Abstract
Copy-number variations (CNVs) are strong risk factors for neurodevelopmental and psychiatric disorders. The 15q13.3 microdeletion syndrome region contains up to ten genes and is associated with numerous conditions, including autism spectrum disorder (ASD), epilepsy, schizophrenia, and intellectual disability; however, the mechanisms underlying the pathogenesis of 15q13.3 microdeletion syndrome remain unknown. We combined whole-genome sequencing, human brain gene expression (proteome and transcriptome), and a mouse model with a syntenic heterozygous deletion (Df(h15q13)/+ mice) and determined that the microdeletion results in abnormal development of cortical dendritic spines and dendrite outgrowth. Analysis of large-scale genomic, transcriptomic, and proteomic data identified OTUD7A as a critical gene for brain function. OTUD7A was found to localize to dendritic and spine compartments in cortical neurons, and its reduced levels in Df(h15q13)/+ cortical neurons contributed to the dendritic spine and dendrite outgrowth deficits. Our results reveal OTUD7A as a major regulatory gene for 15q13.3 microdeletion syndrome phenotypes that contribute to the disease mechanism through abnormal cortical neuron morphological development.
Keywords: 15q13.3 microdeletion syndrome, copy-number variation, OTUD7A, synapse, dendrite, neurodevelopmental disorder, autism spectrum disorder, deubiquitinase, schizophrenia, dendritic spine
Introduction
Neurodevelopmental disorders include various conditions characterized by deficits or delays in typical developmental milestones that appear in childhood. Genetic studies indicate that copy-number variations (CNVs), comprising deletions or duplications of genomic DNA that may directly or indirectly affect the dosage of genes, can be significant risk factors for these disorders. Certain genomic contexts are hotspots for recombination- and replication-based mechanisms that result in microdeletions or microduplications. While 5%–20% of developmentally delayed individuals carry a rare large pathogenic CNV,1 the pathobiology is poorly understood. Furthermore, for most CNVs associated with neurodevelopmental disorders, the underlying genes that contribute to the clinical phenotypes remain unknown. The 22q11.2 and 16p11.2 microdeletions are two specific CNVs associated with schizophrenia (MIM: 181500) and autism spectrum disorder (ASD) (MIM: 209850), respectively, in which substantial progress has been made into the understanding of disease pathophysiology.2, 3, 4, 5, 6, 7, 8, 9 In both, modeling microdeletions in animal models has accelerated the pace at which the underlying biological pathogenic mechanism(s) have been identified and has provided a means to test potential “driver genes” underlying CNV-associated phenotypes.10, 11, 12, 13, 14 Importantly, these studies have led to potential drug therapies that have been tested in preclinical mouse models,5, 7, 15 highlighting the power of this approach.
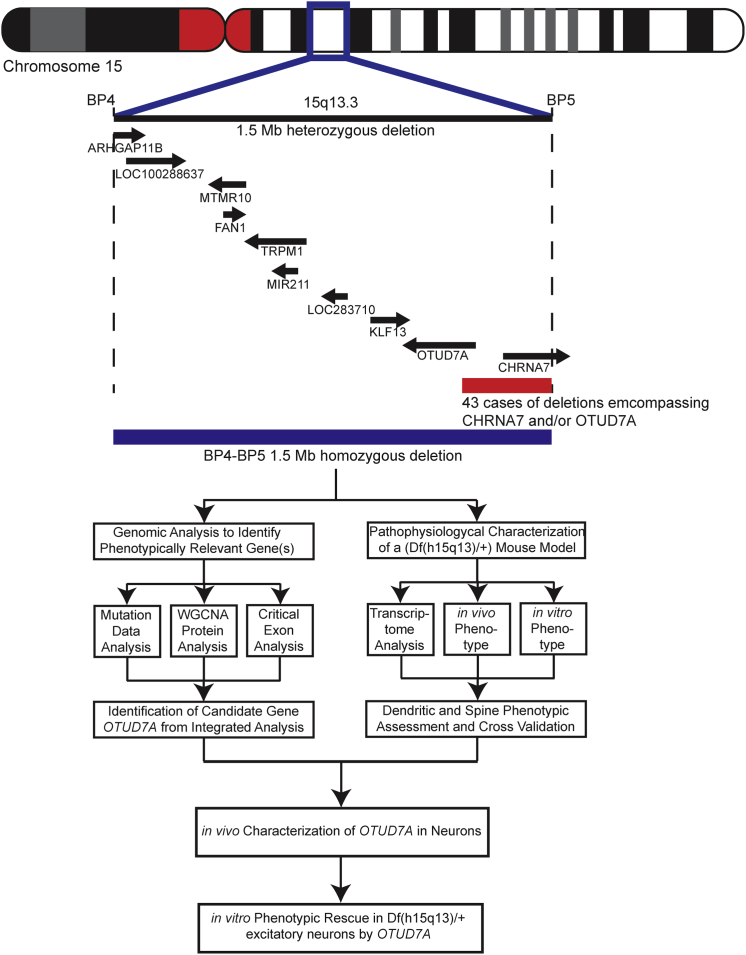
The 15q13.3 1.53 Mb microdeletion syndrome (MIM: 612001) locus (chr15:30,910,306–32,445,407 [hg19]) that resides within breakpoints BP4-BP5 on human chromosome 15 is a recurrent CNV that results in a highly heterogeneous set of phenotypes including intellectual disability (50%–60%), autism spectrum disorder (10%–20%), epilepsy (∼30%) (MIM: 607208), and schizophrenia (10%–20%).16, 17, 18, 19, 20, 21, 22, 23 Among the more than 200 individuals bearing 15q13.3 BP4-BP5 deletions who have been described in the clinical literature, 80% exhibited ascertainment-independent phenotypic manifestations; this implies that one or more genes in the CNV may contribute to disease manifestation, including the incomplete penetrance and variable expressivity observed.24 Individuals are typically heterozygous for the 15q13.3 microdeletion, which encompasses seven protein-coding genes, one microRNA, and two putative pseudogenes (ARHGAP11BI [MIM: 616310], LOC100288637, FAN1 [MIM: 613534], MTMR10, TRPM1 [MIM: 603576], LOC283710, microRNA-211, KLF13 [MIM: 605328], OTUD7A [MIM: 612024], and CHRNA7 [MIM: 118511]). Recently, a heterozygous mouse model of the 15q13.3 microdeletion was generated (Df(h15q13)/+ mice) that contains a deletion of the syntenic murine chromosome region (mouse chromosome 7qC) corresponding to the human CNV. Phenotypic manifestations of this heterozygous mouse model include schizophrenia- and epilepsy-related endophenotypes, notably long-term spatial memory deficits, increased sensitivity to stress, and reductions in auditory-evoked gamma power (similar to schizophrenia patients).25 Many of these observed clinical phenotypes were replicated in another independent 15q13.3 heterozygous microdeletion mouse model.26
In contrast to the heterozygous deletion, the homozygous deletion is extremely rare and only eight case subjects have been reported in the literature.24, 27, 28 All reported case subjects that carry the homozygous deletion manifested severe cognitive and physiological impairments including severe DD/ID, hypotonia, seizures, and visual impairment.24 A recent study analyzed behavioral abnormalities of Df(h15q13)−/− homozygous knock out (KO) mice29 in which both copies of the genes within the locus are deleted.27 While the majority of human case subjects are carriers of a heterozygous microdeletion, the homozygous KO mice displayed more pronounced phenotypes in seizure susceptibility, ASD behavior-related phenotypes, and auditory sensory processing, demonstrating a gene-dosage dependency. While the underlying neurophysiological abnormalities remain unknown, recent studies using resting-state fMRI have revealed that Df(h15q13)/+ mice display altered neuronal firing rates in the prefrontal cortex30 and abnormal brain hyperconnectivity patterns;31 however, the underlying molecular and physiological brain abnormalities contributing to the mouse behavioral, imaging, and electrophysiological phenotypes remain unknown.
The complexities inherent in the identification of the critical driver gene(s) in a given CNV are significant and require evidence from multiple sources. With regard to the 15q13.3 microdeletion, CHRNA7 (encoding a cholinergic receptor) has been proposed as a driver gene, in part because some patients have overlapping deletions that encompass CHRNA7.20 Furthermore, CHRNA7 agonists may improve cognition in humans with schizophrenia32 and abnormal fMRI-derived brain connectivity in Df(h15q13)/+ mice.31 However, Chrna7 KO mice display very mild defects in synaptic function and learning33, 34, 35, 36 and have no consistent behavioral or neurophysiological phenotypes compared to Df(h15q13)/+ mice.37 Furthermore, among clinical case subjects, many of the deletions encompassing CHRNA7 also overlap the adjacent gene OTUD7A.20 One study reported 43 case subjects from the literature that had deletions encompassing both CHRNA7 and the first exon of OTUD7A.24 The Database of Genomic Variants (DGV) includes population control subjects that carry CHRNA7 deletions, bringing into question the penetrance of haplo-insufficiency of CHRNA7.38, 39 Another possible candidate is FAN1, which encodes a DNA repair enzyme. Rare nonsynonymous variants have been found in this gene among individuals with ASD or schizophrenia.40 Consequently, other genes in the 15q13.3 microdeletion could contribute to disease pathogenesis.
Here, we used multiple in silico, in vivo, and in vitro strategies to dissect the cellular phenotypes and critical loci that contribute to the 15q13.3 microdeletion syndrome. By using the Df(h15q13)/+ heterozygous mouse model, RNA sequencing, signaling pathway analysis, and neuronal morphology analysis, we discovered that developing cortical excitatory neurons have deficiencies in dendrite and synapse growth (Figure S2). We concurrently identified candidate gene(s) within this syndromic region contributing to these phenotypes by applying developmental human brain expression (transcriptome and proteome) data analysis together with whole-genome and -exome sequencing data from individuals with neurodevelopmental disorders and ASD.41 These analyses identified a strong candidate gene, OTUD7A, within the 15q13.3 microdeletion syndrome region, which encodes a putative deubiquitinating enzyme that localizes to dendritic spine compartments and has a protein-protein co-expression network that includes synaptic and dendritic signaling pathways. We biologically validated OTUD7A and found that re-expressing OTUD7A WT into cortical neurons from Df(h15q13)/+ mice rescued the dendritic spine defects. However, a mutant OTUD7A harboring an ASD-linked de novo exonic mutation was unable to rescue these defects. Furthermore, we found that both OTUD7A and CHRNA7 contribute to the dendrite outgrowth defects. Our data identify OTUD7A as a candidate gene that contributes to abnormalities in cortical neuron dendritic and spine development and a critical gene in the phenotypic manifestation of the 15q13.3 microdeletion syndrome.
Material and Methods
RNA Sequencing and Differential Expression Analysis
mRNA was extracted from E16, P21, and adult (P63) mouse cortical brain tissue using Trizol LS reagent (ThermoFisher Scientific). Two WT and two Df(h15q13)/+ mice were used for each time point (12 mice total). All postnatal mice used for transcriptome analysis were male whereas sex was undetermined for embryonic mouse brain samples. Quality of total RNA samples was checked on an Agilent Bioanalyzer 2100 RNA Nano chip following Agilent Technologies’ recommendation. We have measured the concentration by Qubit RNA HS Assay on a Qubit fluorometer (ThermoFisher). The library preparation for RNA was performed following the Illumina Stranded mRNA Library Preparation protocol. In brief, 500 ng of total RNA was used as the input material and enriched for poly(A) mRNA, fragmented into the 200- to 300-base range for 4 min at 94°C and converted to double stranded cDNA, end-repaired, and adenylated at the 3′ to create an overhang A to allow for ligation of Illumina adapters with an overhang T; library fragments were amplified under the following conditions: initial denaturation at 98°C for 10 s, followed by 10 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 30 s, and finally an extension step for 5 minutes at 72°C; at the amplification step, each sample was amplified with different barcoded adapters to allow for multiplex sequencing. To check the size fragment, 1 μL of the final RNA libraries was loaded on a Bioanalyzer 2100 DNA High Sensitivity chip (Agilent Technologies); RNA libraries were quantified by qPCR using the Kapa Library Quantification Illumina/ABI Prism Kit protocol (KAPA Biosystems). Libraries were pooled in equimolar quantities and paired-end sequenced on an Illumina HiSeq 2500 platform using a High Throughput Run Mode flowcell and the V4 sequencing chemistry following Illumina’s recommended protocol to generate paired-end reads of 126 bases in length.
