Abstract
The effects of small (~102 Da) and larger (>103 Da) cosolutes on the equilibrium stability of monomeric globular proteins are broadly understood, excluding volume stabilizes proteins and chemical interactions are stabilizing when repulsive, but destabilizing when attractive. Proteins, however, rarely work alone. Here, we investigate the effects of small and large cosolutes on the equilibrium stability of the simplest defined protein–protein interactions, the side-by-side homodimer formed by the A34F variant of the 56-residue B1 domain of protein G. We used 19F nuclear magnetic resonance spectroscopy to quantify the effects of urea, trimethylamine oxide, Ficoll, and more physiologically relevant cosolutes on the dimer dissociation constant. The data reveal the same stabilizing and destabilizing influences from chemical interactions as observed in studies of protein stability. Results with more physiologically relevant molecules such as bovine serum albumin, lysozyme, and reconstituted Escherichia coli cytosol reflect the importance of chemical interactions between these cosolutes and the test protein. Our study serves as a stepping-stone to a more complete understanding of crowding effects on protein–protein interactions.
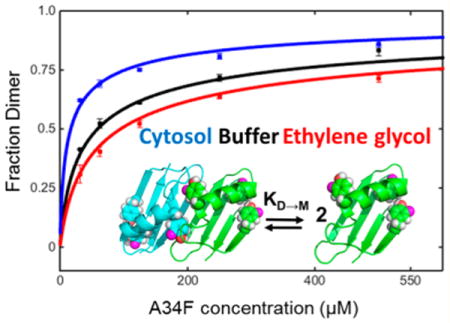
Graphical abstract
The Escherichia coli cytoplasm, which has a macromolecule concentration of greater than 300 g/L,1 is a complex environment crowded with an assortment of proteins, nucleic acids, and small molecules. A reductionist approach featuring simple buffered solutions with macromolecule concentrations of less than 10 g/L, however, is usually utilized to simplify investigations of protein biophysics. Such approaches provide a wealth of knowledge about protein structure, function, and folding, but they neglect the transient interactions that take place in cells brought about by the crowded environment.2,3
To understand how proteins behave in cells, high concentrations of cosolutes are often used to simulate the cellular interior. Cosolutes, from large and supposedly inert macromolecules like polyethylene glycol (PEG) and the cross-linked sucrose polymer Ficoll-70 to globular proteins, have been used to mimic the cellular interior.4–6 In concentrated cosolute solutions, the test protein experiences two interactions between the cosolutes and the test protein that are absent in dilute solution: hard-core repulsions and chemical interactions.7–15 Hard-core repulsions reduce the volume available to the test protein and favor states that occupy the least space.16 Chemical interactions arise from the close proximity of the test protein and the cosolute and include charge–charge,12,17 hydrophobic, and hydrogen bonding interactions. Repulsive chemical interactions between charges of the same sign on the test protein and the cosolute stabilize globular proteins because they enhance the hard-core repulsions. Attractive interactions destabilize globular proteins because protein unfolding exposes additional groups that can form attractive interactions.9,10,14,15,17,18
Environments ranging from small cosolutes to synthetic polymers, globular proteins, cell lysates, and even the cellular interior6–14,17 have been used to explore how crowding affects protein stability and folding. However, proteins rarely act alone, and there are few studies on the effect of crowding on protein–protein interactions.19,20
The B1 domain of protein G (GB1) is one of the most extensively studied globular proteins. This 56-residue molecule adopts a thermally stable 4β+α globular fold, and a large number of variants have been characterized using a variety of biophysical methods.21–23 One variant, A34F, forms a simple side-by-side dimer (Figure 1).24 Such dimers can be thought of as a pair of kissing spheres where the volume of the dimer is approximately twice that of the monomer,25 and there is a small decrease in solvent accessible surface area upon dimer formation. Dimerization of A34F GB1 occurs because the side chain of Phe 34 becomes part of the hydrophobic core, displacing the Tyr 33 side chain at the surface.24 To compensate, the C-terminal end of the sole helix unfolds, forming a pocket for the Tyr 33 side chain of the other GB1 monomer. The dimer is also stabilized by hydrogen bonding along the now adjacent β-sheets.
Figure 1.
