Abstract
Background
Cytochrome P450s form an important group of enzymes involved in xenobiotics degradation and metabolism, both primary and secondary. These enzymes are also useful in industry as biotechnological tools for bioconversion and a few are reported to be involved in pathogenicity. Trichoderma spp. are widely used in industry and agriculture and are known for their biosynthetic potential of a large number of secondary metabolites. For realising the full biosynthetic potential of an organism, it is important to do a genome-wide annotation and cataloguing of these enzymes.
Results
Here, we have studied the genomes of seven species (T. asperellum, T. atroviride, T. citrinoviride, T. longibrachiatum, T. reesei , T. harzianum and T. virens) and identified a total of 477 cytochrome P450s. We present here the classification, evolution and structure as well as predicted function of these proteins. This study would pave the way for functional characterization of these groups of enzymes and will also help in realization of their full economic potential.
Conclusion
Our CYPome annotation and evolutionary studies of the seven Trichoderma species now provides opportunities for exploration of research-driven strategies to select Trichoderma species for various applications especially in relation to secondary metabolism and degradation of environmental pollutants.
Electronic supplementary material
The online version of this article (10.1186/s40694-018-0056-3) contains supplementary material, which is available to authorized users.
Background
Trichoderma (Hypocreales, Ascomycota, Dikarya) species are among the most common fungi frequently isolated as mycotrophs from various fungi and as saprotrophs from free soil, soil litter, dead wood and rhizosphere, and includes more than 256 accepted species [1, 2]. These fungi are economically important due to their ability to produce enzymes of industrial importance, ability to kill/inhibit many plant pathogenic fungi, to boost plant immunity and promote plant growth, in addition to their ability to produce a plethora of secondary metabolites [3, 4]. A few species/strains are known to be opportunistic human pathogens [5]. Trichoderma spp. are thus ideal candidates for genome-wide studies to further augment their biotechnological applications. The first species to be sequenced is Trichoderma reesei, industrial source of cellulases and hemicellulases [6]. This was soon followed by whole genome sequencing of two strongly mycoparasitic species, viz. T. atroviride and T. virens [7]. A comparative analysis of the mycoparasitic species i.e., T. atroviride and T. virens with that of weaker mycoparasitic species T. reesei yielded novel information on the genome-scale differences between these species. In general, the mycoparasitic species are enriched in genes involved mycoparasitism and secondary metabolism [1, 7, 8]. Four more species, i.e., T. asperellum and T. harzianum (biocontrol species) and T. longibrachiatum and T. citrinoviride (opportunistic human pathogens) were subsequently sequenced by US Department of Energy Joint Genome Initiative (Mycocosm [9]; http://jgi.doe.gov/fungi). However, detailed analyses of these four genomes are awaited.
Cytochrome P450 genes (CYPs) are found in the genomes of prokaryotes and lower and higher eukaryotes. CYPs constitute a large superfamily of heme-thiolate proteins involved in the metabolism of a wide variety of both exogenous and endogenous compounds [10]. CYPs are heme b containing monooxygenases which were recognized and defined as a distinct class of hemoproteins [11]. Cyp proteins catalyze the regio-, chemo- and stereospecific oxidation of a vast number of substrates under mild reaction conditions, thus accomplishing chemical transformations. These functions make them important players in xenobiotic degradation and in primary and secondary metabolism. A few such enzymes are also reported to be involved in pathogenicity of plant pathogenic fungi [12–16]. Their diverse functional properties reflect their biological roles and make them important candidates for extensive investigation to explore diverse aspects of P450 functions and regulation as well as for biotechnological applications [17, 18].
Cytochrome P450s are categorized into two main classes, B (initially assigned as Bacterial) and E (initially assigned as Eukaryotic). Bacterial P450s with three component systems [an FAD-containing flavoprotein (NADPH or NADH-dependent reductase), an iron sulphur protein, and the P450 hemeprotein] and the fungal P450 nor (nitric oxide reductase). Clan CYP 55 belong to the ‘B’-class [19]. All the other known P450s from distinct systems, including eukaryotic and bacterial P450s, belong to the ‘E’-class. The eukaryotic microsomal P450 system contains two components, the NADPH:P450 oxidoreductase (POR), a flavoprotein containing both FAD and FMN, and the P450 monooxygenase containing the heme domain. The prokaryotic (bacterial) soluble P450 monooxygenase P450BM3 (Cyp102) exists as a single protein with both heme and flavin functional domains.