We used TopHat42 to align the read and conducted expression analysis for each gene through reads per kilobase of transcript per million mapped reads (rpkm) computation. To compute global differential expression (DE) analysis for each of the three time points—E16, P21, and adult (P63)—we conducted comprehensive DE analysis using a Bioconductor package named edgeR.43 Differential expression was computed against control expression for each of the three time points separately and a gene was considered differentially expressed for a time point only if p < 0.0001 and FDR < 0.05.
Microarray and Sequencing Data
Microarray data were obtained for 38,325 ASD-affected and developmental delay (DD)-affected case subjects from four large sources (Table S3). The first dataset is comprised of 1,026 samples diagnosed primarily with ASD and genotyped with high-resolution microarrays (Illumina 1M). The other three other datasets comprised case subjects with developmental delay. The first of these includes 9,322 samples recruited from USA by a private company (Lineagen Inc.) and the data were generated using high-resolution Affymetrix Cytoscan HD in conjunction with the Chas algorithm. The second dataset comprised 10,619 DD-affected case subjects from two major clinics in Ontario, Canada. For the first and second datasets, ethical clearance was obtained from The Hospital for Sick Children’s institutional ethical board. The third dataset is from a published dataset of 17,358 case subjects with DD in USA, primarily from a private diagnostic company which used the international standards for cytogenomic arrays (ISCA) for variant classification.44 We have preprocessed 22,241 control samples from seven different sources that were recruited as population control subjects.44, 45, 46, 47, 48, 49, 50 For both case and control subjects, we focused on deletions with >50% reciprocal overlap with the 15q13.3 microdeletion region (BP4-BP5). Gene-specific deletions and atypical variants that were larger and overlapped with 15q13.3 were also kept for fine mapping. To identify de novo mutations within the 15q13.3 locus, we analyzed published whole-exome and -genome sequencing data from autism spectrum disorder cohorts51, 52, 53 and a homogeneous population control.54 Variants were validated using Sanger sequencing as described in Yuen et al.53 Consent was obtained from all human participants, as approved by the Research Ethics Boards at The Hospital for Sick Children, McMaster University, and Memorial Hospital.
Critical Exon and Protein Expression Analysis
Whole-genome sequencing data from the 1000 Genomes Project was used to compute the burden of rare missense and loss-of-function mutations for each exon. Furthermore, exon-level expression data from RNA sequencing were obtained for developmental human brain tissues (prenatal and postnatal postmortem donors) from the BrainSpan project and ten other human tissues. The analysis to identify brain-specific critical exons has been described in Uddin et al.41 where 10% of genes within the genome were shown to be brain critical and this specific group of genes is significantly enriched for de novo mutations in autism probands but not in unaffected siblings or in population control subjects. In brief, to classify critical exons, we computed the expression level from RNA-sequencing data from developmental human brain for each exon and classified high and low based on the 75th percentile of the entire dataset. An exon was classified as critical exon if it is highly expressed in a tissue and the burden of deleterious mutation is low. A critical exon was classified as brain specific if critical only for brain and not other tissues. We obtained a brain-specific protein module from Uddin et al.,1 where a comprehensive weighted gene co-expression network analysis (WGCNA) was conducted on the high-resolution protein mass-spectrometry dataset from developmental human tissues (24 different human tissues, each pooled from 3 post-mortem donors which includes 17 adult and 7 prenatal samples). For the 15q13.3 microdeletion syndrome locus, OTUD7A is the only gene within the brain-specific protein module and we have extracted the first-degree neighbors from the module. An over-representation analysis was conducted with gene sets using a Fisher exact test and p value was corrected for multiple tests.
To produce gene level expression information, we normalized the entire protein expression level data from 24 different human tissues and quantified the 75th percentile of the expression value from the entire data. Expression for a gene was considered high if it was above the 75th percentile, and low otherwise. For gene-level RNA-seq expression data, we followed the same protocol as the protein expression data for consistency.
Geneset Enrichment Analysis and Visualization
We conducted a comprehensive geneset enrichment analysis on RNA-seq differentially expressed genes and for the first-degree neighbor proteins for OTUD7A that were extracted from the blue protein module. We manually curated 2,848 gene sets from the gene ontology (GO) (R package, v.2.8.0), pathways from the National Cancer Institute at the National Institutes of Health (NCI-NIH), Kyoto Encyclopedia of Genes and Genomes (KEGG) (May 30, 2013), and Reactome databases. Gene sets were considered significant after Benjamini-Hotchberg false discovery rate of 0.05. To visualize network datasets, we used Cytoscape plugins using v.3.2.0.
Animals
Df(h15q13)/+ mice were generated by Taconic Artemis as described in Fejgin et al.25 Animals were bred, genotyped, and housed at the Central Animal Facility at McMaster University. All procedures received the approval of the Animal Research Ethics Board (AREB). Genotypes were identified during breeding by PCR of ear notches, and three WT females were bred with 1 Df(h15q13)/+ male per breeding cage. The use of only WT females for breeding was performed to minimize effects of potential differences in the embryonic environment and/or mothering of the Df(h15q13)/+ females compared to WT females. To obtain cortical cultures, WT females were timed-bred with Df(h15q13)/+ males and males were removed when a plug was observed, indicating copulation. At E16, mothers were sacrificed and litters were collected. Animals of appropriate genotype were included, and any animals with unclear genotypes were excluded from experiments.
Cell Culture and Transfection
Primary cortical neurons were cultured as follows. Cortices were dissected out of WT and Df(h15q13)/+ mouse embryonic brains at E16. Each brain was cultured individually. Dissociation was aided by incubation in 0.3 mg/mL Papain (Worthington Biochemical)/400 U/mL DNase I (Invitrogen) in 1× Hanks Buffered Saline Solution (HBSS) for 20 min at 37°C, followed by light trituration. Cells were seeded onto 0.1 mg/mL poly-D-lysine (BD Sciences)/3.3 μg/mL Laminin (Sigma)-coated coverslips (Matsunami) in 12-well plates in plating media containing Neurobasal medium, 10% fetal bovine serum, 1% penicillin/streptomycin, and 2 mM L-glutamine (Invitrogen). After 1.5 hr, media was changed to serum-free feeding media containing Neurobasal medium, 2% B27 supplement, 1% penicillin/streptomycin, and 2 mM L-glutamine. At DIV2-4, cultures were treated with 1 μM Cytosine β-D-arabinofuranoside hydrochloride (Ara-C) (Sigma) to inhibit glial cell proliferation. Cultures were maintained at 37°C, 5% CO2. All media components were from GIBCO unless otherwise specified. Transfections were performed at DIV7 using Lipofectamine LTX and Plus reagents (Invitrogen) according to the manufacturer’s instructions. HEK293FT cells (Invitrogen) were grown under standard cell culture conditions and transfected with plasmids using Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen). HEK293FT cells were used for ease of plasmid expression and have not been tested for Mycoplasma contamination.
In Utero Electroporation
Timed-pregnant WT female and Df(h15q13)/+ male mice were anesthetized by isoflurane (gas). The uterine horns were exposed, and FLAG-pcDNA control or FLAG-OTUD7A WT plasmids with pCAG Venus contructs (4:1 ratio) mixed with Fast Green were injected into the lateral ventricles of E16 embryos. 5 current pulses (conditions: 950 ms pulse, 50 ms interval, 36V) were delivered across the head of the embryos. Litters were born and sacrificed by perfusion at P22. Mice were anesthetized by isoflurane gas followed by intraperitoneal injection of sodium pentobarbitol. This was followed by perfusion with lactate ringers solution and then 4% PFA. Following perfusion, brains were harvested and post-fixed in 4% PFA for 48 hr at 4°C. Mice were genotyped as described, and brains were sectioned using a vibratome (Leica) at 50 μm thickness (coronal).
Antibodies and Constructs
The following primary antibodies were used in this study. Mouse anti-FLAG (Sigma F3165; IF 1:2,000; western blotting 1:2,000), rabbit anti-β-actin (Cell Signaling 4970; western blotting 1:2,000), and anti-rabbit PSD95 (Cell Signaling Technologies D27E11; IF 1:200). All secondary antibodies (anti-rabbit cy5, anti-mouse cy3; Jackson ImmunoResearch; IF 1:500, anti-rabbit-HRP, anti-mouse-HRP; GE Life Sciences; IB 1:5,000) were raised in donkey.
pCAGIG-Venus was provided by Dr. Zhigang Xie (Boston University, MA). The FLAG-OTUD7A WT was amplified from a pCMV6_FLAG-WT-OTUD7A construct (Origene, RC213015, GenBank: NM_130901) and cloned into the pcDNA3.3 vector between EcoRI and KpnI. pcDNA3.3_FLAG-OTUD7A p.Asn492_Lys494del plasmid was generated using In-Fusion cloning kit. FLAG-CHRNA7, FLAG-FAN1, and FLAG-KLF13 were amplified from a pCMV6_FLAG-CHRNA7 (Origene, RC221382, GenBank: NM_000746), pCMV6_FLAG-FAN1 (Origene, RC224249, GenBank: NM_014967), and pCMV6_FLAG-KLF13 (Origene, RC200805, GenBank: NM_015995) and cloned into the pcDNA3.3 vector using In-Fusion cloning kit.
Immunocytochemistry
On DIV14, cells on glass coverslips were fixed with 4% formaldehyde in PBS for 20 min at room temperature. Cells were washed in PBS, followed by blocking in blocking/permeabilization solution consisting of 10% Donkey Serum (Millipore) and 0.3% Triton X-100 (Fisher Scientific) in PBS for 1 hr at room temperature. Incubation in primary antibodies was performed at 4°C overnight. Cells were then washed in PBS, followed by incubation with secondary antibodies in 50% blocking/permeabilization solution at room temperature with gentle agitation for 1.5 hr. Cells were then washed in PBS and were mounted on VistaVision glass microscope slides (VWR) using Prolong Gold antifade reagent (Life Technologies). Images were acquired using a Zeiss LSM700 confocal microscope.
Golgi Staining
Golgi staining was performed on mouse brains from P28 male mice using a commercial kit and protocol (FD Rapid GolgiStain Kit, FD NeuroTechnologies, Inc.). Briefly, whole mouse brains were removed and washed with Milli-Q water. The brain was immersed in impregnation solution for 8 days. Following impregnation, brains were rinsed twice with water and incubated for 72 hr with Solution C. Brains were then removed and incubated in 30% sucrose/PBS for 24 hr in the dark. Using a vibratome (Leica), brain slices were cut at 150 μm thickness and mounted onto 3% gelatin-coated SuperFrost Plus microscope slides (ThermoFisher). Sections were incubated in staining solution for 10 min, washed with Milli-Q water, dehydrated with EtOH, and cleared with xylene (Sigma). Stained brain slices were imaged using Zeiss Axiocam ICm1 microscope camera. Z stacks were acquired at 1 μm intervals for sholl analysis and 0.28 μm intervals for spine analysis. Images were processed using ImageJ software (Sholl Analysis plugin).