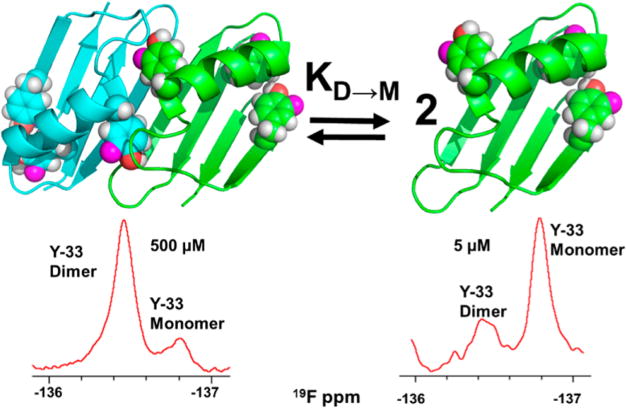
A34F GB1 dimer and monomer and 19F NMR spectra acquired at two concentrations. In each spectrum, the area under a peak is proportional to the concentration of the corresponding state, allowing straightforward quantification of the dissociation constant, KD→M.
For A34F GB1, our calculations show that there is no volume reduction on dimerization; the dimer and monomer molecular volumes are 15 500 Å3 and 7500 Å3, respectively. The change in solvent accessible surface is also small; the area of the dimer, 7023.90 Å2, is only 9% less than that of two monomers (2 × 3924.1 Å2). Scaled-particle theory predicts that hard-core repulsions have a small effect on the stability of side by side dimers,25 making this a good system for focusing on chemical interactions.
As we show, the dissociation constant, KD→M, where D represent the dimer and M the monomer, can be quantified in buffer and buffered solutions containing high concentrations of cosolutes by using 19F nuclear magnetic resonance spectroscopy (NMR). Fluorine-19 is an attractive nucleus because it is 100% abundant, rarely used in biology, has a high NMR sensitivity (83% of protons) and its chemical shift is sensitive to its environment.26–28 Furthermore, the simplicity of one-dimensional 19F spectra allows the acquisition of data in a matter of minutes, even in living cells.29,30 Importantly, Escherichia coli readily incorporate fluorinated aromatic amino acids into recombinant protein.31,32 GB1 contains three tyrosines that are readily labeled with 3-fluorotyrosine. Their resonances have been assigned.33
EXPERIMENTAL PROCEDURES
Vector
The gene for T2Q GB1 in pET11a was used as the wild-type vector. This mutation prevents N-terminal degradation.34 We refer to the T2Q variant as the wild-type protein. The A34F change was made using Agilent’s QuickChange mutagenesis kit.
Expression and Purification
GB1 was expressed in E. coli BL21(DE3) cells and purified using a modified protocol.35 Briefly, a 1-L culture harboring the A34F construct was grown in antibiotic-containing, 15N-enriched M9 media and incubated at 37 °C with shaking (New Brunswick Scientific Innova I26, 225 rpm). When the cells reached an optical density at 600 nm of 0.4, N-phosphonomethylglycine (0.5 g, to inhibit aromatic amino acid synthesis), 3-fluorotyrosine (70 mg), phenylalanine (60 mg), and tryptophan (60 mg) were added.36 Protein expression was induced with isopropyl β-D-1-thiogalactopyranoside (IPTG), at a final concentration of 1 mM, when the culture reached an optical density at 600 nm of 0.6. After 2 h, the cells were pelleted at 1000g for 25 min. The supernatant was discarded, and the pellet was stored at −20 °C. Cells were lysed by sonication (Fischer Scientific Sonic Dismembrator model 500, 15% amplitude, 10 min, 50% duty cycle) in 25 mL of 20 mM tris, pH 7.5, containing a cOmplete protease inhibitor tablet (Roche). The lysate was centrifuged for 45 min at 27000g to remove cell debris, and the supernatant was filtered (0.22 μm). The filtrate was loaded on a 16 mm × 200 mm Q Sepharose anion exchange column attached to a GE AKTA FPLC. The column was eluted with 20 mM tris, pH 7.5 using a gradient from 0 to 1 M NaCl over three column volumes. Fractions were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE) to assess protein content. GB1-containing fractions were concentrated in a 3000-molecular-weight-cutoff Amicon spin concentrator and buffer exchanged into 10 mM potassium phosphate containing 150 mM NaCl (pH 6.0). The final volume was 3 mL. This solution was loaded on a 16 mm × 600 mm GE Superdex-75 gel filtration column and developed over two column volumes of the same buffer. Fractions were subjected SDS PAGE. Fractions containing only GB1 were concentrated and buffer exchanged into filtered, 17 MΩ cm−1 H2O, flash frozen in a CO2(s)/ethanol bath and lyophilized for at least 12 h (Labconco FreeZone). Lyophilized protein was stored at −20 °C. The process was completed in <36 h to avoid aggregation. Protein purity was confirmed by electrospray ionization mass spectrometry (expected 6385 Da, observed 6391 Da, Figure S1), SDS PAGE, and NMR spectroscopy (Figure S2).
