therapies.
INTRODUCTION
Clostridium difficile is a Gram-positive spore-forming obligate anaerobe, first isolated from the stool of newborn infants in 1935 (Hall and O'Toole 1935). Clostridium difficile infection (CDI) is the leading cause of infectious hospital-acquired gastrointestinal illness in the developed world (Musher et al.2006; McFarland 2009). The disease is toxin mediated, ranging in severity from mild diarrhea to toxic megacolon, and can result in death (Bartlett and Gerding 2008). The frequency and severity of CDI are increasing rapidly in the United States and globally (Kelly and LaMont 2008); a recent report estimates that in 2011 CDI was responsible for almost half a million infections and approximately 29 000 deaths in the United States (Lessa et al.2015). Improved methods for preventing and treating CDI will be crucial for reducing morbidity and mortality from this disease.
Bacteria commonly grow as biofilms, surface-associated communities embedded in a polysaccharide-rich substance; this mode of growth is critical to chronic and recurrent infections. Similarities between biofilm infections and CDI suggest that CDI is biofilm mediated. Both biofilm infections and CDIs are surface associated: while biofilms form on surfaces varied as prosthetic implants and the interiors of pipes, C. difficile infects the colon surface. Like many bacteria known to form biofilms, C. difficile employs quorum sensing as a means of cell–cell communication (Davies et al.1998; Carter et al.2005; Lee and Song 2005; Yang et al.2014; Omer Bendori et al.2015). Finally, biofilms are well known to allow their constituents to resist antibiotic treatment (Olsen 2015), and at times CDI does not respond to, or recurs after, appropriate antibiotic therapy. Although the precise mechanisms of CDI treatment resistance and infection relapse are currently unknown, they may be related to the formation of a C. difficile biofilm. Cells growing in a biofilm have a different phenotype compared to their planktonic counterparts. Some cells deep within a biofilm are relatively nutrient deprived and therefore slow or non-growing, which reduces susceptibility to antimicrobials (Becker et al.2001). Determination of the phenotype of C. difficile in CDI may allow for better-targeted therapies. For example, compounds that disrupt the extracellular matrix of biofilms have demonstrated feasibility for reducing bacterial biofilms produced by clinically relevant organisms (Lu and Collins 2007). If C. difficile has growth characteristic of a biofilm in human infection, then study of these compounds could lead to enhancements in the efficacy of antimicrobials. Vaccines based on biofilm-specific antigens have been reported and used successfully in animal models of Staphylococcus aureus osteomyelitis and Haemophilus influenzae otitis media (Brady et al.2011; Novotny et al.2015).
Type IV pili (T4P) are a particular type of bacterial surface appendage that mediate adherence, colonization, biofilm formation, DNA transfer and twitching motility (Craig, Pique and Tainer 2004), among other functions; they have been well characterized in Gram-negative bacteria, and more recently described in Gram-positive bacteria. Gram-positive T4P were first recognized in Ruminococcus albus (Rakotoarivonina et al.2002), and were subsequently observed in C. perfringens (Varga et al.2006). Genes for T4P were identified in the C. difficile genome (Varga et al.2006), although fimbrial structures on C. difficile, resembling T4P, were observed by electron microscopy nearly two decades previously (Borriello, Davies and Barclay 1988). The T4P structural subunits are called pilins; C. difficile T4P are composed primarily of PilA1 pilin (Piepenbrink et al.2015). The C. difficile genome encodes genes for up to nine pilins, as well as the requisite assembly and scaffolding proteins, organized into three gene clusters as previously described by our lab (Maldarelli et al.2014). A complex nanomachine assembles pilin monomers into filaments that can extend several micrometers from the bacterial cell (Craig, Pique and Tainer 2004). In many bacterial species, T4P can also retract; the energy for extension and retraction are provided by distinct ATPases (Whitchurch et al.1991). T4P in Gram-negative organisms, such as enteropathogenic Escherichia coli, Vibrio cholerae and Neisseria meningitidis, are critical for colonization and pathogenesis (Nassif et al.1994; Tacket et al.1998; Humphries et al.2009). In N. meningitidis, the binding of T4P to CD147 facilitates recruitment of beta-adrenergic receptors and leads to passage through the blood–brain barrier, a step absolutely critical to pathogenesis (Nassif et al.1994; Bernard et al.2014).
In addition to their role in colonization, T4P are also critical for development of biofilms in multiple organisms. In Pseudomonas aeruginosa, T4P-mediated motility is critical for early biofilm development (Klausen et al.2003a), particularly in the attachment phase; T4P are also important in developing the architecture of the mature biofilm (Klausen et al.2003b). In an H. influenzae strain, mutation of the major pilin results in thinner and less-stable biofilm as compared to the wild-type strain (Novotny et al.2015). Recent studies also demonstrate the importance of T4P in Gram-positive biofilms. T4P genes are upregulated in C. perfringens biofilms grown at 37°C, and are required for biofilm formation (Obana, Nakamura and Nomura 2014). Recent work demonstrates cyclic-di-GMP regulation of C. difficile T4P gene expression and bacterial aggregation; Bordeleau et al. hypothesize that aggregation regulation is important in the development of C. difficile biofilms (Bordeleau et al.2015; Purcell et al.2016). Biofilm formation in C. difficile has been observed in vitro (Ðapa et al.2013), and putative bacterial aggregates implying biofilm formation have been observed in vivo (Buckley et al.2011). Additionally, examination of the structure of PilA1, the major pilin subunit in C. difficile, shows a strong resemblance to the subunits of T4P known to mediate bacterial self-association, a prerequisite for formation of biofilms that depend on these structures (Piepenbrink et al.2015).
Here, we investigate C. difficile biofilm gene expression and the impact of T4P deficiency, through knockout and complementation of the gene for the major C. difficile pilin, pilA1, on biofilm formation.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The Clostridium difficile R20291 strain, isolated from a 2006 outbreak in the United Kingdom, was chosen for these experiments as its genetic sequence is known and mutants are available. Production of the R20291 pilA1::ermB and R20291 pilA1::ermB ppilA1 C. difficile strains was previously described (Piepenbrink et al.2015). Clostridium difficile was grown at 37°C within an anaerobic chamber (Coy Laboratory Products, Grass Lake, Michigan, US) with 10% H2, 5% CO2 and 85% N2, unless otherwise stated.