The complete CYP complement of one organism, called CYPome, is a collection of CYP genes in the genome of that species [20]. The current state of knowledge on P450 evolution in eukaryotes points to CYP51 as the ancestral P450, which is believed to have led to the evolution of all the present day P450 families [21]. The expansion and diversification of CYPomes may also provide information on fungal evolutionary adaptation to ecological niches. A key development affecting applied P450 research is the need to define and annotate ever-expanding genomic information. Various web-based resources have been developed to probe and assign various orphan CYPs in numerous genomes, owing to the identification of conserved motifs responsible for oxygen and heme-binding. These databases reveal that enormous number of sequence-diverse P450s is yet to be discovered and explored for functions and diverse activities in all kingdoms. One of the most commonly used resources includes the Nelson database (http://drnelson.uthsc.edu/cytochromeP450.html) [21]. The grouping scheme for CYPs is based on amino acid sequence similarity [22]. The original nomenclature for CYPs is based upon amino acid identity where Cyp proteins with at least 40% identity are placed in the same family [22, 23]. However, due to various evolutionary mechanisms, a straight forward nomenclature might be difficult, therefore, family definition is recommended by integrating phylogeny and protein evolution [24]. To each family, Cyp number is designated according to their taxonomic groups. Fungal Cyp families are numbered as Cyp51-Cyp69, Cyp501-Cyp699 and Cyp5001-Cyp6999. With rapid increase in discoveries of new Cyp proteins through genome sequencing, Nelson database lacks efficiency to annotate all Cyp proteins. For higher-level grouping of families identified via the sequence similarity-based scheme, CYP clan system was first developed and then applied to classify metazoan CYPs [25]. The CYP clan approach places all Cyp families with a monophyletic origin into a single clan and has been successfully applied to classify Cyp families in fungi [26]. For example, if new Cyps had equal identity to two or more Cyp families, they can be tentatively assigned to a clan in which these families belong. A site dedicated to filamentous fungi has been developed that includes comprehensive information on P450 clans and families (http://p450.riceblast.snu.ac.kr) [27]. In filamentous fungi, CYPs are involved in various physiological processes including fitness, resistance to xenobiotics and biosynthesis of a vast array of secondary metabolites with applications in biomedical, agricultural and industrial fields [28–31].
Keeping in view the wide spectrum of biotechnological applications of Trichoderma species, and the important role that CYPs play in the biology of fungi, we decided to annotate and make an inventory of the CYPome in the seven species of Trichoderma that have been sequenced by JGI. Annotation of these genes would help in commercial exploitation of these proteins. Earlier, the CYPome of several fungal species have been analysed in detail, e.g., Aspergillus nidulans [29], Phanerochete chrysosporium [32], Mycosphaerella graminicola [33] and Grosmannia clavigera [34]. However, this subject has not been covered in earlier analyses of Trichoderma genomes, except for the inclusion of T. reesei in a broad analysis of fungal CYPomes [35]. Moreover, a detailed phylogenetic analysis of Trichoderma CYPome could advance our understanding of the evolutionary processes of cytochrome P450 proteins in fungi.
Results
CYP proteins in Trichoderma
Trichoderma CYPome embodies a group of cytochrome P450 diverse proteins which are predicted to participate in a spectrum of functions involved in primary, secondary and xenobiotic metabolism. A total of 595 cytochrome P450 proteins have been identified in seven Trichoderma species. These entries were further analysed for the presence of full cytochrome P450 domain which led to the selection of a total 477 Cyp proteins (Table 1) for the detailed study. Entries with incomplete sequences and domains are listed in Additional file 1: Table S1.
Table 1.
Taxonomic distribution of putative CYPs in seven Trichoderma species
| Species | Genome size (Mb) | No. of predicted genes | Total Cyp proteins | Proteins with complete sequences | Clan type | Family type | Families with no FCPD matches |
|---|---|---|---|---|---|---|---|
| T. asperellum | 37.46 | 12,586 | 73 | 62 | 25 | 40 | 7 |
| T. atroviride | 36.10 | 11,863 | 69 | 57 | 22 | 36 | 5 |
| T. citrinoviride | 33.48 | 9397 | 75 | 57 | 23 | 41 | 6 |
| T. longibrachiatum | 32.24 | 10,792 | 68 | 53 | 21 | 38 | 4 |
| T. reesei | 34.10 | 9129 | 70 | 57 | 23 | 42 | 4 |
| T. harzianum | 40.98 | 14,095 | 118 | 101 | 31 | 67 | 12 |
| T. virens | 39.00 | 12,427 | 122 | 90 | 31 | 59 | 12 |
Analysis showed that T. harzianum genome harbours the highest number of Cyps (101), followed by T. virens (90), T. asperellum (62), T. atroviride (57), T. citrinoviride (57), T. reesei (57) and T. longibrachiatum (53). The number of Cyp proteins for families Cyp5080, Cyp52, Cyp534, Cyp535, Cyp541 and Cyp618 were found conserved among seven Trichoderma species. Cytochrome P450 families Cyp504, Cyp505, Cyp5080, Cyp51, Cyp52, Cyp528, Cyp534, Cyp535, Cyp539, Cyp541, Cyp548, Cyp570, Cyp58, Cyp584, Cyp61, Cyp618, Cyp620, Cyp65 and Cyp671 were found ubiquitously present in Trichoderma suggesting a conserved role of these proteins. In Trichoderma, Cyp families Cyp5039, Cyp5044, Cyp5046, Cyp5049, Cyp5055, Cyp5057, Cyp5060, Cyp5128, Cyp5129, Cyp5134, Cyp5168, Cyp5181, Cyp5246, Cyp5262, Cyp5268, Cyp5292, Cyp5296, Cyp5320, Cyp5334, Cyp5390 and Cyp5391 didn’t have any matches in Fungal Cytochrome P450 Database (FCPD). The cytochrome P450 families unique to Trichoderma were identified as Cyp5039, Cyp5049, Cyp5055, Cyp5057, Cyp5128, Cyp5129, Cyp5134, Cyp5268, Cyp5292, Cyp5296, Cyp5390 and Cyp5391. These families were predicted to be involved in both xenobiotic and secondary metabolism (Table 2).