NMR
Fluorine-labeled protein was resuspended at a final concentration of 500 μM in either 20 mM sodium phosphate buffer (pH 7.5, 298 K), or this buffer plus cosolute. Experiments were conducted with a Bruker Avance III HD spectrometer operating at a 19F Larmor frequency of 470 MHz and equipped with a cryogenic QCI probe with an H/F channel. Spectra comprised 31047 points, 128 scans, a delay of 2 s, an acquisition time of 1.4 s, an offset of −100 ppm, and a sweep width of 100 ppm. Samples were internally referenced using a Wilmad coaxial insert containing 0.01% deuterated trifluoroacetic acid (−75.6 ppm) in D2O.
Data were processed using TopSpin 3.2. A line broadening function of 10 Hz was applied to each free-induction decay before Fourier transformation. The resonances from tyrosine 33 corresponding to the monomer and dimer were integrated to obtain the relative populations. The fraction of protein in the dimer, Fd, was calculated by dividing the integral of the dimer peak by the sum of the integrals of the monomer and dimer peaks. KD→M values were obtained as described in Results using MATLAB version R2016A.
Cosolutes
Small cosolutes were weighed, dissolved in 20 mM sodium phosphate buffer, adjusted to pH 7.5, and diluted to the desired concentrations. Lyophilized lysozyme and bovine serum albumin were purchased from Sigma-Aldrich. To prepare these cosolute solutions, protein was weighed and dissolved in 20 mM sodium phosphate buffer (pH 7.5). Concentrations were determined spectrophotometrically with a Nanodrop 1000 spectrophotometer using extinction coefficients of 6700 (L mg−1 cm−1) for bovine serum albumin37 and 26 400 (L mg−1 cm−1) for lysozyme.38 E. coli lysates were prepared as described.39
Volumes and Surface Areas
VADAR40 and POPS41 software were used to calculate the volumes and solvent accessible surface areas, respectively. Volumes were calculated using the standard Voronoi procedure and the PDB structure 2RMM24 for the dimer and 2RMMa and 2RMMb for the monomer. Monomer solvent accessible surface areas were calculated using the “per chain analysis” of 2RMM and 1GB1 with a probe radius of 1.4 Å.
RESULTS
Quantifying Dimerization
A34F GB1 has tyrosines at positions 3, 33, and 45. Its 19F spectrum exhibits six peaks. Tyrosines 3 and 45 are buried in the core. Their rotomers are in slow exchange on the NMR time scale, and, therefore, two resonances are observed for each residue. One resonance is observed for tyrosine 33, suggesting that its rotomers are in fast exchange or have similar chemical shifts. Nevertheless, the shift of the tyrosine 33 peak is affected by dimerization; its resonance exhibits concentration dependence (Figure S3), suggesting that the dimer and the monomer are in slow exchange, in agreement with previous studies.24 We assigned the dimer to the resonance that increases with GB1 concentration. We integrated the dimer and monomer peaks to give their relative populations at five GB1 concentrations using serial dilution. Experiments were performed in triplicate. The data were fitted to eq 1,42 where PT is the total GB1 concentration, to yield a KD→M at 298 K and pH 7.5 of 59 ± 2 μM, where the uncertainty is the standard deviation of the mean. Our measured dissociation constant is approximately twice that determined at pH 5.5 in 50 mM sodium phosphate (27 ± 4 μM).24
| (1) |
The near identity of 15N−1H HSQC spectra shows that fluorine labeling has a small effect on the structure of A34F (Figure S4) and KD→M, because equilibrium analytical ultracentrifugation of the unlabeled protein gives the same value (Figured S5). In summary, 19F NMR permits facile and precise quantification of KD→M for the GB1 A34F side-by-side dimer from five A34F concentrations in less than 2 h.
Small Molecule and Synthetic Polymer Cosolutes
Two osmolytes, TMAO (75 Da) and urea (60 Da), were tested. TMAO (38 g/L) stabilized the dimer, reducing KD→M to 25 ± 2 μM. Urea destabilized the dimer, increasing KD→M to 90 ± 5 μM. The effects of polyethylene glycol (PEG, 8 kDa) its monomer [ethylene glycol (200 g/L)], Ficoll-70 (70 kDa) and its monomer [sucrose (300 g/L)] were examined at the highest concentrations consistent with acquiring high quality NMR data. PEG and ethylene glycol destabilized the dimer, giving KD→M values of 85 ± 5 μM and 95 ± 6 μM, respectively. Ficoll weakly stabilized the dimer yielding a KD→M of 45 ± 2 μM, but its monomer, sucrose, at the same g/L concentration, had a small effect compared to buffer alone (KD→M = 55 ± 2 μM). The results are summarized in Table 1.