RNA sample growth
For planktonic samples, overnight culture in brain heart infusion (BHI) media (Sigma-Aldrich, Saint Louis, Missouri, US) supplemented with 0.5% yeast extract and 0.1% L-cysteine brain heart infusion-supplemented (BHIS) was mixed 1:10 with fresh BHIS and grown with shaking for 6 h before mixing 1:1 with RNAProtect (Qiagen, Hilden, Germany). Biofilm samples were grown for 1 week in glass jars containing 8 mm glass beads. The glass beads were added in order to increase surface area and maximize RNA yields. BHIS medium was changed daily and 6 h prior to cell harvest. To harvest adherent cells (biofilm), media was removed and the beads rinsed with fresh media before covering with RNAProtect and subjecting to vigorous shaking for 2 min. For plate growth samples, overnight culture was streaked onto Columbia blood agar plates (BD Biosciences, San Jose, California, US) and kept in the anaerobic chamber for 24 h prior to a 3-day room temperature growth in an anaerobic GasPak container (BD Biosciences). Plates were returned to the anaerobic chamber for cell harvesting and placement into RNAprotect. All samples were stored at −80°C until used for RNA sequencing.
Preparation of biofilm for imaging
Glass coverslips were placed in six-well (Costar, Corning, New York, US) tissue culture plates, and were inoculated with turbid overnight culture diluted to OD600 = 0.125 in pre-reduced BHI broth to a final volume of 4 mL, supplemented with thiamphenicol as needed for plasmid maintenance. Media was changed every other day. At day 1 or day 7, coverslips were washed with sterile PBS and stained with BacLight Live/Dead stain (Life Technologies, Carlsbad, California, US) according to manufacturer's instructions. Slides were fixed by incubation with 4% paraformaldehyde in PBS, mounted on glass slides, and stored in the dark at −20°C until imaged. Three slides per strain were grown for each strain per time point; two growths were conducted.
RNA sequencing
After thawing, cells were pelleted and RNAprotect removed. Cells were resuspended in 100 uL TES buffer with 35 000 units of Ready-Lyse lysozyme solution (Epicentre, Madison, Wisconsin, US) and incubated with shaking at room temperature for 90 min. The RNeasy RNA purification kit (Qiagen) was then used as per the manufacturer's instructions. Total RNA samples were treated with DNase I (Invitrogen, Carlsbad, California, US). The level of ribosomal RNA present in total RNA samples was reduced prior to library construction using the Ribo-Zero Gram-Positive Bacteria rRNA Removal Kit (Epicentre). Illumina RNAseq libraries were prepared with the TruSeq RNA Sample Prep kit (Illumina, San Diego, California, US), omitting the poly-A selection steps. Adapters containing six nucleotide indices were ligated to the double-stranded cDNA. The DNA was purified between enzymatic reactions, and the size selection of the library was performed with AMPure XT beads (Beckman Coulter Genomics, Danvers, Massachusetts, US). The libraries were sequenced on a 101 bp paired end run on the HiSeq 2000 (Illumina). Reads were processed for quality in Trimmomatic version 0.30 (Bolger, Lohse and Usadel 2014) using a leading and trailing minimum score of 10 and a four-base sliding window minimum score of 15; on average for all libraries 99.9% of reads survived. Bowtie2 version 2.1.0 (Langmead and Salzberg 2012) was used to map an average of 23.6 million paired reads to the C. difficile genome (NC_013316.1, NCBI) with an overall alignment rate of 96.2%. Samtools version 0.1.18 (Li et al.2009) was used for file conversion to the binary BAM format. Transcript abundances were evaluated in Cufflinks version 2.1.1 (Li et al.2009) using a ribosomal masking file for all 5S, 16S and 23S loci (NC_013316.gff annotation, NCBI), and fragments per kilobase per million were reported (http://cufflinks.cbcb.umd.edu) (Trapnell et al.2010). For comparative analysis of biofilm, plate and planktonic conditions, the Cuffdiff package in Cufflinks was used, and significant genes were reported. A cutoff false discovery rate-adjusted p-value (or q-value) of less than 0.05 was used to select significant differentially expressed genes. Heatmaps were generated using the gplots version 2.17.0 package (Warnes et al.2015) in R (Team RC 2014). Mapped RNA-Seq reads were visualized using the Integrative Genomics Viewer (Robinson et al.2011; Thorvaldsdóttir, Robinson and Mesirov et al.2013).
RNA-sequencing data accession number
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar, Domrachev and Lash 2002) and are accessible through GEO Series accession number GSE69001 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE69001).
Confocal scanning laser microscopy (CLSM)
Biofilms were grown on 18×18 mm glass coverslips and prepared as above. Slides were read using a Zeiss 510 Meta confocal laser scanning microscope and accompanying software (Carl Zeiss AG, Oberkochen, Germany). Ten fields of view were imaged per slide. Live cells per field of view were enumerated with CellProfiler (Kamentsky et al.2011). Z-stacks were acquired for a subset of these images; at least six z-stacks were analyzed per strain per time point. The structural organization of the biofilms was analyzed using the Comstat2 software package (http://www.comstat.dk) (Heydorn and Nielsen 2000; Vorregaard 2008). The 3D representations of the biofilms were generated using the 3D viewer plugin for ImageJ (http://3dviewer.neurofly.de) (Schmid et al.2010).
Scanning electron microscopy (SEM)
Biofilms were grown on glass coverslips as described above and fixed in 100% ethanol. The samples were shipped to the Image and Chemical Analysis Laboratory, Department of Physics, Montana State University, where they were allowed to air dry and then taped to an SEM mount using double-sided conductive carbon tape. The samples were coated with iridium and then imaged with a Zeiss Supra 55VP Field Emission Scanning Electron Microscope using a 1 keV electron beam.