Morphological Analyses
Images were processed and analyzed with ImageJ 1.44 software. Sholl analysis was performed using the Sholl analysis plugin in ImageJ. This plugin was used to make concentric circles increasing at a constant radius of 10 μm and to count the number of dendritic intersections. Neuron tracing was performed using the NeuronJ plugin for ImageJ 1.44. Spine density was calculated by visually counting all protrusions from a dendrite within a 15–25 μm distance starting at a secondary branch point. One to three dendritic segments were analyzed per neuron. Maximal spine head width (HW), neck width (NW), length (L), and neck length (NL) were measured for each dendritic protrusion using the segmented line tool in ImageJ. Spines were defined as follows: stubby (L < 1 μm), mushroom (1 ≤ L ≤ 5 μm; HW ≤ 2× NW), or thin (1 ≤ L ≤ 5 μm; WH ≤ 2× NW). The proportion of each spine type was calculated by dividing the number of spines in each spine category (mushroom, thin, and stubby) by the total number of spines within the dendritic segment. For analyses on Golgi-stained brains, 40 neurons from 4 brains per condition were used for sholl analysis and 40 dendritic segments from 4 mouse brains per condition were used for spine analysis. For IUE experiments, 21–23 neurons from 3 brains per condition were used for sholl analysis, and 30 dendritic segments from 3 brains per condition were used for spine analysis. For in vitro analyses, 21–50 neurons were used from 3–4 biological replications (mouse litters) for sholl analysis, and 20–36 dendritic segments from 3–5 biological replications (mouse litters) for spine analysis.
Quantitative PCR
To measure postnatal developmental RNA expression of OTUD7A in the mouse brain, we obtained whole-brain samples of P0, P7, P14, P21, P28, and P63 WT C57BL/6 mice. Total RNA was extracted using TRIzol LS Reagent (Thermo Fisher Scientific), followed by cDNA synthesis using the qScript cDNA synthesis kit according to manufacturer instructions (Quanta Biosciences). Primers and probes were designed and selected using the Universal Probe Library (UPL) Probefinder software for mouse (Roche). Quantitative PCR was performed using the Perfecta qPCR Supermix (Quanta Biosciences) and UPL probe #50 (OTUD7A) or UPL probe #52 (GAPDH). Data represented as levels of OTUD7A normalized to GAPDH levels.
Western Blotting
Western blotting was performed using standard protocols. Briefly, HEK293FT cells were lysed in lysis buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris-Cl, and cOmplete mini protease inhibitor cocktail (Roche). 20–30 μg of sample was loaded into a 10% Tris-Glycine gel and transferred to a PVDF membrane (Bio-Rad). Membranes were blocked for 1 hr in 3% milk in 1× TBST, incubated with primary antibody overnight at 4°C, then with secondary antibody (donkey anti-mouse or anti-rabbit HRP, GE Healthcare) for 1 hr at room temperature before exposure using a ChemiDoc MP system (Bio-Rad).
Statistical Analysis
Data are expressed as mean ± SEM. A minimum of three mice per condition or three mouse litters for in vitro culture experiments, was used for statistical analysis. Blinding was not performed. We used the Student’s t test, one-way ANOVA with post hoc Tukey’s test, and two-way ANOVA in GraphPad Prism 6 statistical software for statistical analyses. p values in the figure legends are from the specified tests, and p < 0.05 was considered statistically significant.
Results
Df(h15q13)/+ Mice Show Defects in Forebrain Development Including Abnormalities in Dendritic Spines and Dendrite Growth
To study the 15q13.3 microdeletion (Figure 1), we utilized the heterozygous BP4-BP5 mouse model (Df(h15q13)/+).25 We examined temporal gene expression (using RNA sequencing) in cortical brain tissue from Df(h15q13)/+ and WT mice at three stages: prenatal (embryonic day 16; E16), early postnatal (postnatal day 21; P21), and adult (P63) (Figures S1A and S1B and Table S1). We identified 66 genes with significant (p < 0.0001 and FDR < 0.05) differential expression from the three developmental time points. Mtmr10, Otud7a, Klf13, Fan1, and Chrna7 displayed significant differential expression (DE) in all periods (Trpm1 is not expressed in the brain, and miR211 was not detected because samples were not enriched for small RNAs during sample preparation) (Figure S1A), demonstrating that the heterozygosity of gene expression persists into adulthood. Interestingly, we found that differential gene expression was most pronounced (∼2-fold) at postnatal time points, P21, and adulthood (when spine and synapse development is peaking). We conducted extensive gene enrichment analyses for all differentially expressed genes by using 2,848 gene sets with defined biological pathways in KEGG, GO, NCI-NIH, and Reactome databases. The most significant enrichment across all time points was observed for prenatal organ development (p < 2.9 × 10−05, significant after Benjamini-Hochberg correction) and forebrain development (p < 6.8 × 10−5) (Figure S1C and Table S2). The significant enrichment of the forebrain development pathway in the WT versus Df(h15q13)/+ comparison suggests that cortical development is disrupted by the microdeletion.
Figure 1.
Overview of the 15q13.3 Microdeletion and CHRNA7/OTUD7A Overlapping Deletion
Schematic diagram showing the location of the human 15q13.3 locus and all ten genes within the 1.5 Mb BP4-BP5 deletion. Smaller deletions (red bar) are found within the BP4-BP5 region overlapping CHRNA7 and/or OTUD7A, modified from Lowther et al.24 The experimental workflow of the current study is presented below the schematic.
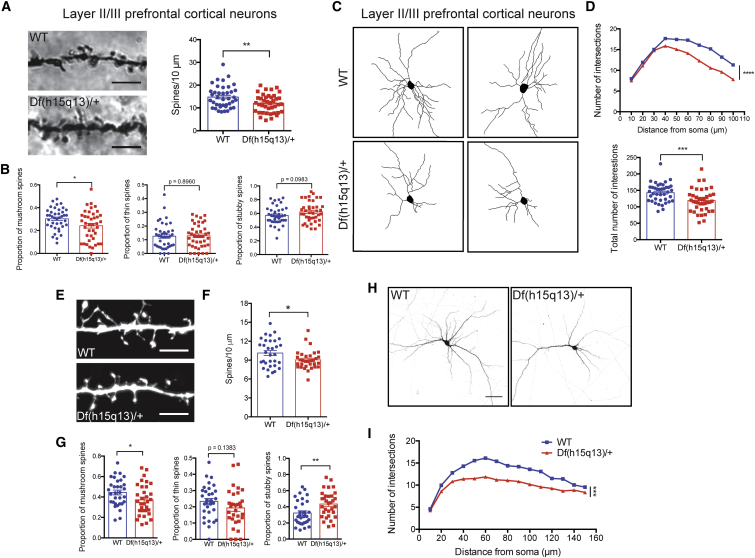
We next focused on postnatal cortical (forebrain) development in Df(h15q13)/+ mice because of our transcriptional data and previous studies that identified abnormalities in excitatory neuron development.55, 56, 57, 58 Therefore, we measured dendritic spine morphology and dendrite growth in vivo on Golgi-Cox-stained WT and Df(h15q13)/+ mouse brains. We analyzed cortical neurons in layers 2/3 of the prefrontal cortex (PFC) of male P28 Df(h15q13)/+ and age/sex-matched WT mice. We found that in Df(h15q13)/+ prefrontal cortex, there was a small but significant (p < 0.01) reduction in spine density and mature mushroom-shaped spines (Figures 2A and 2B). Df(h15q13)/+ mice also displayed a significant (p < 0.05) reduction in spine length and spine neck length (Figure S2A). Dendritic arborization was measured using Sholl analysis59 and revealed that Df(h15q13)/+ mice showed decreased arborization in layer 2/3 PFC neurons compared to WT mice (Figures 2C and 2D). Since these findings were obtained in vivo, it is difficult to determine whether they are cell autonomous. Therefore, we cultured cortical neurons from littermate Df(h15q13)/+ and WT mice, transfected with a Venus fluorescent reporter, and analyzed neurons at 14 days in vitro (DIV14). Df(h15q13)/+ neurons also showed a significant reduction in the density of dendritic spines and spine length (Figures 2E, 2F, and S2B), as well as a decrease in mature mushroom-type spines (p < 0.05) and increase in immature stubby-type spines (p < 0.01) (Figure 2G), replicating many of the in vivo phenotypes. In addition, there was a significant decrease in dendritic arborization in Df(h15q13)/+ neurons compared to WT littermates (Figures 2H and 2I). The spine and dendrite phenotypes in Df(h15q13)/+ observed in vitro were stronger than in vivo, suggesting some level of compensation, similar to previous reports.3, 60
Figure 2.
Df(h15q13)/+ Mice Have Defects in Dendritic Spine Development and Neuronal Morphology
(A) Golgi-stained dendritic spine images (left) and spine density analysis (right). Df(h15q13)/+ mice show a decrease in spine density in layer II/III PFC neurons. WT, n = 40 dendritic segments, 22 neurons; Df(h15q13)/+, n = 40 dendritic segments, 26 neurons, 4 mice per condition, Student’s t test, ∗∗p < 0.01, t(78) = 2.846. Scale bar = 5 μm.
(B) Df(h15q13)/+ mice show a decrease in PFC mushroom spines (left). WT, n = 40 dendritic segments, 22 neurons; Df(h15q13)/+, n = 40 dendritic segments, 26 neurons, 4 mice per condition, Student’s t test, ∗p < 0.05, t(78) = 2.403, t(78) = 0.1312, t(78) = 1.673.
(C) Traces of P28 WT and Df(h15q13)/+ Golgi-stained layer II/III prefrontal cortical (PFC) neurons.
(D) Sholl analysis (left) and the total number of intersections (right). Df(h15q13)/+ mice show a decrease in dendrite growth in layer II/III PFC neurons. n = 40 neurons, 4 mice per condition, two-way ANOVA followed by Tukey’s post hoc test, ∗∗∗p < 0.001, F(9, 780) = 109.2, t(78) = 3.449.
(E) Dendritic spine images from WT and Df(h15q13)/+ cultured neurons (scale bar = 5 μm).
(F) Spine density measurements in cultured neurons. Spine density is decreased in Df(h15q13)/+ cultured cortical neurons. WT, n = 32 dendritic segments from 12 neurons; Df(h15q13)/+, n = 32 dendritic segments from 15 neurons, 3 cultures, Student’s t test, ∗p < 0.05, t(62) = 2.311.
(G) Dendritic spine classification in WT and Df(h15q13)/+ neurons. In Df(h15q13)/+ cultured cortical neurons, the proportion of mushroom-type spines is decreased (left) and the proportion of stubby type spines is increased (right). WT, n = 32 dendritic segments from 12 neurons; Df(h15q13)/+, n = 32 dendritic segments from 15 neurons, 3 cultures, Student’s t test, ∗p < 0.05, ∗∗p < 0.01, t(62) = 2.007, t(62) = 0.1383, t(62) = 2.875.
(H) DIV14 WT and Df(h15q13)/+ cultured cortical neurons expressing Venus (scale bar = 50 μm).
(I) Sholl analysis of cultured cortical neurons. Dendrite growth is decreased in Df(h15q13)/+ neurons. n = 50 neurons, 3 cultures, two-way ANOVA followed by Sidak post hoc test, ∗∗∗p < 0.001, F(14, 1470) = 85.55. Error bars represent SEM.