Table 2.
Phylogenetic clustering of Trichoderma CYP families and clans
| Phylogenetic group ID | Total entries | CYP family | CYP Clan | Putative functions |
|---|---|---|---|---|
| 1 | 33 | Cyp5044a, Cyp5078, Cyp5080, Cyp5104, Cyp528, Cyp531, Cyp532, Cyp5320a, Cyp631 | CYP528, CYP531, CYP532 | Xenobiotic metabolism |
| 2 | 19 | Cyp535, Cyp570 | CYP507 | Xenobiotic metabolism |
| 3 | 3 | Cyp673 | CYP673 | |
| 4 | 11 | Cyp5055a, Cyp5057a, Cyp5262a, Cyp537, Cyp62, Cyp684 | CYP537, CYP62 | Xenobiotic metabolism Secondary metabolism |
| 5 | 35 | Cyp5039a, Cyp5094, Cyp5128a, Cyp5129a, Cyp5292a, Cyp551, Cyp552, Cyp58, Cyp677, Cyp680, Cyp682 | CYP58, CYP677 | Secondary metabolism Xenobiotic metabolism |
| 6 | 10 | Cyp5246a, Cyp53 | CYP53 | Xenobiotic metabolism |
| 7 | 3 | Cyp630 | CYP630 | Primary metabolism |
| 8 | 23 | Cyp574, Cyp5076, Cyp5168a, Cyp671 | CYP574 | Secondary metabolism |
| 9 | 17 | Cyp548 | CYP548 | Xenobiotic metabolism |
| 10 | 56 | Cyp5117, Cyp561, Cyp563, Cyp65 | CYP65 | Secondary metabolism |
| 11 | 3 | Cyp627 | CYP627 | |
| 12 | 62 | Cyp5049a, Cyp52, Cyp5296a, Cyp538, Cyp539, Cyp584, Cyp587, Cyp655 | CYP52, CYP59 | Xenobiotic metabolism |
| 13 | 19 | Cyp5181a, Cyp5334a, Cyp534, Cyp613, Cyp685 | CYP534, CYP613 | Xenobiotic metabolism |
| 14 | 39 | Cyp5134a, Cyp526, Cyp5390a, Cyp617, Cyp618 | CYP526, CYP547 | Secondary metabolism |
| 15 | 36 | Cyp505, Cyp5099, Cyp540, Cyp541 | CYP505, CYP540, CYP56 | Primary metabolism |
| 16 | 9 | Cyp504 | CYP504 | Xenobiotic metabolism |
| 17 | 51 | Cyp5046a, Cyp5068, Cyp5268a, Cyp530, Cyp5391a, Cyp620, Cyp621 | CYP530, CYP533 | Xenobiotic metabolism |
| 18 | 1 | Cyp5042 | CYP5042 | |
| 19 | 18 | Cyp503, Cyp5090, Cyp559, Cyp611, Cyp635, Cyp636, Cyp641, Cyp642 | CYP54, CYP550, CYP559, CYP642, CYP657, CYP659 | Secondary metabolism |
| 20 | 29 | Cyp5060a, Cyp51, Cyp55, Cyp61 | CYP51, CYP55, CYP61 | Primary metabolism |
aCorresponding clans for these families are absent in FCPD
Abundance and diversity of cytochrome P450 family/clan
Identified cytochrome P450 proteins were annotated and classified into 85 families (Fig. 1) and 37 clans (Fig. 2). Trichoderma species showed diversity in the number of annotated Cyp families (Table 1, Figs. 1, 2, 3). The numbers of annotated Cyp families among Trichoderma species ranged from 36 (T. atroviride) to 67 (T. harzianum). Annotated CYP clans were also found to be diverse in Trichoderma (Fig. 3). The highest numbers of CYP clans were identified in T. harzianum (31) and T. virens (31). T. asperellum and T. atroviride contained 25 and 22 clan types respectively. Clans CYP52 and CYP65 were found to be most abundant with 55 and 56 protein entries, respectively (Fig. 3). The number of proteins in the most abundant clans CYP52 and CYP65 ranged from 6 to 12 among Trichoderma species. Clan CYP673 was identified only in T. virens and T. harzianum, containing 1 and 2 members respectively, and was found to be absent in other five species. Similarly, clan CYP56 proteins were found to be unique to T. asperellum, T. harzianum and T. virens with single entries in each species. Clan540 proteins were found absent in T. citrinoviride, T. longibrachiatum and T. reesei. Clans CYP5042, 642, 659 and 677 were identified only in T. virens and were absent in all other species.