RESULTS
RNA sequencing reveals a unique Clostridium difficile transcriptome during biofilm growth
We performed RNA-Seq on C. difficile grown on plates under conditions favoring expression of PilA1 pili, on planktonic bacteria from BHIS broth culture, and on bacteria grown for 1 week in BHIS as a biofilm on glass beads. Of the 3652 C. difficile R20291 genes, 1604 were significantly differentially expressed in biofilm samples when compared to planktonic samples, and 2105 were significantly differentially expressed in biofilm samples when compared to plate-growth samples. We evaluated specific genes suspected to be growth phenotype related (Table 1). Not surprisingly, the flagellin gene fliC was significantly decreased in biofilm samples relative to planktonic samples, while there was no significant difference between biofilm and plate samples. Suspecting adhesins important in biofilm formation, we interrogated the adhesin genes cwp66 (Waligora et al.2001; Janoir et al.2007) and CD0802, the equivalent of CD0873 in strain 630 (Kovacs-Simon et al.2014). Unexpectedly, we found decreased expression of both in biofilm samples when compared to planktonic samples, although only CD0802 was significantly different. When comparing biofilm- and plate-growth samples, we found significantly decreased expression of cwp66 and significantly increased expression of CD0802 in biofilm samples. Another adhesin, cbpA, encoding a collagen-binding adhesin, was recently identified in strain 630 (Tulli et al.2013), but the gene is not present in strain R20291. cwp84 encodes a cysteine protease that cleaves the highly expressed precursor surface layer protein SlpA into high-molecular-weight (HMW) and low-MW constituents; of the two, the HMW protein is more important for binding to human gastrointestinal tissues (Calabi et al.2002). Interestingly, we find no differences in cwp84 expression between all three growth conditions. This may reflect that our RNA-Seq biofilm samples were obtained after 1 week of growth, as a prior report documents that a cwp84 mutant has a more dramatic decrease in biofilm formation on day 1 compared to days 3 and 5 (Ðapa et al.2013).
Table 1.
Differential expression of specific C. difficile genes during biofilm growth (pairwise comparison).
| Gene | Description | Biofilm/plate differential expression log2(fold change) | Biofilm/planktonic differential expression log2(fold change) |
|---|---|---|---|
| pilA1 | Major pilin | −0.90 | 0.87* |
| pilA2 | Predicted pilin | −3.12* | −0.41 |
| pilA3 | Predicted pilin | −2.17 | 0.37 |
| pilJ | Minor pilin | 0.43 | 0.07 |
| pilK | Pilin-like protein | 1.02* | 1.18* |
| pilU | Pilin-like protein | 1.04* | 1.08* |
| pilV | Pilin-like protein | 1.60* | 0.96* |
| pilW | Pilin-like protein | −0.27 | −0.60* |
| pilX | Pilin-like protein | −2.21 | −0.26 |
| fliC | Flagellin | −0.25 | −1.35* |
| cwp66 | Adhesin (cell-wall protein) | −1.73* | −0.18 |
| cwp84 | Cysteine protease important for surface layer maturation | 0.37 | −0.31 |
| CD0802 | Adhesin (lipoprotein) | 4.87* | −0.87* |
| luxS | Quorum-sensing regulator | 1.99 | 0.12 |
| Spo0A | Transcription factor controlling spore formation | −1.35 | −0.23 |
| tcdA | Toxin A | −1.70* | 0.02 |
| tcdB | Toxin B | −1.53* | 0.59* |
| sigB | Housekeeping gene | −0.26 | −0.34 |
Statistically significant (q-value < 0.05).
Cell-to-cell communication via a quorum-sensing system is vital for biofilm formation in multiple organisms (Davies et al.1998; Yang et al.2014; Omer Bendori et al.2015). Clostridium difficile encodes the quorum-sensing regulator luxS, which is shown to regulate the expression of the genes encoding C. difficile toxin A and toxin B (Carter et al.2005; Lee and Song, 2005). Mutation of C. difficile luxS results in a dramatic deficiency in biofilm formation, with inability to form even a bacterial monolayer on glass surfaces (Ðapa et al.2013). We observed no significant differences in expression of luxS between the three growth phenotypes examined. We did, however, find differential expression of toxin genes, suggesting that factors other than quorum sensing via the luxS regulator are important in regulation of toxin gene expression. The toxin B gene (tcdB), but not the toxin A gene (tcdA), was significantly increased in biofilm samples when compared to planktonic samples, while both tcdA and tcdB were significantly decreased in biofilm samples in comparison to plate-growth samples.
RNA sequencing shows differences in pilin gene expression
We previously demonstrated that there are far more sequence reads from pilA1 than any other pilin gene when C. difficile is grown on blood agar under conditions that enhance the expression of pili (Fig. 1C and D), and that pilA1 encodes the major pilin (Piepenbrink et al.2015). Here, we also show that pilA1 is the most highly expressed pilin gene during biofilm and planktonic growth (Fig. 1A, B and D), further supporting the designation of pilA1as the major pilin gene. We also find pilJ is the second most highly expressed pilin gene in all growth conditions. This is consistent with our prior work demonstrating PilJ incorporation into pili, indicating that PilJ is a minor pilin (Piepenbrink et al.2014). When growth conditions are compared, significant expression differences are seen for multiple pilin genes (Fig. 1E). Expression of pilA1 is significantly greater on plates than under planktonic conditions in broth (q < 0.05), as was expected from prior experience examining pili by electron microscopy (Piepenbrink et al.2015). Relative expression of pilA1 is also significantly greater when grown in BHIS as a biofilm on glass beads than when grown as planktonic cells. This finding is consistent with recent work using quantitative reverse transcriptase PCR that found a significant increase in pilA1 expression in 48 h R20291 biofilms grown on plastic and compared to planktonic growth (Purcell et al.2016). Although they reside in the same gene cluster as pilA1, our data indicate that expression of the pilin-like protein genes pilK, pilU and pilV may have different regulation. All three of these RNAs are expressed at significantly higher levels in biofilm conditions when compared to plate or planktonic growth, while pilA1 is expressed at higher levels in biofilm relative to planktonic growth, but not relative to plate growth (Fig. 1E). Furthermore, while pilA1 is expressed at a significantly higher level in plate growth in comparison to planktonic growth, this is not the case for pilK, pilU and pilV (Fig. 1E). Recent data demonstrate a predicted transcription terminator 36 bp downstream from the pilA1 stop codon (Bordeleau et al.2015), consistent with our finding of greater pilA1 expression relative to pil K, pil U and pilV expression. Close inspection of the RNA-Seq data does not reveal evidence of a separate promoter or transcriptional terminator in this gene cluster. In contrast to the pilins from the largest gene cluster, pilJ, which is unlinked to the pilA1 gene cluster, is expressed at equivalent levels under all three conditions.