OTUD7A Is a Candidate Driver Gene in the 15q13.3 Microdeletion
We further assessed the pathogenicity of the 15q13.3 microdeletion by using large datasets to compile all available published and unpublished case subjects (Table S3). In addition, clinical microarray data from 38,325 case subjects with a range of neurodevelopmental disorders were examined to identify the frequency of 15q13.3 deletions (chr15: 30910306–32445407 [hg19], encapsulating BP4 and BP5). Stringent CNV calling parameters were applied (minimum of 5 probes, minimum of 2 algorithms, >30 kb in length) on the clinical microarray data, facilitating the discovery of 156 individuals with 15q13.3 microdeletions impacting the typical BP4-BP5. Among these microdeletions, 148 were detected in case subjects with broad developmental phenotypes and 8 in case subjects with a primary diagnosis of ASD (Figure S3A). In contrast, we identified only one BP4-BP5 deletion within a control population of 22,241 individuals (Figure S3A). The enrichment of 15q13.3 microdeletions within neurodevelopmental disorder-affected and ASD-affected case subjects (0.4% combined in NDD and ASD case subjects; 0.004% in control subjects) is highly significant (p < 1.30 × 10−29). Together, these data re-confirm that the 15q13.3 microdeletion is strongly associated with neurodevelopmental disorders.16, 61
We then applied three approaches to narrow down the potential contributing gene(s) from the ten genes present in BP4-BP5. First, we determined whether there are smaller deletions within BP4-BP5 that are associated with neurodevelopmental disorders. We delineated the minimal region (530 kb in size) of overlap from our clinical microarray sample and found that only two genes (CHRNA7 and OTUD7A) are associated with an atypical smaller CNV. This has been reported previously, but it is particularly noteworthy that at least 43 overlapping deletions encompassing CHRNA7 also impacted OTUD7A20, 24 (Figure 1). Furthermore, since 52 microdeletions encompassing CHRNA7 are found within our control samples (compared to zero smaller deletions in controls within OTUD7A), this also implicates other genes within the CNV. In further support of this, we identified a 5-year-old female, previously unreported in the literature, with global developmental delay who possessed a genetic deletion that spanned BP4-BP5 including OTUD7A but not CHRNA7 (Figure S3B). Similarly, a duplication breakpoint that does not involve CHRNA7 has also been recently reported in a female patient with atypical Rett syndrome who is negative for an MECP2 mutation.62 Taken together, these data suggest that genes other than CHRNA7, such as OTUD7A, may contribute to the 15q13.3 microdeletion syndrome.
We next investigated DNA sequence-level mutations in the ten genes within the microdeletion among individuals with a neurodevelopmental disorder but who did not have a 15q13.3 microdeletion. We focused on de novo mutations, given their established role in pathogenicity.41, 51, 52, 53, 63, 64, 65, 66 We investigated whole-genome sequencing data (n = 84 ASD quad families53) and exome-sequencing data (n = 5,953 ASD trios51, 52) and discovered 8 de novo mutations impacting regions in the BP4-BP5 interval. Three of these mutations impacted OTUD7A, one impacted MTMR10, one impacted TRPM1 (Table 1, case 2, GenBank: NM_001252024.1; c.2382C>G [p.Phe794Leu]), and the remainder were intergenic de novo mutations. Among the de novo OTUD7A mutations specifically, the first is an exonic de novo 9 bp non-frameshift deletion (Table 1, case 3, GenBank: NM_130901.2; c.1474_1482del [p.Asn492_Lys494del]), in an ASD-affected proband and their affected sibling (Figures 3A, S3D, and S3E, and Table 1). The second and third are both intronic, one a de novo indel (2 bp deletion) observed in an ASD-affected proband and the other an intronic de novo single-nucleotide mutation in an ASD-affected proband (Table 1). One of the intergenic de novo mutations was identified in two ASD-affected siblings between OTUD7A and CHRNA7 (cases 5 and 6 in Table 1); genome-wide, no other pathogenic exonic mutations or CNVs were identified in either of these affected siblings. Interestingly, the intergenic mutation between OTUD7A and CHRNA7 occurs in the binding site of the transcription factor, enhancer of zeste 2 polycomb repressive complex subunit 2 (EZH2) (Figure S4). EZH2 (MIM: 601573) has previously been implicated in dendrite growth,67 which raises the hypothesis that EZH2 regulates OTUD7A expression; however, further investigation is required to investigate the intergenic mutations. In contrast, no de novo mutations within OTUD7A were identified from whole-genome sequencing of 250 Dutch trio control samples54 or exome sequencing of the unaffected siblings of ASD-affected probands (from a family dataset of 5,205 individuals).52 The identification of de novo mutations either within or near OTUD7A is further evidence in support of OTUD7A as a major critical gene in the 15q13.3 microdeletion syndrome.
Table 1.
Individual ASD-Affected Case Subjects with De Novo Mutations Found near or within OTUD7A
| ASD Cases | Chromosome | Nucleotide-Level Change-HGVS Format | Protein-Level Change | Seq-Type Variants | Gene/Nearby Genes |
|---|---|---|---|---|---|
| Case 1 | 15 | XM_005254506.1; c.936−17A>G | – | intronic | MTMR10 |
| Case 2 | 15 | NM_001252024.1; c.2382C>G | p.Phe794Leu | missense | TRPM1 |
| Case 3 | 15 | NM_130901.2; c.1474_1482del | p.Asn492_Lys494del | exonic-indel | OTUD7A |
| Case 4 | 15 | NC_000015.9; g.31557488A>C | – | intergenic | KLF13 |
| Case 5 | 15 | NC_000015.9; g.32309599_32309610del | – | intergenic-indel | OTUD7A, CHRNA7 |
| Case 6 | 15 | NC_000015.9; g.32309599_32309610del | – | intergenic-indel | OTUD7A, CHRNA7 |
| Case 7 | 15 | XM_005254201.1; c.−223+11014A>G | – | intronic | OTUD7A |
| Case 8 | 15 | NM_130901.1; c.1150+935del | – | intronic-indel | OTUD7A |
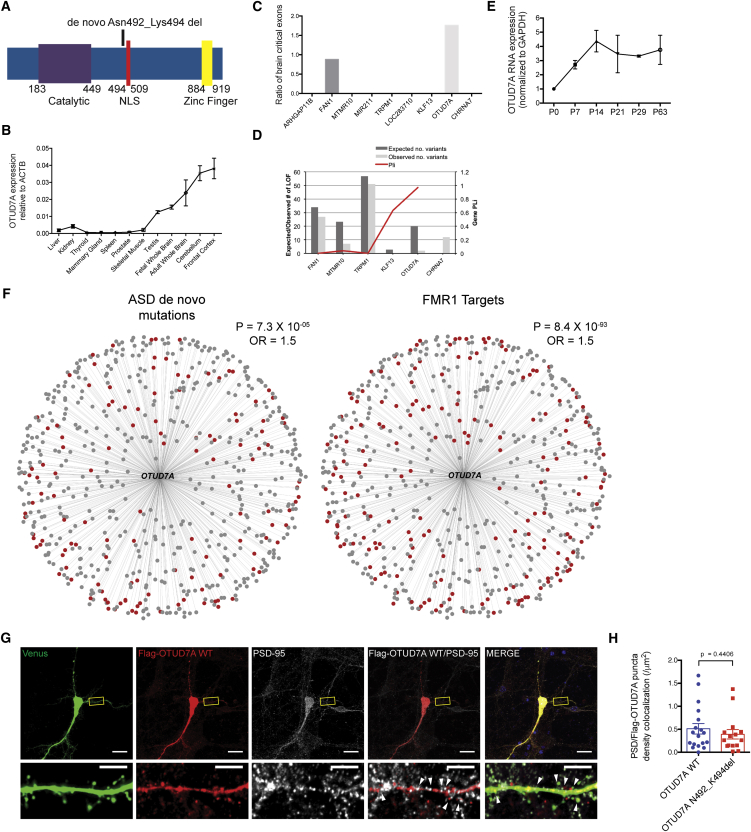
Figure 3.
Identification of OTUD7A as a Driver Gene of the 15q13.3 Microdeletion
(A) Schematic of the human OTUD7A protein showing the exonic 9 bp mutation (de novo p.Asn492_Lys494 deletion in the protein) found in an ASD proband and affected sibling (proband is case 3 in Table 1). The mutation is located in the nuclear localization sequence (NLS) of OTUD7A.
(B) OTUD7A mRNA expression relative to ACTB in various human tissues. OTUD7A is enriched in the brain, with highest expression in the frontal cortex.
(C) Ratio of brain critical exons in nine genes within the 15q13.3 microdeletion region. FAN1 and OTUD7A contain brain critical exons, with OTUD7A showing a higher ratio than that of FAN1.
(D) OTUD7A has the highest pLI (a score that indicates the probability that a gene is intolerant to a loss-of-function mutation) value compared to the other genes in the 15q13.3 critical region. The left y axis represents the number of observed LOF mutations within the ExAC population scale dataset and the right y axis shows the computed pLI score.
(E) OTUD7A mRNA expression in the mouse brain is low in early postnatal life, increases into adolescence, and remains stable into adulthood (n = 2 technical replicates).
(F) OTUD7A protein co-expression network module. Weighted gene correlation network analysis of OTUD7A from human proteome data shows that 616 genes are highly co-expressed with OTUD7A (top 25th percentile). 21% of genes highly co-expressed with OTUD7A harbor known ASD de novo mutations (left, red dots) and 30% of highly co-expressed genes are targets of FMR1 (right, red dots).
(G) DIV14 WT mouse cortical neurons co-expressing Venus and FLAG-OTUD7A WT and co-stained for FLAG and PSD95. OTUD7A is co-localized with PSD95 in dendrites and dendritic spines. Arrows indicate co-localized puncta located in dendritic spines (scale bars = 20 μm, top; 5 μm, bottom).
(H) Quantification of FLAG-OTUD7A and PSD95 puncta co-localization showed no changes between OTUD7A WT and OTUD7A p.Asn492_Lys494del overexpression (OTUD7A WT, n = 18 neurons; OTUD7A p.Asn492_Lys494del, n = 15 neurons, 2 cultures, Student’s t test, t(31) = 0.7813). Error bars represent SEM.
In our final approach, we assessed the exons of human genes for relevancy in neurodevelopment according to their population-level deleterious mutational burden in concert with brain mRNA expression (from RNA-sequencing data), earmarking those exons with a low mutational burden and high mRNA expression only in the developing human brain as being “brain critical.”1, 41 This analysis identified FAN1 and OTUD7A as genes bearing critical exons, with OUTD7A having a higher ratio of brain critical exons than FAN1 (Figure 3C). FAN1 was recently suggested to be a susceptibility gene in the 15q13.3 microdeletion based on the identification of an enrichment of small nucleotide variants (SNVs) in ASD and SCZ populations.40 However, in our analysis, we did not identify de novo mutations in FAN1 or overlapping deletions containing this gene and therefore did not pursue it further. To complement the critical exon analysis, we also examined the tolerance of loss-of-function (pLI) mutations of each of the 15q13.3 genes (a measure of the probability that a given gene is extremely intolerant of loss-of-function variation) from the exome aggregation consortium (ExAC) database.68 Comparison of the genes in the 15q13.3 microdeletion (no data were available for CHRNA7) revealed that OTUD7A possessed the most constrained gene scores (pLI = 0.97 and Z = 5.92) (Figure 3D), supporting its important evolutionary role in development.
To investigate the expression patterns of the genes within the BP4-BP5 deletion in the brain, we conducted qRT-PCR analysis of 8 genes (except KLF13 and miR-211; LOC100288637 was not detected) in 12 human tissues. For each gene, primers were designed targeting the exons that are common in all transcripts and our qRT-PCR analysis revealed that OTUD7A was the only gene that displayed a brain-specific mRNA expression pattern compared to other tissues (Figures 3B and S3C). This is similar to the expression profile of OTUD7A in the GTeX database. Furthermore, the BrainSpan database for human developmental brain expression shows that many OTUD7A exons are expressed above the 75th percentile of the total brain expression dataset (Figure S3F). KLF13 is notable because it has higher brain expression than OTUD7A in these databases; however, upon further examination it is largely driven by one exon (Figure S3F), and its expression is not brain specific, which is evident in the critical-exon analysis.