Table 1.
KD→M and ΔGo′D→M values for the A34F GB1 Dimer at pH 7.5, 298 Ka
| condition |
μM
|
kcal/mol
|
|
|---|---|---|---|
| KD→M | ΔG°′D→M | ΔΔG°′D→M | |
| buffer (20 mM NaPO4) | 59 ± 2 | 5.75 ± 0.03 | N/A |
| ethylene glycol, 200 g/L | 95 ± 6 | 5.46 ± 0.04 | −0.29 ± 0.05 |
| urea, 100 g/L | 90 ± 5 | 5.50 ± 0.03 | −0.25 ± 0.04 |
| 8 kDa PEG, 200 g/L | 85 ± 5 | 5.53 ± 0.03 | −0.22 ± 0.03 |
| lysozyme, 50 g/L | 80 ± 5 | 5.57 ± 0.04 | −0.18 ± 0.05 |
| sucrose, 300 g/L | 55 ± 3 | 5.78 ± 0.03 | 0.03 ± 0.04 |
| Ficoll-70, 300 g/L | 45 ± 2 | 5.90 ± 0.03 | 0.15 ± 0.04 |
| BSA, 100 g/L | 26 ± 2 | 6.23 ± 0.05 | 0.48 ± 0.06 |
| TMAO, 38 g/L | 25 ± 2 | 6.25 ± 0.05 | 0.50 ± 0.06 |
| cytosol, 75 g/L | 17 ± 2 | 6.47 ± 0.07 | 0.72 ± 0.08 |
Values are averages. Uncertainties are standard deviations of the mean from three trials.
Proteins and Freeze-Dried Cytosol
Two protein cosolutes, lysozyme (14 kDa, pI 11.4) and bovine serum albumin (BSA, 68 kDa, pI 4.5) and freeze-dried E. coli cytosol39 (75 g/L) were tested. BSA at 100 g/L stabilized the dimer (KD→M 26 ± 2 μM), but 50 g/L lysozyme destabilized the dimer (KD→M 80 ± 5 μM). The cosolutes were used at the highest concentrations that give interpretable NMR spectra (higher concentrations caused excessive broadening). Cytosol had the largest stabilizing effect of any cosolute (KD→M 17 ± 2 μM). The results are summarized in Table 1.
DISCUSSION
Dissociation constants (KD→M) and modified standard state dissociation free energies (ΔG°′D→M) with respect to buffer were compared at pH 7.5 and 298 K (Figure 3, Table 1).
Figure 3.
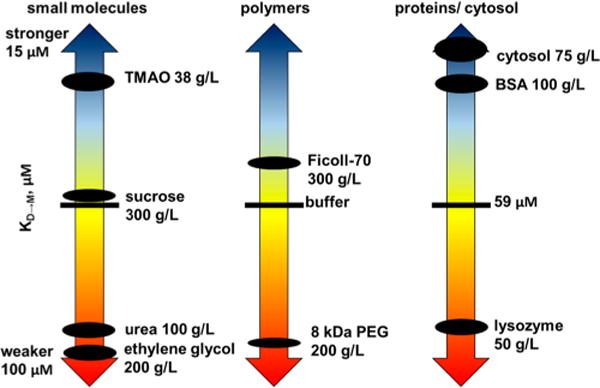
A34F dissociation constants (KD→M) in buffer, osmolytes, synthetic polymers, their monomers, and protein crowders. The size of the dots reflects the uncertainty.
Small Molecule Cosolutes
Urea and ethylene glycol interact favorably with the protein surface, favoring states with the most surface because expansion maximizes interactions between the protein and the cosolute.43–45 Consistent with this idea, urea and ethylene glycol destabilize the dimer by 0.25 ± 0.04 kcal/mol and 0.29 ± 0.05 kcal/mol, respectively. TMAO and sucrose work in the opposite manner, interacting unfavorably with the protein, favoring compact, over expanded, states.46,47 As expected, TMAO stabilizes the dimer by 0.50 ± 0.06 kcal/mol. It is unclear why sucrose has an almost negligible effect, even though it stabilizes proteins. Perhaps the result arises from a balancing of repulsive interactions from the backbone and attractive interactions from the exposed residues on the protein surface.47 Formal proof of chemical interactions, however, must await determination of dissociation enthalpies.18,48–51
Synthetic Polymers and Their Monomers
Both PEG and ethylene glycol destabilize the dimer, but PEG is less destabilizing. Sucrose has almost no effect, but its polymer Ficoll is stabilizing. An explanation for both observations is that there is a small stabilizing macromolecular effect. However, the small net stabilizing effect could also arise from polymer induced shielding of destabilizing attractive interactions by the polymer, as suggested by Knowles et al.44
Proteins
To gain more biologically relevant information, we turned to globular proteins as cosolutes and examined the effects of lysozyme (14 kDa, pI 9.7, 50 g/L) and BSA (68 kDa, pI 4.5, 100 g/L). At pH 7.5, both are polyanions. The GB1 monomer has a net charge of approximately −4.3, and BSA has a net charge of approximately −18. The resulting charge–charge repulsion between BSA and GB1 should favor dimerization. This prediction is borne out; BSA stabilizes the dimer by 0.48 ± 0.06 kcal/mol.