Figure 1.
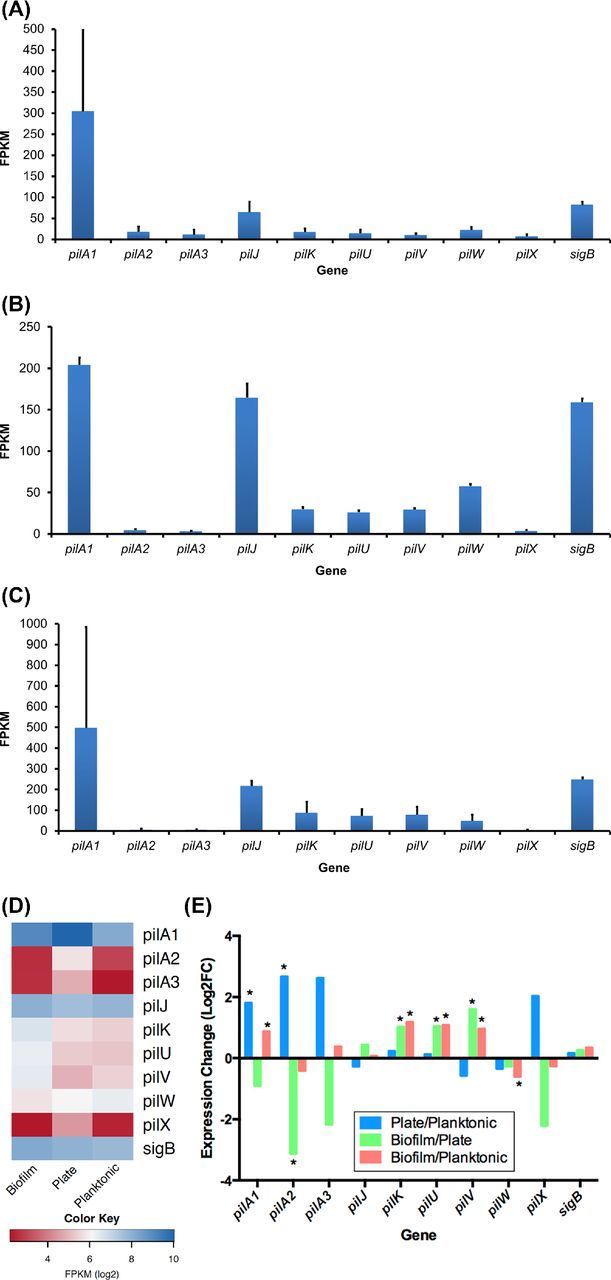
Pilin gene expression as determined by RNA sequencing of C. difficile samples grown on blood agar under conditions that express pili (A) as planktonic culture (B), and as a biofilm on glass beads (C). Using a three-way comparison, pilin gene expression is shown as a heatmap (D), and differential expression between the three growth conditions is shown using pairwise comparisons (E). *q value < 0.05. Please note that data included in panel A were previously published (Piepenbrink et al.2015).
A non-piliated mutant is deficient in initial biofilm formation
We previously demonstrated that PilA1 is the major C. difficile pilin, that the pilA1 mutant produces no visible pili in conditions known to induce piliation in the wild-type strain and that piliation is restored with addition of a complementation plasmid (Piepenbrink et al.2015). To investigate the role of T4P in biofilm formation, we grew wild-type C. difficile, the T4P-deficient mutant and the complemented mutant under biofilm-producing conditions.
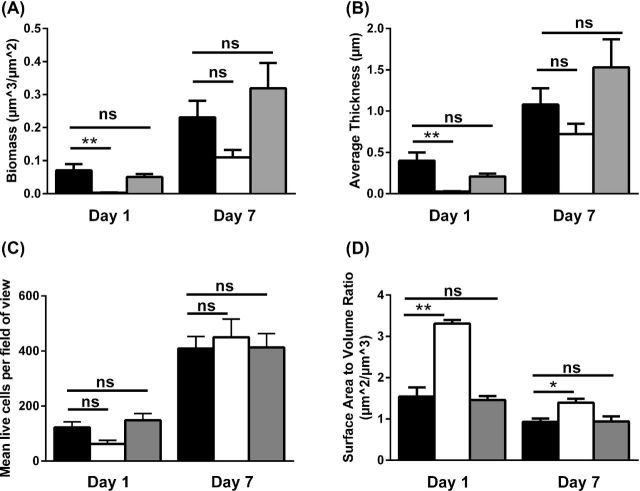
If T4P are important in biofilm formation, we would expect a non-piliated C. difficile mutant to be delayed in biofilm formation as indicated by decreased biomass and decreased live cell count in early biofilms as compared to wild-type bacteria; indeed, we observe this to be the case. Analysis of the 3D biofilm structure demonstrates that biofilms formed by the pilA1 mutant accumulate significantly less biomass at day 1 than the wild-type strain or the complemented mutant (Fig. 2A); correspondingly, the wild-type and the complemented pilA1 mutant form significantly thicker biofilms than the pilA1 mutant at day 1 (Fig. 2B). Quantification of CLSM images demonstrates that after 1 day of growth pilA1 mutant biofilm appears to have fewer live cells present per field of view than the parent wild-type strain (Figs 2C and 3A–C), although this finding is not statistically significant. The 3D reconstructions of biofilms illustrate the thicker biofilms formed by the wild-type and complemented mutant as compared to the pilA1 mutant (Fig. 3G–J) Complementation of pilA1 restores the count of live cells per field of view to levels slightly higher than seen in the wild-type strain.