We also determined the protein co-expression network for OTUD7A in the brain using recently published human tissue protein-level data.69 To identify a core set of proteins having an important role in brain development, we analyzed protein data from 24 human tissues (each pooled from 3 post-mortem samples) including 17 adult and 7 prenatal samples that were run through high-resolution Fourier transform mass spectrometers for fragmentation.69 Weighted gene co-expression network analysis (WGCNA) analysis was used to identify 23 unbiased and independent protein modules. After rigorous gene set enrichment analysis (see STAR Methods), one of the modules was chosen for further analysis, as it was brain-specific and significantly associated with neurodevelopment and comprised of 2,484 proteins.70 Strikingly, WGCNA analysis identified OTUD7A as being the only gene within 15q13.3 microdeletion that was part of the brain-specific protein module. This is because the other 15q13.3 genes’ protein products did not have brain-specific expression. We identified 616 strongly connected (top 25th percentile) first-degree neighbor genes for OTUD7A within the brain-specific protein module. A comprehensive enrichment analysis was conducted, which determined that these OTUD7A neighboring genes are highly enriched for targets of FMR1 (Fragile X Mental Retardation 1), many of which are known to play a role at the synapse, as well as genes harboring ASD de novo mutations (Figure 3F). We performed a comprehensive gene set enrichment analysis to decipher the pathways and biological relevance of OTUD7A neighbor genes. The three most significant associations were observed with synapse (GO:0045202) (p < 1.1 × 10−82, Benjamin-Hochberg corrected), synaptic component (GO:0044456) (p < 9.92 × 10−72), and neuron projection (GO: 0043005) (p < 9.96 × 10−71) (Table S4). These co-expression networks align with the potential involvement of OTUD7A in the spine and dendrite abnormalities observed in Df(h15q13)/+ mice. Furthermore, the collective data indicate that OTUD7A is the only gene in the 15q13.3 microdeletion that has brain-specific expression, contains critical exons, and is interconnected with a brain-specific synaptic signaling network linked to ASD.
Expression and Localization of OTUD7A in Neurons
We utilized WT and Df(h15q13)/+ mice to investigate the biological function of Otud7a in the brain and to test our hypothesis that OTUD7A is a critical gene in the 15q13.3 microdeletion. We first analyzed the expression of Otud7a mRNA in the developing WT mouse brain and observed an expression profile similar to the developing human brain, with an increase in expression during early postnatal stages when dendrite and spine development are important (Figure 3E) and similar to the RNA-sequencing data (Figure S1A). To determine the localization of OTUD7A in neurons, we transfected a FLAG-tagged human OTUD7A (FLAG-OTUD7A WT) construct in cultured WT mouse cortical neurons and analyzed FLAG levels at DIV14, since no validated antibodies against OTUD7A are available. This experiment revealed that OTUD7A has a punctate localization pattern and is restricted to the soma and dendrites in Venus-transfected neurons (Venus is used for visualization) (Figure 3G). A very similar protein localization pattern was also independently reported by a companion study in this issue of AJHG.37 Furthermore, we detected a fraction of FLAG-OTUD7A puncta in dendritic spines that co-localized with PSD95, a postsynaptic marker of mature excitatory synapses (Figures 3G and 3H). These data reveal that Otud7a is expressed in the brain and can localize to dendritic and synaptic compartments in cortical neurons.
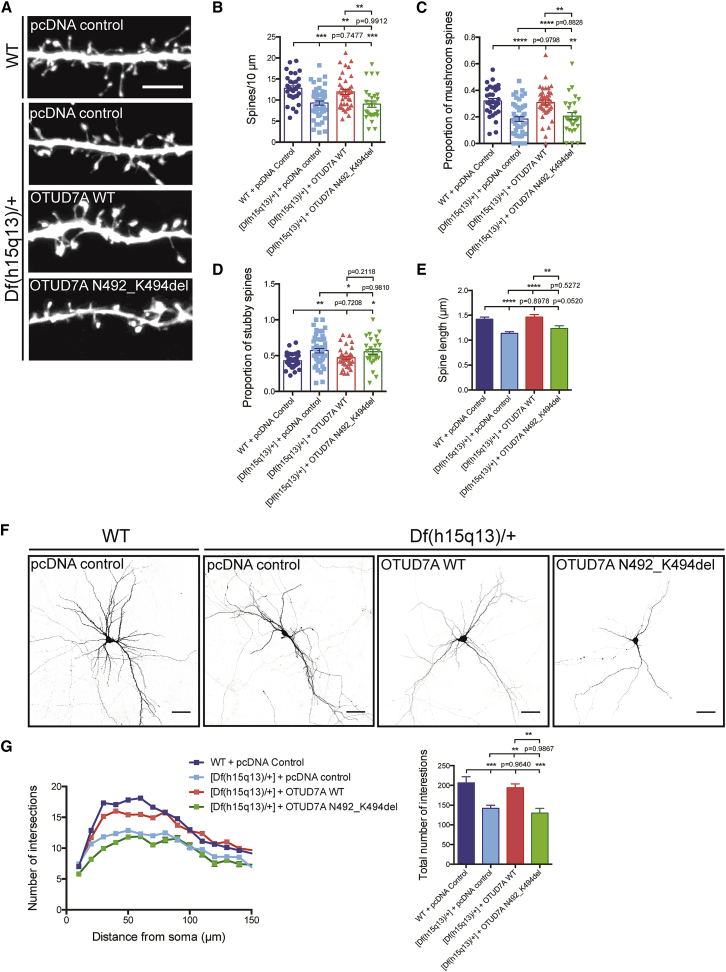
Rescue of Spine and Dendrite Phenotypes in Df(h15q13)/+ Excitatory Neurons by OTUD7A
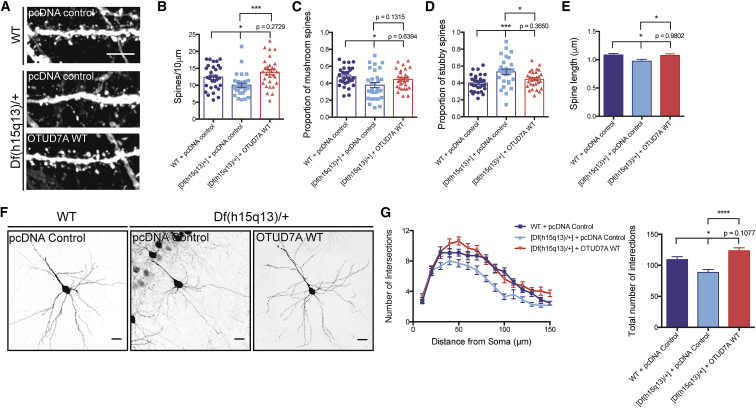
We hypothesized that the reduced expression of Otud7a in Df(h15q13)/+ heterozygous neurons mediated the dendritic spine and dendrite growth phenotypes, and increasing OTUD7A expression in these neurons would rescue the deficit. To test this, we co-transfected Venus and FLAG-OTUD7A WT (or a control plasmid) into littermate WT or Df(h15q13)/+ E16 mice using in utero electroporation to label layer 2/3 prefrontal cortical neurons. We examined dendritic spines and dendrite growth in brain slices at P22. We found that expression of OTUD7A WT in Df(h15q13)/+ neurons completely rescued the deficit in spine density, the proportion of mushroom and stubby type spines, and spine length compared to WT conditions (Figures 4A–4E). We also found that expression of OTUD7A WT in Df(h15q13)/+ neurons rescued the deficit in dendrite branching, similar to WT conditions (Figures 4F and 4G). We then tested whether the rescue by OTUD7A is recapitulated in vitro. We cultured cortical neurons from littermate WT and Df(h15q13)/+ mice at E16 and co-transfected FLAG-OTUD7A WT and Venus at DIV7. We evaluated FLAG/Venus double-positive neurons at DIV14 to visualize and quantify dendrites and spines expressing OTUD7A WT. We found that expression of OTUD7A WT in cultured Df(h15q13)/+ cortical neurons rescued the reduction in spine density, spine length, and the proportion of mushroom and stubby spines but had no effect on the proportion of thin spines or spine head width, neck width, or neck length (Figures 5A–5E, S5B, and S5C). OTUD7A also rescued dendritic branching compared to WT controls as measured by Sholl analysis and the total number of dendritic intersections (Figures 5F and 5G). Together, these data indicate that OTUD7A contributes to spine and dendrite deficits caused by the 15q13.3 microdeletion.
Figure 4.
Reduced Expression of OTUD7A Contributes to Spine and Dendrite Defects in Df(h15q13)/+ Mice
(A) Representative images of Venus-expressing dendritic spines from P22 WT or Df(h15q13)/+ neurons co-expressing Venus and pcDNA control or FLAG-OTUD7A WT (scale bar = 5 μm).
(B) Expression of OTUD7A WT rescues dendrite spine density defects in Df(h15q13)/+ cortical neurons. WT + pcDNA control, n = 30 dendrite segments; [Df(h15q13)/+] + pcDNA, n = 30 dendrite segments; [Df(h15q13)/+] + OTUD7A WT, n = 30 dendrite segments; 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, ∗∗∗p < 0.001, F(2, 87) = 8.049.
(C and D) Df(h15q13)/+ neurons show a decrease in mushroom spines and an increase of stubby spines compared to WT neurons. Expression of OTUD7A WT in Df(h15q13)/+ neurons decreases the proportion of stubby spines compared to Df(h15q13)/+ neurons. WT + pcDNA control, n = 30 dendrite segments; [Df(h15q13)/+] + pcDNA control, n = 30 dendrite segments; [Df(h15q13)/+] + OTUD7A WT, n = 30 dendrite segments; 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, ∗∗∗p < 0.001, F(2, 87) = 4.249, F(2, 87) = 8.636.
(E) Df(h15q13)/+ neurons showed a decrease in spine length compared to WT neurons, and expression of OTUD7A WT in Df(h15q13)/+ neurons increased spine length. WT + pcDNA control, n = 586 spines; [Df(h15q13)/+] + pcDNA control, n = 475 spines; [Df(h15q13)/+] + OTUD7A WT, n = 664 spines; 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, ∗∗∗p < 0.001, F(2, 1722) = 3.956.
(F) Representative images of littermate P22 WT and Df(h15q13)/+ neurons expressing Venus and pcDNA control or OTUD7A WT (scale bar = 50 μm).
(G) Expression of OTUD7A WT rescues dendrite growth defects (number and total number of intersections) in Df(h15q13)/+ cortical neurons. WT + pcDNA control, n = 28 neurons, [Df(h15q13)/+] + pcDNA control, n = 24; [Df(h15q13)/+] + OTUD7A WT, n = 28 neurons; 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, ∗∗∗∗p < 0.0001, F(2, 61) = 13.53. Error bars represent SEM.
Figure 5.
An Autism Spectrum Disorder-Linked De Novo Mutation in OTUD7A Disrupts Dendritic Spine Development and Neuronal Morphology
(A) Dendritic spines from DIV14 WT and Df(h15q13)/+ cultured cortical neurons co-expressing Venus, and pcDNA control, FLAG-OTUD7A WT, or FLAG-OTUD7A p.Asn492_Lys494del (scale bar = 5 μm).
(B) In Df(h15q13)/+ neurons, expression of OTUD7A p.Asn492_Lys494del does not rescue the reduction of dendritic spine density. WT + pcDNA, n = 32 dendritic segments, 18 neurons; pcDNA control, n = 48 dendritic segments, 30 neurons; OTUD7A WT, n = 38 dendritic segments, 21 neurons; OTUD7A p.Asn492_Lys494del, n = 28 dendritic segments, 19 neurons, 5 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗∗p < 0.01, ∗∗∗p < 0.001, F(3, 142) = 9.422.
(C) Expression of OTUD7A p.Asn492_Lys494del does not increase the reduced proportion of mushroom spines in Df(h15q13)/+ neurons. n = same as (B), one-way ANOVA followed by Tukey’s post hoc test, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, F(3, 141) = 11.99.
(D) Expression of OTUD7A p.Asn492_Lys494del does not significantly decrease the increased proportion of stubby spines in Df(h15q13)/+ neurons. n = same as (B), one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, ∗∗p < 0.01, F(3, 141) = 5.751.
(E) Expression of OTUD7A p.Asn492_Lys494del is unable to increase the reduction in spine length in Df(h15q13)/+ neurons. WT + pcDNA, n = 653 spines; [Df(h15q13)/+] + pcDNA control, n = 770 spines; [Df(h15q13)/+] + OTUD7A WT, n = 712 spines; [Df(h15q13)/+] + OTUD7A p.Asn492_Lys393del, n = 365 spines, 5 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, F(3, 2496) = 13.20.