Lysozyme is expected to have the opposite effect, because it is a polycation with a charge of +8 at pH 7.5. The resulting attraction between lysozyme and GB1 should favor the monomer because it has more charged surface to interact with lysozyme. As predicted, lysozyme destabilizes the dimer by 0.18 ± 0.05 kcal/mol. The effect would have been larger, but lysozyme concentrations of >50 g/L give poor quality spectra, again consistent with idea that its attractive interactions with GB1 increase its effective molecular weight, broadening the resonances.
Freeze-Dried Lysate
Proteins comprise ~55% of the dry weight of the E. coli cytoplasm,52 and the molecular masses and isoelectric points of the proteome range from ~5 kDa to >200 kDa and from 4 to 12, respectively.14,15 The E. coli proteome has an abundance of acidic proteins, and GB1 also has a net negative charge at the pH studied here. Therefore, we expect a stabilizing effect from the cellular environment. To test this hypothesis we examined the effect of freeze-dried E. coli lysates39 at 75 g/L. These lysates comprise only proteins and nucleic acids, because small molecules were removed during preparation. As predicted, lysate increases dimer stability by 0.72 ± 0.08 kcal/mol. It is difficult to parse this stability increase between macromolecular effects and repulsive charge–charge interactions, but the charge–charge portion arises from a combination of protein charge and nucleic acids charge.
The protein stabilizing or destabilizing effect of most cosolutes,47,53 including reconstituted lysates,14 increases with increasing concentration. We anticipate, therefore, that lysates with protein concentrations approaching those found in cells1 will increase stabilization and that the intracellular environment might have an even larger effect on protein–protein interactions than we observe in these model studies.
CONCLUSIONS
A recent review of macromolecular crowding highlighted the need for studies of protein association under crowded conditions.54 We undertook this challenge using a simple homodimeric system. We find that the attractive and repulsive interactions that govern the effects of cosolutes on protein stability also govern their effects on this protein–protein interaction. Polymer crowders, which are believed to stabilize proteins through hard-core repulsions, did not show a large stabilizing effect. Reconstituted E. coli cytosol was the most stabilizing cosolute, which highlights the important differences between physiologically relevant cosolutes and synthetic polymer crowders, a difference also noted for effects on protein stability.2 Our results highlight the importance of chemical interactions as a mechanism for regulating protein complex stability. This study lays the foundation for defining the role of chemical interactions in protein–protein interactions.
Supplementary Material
Figure 2.
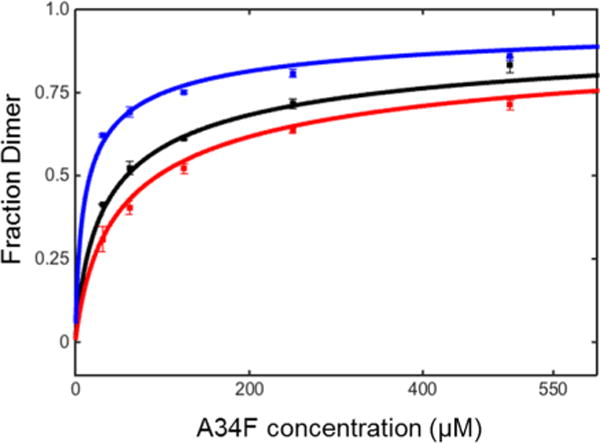
Binding isotherms for 3-fluorotyrosine labeled A34F in solutions of cytosol (75 g/L blue), buffer (black), and ethylene glycol (200 g/L, red) at pH 7.5, 298 K.
Acknowledgments
The authors thank the members of the Pielak lab for insightful discussions, Greg Young for assistance with NMR, Ashutosh Tripathy for assistance with analytical ultracentrifugation, Brandie Ehrmann for assistance with mass spectrometry, and Elizabeth Pielak for comments on the manuscript.
Funding
This work was supported by grants from the National Science Foundation (MCB-1410854 and CHE-1607359) and was performed in facilities supported by the National Cancer Institute (P30 CA016086).