Figure 2.
Assessment of lack of piliation on C. difficile biofilm formation. Biofilm biomass (A), thickness (B), live cell count (C) and surface-area-to-volume ratio (D) were calculated for wild-type C. difficile (black bars), the pilA1 mutant strain (white bars) and the complemented mutant (gray bars) at 1 and 7 days of growth. After 1 day of growth, the pilA1 mutant forms biofilms of significantly reduced biomass and thickness that contain fewer live cells as compared to the parent wild-type strain; the complemented mutant resembles the wild-type strain. These differences are still apparent after 7 days of growth, though these differences are no longer significant. The pilA1 mutant strain also has a significantly elevated surface-to-volume ratio as compared to the wild type at 1 and 7 days of growth. * indicates P < 0.01, ** indicates p < 0.001. Groups were compared using a one-way ANOVA with Dunnett's post-test. Error bars indicate SEM.
Figure 3.
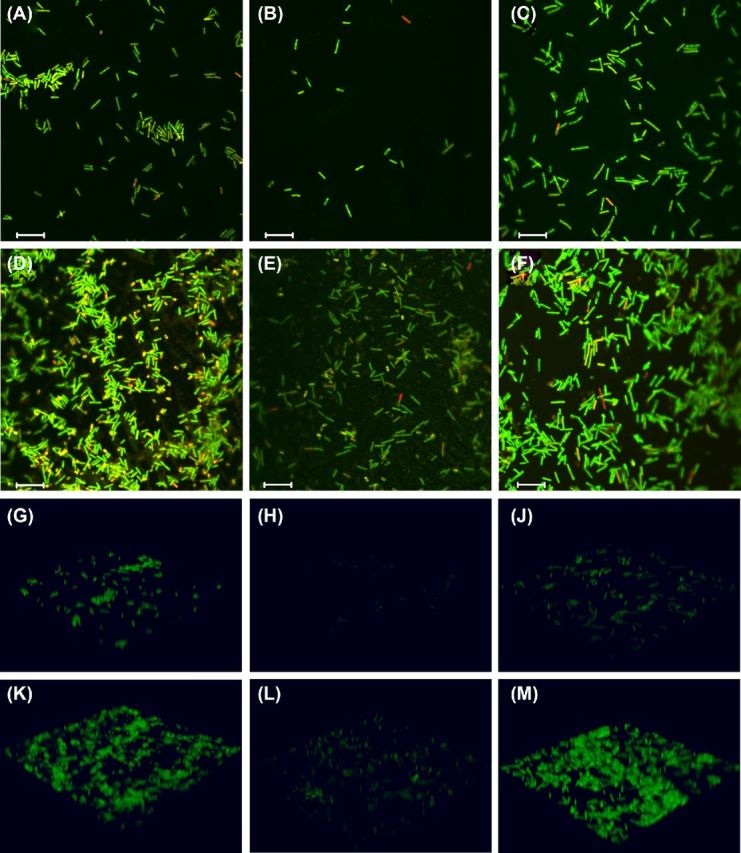
Visualization of C. difficile biofilms by CLSM. Representative images from each strain at both time points demonstrate the reduced biomass and thickness of the pilA1 mutant biofilms as compared to the wild-type strain. Complementation of the mutation restores the wild-type phenotype. The 3D reconstructions of the imaged biofilms further illustrate these observations. Images demonstrate 1 day (A-C, G-J) and 7 days (D–F, K-M) of growth for wild type (A, D, G and K), the pilA1 mutant (B, E, H and L) and the complemented mutant (C, F, J and M). Live cells are stained green, dead cells are stained red. The scale bar for images A–F indicates 10 μm.
The pilA1 mutant strain's biofilm formation deficiency is not simply due to differences in growth rates between the wild-type and mutant strains. In broth culture, the pilA1 mutant demonstrates similar growth as compared to the parent wild-type strain, indicating that the absence of T4P is the principal reason for the deficit in biofilm formation in the pilA1 mutant strain. While the non-piliated pilA1 mutant shows fewer live cells per field of view than the parent wild-type strain after 24 h of growth, this difference disappears by day 7 (Fig. 2C and H–J). As above, the 3D biofilm reconstructions demonstrate this phenotype (Fig. 3K–M). This phenotype, that of an early deficiency in biofilm formation, is consistent with previous observations of T4P-associated biofilms in C. perfringens and non-typeable Haemophilus influenzae (NTHI) (Carruthers et al.2012; Novotny et al.2015).
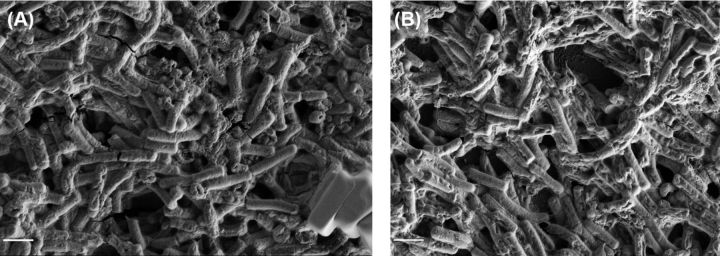
Along with total biomass and biofilm thickness, the measured surface-area-to-volume ratio provides a useful descriptor of a strain's tendency to form clumps or aggregates of bacteria; a lower surface-area-to-volume ratio indicates fewer isolated bacteria and more aggregates. Given that T4P are required for aggregation or microcolony formation by other bacteria, we would anticipate an increased surface-area-to-volume ratio in the pilA1 mutant relative to the wild-type strain. Indeed, we find that the pilA1 mutant has an increased ratio relative to the wild-type strain, and complementation restores this ratio to wild-type levels (Fig. 2D). Importantly, these results clearly indicate a role for T4P in promoting bacterial aggregation rather than simply increasing adherence to an abiotic surface. However, CLSM images show aggregates of bacteria in all strains at both time points (Fig. 3A–G), although surface-area-to-volume ratios demonstrate that the wild-type strain has a greater propensity toward aggregate formation than the pilA1 mutant (Fig. 3K–M). Similarly, SEM data show that both wild-type and pilA1 mutants are capable of forming aggregates with an extracellular matrix (Fig. 4).