(F) DIV14 WT and Df(h15q13)/+ cortical neurons expressing Venus and pcDNA control, FLAG-OTUD7A WT, or FLAG-OTUD7A p.Asn492_Lys494del (scale bar = 50 μm).
(G) In Df(h15q13)/+ neurons, expression of OTUD7A p.Asn492_Lys494del does not rescue the reduction of dendrite growth. WT + pcDNA, n = 31 neurons; pcDNA control, n = 23 neurons; OTUD7A WT, n = 26 neurons; OTUD7A p.Asn492_Lys393del, n = 15 neurons, 5 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗∗p < 0.01, ∗∗∗p < 0.001, F(3, 133) = 9.613. Error bars represent SEM.
An ASD-Associated De Novo Mutation in OTUD7A Impairs Dendrite and Spine Growth
We next examined whether the ASD-linked de novo mutation in OTUD7A (p.Asn492_Lys494del; case 3, Table 1) has a functional effect on dendritic and spine morphology. We generated a mutant OTUD7A (encoding p.Asn492_Lys494del) in the same FLAG-tagged plasmid as OTUD7A WT and found no overt difference in protein levels or localization between the WT and the mutant form of OTUD7A (Figures S5A, S6L, and S6M). Furthermore, OTUD7A p.Asn492_Lys494del failed to rescue the defects in dendritic spine density, morphology, or spine length, spine neck length, spine head width, or neck width in Df(h15q13)/+ neurons (Figures 5A–5E, S5B, and S5C). Compared to WT control neurons and OTUD7A WT, the expression of OTUD7A p.Asn492_Lys494del in Df(h15q13)/+ cortical neurons failed to rescue dendritic arborization defects (Figures 5F and 5G). We also investigated whether OTUD7A p.Asn492_Lys494del has a dominant-negative effect when expressed in WT cortical neurons. We co-transfected Venus with OTUD7A WT or OTUD7A p.Asn492_Lys494del in cultured WT neurons and determined that expression of OTUD7A p.Asn492_Lys494del had a mild dominant-negative effect, significantly reducing dendritic spine length (Figures S6A and S6F) and arborization (Figures S6J and S6K) compared to the pcDNA control but had no effect on spine density, spine type, or length (Figures S6A–S6I). These experiments reveal that a de novo mutation in OTUD7A is sufficient to impair its function during dendritic spine and dendrite development.
OTUD7A Is the Predominant Gene Regulating Dendrite Spine Development
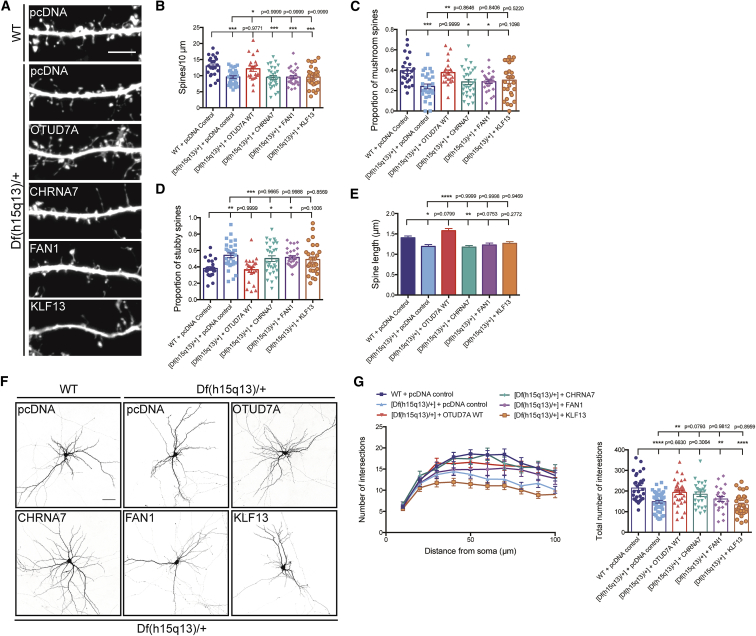
Finally, given that other genes in the 15q13.3 CNV have been implicated (CHRNA7 and FAN1), we tested the function of OTUD7A compared to other 15q13.3 genes. In addition to CHRNA7 and FAN1, KLF13 was included in our analysis due to its high expression (Figure S3F). We created FLAG-tagged CHRNA7, KLF13, or FAN1 constructs to determine whether they could rescue the cellular phenotypes. Analysis of spine density and morphology showed that OTUD7A was the only gene tested that could rescue spine density, length, and the proportion of mushroom and stubby spines, whereas CHRNA7, KLF13, or FAN1 were unable to (Figures 6A–6E). For dendrite outgrowth, we found that KLF13 and FAN1 were unable to rescue the defect in dendrite growth, further suggesting that these genes have a minimal role the 15q13.3 microdeletion syndrome (Figures 6F and 6G). Interestingly, expression of CHRNA7 had a similar rescue effect as OTUD7A in dendrite outgrowth, suggesting that heterozygous loss of both genes contribute, possibly together, to the dendrite growth defects in Df(h15q13)/+ mice. These data indicate that OTUD7A is the only gene in the BP4-BP5 interval regulating dendritic spine developmental phenotypes. A companion study by Yin et al. shows that Otud7a KO mice recapitulate many phenotypes of the 15q13.3 microdeletion syndrome including defective spine-synapse development, whereas a previous study from this laboratory showed no consistent behavioral phenotypes in a Chrna7 KO mouse model.37, 71 Our study, along with the companion study, provides evidence that OTUD7A is a critical gene in the 15q13.3 microdeletion syndrome.
Figure 6.
OTUD7A Is the Predominant Gene in the 15q13.3 Microdeletion Regulating Dendritic Spine Development
(A) Dendritic spines from DIV14 WT and Df(h15q13)/+ cortical neurons co-expressing Venus and the indicated construct (scale bar = 5 μm).
(B–E) Expression of CHRNA7, FAN1, or KLF13 in Df(h15q13)/+ neurons did not rescue the defects in certain phenotypes.
(B) Dendritic spine density. WT + pcDNA control, n = 23 dendritic segments; [Df(h15q13)/+] + pcDNA control, n = 24 dendritic segments; [Df(h15q13)/+] + OTUD7A WT, n = 22 dendritic segments; [Df(h15q13)/+] + CHRNA7, n = 30 dendritic segments; [Df(h15q13)/+] + FAN1, n = 24 dendritic spines; [Df(h15q13)/+] + KLF13, n = 30 dendritic segments; 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, F(5, 157) = 6.907.
(C and D) Proportion of mushroom spines (C) and the proportion of stubby spines (D). WT + pcDNA control, n = 23 dendritic segments; [Df(h15q13)/+] + pcDNA control, n = 24 dendritic segments; [Df(h15q13)/+] + OTUD7A WT, n = 22 dendritic segments; [Df(h15q13)/+] + CHRNA7, n = 30 dendritic segments; [Df(h15q13)/+] + FAN1, n = 24 dendritic spines; [Df(h15q13)/+] + KLF13, n = 30 dendritic segments; 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, ∗∗p < 0.01, F(5, 156) = 5.045, F(5, 157) = 5.666.
(E) Spine length. WT + pcDNA control, n = 484 spines; [Df(h15q13)/+] + pcDNA control, n = 379 spines; [Df(h15q13)/+] + OTUD7A WT, n = 444 spines; [Df(h15q13)/+] + CHRNA7, n = 454 spines; [Df(h15q13)/+] + FAN1, n = 374 spines; [Df(h15q13)/+] + KLF13, n = 457 spines; 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, F(5, 2728) = 10.64.
(F) DIV14 WT and Df(h15q13)/+ cortical neurons co-expressing Venus and the indicated construct (scale bar = 50 μm).
(G) Expression of OTUD7A WT or CHRNA7 in Df(h15q13)/+ cortical neurons rescues dendritic arborization defects, whereas FAN1 and KLF13 did not. WT + pcDNA, n = 30 neurons, [Df(h15q13)/+] + pcDNA control, n = 40 neurons; [Df(h15q13)/+] + OTUD7A WT, n = 35 neurons; [Df(h15q13)/+] + CHRNA7, n = 26 neurons; [Df(h15q13)/+] + FAN1, n = 23 neurons; [Df(h15q13)/+] + KLF13, n = 33 neurons, 3 cultures, one-way ANOVA followed by Tukey’s post hoc test, ∗p < 0.05, F(5, 181) = 6.873. Error bars represent SEM.
Discussion
Our data support an important and previously unidentified role for OTUD7A in the 15q13.3 microdeletion syndrome. Specifically, we identified that OTUD7A is a critical gene for brain development and an important contributing gene in the 15q13.3 microdeletion syndrome. We also biologically validated that Otud7a was the only gene in the interval that we tested that regulates the dendritic spine defects caused by the BP4-BP5 microdeletion in a validated mouse model. Our identification of OTUD7A as a critical component of brain protein-signaling networks that localizes to PSD95-positive spine synapses and modulates spine morphology and dendrite growth leads us to conclude that OTUD7A is a critical driver gene of the 15q13.3 microdeletion.
Our findings are overall consistent with a companion study by Yin et al. showing that Otud7a localizes to dendritic spines and that Otud7a KO mice have deficits in cortical neuron dendritic spine density and glutamatergic transmission.37 One difference is the lack of a dendrite outgrowth phenotype in Otud7a KO mice. However, given that Yin et al.37 demonstrated that expression of OTUD7A increased dendrite branching, this suggests that OTUD7A expression is able to stimulate dendrite development, which is consistent with our rescue findings. Moreover, we found that Otud7a and Chrna7 both contribute to dendrite growth defects in Df(h15q13)/+ mice. Therefore, CHRNA7 and OTUD7A could regulate dendrite growth together in excitatory cortical neurons or alternatively, CHRNA7 and OTUD7A may regulate dendritic development at different times and/or through different cell types, since CHRNA7 is also expressed in inhibitory neurons.72 Since OTUD7A was the only gene able to rescue spine deficits in Df(h15q13)/+ neurons, this indicates a unique and important role for it at the synapse during development. Although we have observed a strong and cross-validated OTUD7A phenotype, our results indicate a multi-hit hypothesis where more than one gene contributes to the etiology of 15q13.3 microdeletion syndrome, which is in line with other CNVs. For example, DGCR8 and ZDHHC8 are proposed driver genes in the 22q11.2 microdeletion, UBE3A and MAGEL2 for the 15q11-13 microdeletion region,73, 74 and GTF2I and FZD9 for the 7q11.23 region.75, 76, 77 For the 15q13.3 CNV, future experiments are needed using OTUD7A and CHRNA7 single and double KO cellular models to compare to the 15q13.3 microdeletion to determine the unique and shared contribution of each gene to disease pathogenesis.
It is also possible that while the heterozygous loss of the driver genes in the 15q13.3 region results in a core set of neurodevelopmental deficiencies, the heterogeneity of genetic background of the deletion carrier might contribute to the heterogeneous clinical presentation. Interestingly, such a mechanism has been shown to be at play in the 22q11.2 microdeletion syndrome,78 suggesting that it could be the case for the 15q13.3 microdeletion.