ABBREVIATIONS
- GB1
B1 domain of protein G
- IPTG
isopropyl β-D-1-thiogalactopyr-anoside
- NMR
nuclear magnetic resonance
- PEG
polyethylene glycol
- SDS PAGE
sodium dodecyl sulfate poly acrylamide gel electrophoresis
- TMAO
trimethylamine oxide
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.6b01251.
Figures showing 19F incorporation efficiency by mass spectrometry, monomer–dimer HSQC spectra, 19F tyrosine assignments, fluorinated and HSQC spectra of nonfluorinated A34F GB1, and analytical ultracentrifugation profiles of fluorinated and nonfluorinated A34F GB1 (PDF)
ORCID
Gary J. Pielak: 0000-0001-6307-542X
Notes
The authors declare no competing financial interest.
References
- 1.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar M, Li C, Pielak GJ. Soft interactions and crowding. Biophys Rev. 2013;5:187–194. doi: 10.1007/s12551-013-0104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen RD, Pielak GJ. A cell is more than the sum of its (dilute) parts: A brief history of quinary structure. Protein Sci. 2016 doi: 10.1002/pro.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McPhie P, Ni Y, Minton AP. Macromolecular crowding stabilizes the molten globule form of apomyoglobin with respect to both cold and heat unfolding. J Mol Biol. 2006;361:7–10. doi: 10.1016/j.jmb.2006.05.075. [DOI] [PubMed] [Google Scholar]
- 5.Hong J, Gierasch LM. Macromolecular crowding remodels the energy landscape of a protein by favoring a more compact unfolded state. J Am Chem Soc. 2010;132:10445–10452. doi: 10.1021/ja103166y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christiansen A, Wittung-Stafshede P. Quantification of excluded volume effects on the folding landscape of Pseudomonas aeruginosa apoazurin in vitro. Biophys J. 2013;105:1689–1699. doi: 10.1016/j.bpj.2013.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miklos AC, Sarkar M, Wang Y, Pielak GJ. Protein crowding tunes protein stability. J Am Chem Soc. 2011;133:7116–7120. doi: 10.1021/ja200067p. [DOI] [PubMed] [Google Scholar]
- 8.Gnutt D, Gao M, Brylski O, Heyden M, Ebbinghaus S. Excluded-volume effects in living cells. Angew Chem, Int Ed. 2015;54:2548–2551. doi: 10.1002/anie.201409847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danielsson J, Mu X, Lang L, Wang H, Binolfi A, Theillet FX, Bekei B, Logan DT, Selenko P, Wennerström H, Oliveberg M. Thermodynamics of protein destabilization in live cells. Proc Natl Acad Sci U S A. 2015;112:12402–12407. doi: 10.1073/pnas.1511308112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monteith WB, Cohen RD, Smith AE, Guzman-Cisneros E, Pielak GJ. Quinary structure modulates protein stability in cells. Proc Natl Acad Sci U S A. 2015;112:1739–1742. doi: 10.1073/pnas.1417415112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Zhuravleva A, Gierasch LM. Exploring weak, transient protein–protein interactions in crowded in vivo environments by in-cell nuclear magnetic resonance spectroscopy. Biochemistry. 2011;50:9225–9236. doi: 10.1021/bi201287e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen RD, Guseman AJ, Pielak GJ. Intracellular pH modulates quinary structure. Protein Sci. 2015;24:1748–1755. doi: 10.1002/pro.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crowley PB, Chow E, Papkovskaia T. Protein interactions in the Escherichia coli cytosol: An impediment to in-cell NMR spectroscopy. ChemBioChem. 2011;12:1043–1048. doi: 10.1002/cbic.201100063. [DOI] [PubMed] [Google Scholar]
- 14.Sarkar M, Smith AE, Pielak GJ. Impact of reconstituted cytosol on protein stability. Proc Natl Acad Sci U S A. 2013;110:19342–19347. doi: 10.1073/pnas.1312678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarkar M, Lu J, Pielak GJ. Protein crowder charge and protein stability. Biochemistry. 2014;53:1601–1606. doi: 10.1021/bi4016346. [DOI] [PubMed] [Google Scholar]
- 16.Minton AP. Excluded volume as a determinant of macromolecular structure and reactivity. Biopolymers. 1981;20:2093–2120. [Google Scholar]
- 17.Cohen RD, Pielak GJ. Electrostatic contributions to protein quinary structure. J Am Chem Soc. 2016;138:13139–13142. doi: 10.1021/jacs.6b07323. [DOI] [PubMed] [Google Scholar]