Figure 4.
Formation of biofilm by C. difficile. SEM imaging demonstrates formation of cell aggregates linked by matrix by both wild-type (A) and pilA1 mutant (B) strains of C. difficile after 7 days of growth. Scale bar indicates 2 μm.
Given the presence of these aggregates in the pilA1 mutant, these aggregates can form in the absence of T4P composed of PilA1. Thus, other adhesive proteins, for example CbpA or SlpA (Merrigan et al.2013; Tulli et al.2013), may be involved in cell–cell adhesion alongside T4P; alternately, T4P composed of PilA2 or PilA3 may be involved in aggregate formation. As shown above, pilA2 and pilA3 are expressed at low levels in the C. difficile biofilm; however, no pili were observed on the pilA1 mutant, suggesting their role, if any, is a small one.
In sum, T4P genes are differentially expressed during biofilm growth and critical for early biofilm formation in C. difficile: a non-piliated strain is deficient in initial biofilm formation, forms a biofilm of reduced thickness and biomass and has decreased tendency to aggregate in comparison to the parent wild-type strain. Complementation of the mutant restores the above measures to wild-type levels.
DISCUSSION
Despite the human and medical costs of CDI, its mechanisms of colon colonization are poorly understood. Previous research demonstrates that C. difficile forms biofilms and expresses T4P (Ðapa et al.2013; Maldarelli et al.2014; Bordeleau et al.2015; Purcell et al.2016). In this work, we sought to elucidate the role of T4P in C. difficile biofilm formation. We demonstrate major differences in gene expression when C. difficile is grown as a biofilm as compared to planktonic or plate growth, including differences in pilin gene expression. We also demonstrate the role of T4P in biofilm generation: a non-piliated mutant is slower to form biofilm and less likely to form aggregates than the parent wild-type strain. Together, our results demonstrate that biofilm is a distinct mode of C. difficile growth, one in which T4P play an important role. Furthermore, these data reinforce earlier work (Ðapa et al.2013) that emphasizes the importance of studying biofilms in this clinically relevant organism.
Our RNA-Seq studies provide valuable information regarding gene expression in C. difficile biofilms. We find that about half of all genes in our biofilm-growth samples are significantly differentially expressed when compared to either planktonic- or plate-growth samples. When comparing biofilm and planktonic gene expression, we expected to see increased expression of adhesin genes in the biofilm samples; however, we saw significantly lower expression of CD0802 and insignificantly lower expression of cwp66. These data lead us to hypothesize that these adhesins are not expressed at high levels once a biofilm is established, and this finding may reflect that samples were grown for 1 week before RNA extraction. Alternatively, as the ligands for these adhesins are unknown, and may be present on host tissue but not the surface of C. difficile, increased expression of these adhesins may rely on the presence of the host. Although others have shown a role for the quorum-sensing regulator luxS, the sporulation transcription factor spo0A and cysteine protease cwp84 in biofilm formation (Ðapa et al.2013), we found no significant differences in expression of any of these genes in our samples. One limitation of our study is that we only have expression data for 1-week biofilm samples, and it is quite possible that differences occur at earlier time points.
Using quantitative biofilm formation assays, we show that a non-piliated mutant is deficient in early biofilm formation, which supports our hypothesis. The trend toward a decreased day 1 live-cell count in the non-piliated strain suggests that T4P are important in surface attachment, the first step of biofilm formation. While pili primarily composed of PilA2 and PilA3 may be expressed under other conditions, the genes encoding those proteins may be remnants of other complete T4P gene clusters, and may no longer be expressed. Our RNA-Seq data suggest that pilA2 and pilA3 are poorly expressed under the conditions tested here, and no pili were observed in the pilA1 mutant, indicating that, under the conditions used here, PilA2 and PilA3 pili are unlikely to contribute to biofilm formation. Deficiency in biofilm formation upon loss of piliation is common to other T4P-expressing, biofilm-forming organisms, including NTHI and Pseudomonas aeruginosa. NTHI strains lacking expression of the major pilin pilA form biofilms of reduced thickness and reduced biomass after 24 h of growth (Carruthers et al.2012), results consistent with our findings in C. difficile. Similarly, experiments with cocultures of wild-type and non-piliated mutant P. aeruginosa strains demonstrate that non-piliated bacteria are less able to move between microcolonies, and consequently form less regular biofilms (Klausen et al.2003a). The two T4P systems of the aquatic pathogen Vibrio parahaemolyticus have discrete roles in biofilm formation; the mannose-sensitive hemagglutinin pilus is involved in bacterial adhesion to surfaces, whereas the chitin-regulated pilus plays a role in cell–cell adhesion (Shime-Hattori et al.2006). T4P in C. difficile may serve similar purposes, allowing bacteria in the nascent biofilm to adhere to each other, as well as to surfaces. Clostridium difficile encodes three putative major pilins, pilA1, pilA2 and pilA3 (Merrigan et al.2013). We demonstrate here that pili composed of PilA1 are involved in early biofilm formation, although this result does not exclude the possibility that pili composed primarily of PilA2 or PilA3 also contribute to biofilm formation and maintenance. As in V. parahaemolyticus, multiple types of pili may be involved in aggregation and biofilm formation; other mechanisms unrelated to pili are most likely involved as well.
Our observations regarding increased surface-area-to-volume ratio in the pilA1 mutant relative to the wild-type strain support the hypothesis that T4P aid the bacteria in forming aggregates or microcolonies, as a lower relative surface-to-volume ratio is indicative of more cell aggregates. Microcolonies are a critical part of P. aeruginosa biofilm initiation, and in Neisseria meningitidis, T4P-reliant microcolony formation is critical for bacterial passage across the blood–brain barrier. Our results provide a potential explanation for C. difficile cell aggregates, that is linkage by hair-like structures, which have been seen by SEM in the ceca and colons of C. difficile-infected hamsters 24 and 36 h post-infection (Buckley et al.2011). Taken together, our findings and the images acquired by Buckley and colleagues suggest a possible mechanism of colonic C. difficile colonization: T4P facilitate initial attachment to the colonic epithelium and microcolony formation, which in turn leads to progression of disease. The involvement of T4P in initial C. difficile biofilm formation is dependent upon c-di-GMP levels, as was recently demonstrated by Purcell et al. (2016).