OTUD7A is part of a family of deubquitinases (DUBs) that are important for removing ubiquitin molecules from proteins that are targeted for degradation.79, 80 While we did not investigate the mechanistic action of OTUD7A, its localization to the postsynaptic density suggests that it could regulate key proteins involved in synapse maturation. Given the limited information on the role of OTUD7A, its protein-interaction network in neurons needs to be identified. Interestingly, a recent study identified that biallelic variants in OTUD6B, a family member, causes a novel syndrome characterized by intellectual disability, dysmorphic features, and seizures,81 indicating that DUBs could play a broader role in NDDs. Important questions regarding OTUD7A remain, such as: what specific proteins does OTUD7A regulate and interact with at the synapse? Which stages of synapse development are impaired by a lack of OTUD7A and how does this impact synaptic physiology and the epilepsy phenotypes observed in this microdeletion? It should also be noted that the dysregulation of protein turnover and the ubiquitin pathway has been implicated in other neurodevelopmental disorder-related CNVs. For instance, at the 15q11–13 locus, UBE3A has been reported to ubiquitinate key synaptic proteins.82, 83 Furthermore, defects in the regulation of synaptic protein turnover (i.e., synaptic protein synthesis) have also been observed in the 16p11.2 deletion and single-gene disorders such as fragile X syndrome (FXS) and tuberous sclerosis complex (TSC).55, 84 Thus, if OTUD7A is involved in protein turnover, particularly at the synapse, this would add to growing evidence that this is a common pathological mechanism for neurodevelopmental disorders involving defective synaptic signaling.
Acknowledgments
This work was supported by grants to K.K.S. (Canadian Institutes of Health Research [CIHR], Natural Sciences and Engineering Research Council, Brain Canada Platform Support Grant, and the Ontario Brain Institute-POND study), S.W.S. (OB, CIHR, and Autism Speaks), K.J.H. (CIHR and the Ontario Institute of Cancer Research), and B.W.D. (CIHR).
Published: February 1, 2018
Footnotes
Supplemental Data include six figures and four tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.01.006.
Contributor Information
Stephen W. Scherer, Email: stephen.scherer@sickkids.ca.
Karun K. Singh, Email: singhk2@mcmaster.ca.
Accession Numbers
Whole-genome datasets generated during and analyzed during the current study are available at the Autism Speaks MSSNG repository.85 Microarray data has been submitted to dbGAP under accession number phs001154.v1.p1.
Web Resources
Allen Brain Atlas, http://www.brain-map.org/
Autism Speaks MSSNG Project, https://www.mss.ng
Database of Genomic Variants (DGV), http://dgv.tcag.ca/dgv/app/home
Human ProteomeMap, http://www.humanproteomemap.org/
OMIM, http://www.omim.org/
Supplemental Data
List for each developmental time point is given in separate columns.
The list comprised of results from gene enrichment analysis for the differentially expressed genes.
References
- 1.Uddin M., Pellecchia G., Thiruvahindrapuram B., D’Abate L., Merico D., Chan A., Zarrei M., Tammimies K., Walker S., Gazzellone M.J. Indexing effects of copy number variation on genes involved in developmental delay. Sci. Rep. 2016;6:28663. doi: 10.1038/srep28663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H., Abecasis G.R., Heath S.C., Knowles A., Demars S., Chen Y.J., Roos J.L., Rapoport J.L., Gogos J.A., Karayiorgou M. Genetic variation in the 22q11 locus and susceptibility to schizophrenia. Proc. Natl. Acad. Sci. USA. 2002;99:16859–16864. doi: 10.1073/pnas.232186099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukai J., Dhilla A., Drew L.J., Stark K.L., Cao L., MacDermott A.B., Karayiorgou M., Gogos J.A. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat. Neurosci. 2008;11:1302–1310. doi: 10.1038/nn.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Portmann T., Yang M., Mao R., Panagiotakos G., Ellegood J., Dolen G., Bader P.L., Grueter B.A., Goold C., Fisher E. Behavioral abnormalities and circuit defects in the basal ganglia of a mouse model of 16p11.2 deletion syndrome. Cell Rep. 2014;7:1077–1092. doi: 10.1016/j.celrep.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blizinsky K.D., Diaz-Castro B., Forrest M.P., Schürmann B., Bach A.P., Martin-de-Saavedra M.D., Wang L., Csernansky J.G., Duan J., Penzes P. Reversal of dendritic phenotypes in 16p11.2 microduplication mouse model neurons by pharmacological targeting of a network hub. Proc. Natl. Acad. Sci. USA. 2016;113:8520–8525. doi: 10.1073/pnas.1607014113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Migliavacca E., Golzio C., Männik K., Blumenthal I., Oh E.C., Harewood L., Kosmicki J.A., Loviglio M.N., Giannuzzi G., Hippolyte L., 16p11.2 European Consortium A potential contributory role for ciliary dysfunction in the 16p11.2 600 kb BP4-BP5 pathology. Am. J. Hum. Genet. 2015;96:784–796. doi: 10.1016/j.ajhg.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pucilowska J., Vithayathil J., Tavares E.J., Kelly C., Karlo J.C., Landreth G.E. The 16p11.2 deletion mouse model of autism exhibits altered cortical progenitor proliferation and brain cytoarchitecture linked to the ERK MAPK pathway. J. Neurosci. 2015;35:3190–3200. doi: 10.1523/JNEUROSCI.4864-13.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumenthal I., Ragavendran A., Erdin S., Klei L., Sugathan A., Guide J.R., Manavalan P., Zhou J.Q., Wheeler V.C., Levin J.Z. Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am. J. Hum. Genet. 2014;94:870–883. doi: 10.1016/j.ajhg.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golzio C., Willer J., Talkowski M.E., Oh E.C., Taniguchi Y., Jacquemont S., Reymond A., Sun M., Sawa A., Gusella J.F. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485:363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stark K.L., Xu B., Bagchi A., Lai W.S., Liu H., Hsu R., Wan X., Pavlidis P., Mills A.A., Karayiorgou M., Gogos J.A. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 11.Fénelon K., Mukai J., Xu B., Hsu P.K., Drew L.J., Karayiorgou M., Fischbach G.D., Macdermott A.B., Gogos J.A. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc. Natl. Acad. Sci. USA. 2011;108:4447–4452. doi: 10.1073/pnas.1101219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu B., Hsu P.K., Stark K.L., Karayiorgou M., Gogos J.A. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sigurdsson T., Stark K.L., Karayiorgou M., Gogos J.A., Gordon J.A. Impaired hippocampal-prefrontal synchrony in a genetic mouse model of schizophrenia. Nature. 2010;464:763–767. doi: 10.1038/nature08855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian D., Stoppel L.J., Heynen A.J., Lindemann L., Jaeschke G., Mills A.A., Bear M.F. Contribution of mGluR5 to pathophysiology in a mouse model of human chromosome 16p11.2 microdeletion. Nat. Neurosci. 2015;18:182–184. doi: 10.1038/nn.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura M., Mukai J., Gordon J.A., Gogos J.A. Developmental inhibition of Gsk3 rescues behavioral and neurophysiological deficits in a mouse model of schizophrenia predisposition. Neuron. 2016;89:1100–1109. doi: 10.1016/j.neuron.2016.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharp A.J., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helbig I., Mefford H.C., Sharp A.J., Guipponi M., Fichera M., Franke A., Muhle H., de Kovel C., Baker C., von Spiczak S. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonacci F., Dennis M.Y., Huddleston J., Sudmant P.H., Steinberg K.M., Rosenfeld J.A., Miroballo M., Graves T.A., Vives L., Malig M. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat. Genet. 2014;46:1293–1302. doi: 10.1038/ng.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziats M.N., Goin-Kochel R.P., Berry L.N., Ali M., Ge J., Guffey D., Rosenfeld J.A., Bader P., Gambello M.J., Wolf V. The complex behavioral phenotype of 15q13.3 microdeletion syndrome. Genet. Med. 2016;18:1111–1118. doi: 10.1038/gim.2016.9. [DOI] [PubMed] [Google Scholar]
- 20.Shinawi M., Schaaf C.P., Bhatt S.S., Xia Z., Patel A., Cheung S.W., Lanpher B., Nagl S., Herding H.S., Nevinny-Stickel C. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat. Genet. 2009;41:1269–1271. doi: 10.1038/ng.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefansson H., Rujescu D., Cichon S., Pietiläinen O.P., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E., GROUP Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagnamenta A.T., Wing K., Sadighi Akha E., Knight S.J., Bölte S., Schmötzer G., Duketis E., Poustka F., Klauck S.M., Poustka A., International Molecular Genetic Study of Autism Consortium A 15q13.3 microdeletion segregating with autism. Eur. J. Hum. Genet. 2009;17:687–692. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowther C., Costain G., Stavropoulos D.J., Melvin R., Silversides C.K., Andrade D.M., So J., Faghfoury H., Lionel A.C., Marshall C.R. Delineating the 15q13.3 microdeletion phenotype: a case series and comprehensive review of the literature. Genet. Med. 2015;17:149–157. doi: 10.1038/gim.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fejgin K., Nielsen J., Birknow M.R., Bastlund J.F., Nielsen V., Lauridsen J.B., Stefansson H., Steinberg S., Sorensen H.B., Mortensen T.E. A mouse model that recapitulates cardinal features of the 15q13.3 microdeletion syndrome including schizophrenia- and epilepsy-related alterations. Biol. Psychiatry. 2014;76:128–137. doi: 10.1016/j.biopsych.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Kogan J.H., Gross A.K., Featherstone R.E., Shin R., Chen Q., Heusner C.L., Adachi M., Lin A., Walton N.M., Miyoshi S. Mouse model of chromosome 15q13.3 microdeletion syndrome demonstrates features related to autism spectrum disorder. J. Neurosci. 2015;35:16282–16294. doi: 10.1523/JNEUROSCI.3967-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masurel-Paulet A., Drumare I., Holder M., Cuisset J.M., Vallée L., Defoort S., Bourgois B., Pernes P., Cuvellier J.C., Huet F. Further delineation of eye manifestations in homozygous 15q13.3 microdeletions including TRPM1: a differential diagnosis of ceroid lipofuscinosis. Am. J. Med. Genet. A. 2014;164A:1537–1544. doi: 10.1002/ajmg.a.36471. [DOI] [PubMed] [Google Scholar]
- 28.Spielmann M., Reichelt G., Hertzberg C., Trimborn M., Mundlos S., Horn D., Klopocki E. Homozygous deletion of chromosome 15q13.3 including CHRNA7 causes severe mental retardation, seizures, muscular hypotonia, and the loss of KLF13 and TRPM1 potentially cause macrocytosis and congenital retinal dysfunction in siblings. Eur. J. Med. Genet. 2011;54:e441–e445. doi: 10.1016/j.ejmg.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 29.Forsingdal A., Fejgin K., Nielsen V., Werge T., Nielsen J. 15q13.3 homozygous knockout mouse model display epilepsy-, autism- and schizophrenia-related phenotypes. Transl. Psychiatry. 2016;6:e860. doi: 10.1038/tp.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nilsson S.R., Celada P., Fejgin K., Thelin J., Nielsen J., Santana N., Heath C.J., Larsen P.H., Nielsen V., Kent B.A. A mouse model of the 15q13.3 microdeletion syndrome shows prefrontal neurophysiological dysfunctions and attentional impairment. Psychopharmacology (Berl.) 2016;233:2151–2163. doi: 10.1007/s00213-016-4265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gass N., Weber-Fahr W., Sartorius A., Becker R., Didriksen M., Stensbøl T.B., Bastlund J.F., Meyer-Lindenberg A., Schwarz A.J. An acetylcholine alpha7 positive allosteric modulator rescues a schizophrenia-associated brain endophenotype in the 15q13.3 microdeletion, encompassing CHRNA7. Eur. Neuropsychopharmacol. 2016;26:1150–1160. doi: 10.1016/j.euroneuro.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 32.Freedman R. α7-nicotinic acetylcholine receptor agonists for cognitive enhancement in schizophrenia. Annu. Rev. Med. 2014;65:245–261. doi: 10.1146/annurev-med-092112-142937. [DOI] [PubMed] [Google Scholar]