- 18.Benton LA, Smith AE, Young GB, Pielak GJ. Unexpected effects of macromolecular crowding on protein stability. Biochemistry. 2012;51:9773–9775. doi: 10.1021/bi300909q. [DOI] [PubMed] [Google Scholar]
- 19.Phillip Y, Sherman E, Haran G, Schreiber G. Common crowding agents have only a small effect on protein-protein interactions. Biophys J. 2009;97:875–885. doi: 10.1016/j.bpj.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiao M, Li HT, Chen J, Minton AP, Liang Y. Attractive protein-polymer interactions markedly alter the effect of macromolecular crowding on protein association equilibria. Biophys J. 2010;99:914–923. doi: 10.1016/j.bpj.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gronenborn A, Filpula D, Essig N, Achari A, Whitlow M, Wingfield P, Clore G. A novel, highly stable fold of the immunoglobulin binding domain of streptococcal protein G. Science. 1991;253:657–661. doi: 10.1126/science.1871600. [DOI] [PubMed] [Google Scholar]
- 22.Smith CK, Bu Z, Engelman DM, Regan L, Anderson KS, Sturtevant JM. Surface point mutations that significantly alter the structure and stability of a protein’s denatured state. Protein Sci. 1996;5:2009–2019. doi: 10.1002/pro.5560051007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honda S, Kobayashi N, Munekata E. Thermodynamics of a β-hairpin structure: Evidence for cooperative formation of folding nucleus. J Mol Biol. 2000;295:269–278. doi: 10.1006/jmbi.1999.3346. [DOI] [PubMed] [Google Scholar]
- 24.Jee J, Byeon IJL, Louis JM, Gronenborn AM. The point mutation A34F causes dimerization of GB1. Proteins: Struct, Funct, Genet. 2008;71:1420–1431. doi: 10.1002/prot.21831. [DOI] [PubMed] [Google Scholar]
- 25.Berg OG. The influence of macromolecular crowding on thermodynamic activity: Solubility and dimerization constants for spherical and dumbbell-shaped molecules in a hard-sphere mixture. Biopolymers. 1990;30:1027–1037. doi: 10.1002/bip.360301104. [DOI] [PubMed] [Google Scholar]
- 26.Harper DB, O’Hagan D. The fluorinated natural products. Nat Prod Rep. 1994;11:123–133. doi: 10.1039/np9941100123. [DOI] [PubMed] [Google Scholar]
- 27.O’Hagan D, Harper DB. Fluorine-containing natural products. J Fluorine Chem. 1999;100:127–133. [Google Scholar]
- 28.Chen H, Viel S, Ziarelli F, Peng L. 19F NMR: A valuable tool for studying biological events. Chem Soc Rev. 2013;42:7971–7982. doi: 10.1039/c3cs60129c. [DOI] [PubMed] [Google Scholar]
- 29.Li C, Wang GF, Wang Y, Creager-Allen R, Lutz EA, Scronce H, Slade KM, Ruf RAS, Mehl RA, Pielak GJ. Protein 19F NMR in Escherichia coli. J Am Chem Soc. 2010;132:321–327. doi: 10.1021/ja907966n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith AE, Zhou LZ, Gorensek AH, Senske M, Pielak GJ. In-cell thermodynamics and a new role for protein surfaces. Proc Natl Acad Sci U S A. 2016;113:1725–1730. doi: 10.1073/pnas.1518620113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campos-Olivas R, Aziz R, Helms GL, Evans JNS, Gronenborn AM. Placement of 19F into the center of GB1: Effects on structure and stability. FEBS Lett. 2002;517:55–60. doi: 10.1016/s0014-5793(02)02577-2. [DOI] [PubMed] [Google Scholar]
- 32.Crowley PB, Kyne C, Monteith WB. Simple and inexpensive incorporation of 19F-tryptophan for protein NMR spectroscopy. Chem Commun. 2012;48:10681–10683. doi: 10.1039/c2cc35347d. [DOI] [PubMed] [Google Scholar]
- 33.Ye Y, Liu X, Zhang Z, Wu Q, Jiang B, Jiang L, Zhang X, Liu M, Pielak GJ, Li C. 19F NMR spectroscopy as a probe of cytoplasmic viscosity and weak protein interactions in living cells. Chem - Eur J. 2013;19:12705–12710. doi: 10.1002/chem.201301657. [DOI] [PubMed] [Google Scholar]
- 34.Smith CK, Withka JM, Regan L. A thermodynamic scale for the β sheet forming tendencies of the amino acids. Biochemistry. 1994;33:5510–5517. doi: 10.1021/bi00184a020. [DOI] [PubMed] [Google Scholar]
- 35.Monteith WB, Pielak GJ. Residue level quantification of protein stability in living cells. Proc Natl Acad Sci U S A. 2014;111:11335–11340. doi: 10.1073/pnas.1406845111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan F, Kuprov I, Craggs TD, Hore PJ, Jackson SE. 19F NMR studies of the native and denatured states of green fluorescent protein. J Am Chem Soc. 2006;128:10729–10737. doi: 10.1021/ja060618u. [DOI] [PubMed] [Google Scholar]