The ability to form biofilms may provide C. difficile a selective advantage. If T4P facilitate C. difficile adherence (both bacterium to bacterium and bacterium to epithelium), T4P-mediated adhesion could allow the bacteria to persist in the colon. Although spore formation may be the predominant mechanism underlying recurrent CDI, the presence of sessile communities in the colon may help to explain why symptoms of CDI can persist in some individuals despite recommended therapy and why infection recurs with high frequency. Bacteria in the biofilm mode of growth are more resistant to antimicrobials than bacteria grown on plates or in broth (Folsom et al.2010). This phenomenon has been demonstrated in vitro in C. difficile; bacteria grown as a biofilm were demonstrably more resistant to killing by super-MIC concentrations of vancomycin when compared to equivalent planktonic cultures (Ðapa et al.2013). This advantage at super-MIC levels of vancomycin may indicate one mechanism by which C. difficile evades eradication during oral vancomycin treatment.
In conclusion, we demonstrate that the RNA expression profile of C. difficile in biofilm is distinct from that of C. difficile grown in broth or on agar plates, including significant differences in pilin, adhesin and toxin gene expression. We find that bacteria unable to produce T4P form thinner biofilms with decreased biomass and a trend toward fewer live cells than biofilms formed by T4P-producing, wild-type bacteria. This work leads us to a more general understanding of biofilm development and the role of T4P in that development.
FUNDING
This work was supported by the National Institutes of Health training fellowship F30 DK105539 (GAM), National Institutes of Health training grant T32 AI 095190 (KHP), National Institutes of Health training fellowship F32 AI 110045 (KHP), National Institutes of Health grant R21 AI105881 (MSD), National Institutes of Health grant R01 AI114902 (EJS), and a pilot project grant from the Baltimore Research and Education Foundation (ECvR).
Conflict of interest. None declared.
REFERENCES
- Bartlett JG, Gerding DN. Clinical recognition and diagnosis of Clostridium difficile infection. Clin Infect Dis. 2008;46:S12–8. doi: 10.1086/521863. [DOI] [PubMed] [Google Scholar]
- Becker P, Hufnagle W, Peters G, et al. Detection of differential gene expression in biofilm-forming versus planktonic populations of Staphylococcus aureus using micro-representational-difference analysis. Appl Environ Microb. 2001;67:2958–65. doi: 10.1128/AEM.67.7.2958-2965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard SC, Simpson N, Join-Lambert O, et al. Pathogenic Neisseria meningitidis utilizes CD147 for vascular colonization. Nat Med. 2014;20:725–31. doi: 10.1038/nm.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau E, Purcell EB, Lafontaine DA, et al. Cyclic di-GMP riboswitch-regulated type IV pili contribute to aggregation of Clostridium difficile. J Bacteriol. 2015;197:819–32. doi: 10.1128/JB.02340-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borriello SP, Davies HA, Barclay FE. Detection of fimbriae amongst strains of Clostridium difficile. FEMS Microbiol Lett. 1988;49:65–7. [Google Scholar]
- Brady RA, O'May GA, Leid JG, et al. Resolution of Staphylococcus aureus biofilm infection using vaccination and antibiotic treatment. Infect Immun. 2011;79:1797–803. doi: 10.1128/IAI.00451-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley AM, Spencer J, Candlish D, et al. Infection of hamsters with the UK Clostridium difficile ribotype 027 outbreak strain R20291. J Med Microbiol. 2011;60:1174–80. doi: 10.1099/jmm.0.028514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabi E, Calabi F, Phillips AD, et al. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect Immun. 2002;70:5770–8. doi: 10.1128/IAI.70.10.5770-5778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carruthers MD, Tracy EN, Dickson AC, et al. Biological roles of nontypeable Haemophilus influenzae type IV pilus proteins encoded by the pil and com operons. J Bacteriol. 2012;194:1927–33. doi: 10.1128/JB.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter GP, Purdy D, Williams P, et al. Quorum sensing in Clostridium difficile: analysis of a luxS-type signalling system. J Med Microbiol. 2005;54:119–27. doi: 10.1099/jmm.0.45817-0. [DOI] [PubMed] [Google Scholar]
- Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–78. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- Ðapa T, Leuzzi R, Ng YK, et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol. 2013;195:545–55. doi: 10.1128/JB.01980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–8. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsom JP, Richards L, Pitts B, et al. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol. 2010;10:294–310. doi: 10.1186/1471-2180-10-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IC, O'Toole E. Intestinal flora in new-born infants: With a description of a new pathogenic anaerobe, Bacillus difficilis. Am J Dis Child. 1935;49:390–402. [Google Scholar]
- Heydorn A, Nielsen AT. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Humphries RM, Waterhouse CCM, Mulvey G, et al. Interactions of enteropathogenic Escherichia coli with pediatric and adult intestinal biopsy specimens during early adherence. Infect Immun. 2009;77:4463–8. doi: 10.1128/IAI.00686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoir C, Péchiné S, Grosdidier C, et al. Cwp84, a surface-associated protein of Clostridium difficile, is a cysteine protease with degrading activity on extracellular matrix proteins. J Bacteriol. 2007;189:7174–80. doi: 10.1128/JB.00578-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamentsky L, Jones TR, Fraser A, et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27:1179. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CP, LaMont JT. Clostridium difficile — more difficult than ever. N Engl J Med. 2008;359:1932–40. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- Klausen M, Aaes-Jørgensen A, Molin S, et al. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol. 2003a;50:61. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- Klausen M, Heydorn A, Ragas P, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol. 2003b;48:1511–24. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- Kovacs-Simon A, Leuzzi R, Kasendra M, et al. Lipoprotein CD0873 is a novel adhesin of Clostridium difficile. J Infect Dis. 2014;210:274–84. doi: 10.1093/infdis/jiu070. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASY, Song KP. LuxS/autoinducer-2 quorum sensing molecule regulates transcriptional virulence gene expression in Clostridium difficile. Biochem Bioph Res Co. 2005;335:659–66. doi: 10.1016/j.bbrc.2005.07.131. [DOI] [PubMed] [Google Scholar]