- 33.Young J.W., Meves J.M., Tarantino I.S., Caldwell S., Geyer M.A. Delayed procedural learning in α7-nicotinic acetylcholine receptor knockout mice. Genes Brain Behav. 2011;10:720–733. doi: 10.1111/j.1601-183X.2011.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Azzopardi E., Typlt M., Jenkins B., Schmid S. Sensorimotor gating and spatial learning in α7-nicotinic receptor knockout mice. Genes Brain Behav. 2013;12:414–423. doi: 10.1111/gbb.12038. [DOI] [PubMed] [Google Scholar]
- 35.Lin H., Hsu F.C., Baumann B.H., Coulter D.A., Lynch D.R. Cortical synaptic NMDA receptor deficits in α7 nicotinic acetylcholine receptor gene deletion models: implications for neuropsychiatric diseases. Neurobiol. Dis. 2014;63:129–140. doi: 10.1016/j.nbd.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orr-Urtreger A., Göldner F.M., Saeki M., Lorenzo I., Goldberg L., De Biasi M., Dani J.A., Patrick J.W., Beaudet A.L. Mice deficient in the alpha7 neuronal nicotinic acetylcholine receptor lack alpha-bungarotoxin binding sites and hippocampal fast nicotinic currents. J. Neurosci. 1997;17:9165–9171. doi: 10.1523/JNEUROSCI.17-23-09165.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin J., Wu C., Chao E.S., Soriano S., Wang L., Wang W., Cummock S.E., Tao H., Pang K., Liu Z. Otud7a knockout mice recapitulate many neurological features of 15q13.3 microdeletion syndrome. Am. J. Hum. Genet. 2018;102:296–308. doi: 10.1016/j.ajhg.2018.01.005. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Locke D.P., Sharp A.J., McCarroll S.A., McGrath S.D., Newman T.L., Cheng Z., Schwartz S., Albertson D.G., Pinkel D., Altshuler D.M., Eichler E.E. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ionita-Laza I., Xu B., Makarov V., Buxbaum J.D., Roos J.L., Gogos J.A., Karayiorgou M. Scan statistic-based analysis of exome sequencing data identifies FAN1 at 15q13.3 as a susceptibility gene for schizophrenia and autism. Proc. Natl. Acad. Sci. USA. 2014;111:343–348. doi: 10.1073/pnas.1309475110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uddin M., Tammimies K., Pellecchia G., Alipanahi B., Hu P., Wang Z., Pinto D., Lau L., Nalpathamkalam T., Marshall C.R. Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nat. Genet. 2014;46:742–747. doi: 10.1038/ng.2980. [DOI] [PubMed] [Google Scholar]
- 42.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coe B.P., Witherspoon K., Rosenfeld J.A., van Bon B.W., Vulto-van Silfhout A.T., Bosco P., Friend K.L., Baker C., Buono S., Vissers L.E. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bierut L.J., Agrawal A., Bucholz K.K., Doheny K.F., Laurie C., Pugh E., Fisher S., Fox L., Howells W., Bertelsen S., Gene, Environment Association Studies Consortium A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. USA. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coviello A.D., Haring R., Wellons M., Vaidya D., Lehtimäki T., Keildson S., Lunetta K.L., He C., Fornage M., Lagou V. A genome-wide association meta-analysis of circulating sex hormone-binding globulin reveals multiple Loci implicated in sex steroid hormone regulation. PLoS Genet. 2012;8:e1002805. doi: 10.1371/journal.pgen.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bierut L.J., Madden P.A., Breslau N., Johnson E.O., Hatsukami D., Pomerleau O.F., Swan G.E., Rutter J., Bertelsen S., Fox L. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum. Mol. Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verhoeven V.J., Hysi P.G., Wojciechowski R., Fan Q., Guggenheim J.A., Höhn R., MacGregor S., Hewitt A.W., Nag A., Cheng C.Y., Consortium for Refractive Error and Myopia (CREAM) Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) Research Group. Wellcome Trust Case Control Consortium 2 (WTCCC2) Fuchs’ Genetics Multi-Center Study Group Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat. Genet. 2013;45:314–318. doi: 10.1038/ng.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart A.F., Dandona S., Chen L., Assogba O., Belanger M., Ewart G., LaRose R., Doelle H., Williams K., Wells G.A. Kinesin family member 6 variant Trp719Arg does not associate with angiographically defined coronary artery disease in the Ottawa Heart Genomics Study. J. Am. Coll. Cardiol. 2009;53:1471–1472. doi: 10.1016/j.jacc.2008.12.051. [DOI] [PubMed] [Google Scholar]
- 50.Krawczak M., Nikolaus S., von Eberstein H., Croucher P.J., El Mokhtari N.E., Schreiber S. PopGen: population-based recruitment of patients and controls for the analysis of complex genotype-phenotype relationships. Community Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 51.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuen R.K., Thiruvahindrapuram B., Merico D., Walker S., Tammimies K., Hoang N., Chrysler C., Nalpathamkalam T., Pellecchia G., Liu Y. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat. Med. 2015;21:185–191. doi: 10.1038/nm.3792. [DOI] [PubMed] [Google Scholar]
- 54.Genome of the Netherlands Consortium Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 55.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci. 2015;16:551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- 56.Lee E., Lee J., Kim E. Excitation/inhibition imbalance in animal models of autism spectrum disorders. Biol. Psychiatry. 2017;81:838–847. doi: 10.1016/j.biopsych.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 57.de la Torre-Ubieta L., Won H., Stein J.L., Geschwind D.H. Advancing the understanding of autism disease mechanisms through genetics. Nat. Med. 2016;22:345–361. doi: 10.1038/nm.4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J.A., Peñagarikano O., Belgard T.G., Swarup V., Geschwind D.H. The emerging picture of autism spectrum disorder: genetics and pathology. Annu. Rev. Pathol. 2015;10:111–144. doi: 10.1146/annurev-pathol-012414-040405. [DOI] [PubMed] [Google Scholar]
- 59.Kutzing M.K., Langhammer C.G., Luo V., Lakdawala H., Firestein B.L. Automated Sholl analysis of digitized neuronal morphology at multiple scales. J. Vis. Exp. 2010;45:2354. doi: 10.3791/2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwan V., Meka D.P., White S.H., Hung C.L., Holzapfel N.T., Walker S., Murtaza N., Unda B.K., Schwanke B., Yuen R.K.C. DIXDC1 phosphorylation and control of dendritic morphology are impaired by rare genetic variants. Cell Rep. 2016;17:1892–1904. doi: 10.1016/j.celrep.2016.10.047. [DOI] [PubMed] [Google Scholar]
- 61.Cooper G.M., Coe B.P., Girirajan S., Rosenfeld J.A., Vu T.H., Baker C., Williams C., Stalker H., Hamid R., Hannig V. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sajan S.A., Jhangiani S.N., Muzny D.M., Gibbs R.A., Lupski J.R., Glaze D.G., Kaufmann W.E., Skinner S.A., Annese F., Friez M.J. Enrichment of mutations in chromatin regulators in people with Rett syndrome lacking mutations in MECP2. Genet. Med. 2017;19:13–19. doi: 10.1038/gim.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Roak B.J., Stessman H.A., Boyle E.A., Witherspoon K.T., Martin B., Lee C., Vives L., Baker C., Hiatt J.B., Nickerson D.A. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat. Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Helsmoortel C., Vulto-van Silfhout A.T., Coe B.P., Vandeweyer G., Rooms L., van den Ende J., Schuurs-Hoeijmakers J.H., Marcelis C.L., Willemsen M.H., Vissers L.E. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat. Genet. 2014;46:380–384. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O’Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qi C., Liu S., Qin R., Zhang Y., Wang G., Shang Y., Wang Y., Liang J. Coordinated regulation of dendrite arborization by epigenetic factors CDYL and EZH2. J. Neurosci. 2014;34:4494–4508. doi: 10.1523/JNEUROSCI.3647-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin J., Chen W., Yang H., Xue M., Schaaf C.P. Chrna7 deficient mice manifest no consistent neuropsychiatric and behavioral phenotypes. Sci. Rep. 2017;7:39941. doi: 10.1038/srep39941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adams C.E., Yonchek J.C., Schulz K.M., Graw S.L., Stitzel J., Teschke P.U., Stevens K.E. Reduced Chrna7 expression in mice is associated with decreases in hippocampal markers of inhibitory function: implications for neuropsychiatric diseases. Neuroscience. 2012;207:274–282. doi: 10.1016/j.neuroscience.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fountain M.D., Tao H., Chen C.A., Yin J., Schaaf C.P. Magel2 knockout mice manifest altered social phenotypes and a deficit in preference for social novelty. Genes Brain Behav. 2017;16:592–600. doi: 10.1111/gbb.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schaaf C.P., Gonzalez-Garay M.L., Xia F., Potocki L., Gripp K.W., Zhang B., Peters B.A., McElwain M.A., Drmanac R., Beaudet A.L. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat. Genet. 2013;45:1405–1408. doi: 10.1038/ng.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adamo A., Atashpaz S., Germain P.L., Zanella M., D’Agostino G., Albertin V., Chenoweth J., Micale L., Fusco C., Unger C. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat. Genet. 2015;47:132–141. doi: 10.1038/ng.3169. [DOI] [PubMed] [Google Scholar]
- 76.Sakurai T., Dorr N.P., Takahashi N., McInnes L.A., Elder G.A., Buxbaum J.D. Haploinsufficiency of Gtf2i, a gene deleted in Williams Syndrome, leads to increases in social interactions. Autism Res. 2011;4:28–39. doi: 10.1002/aur.169. [DOI] [PubMed] [Google Scholar]
- 77.Chailangkarn T., Trujillo C.A., Freitas B.C., Hrvoj-Mihic B., Herai R.H., Yu D.X., Brown T.T., Marchetto M.C., Bardy C., McHenry L. A human neurodevelopmental model for Williams syndrome. Nature. 2016;536:338–343. doi: 10.1038/nature19067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Merico D., Zarrei M., Costain G., Ogura L., Alipanahi B., Gazzellone M.J., Butcher N.J., Thiruvahindrapuram B., Nalpathamkalam T., Chow E.W. Whole-genome sequencing suggests schizophrenia risk mechanisms in humans with 22q11.2 deletion syndrome. G3 (Bethesda) 2015;5:2453–2461. doi: 10.1534/g3.115.021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mevissen T.E.T., Kulathu Y., Mulder M.P.C., Geurink P.P., Maslen S.L., Gersch M., Elliott P.R., Burke J.E., van Tol B.D.M., Akutsu M. Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne. Nature. 2016;538:402–405. doi: 10.1038/nature19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mevissen T.E., Hospenthal M.K., Geurink P.P., Elliott P.R., Akutsu M., Arnaudo N., Ekkebus R., Kulathu Y., Wauer T., El Oualid F. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santiago-Sim T., Burrage L.C., Ebstein F., Tokita M.J., Miller M., Bi W., Braxton A.A., Rosenfeld J.A., Shahrour M., Lehmann A., EuroEPINOMICS RES Consortium Autosomal Recessive working group, S. Hande Caglayan Biallelic variants in OTUD6B cause an intellectual disability syndrome associated with seizures and dysmorphic features. Am. J. Hum. Genet. 2017;100:676–688. doi: 10.1016/j.ajhg.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Greer P.L., Hanayama R., Bloodgood B.L., Mardinly A.R., Lipton D.M., Flavell S.W., Kim T.K., Griffith E.C., Waldon Z., Maehr R. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kühnle S., Mothes B., Matentzoglu K., Scheffner M. Role of the ubiquitin ligase E6AP/UBE3A in controlling levels of the synaptic protein Arc. Proc. Natl. Acad. Sci. USA. 2013;110:8888–8893. doi: 10.1073/pnas.1302792110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ebrahimi-Fakhari D., Sahin M. Autism and the synapse: emerging mechanisms and mechanism-based therapies. Curr. Opin. Neurol. 2015;28:91–102. doi: 10.1097/WCO.0000000000000186. [DOI] [PubMed] [Google Scholar]
- 85.Yuen R.K.C., Merico D., Bookman M., Howe J.L., Thiruvahindrapuram B., Patel R.V., Whitney J., Deflaux N., Bingham J., Wang Z. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017;20:602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List for each developmental time point is given in separate columns.
The list comprised of results from gene enrichment analysis for the differentially expressed genes.