- 37.Putnam F. Progress in plasma proteins. In: Putnam F, editor. The Plasma Proteins. 2nd. Academic Press; New York: 1984. pp. 1–44. [Google Scholar]
- 38.Aune KC, Tanford C. Thermodynamics of the denaturation of lysozyme by guanidine hydrochloride. I. Dependence on pH at 25°. Biochemistry. 1969;8:4579–4585. doi: 10.1021/bi00839a052. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Li C, Pielak GJ. Effects of proteins on protein diffusion. J Am Chem Soc. 2010;132:9392–9397. doi: 10.1021/ja102296k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS. VADAR: A web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 2003;31:3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavallo L, Kleinjung J, Fraternali F. POPS: A fast algorithm for solvent accessible surface areas at atomic and residue level. Nucleic Acids Res. 2003;31:3364–3366. doi: 10.1093/nar/gkg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aramini J, Hamilton K, Ma LC, Swapna GVT, Leonard P, Ladbury J, Krug R, Montelione G. 19F NMR reveals multiple conformations at the dimer interface of the Nonstructural Protein 1 effector domain from Influenza A virus. Structure. 2014;22:515–525. doi: 10.1016/j.str.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guinn EJ, Pegram LM, Capp MW, Pollock MN, Record MT. Quantifying why urea is a protein denaturant, whereas glycine betaine is a protein stabilizer. Proc Natl Acad Sci U S A. 2011;108:16932–16937. doi: 10.1073/pnas.1109372108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knowles DB, Shkel IA, Phan NM, Sternke M, Lingeman E, Cheng X, Cheng L, O’Connor K, Record MT. Chemical interactions of polyethylene glycols (PEGs) and glycerol with protein functional groups: Applications to effects of PEG and glycerol on protein processes. Biochemistry. 2015;54:3528–3542. doi: 10.1021/acs.biochem.5b00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shkel IA, Knowles DB, Record MT. Separating chemical and excluded volume interactions of polyethylene glycols with native proteins: Comparison with PEG effects on DNA helix formation. Biopolymers. 2015;103:517–527. doi: 10.1002/bip.22662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Street TO, Bolen DW, Rose GD. A molecular mechanism for osmolyte-induced protein stability. Proc Natl Acad Sci U S A. 2006;103:13997–14002. doi: 10.1073/pnas.0606236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Auton M, Rösgen J, Sinev M, Holthauzen LMF, Bolen DW. Osmolyte effects on protein stability and solubility: A balancing act between backbone and side-chains. Biophys Chem. 2011;159:90–99. doi: 10.1016/j.bpc.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Sarkar M, Smith AE, Krois AS, Pielak GJ. Macromolecular crowding and protein stability. J Am Chem Soc. 2012;134:16614–16618. doi: 10.1021/ja305300m. [DOI] [PubMed] [Google Scholar]
- 49.Senske M, Törk L, Born B, Havenith M, Herrmann C, Ebbinghaus S. Protein stabilization by macromolecular crowding through enthalpy rather than entropy. J Am Chem Soc. 2014;136:9036–9041. doi: 10.1021/ja503205y. [DOI] [PubMed] [Google Scholar]
- 50.Politi R, Harries D. Enthalpically driven peptide stabilization by protective osmolytes. Chem Commun. 2010;46:6449–6451. doi: 10.1039/c0cc01763a. [DOI] [PubMed] [Google Scholar]
- 51.Sapir L, Harries D. Is the depletion force entropic? Molecular crowding beyond steric interactions. Curr Opin Colloid Interface Sci. 2015;20:3–10. [Google Scholar]
- 52.Neidhardt FC. Chemical composition of Escherichia coli. In: Frederick C, Neidhardt JLI, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium. American Society for Microbiology; Washington, DC: 1987. pp. 3–5. [Google Scholar]
- 53.Schellman JA. A simple model for solvation in mixed solvents. Applications to the stabilization and destabilization of macromolecular structures. Biophys Chem. 1990;37:121–140. doi: 10.1016/0301-4622(90)88013-i. [DOI] [PubMed] [Google Scholar]
- 54.Rivas G, Minton AP. Macromolecular crowding in vitro, in vivo, and in between. Trends Biochem Sci. 2016;41:970–981. doi: 10.1016/j.tibs.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