Fig. 1.
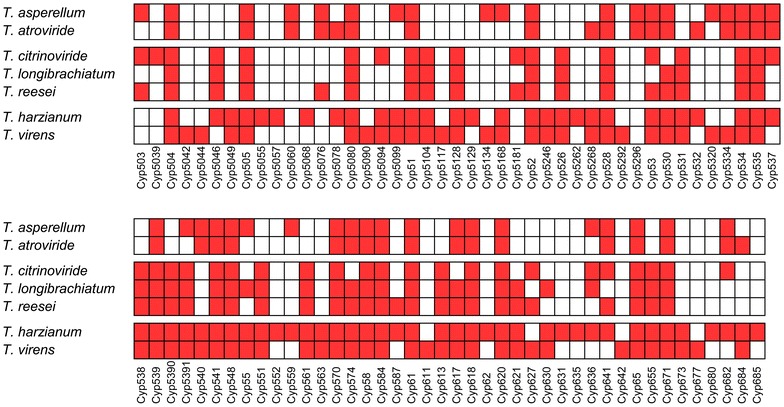
Cytochrome P450 families identified in Trichoderma
Fig. 2.
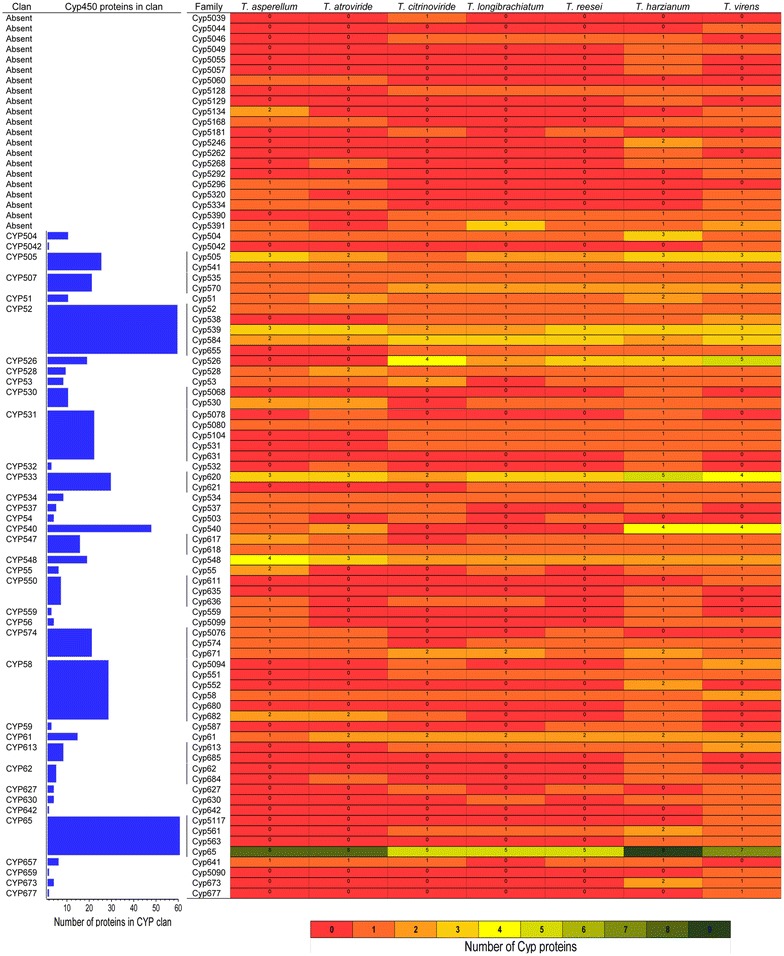
Abundance of cytochrome P450 families and clans in Trichoderma. The heatmap displays the abundance of Cyp protein families among Trichoderma species. Blue bar represent the number of Cyp proteins in a clan
Fig. 3.