- Lessa FC, Mu Y, Bamberg WM, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825–34. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsacker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. P Natl Acad Sci USA. 2007;104:11197–202. doi: 10.1073/pnas.0704624104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland LV. Renewed interest in a difficult disease: Clostridium difficile infections – epidemiology and current treatment strategies. Curr Opin Gastroen. 2009;25:24–35. doi: 10.1097/MOG.0b013e32831da7c4. [DOI] [PubMed] [Google Scholar]
- Maldarelli GA, De Masi L, von Rosenvinge EC, et al. Identification, immunogenicity, and cross-reactivity of type IV pilin and pilin-like proteins from Clostridium difficile. Pathog Dis. 2014;71:302–14. doi: 10.1111/2049-632X.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrigan MM, Venugopal A, Roxas JL, et al. Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLoS One. 2013;8:1–12. doi: 10.1371/journal.pone.0078404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musher DM, Logan N, Hamill RJ, et al. Nitazoxanide for the treatment of Clostridium difficile colitis. Clin Infect Dis. 2006;43:421–7. doi: 10.1086/506351. [DOI] [PubMed] [Google Scholar]
- Nassif X, Beretti J.-L, Lowy J, et al. Roles of pilin and PilC in adhesion of Neisseria meningitidis to human epithelial and endothelial cells. P Natl Acad Sci USA. 1994;91:3769–73. doi: 10.1073/pnas.91.9.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny LA, Jurcisek JA, Ward MO, et al. Antibodies against the majority subunit of type IV pili disperse nontypeable Haemophilus influenzae biofilms in a LuxS-dependent manner and confer therapeutic resolution of experimental otitis media. Mol Microbiol. 2015;96:276–92. doi: 10.1111/mmi.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obana N, Nakamura K, Nomura N. A sporulation factor is involved in the morphological change of Clostridium perfringens biofilms in response to temperature. J Bacteriol. 2014;196:1540–50. doi: 10.1128/JB.01444-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen I. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol. 2015;34:877–86. doi: 10.1007/s10096-015-2323-z. [DOI] [PubMed] [Google Scholar]
- Omer Bendori S, Pollak S, Hizi D, et al. The RapP-PhrP quorum-sensing system of Bacillus subtilis strain NCIB3610 affects biofilm formation through multiple targets, due to an atypical signal-insensitive allele of RapP. J Bacteriol. 2015;197:592–602. doi: 10.1128/JB.02382-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenbrink KH, Maldarelli GA, de la Peña CFM, et al. Structure of Clostridium difficile PilJ exhibits unprecedented divergence from known type IV pilins. J Biol Chem. 2014;289:4334–45. doi: 10.1074/jbc.M113.534404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenbrink KH, Maldarelli GA, Martinez de la Peña CF, et al. Article: structural and evolutionary analyses show unique stabilization strategies in the type IV pili of Clostridium difficile. Structure. 2015;23:385–96. doi: 10.1016/j.str.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell EB, McKee RW, Bordeleau E, et al. Regulation of type IV pili contributes to surface behaviors of historical and epidemic strains of Clostridium difficile. J Bacteriol. 2016;198:565–77. doi: 10.1128/JB.00816-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakotoarivonina H, Jubelin G, Hebraud M, et al. Adhesion to cellulose of the Gram-positive bacterium Ruminococcus albus involves type IV pili. Microbiol. 2002;148:1871–80. doi: 10.1099/00221287-148-6-1871. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative Genomics Viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B, Schindelin J, Cardona A, et al. A high-level 3D visualization API for Java and ImageJ. BMC Bioinformatics. 2010;11:1–7. doi: 10.1186/1471-2105-11-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shime-Hattori A, Iida T, Arita M, et al. Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol Lett. 2006;264:89–97. doi: 10.1111/j.1574-6968.2006.00438.x. [DOI] [PubMed] [Google Scholar]
- Tacket CO, Taylor RK, Losonsky G, et al. Investigation of the roles of toxin-coregulated pili and mannose-sensitive hemagglutinin pili in the pathogenesis of Vibrio cholerae O139 infection. Infect Immun. 1998;66:692–5. doi: 10.1128/iai.66.2.692-695.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team RC, et al. R: The R Project for Statistical Computing [WWW Document] 2014 https://www.r-project.org/ (3 May 2016, date last accessed) [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulli L, Marchi S, Petracca R, et al. CbpA: a novel surface exposed adhesin of Clostridium difficile targeting human collagen. Cell Microbiol. 2013;15:1674–87. doi: 10.1111/cmi.12139. [DOI] [PubMed] [Google Scholar]
- Varga JJ, Nguyen V, O'Brien DK, et al. Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other Clostridia. Mol Microbiol. 2006;62:680–94. doi: 10.1111/j.1365-2958.2006.05414.x. [DOI] [PubMed] [Google Scholar]
- Vorregaard M. Comstat2 - A Modern 3D Image Analysis Environment for Biofilms (Master's) Kongens Lyngby, Denmark: Technical University of Denmark (DTU), Informatics and Mathematical Modeling; 2008. [Google Scholar]
- Waligora AJ, Hennequin C, Mullany P, et al. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect Immun. 2001;69:2144–53. doi: 10.1128/IAI.69.4.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, et al. gplots: Various R Programming Tools for Plotting Data. 2015. R package version 2.17.0. http://CRAN.R-project.org/package=gplots. [Google Scholar]
- Whitchurch CB, Hobbs M, Livingston SP, et al. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- Yang K, Meng J, Huang Y, et al. The role of the QseC quorum-sensing sensor kinase in epinephrine-enhanced motility and biofilm formation by Escherichia coli. Cell Biochem Biophys. 2014;70:391–8. doi: 10.1007/s12013-014-9924-5. [DOI] [PubMed] [Google Scholar]