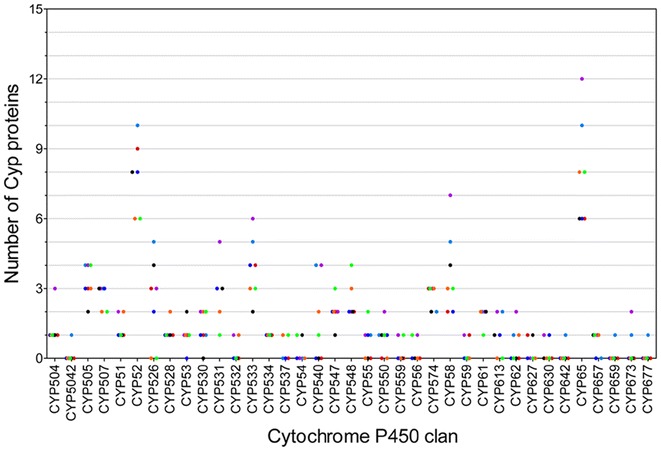
Diversity of cytochrome P450 clans among Trichoderma species. Scatter-plot presents number of proteins in CYP clans in T. asperellum (orange), T. atroviride (green), T. citrinoviride (black), T. harzianum (purple), T. longibrachiatum (dark blue), T. reesei (red) and T. virens (light blue)
Phyletic distribution of CYP families and clans in Trichoderma
The genome-wide comparisons and annotations of P450s have allowed us to further develop the relationships among Cyp families in different Trichoderma species. To demonstrate the divergence of the primary sequences and evolutionary relationships of cytochrome P450 families in Trichoderma, a detailed phylogenetic analysis was carried out using 477 aligned Cyp protein sequences. The phylogenetic tree depicting evolutionary relationships among Trichoderma cytochrome P450 proteins are illustrated in Fig. 4. Further, the distribution of different CYP clans and families in 20 phylogenetic groups with their putative functions are summarized in Table 2.
Fig. 4.
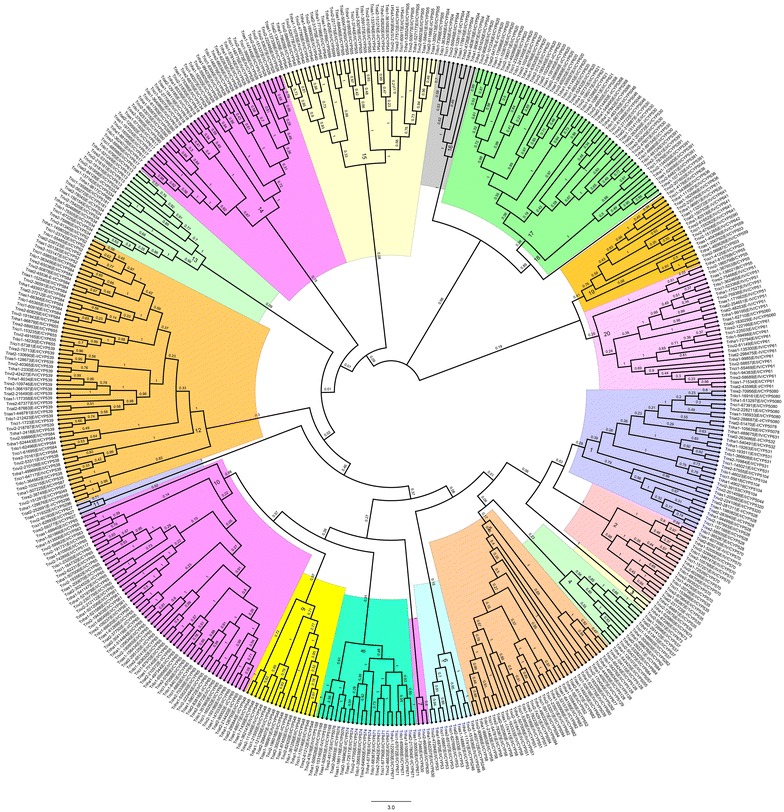
Evolutionary relationships of cytochrome P450 proteins among Trichoderma species. Phylogenetic tree was constructed inferred using the minimum evolution method72 using MEGA5 software. Phylogenetic groups (1–20) and bootstrap frequencies are shown in the tree. Tree includes Cyp proteins from all seven Trichoderma species including T. asperellum, T. atroviride, T. citrinoviride, T. harzianum, T. longibrachiatum, T. reesei and T. virens. Each phylogenetic group is indicated by a specific color
Evolutionary studies differentiated 477 cytochrome P450 proteins from 7 Trichoderma species into 20 phylogenetic groups (Fig. 4). Group 1 consisted of a total of 33 Cyp proteins from clans CYP528, CYP531 and CYP532. In Trichoderma, clan CYP531 consists of five Cyp families including Cyp5078, Cyp5080, Cyp5104, Cyp531 and Cyp631. Group 2 consisted of total 19 protein members belonging to clan CYP507. Members of clan CYP507 have been predicted to be involved in xenobiotic metabolism in Pezizomycotina [36]. In FCPD, clan CYP507 consists of four Cyp families including Cyp 507, Cyp525, Cyp535 and Cyp570. Of these four families, only Cyp535 and Cyp570 families are present in Trichoderma species. Group 2 containing clan CYP507 proteins was further differentiated into two sub-groups containing families Cyp535 (7 proteins) and Cyp570 (12 proteins) respectively. In Trichoderma, all 19 proteins belonging to clan 507 are grouped together in group 2 suggesting conserved putative role of Cyp535 and Cyp570 in xenobiotics metabolism. Clan CYP673 in group 3 consists of only three members-two from T. harzianum and one from T. virens.
Group 4 consists of 11 proteins from 2 clans (CYP537 and CYP62). In FCPD, clan CYP537 consists of two families: Cyp537 and Cyp577. In Trichoderma, Cyp577 family is absent and Cyp537 proteins are present only in T. asperellum, T. atroviride, T. citrinoviride and T. harzianum. In group 4, all identified members of clan CYP62 grouped together. Clan CYP62 in FCPD consists of three CYP families including CYP62, CYP626 and CYP684. In Trichoderma, one Cyp62 (T. harzianum) and three Cyp684 proteins (one each) were identified in T. atroviride, T. harzianum and T. virens. Group 4 also contained Cyp50555, Cyp5057 and Cyp5262 proteins. The corresponding clans for these three families are absent in FCPD. Protein Cyp5262 was grouped together with members of clan CYP537, whereas Cyp50555 and Cyp5057 proteins formed a separate subgroup in Group 4. Group 5 contained total 35 proteins belonging to clans CYP58 and CYP677, which includes diverse Cyp families Cyp5039, Cyp5094, Cyp5128, Cyp5129, Cyp5292, Cyp551, Cyp552, Cyp58, Cyp677, Cyp680 and Cyp682. Clans for Cyp families 5039, 5128, 5129 and 5292 are not available in FCPD. Trichoderma has only one Cyp677 protein i.e., in T. virens which was grouped closely with Cyp5292 in phyletic Group 5. A total of 26 proteins belonging to clan CYP58 are identified in Trichoderma. Cyp58 family had a single member in all Trichoderma species analysed except in T. virens (2 proteins). All 7 members of CYP53 clan were grouped together in Group 6. Cyp53 family was found in all Trichoderma species except T. longibrachiatum. These proteins are involved in xenobiotic metabolism. The group 6 also contained three Cyp5246 proteins, clan for this family is absent in FCPD. Family Cyp5246 is present only in T. harzianum and T. virens. Members of Cyp53 and Cyp5246 families were differentiated in two clear sub-groups. Group 7 consists of only three proteins belonging to clan CYP630; one each from T. harzianum, T. longibrachiatum and T. virens. The group 8 consists of 23 proteins from clan CYP574 including families Cyp5076, Cyp574 and Cyp671. Four members of Cyp5168 family were also clustered in group 8.
Group 9 consists of all 17 proteins of clan CYP548. In Trichoderma, Cyp548 family is ubiquitously present in all seven species, where T. asperellum and T. atroviride contained four and three proteins respectively followed by two each in T. citrinoviride, T. harzianum, T. longibrachiatum, T. reesei and T. virens. These proteins are known to be involved in xenobiotic metabolism. The second largest phylogenetic Group 10 has 56 Cyps from the clan CYP65 which are involved in secondary metabolism. It comprised of families Cyp5117, Cyp561, Cyp563 and Cyp65. Group 11 consists of three Cyp627 proteins.
In Trichoderma, group 12 is the largest with 62 Cyp proteins. These Cyps from clans CYP52 and CYP59 were differentiated separately into two sub-groups. Clans CYP52 and CYP59 involve members of Cyp52, Cyp538, Cyp539, Cyp584, Cyp587 and Cyp655 families. Two entries of Cyp587 family belonging to clan CYP59 were grouped together with two proteins each from Cyp5049 and Cyp5296 families. The corresponding clan for Cyp5049 and Cyp5296 families were found to be absent in FCPD. In group 12, Cyp proteins of clan CYP52 were grouped together in the separate sub-group. Group 13 contained 19 Cyps belonging to clans CYP534 and CYP613. Two Cyp proteins belonging to family Cyp5181 were also present in group 13. Protein members of groups 12 and 13 were predicted to be involved in xenobiotic metabolism (Table 2).
Group 14 consists of proteins belonging to clans CYP526 and CYP547 which were differentiated separately into two sub-groups. In Trichoderma, 2 Cyp families of clan CYP547 were identified that includes Cyp617 (7) and Cyp618 (7). Cyp5134 proteins were grouped together in sub-group containing clan CYP526 proteins. Group 15 consists of 36 proteins involved in primary metabolism that includes members of clans CYP505 (15), CYP540 (11), CYP541 (7) and Cyp5099 (3). All three Cyp5099 proteins belonging to clan CYP56 family were included in this group. These proteins were identified only in T. asperellum, T. harzianum and T. virens. Cyp5099 proteins were found closely related to Cyp540 proteins and together formed a separate sub-group. Another sub-group contained all proteins belonging to clan CYP505 which includes Cyp505 and Cyp541 families. All nine protein members of clan CYP504 were clustered together in group 16. These proteins are known to be involved in xenobiotic metabolism. Trichoderma species contain single copy of Cyp504 protein except T. harzianum which contains three copies of Cyp504 protein involved in phenylacetate catabolism [37].
Group 17 is the third largest Cyp group consisting of 51 Cyps from clans CYP530 and CYP533. In this group, CYP533 is the most dominant clan followed by CYP530. Clans CYP530 and CYP533 include Cyp families Cyp530 (8 proteins) and Cyp5068 (1 protein), and Cyp620 (23 proteins) and Cyp621 (4 proteins) respectively. This group also contained Cyp5046 (4), Cyp5391 (8) and Cyp5268 (3) proteins. The corresponding clans for these families are absent in FCPD. Group 18 contains one Cyp5042 protein of T. virens. Group19 includes 18 proteins belonging to clans CYP54, CYP550, CYP559, CYP642, CYP657 and CYP659. These clans are involved in secondary metabolism. A total of 29 proteins from 7 Trichoderma species corresponding to three clans including CYP51, CYP55 and CYP61 were clustered together in group 20. These are known to be involved in primary metabolism. In this group, CYP51 and CYP61 families dominate with 9 and 15 members respectively. Further, all proteins belonging to Cyp51 were grouped together in group 20. This suggests that Cyp51 protein which is involved in primary metabolism (sterol biosynthesis) is diversified only to a lesser extent in Trichoderma. In comparison to some of the ascomycetous fungi, which carry multiple CYP51 proteins, T. atroviride and T. harzianum contained two copies each, whereas T. asperellum, T. citrinoviride, T. longibrachiatum, T. reesei and T. virens contained only single copy of Cyp51 protein.
Characteristic motifs of the Trichoderma CYP families
Several signature motifs are conserved in fungal Cyp proteins as per pervious findings [26, 35, 37, 38]. In Trichoderma, we identified the characteristic signature motifs of CYP super family AGXDTT, EXXR, PERW and FXXGXRXCXG for each phylogenetic group (Fig. 5). These motifs are functionally essential for the Cyp proteins. Conserved motif FXXGXRXCXG (also known as CXG) is designated as a heme-binding domain [26, 29, 39] and includes a conserved cysteine residue that binds to the Fe of the heme. In Trichoderma, the cysteine residue of the P450 signature CXG motif is invariantly conserved in all P450s, whereas two glycine and one phenylalanine residues were also found to be conserved among majority of phylogenetic groups, which are in accordance with previous reports [37, 40]. In phylogenetic groups 13, 16 and 19, Cyp proteins contain glutamate/aspartate, tyrosine and glycine respectively instead of a phenylalanine residue. Another variant of FXXGXRXCXG motif was found in groups 1, 6 and 20 where first amino acid residue of the motif was either phenylalanine or tryptophan. Further, in groups 5, 8, 12 and 20, FXXGXRXCXG and FXXGXRXCXA variants were identified. Conserved motif EXXR is present in helix K, on the proximal side of heme and probably is involved in the stabilization of the core structure of Cyp proteins [26, 35, 39]. In Trichoderma motif EXXR, glutamic acid and arginine residues were found to be highly conserved, whereas two middle ‘XX’ residues were found to be highly variable. These results are in concurrence with previously reported literature for fungal cytochrome P450 proteins [35, 37, 40, 41]. Another conserved motif of cytochrome P450 protein family is PERW (known as PER) which forms E-R-R triad in Cyp proteins [26]. In Trichoderma, we found PERW as the predominant signature, in accordance with previous reports in fungi [27, 35]. Motif PERW was found to be relatively conserved in Trichoderma with few exceptions that mainly includes phylogenetic groups 19 and 20. Group 19 consists of Cyp proteins from clans CYP54, CYP550, CYP559, CYP642, CYP657 and CYP659, which have been predicted to play role in secondary metabolism. High diversity of PER motif of this group could be attributed to the evolving functions of Cyp P450 protein members. Phylogenetic group 20 consisting of clans CYP51, CYP61 and CYP55 includes Cyp proteins belonging to both class E (CYP51 and CYP61) and B (CYP55). In this group, variant of PERW motif was identified where clan CYP55 proteins (class B) contained amino acid residues K/E/Q between PER and W/Y. The absence of an amino acid residue between arginine and tryptophan residues in “PERW” motif in all class E Cyp proteins indicate the early functional divergence of PERW motif in class B and E cytochrome P450 proteins. These results provide an insight on the structure–function relationships in such a diverse and complex Cyp protein families. Further, we also identified conserved motif, AGXDTT in Trichoderma cytochrome P450 proteins. Motif AGXDTT contributes to oxygen binding and activation [35]. The oxygen-binding domain (AGXDTT) was found to be highly variable in Trichoderma cytochrome P450 proteins. The terminal threonine residue in AGXDTT motif involved in the formation of the enzyme’s critical oxygen-binding pocket was found to be replaced predominately by valine in phylogenetic group 16. Other amino acid residues that replaced terminal threonine in different groups included serine or methionine. For motifs AGXDTT and CXG, Cyp proteins in phyletic groups 13, 19 and 20 (Table 2) were relatively less conserved, suggesting divergence of these Cyp protein sequences and their functions in Trichoderma. We found that the conserved signature motifs and their variants identified in Trichoderma showed few exceptions to previous reports. These results suggest Cyp signature motifs have evolved in Trichoderma to accommodate enormously wide range of substrate specificities and their substrate-binding regions.
Fig. 5.

Conserved signature motifs of Trichoderma Cyp proteins for 20 phylogenetic groups
Cytochrome P450s associated with secondary metabolism related gene clusters
A survey of the genomes of seven Trichoderma spp. revealed that of the 477 cytochrome P450 genes present in the seven genomes, as many as 100 genes are associated with putative secondary metabolism related gene clusters namely NRPS, PKS, NRPS-PKS, NRPS-like, and terpene cyclase clusters (Additional file 1: Table S2).
Discussion
Trichoderma species are the champions of opportunistic success [1]. They can be found virtually in all ecological niches, both terrestrial and aquatic. These fungi are capable of parasitizing a wide range of fungal and oomycetes species. Many species are known to colonize the rhizosphere and roots, both externally and internally [3]. Some are reported to be endophytes [42] while a few are aggressive parasites on cultivated mushrooms [43]. A few species are known to be opportunistic human pathogens while some strains are nematode-parasite, demonstrating their ability to parasitize members of animal kingdom [1]. Several Trichoderma strains are plant growth enhancers and some can colonize composts [44]. A few strains are known to be xenobiotics degraders. Most species are prolific producers of a wide range of secondary metabolites, with a total of more than 1000 compounds chemically characterized [45]. Cytochrome P450s are important for cells to perform a wide variety functions like primary and secondary metabolism, xenobiotic degradation and cellular defence (e.g., in interaction with other fungi). Recently, a T. virens P450 (TvCyt2; Protein Id. 190045) has been shown to be involved in biocontrol and plant growth promotion [46]. Basidiomycetes are capable of metabolizing a wide range of endogenous and exogenous compounds by using cytochrome P450s [47]. Great deal of information is available on the role of P450s in degradation of lignins and polyaromatic hydrocarbons by white rot fungus Phanerochaete chrysosporium and brown-rot fungus Postia placenta, as well as medicinal mushrooms like Coriolus versicolor and Lentinula edodes [48–51]. Role of P450s in colonization of living wood by the plant pathogen Heterobasidion irregulare is also well established [52].
In the present study, Trichoderma CYPome from seven Trichoderma species viz. T. asperellum, T. atroviride, T. citrinoviride, T. harzianum, T. longibrachiatum, T. reesei and T. virens is annotated. Overall, our analysis identified a total of 477 CYPs in these genomes. To provide support for the annotation process, the identified CYPs were also examined for conserved CYP domain. Our analysis of the CYPome has identified 12 families unique to Trichoderma. All the Trichoderma species examined are a rich source of Cyp proteins (55 in T. longibrachiatum to 100 in T. harzianum).
In Trichoderma, clan CYP52 consisted of families Cyp52, Cyp538, Cyp539, Cyp584 and Cyp655. Cyp52 family is found only amongst Candida-related species of fungi and these proteins catalyze the conversion of fatty acids and alkanes to alpha, omega-dicarboxylic acids [53]. The number of Cyp61 proteins was conserved in all Trichoderma species and these proteins were also grouped together in Group 20. Cyp61 proteins are involved in primary metabolism. In Saccharomyces cerevisiae, CYP61 codes for sterol 22 desaturase [54], which is involved in later stages of the ergosterol pathway in metabolizing Ergosta-5,7,24(28)-trienol to Ergosta-5,7,22,24(28)-tetraenol by introducing a C-22(23) double bond in the sterol side chain. Since Cyp61 is involved in the later stages of ergosterol pathway, it is considered to have evolved as a result of duplication and diversification of the CYP51 gene. In ascomycetes and basidiomycetes, clan CYP51 is involved in sterol biosynthesis and is recognized as the housekeeping CYP, and has been a popular antifungal target for the control of fungal diseases in humans and crop plants [29–31, 55]. In comparison to some of the ascomycetous fungi, which carry multiple CYP51 genes, T. atroviride and T. harzianum contained two copies whereas T. asperellum, T. citrinoviride, T. longibrachiatum, T. reesei and T. virens contained only single copy of Cyp51 protein. In addition, all members of both clans CYP51 and CYP61 which are involved in primary metabolism (sterol biosynthesis) are grouped together in group 20, suggesting that both Cyp51 and Cyp61 proteins are diversified only to a lesser extent in Trichoderma.
Motif analysis led to the identification of four signature motifs in phylogenetic groups, which correspond to the conserved tertiary structure and enzyme functions in spite of the wide sequence diversity and functions of Cyp proteins. Modifications found in the heme-binding domain FXXGXRXCXG are more frequently found in CYPs with catalytic activity, often not requiring oxygen [29]. These results indicate Cyp members of groups 5, 8, 12, 13, 16, 19 and 20 may have novel catalytic activities in Trichoderma. Some P450s showed variations of the signature motifs mainly in AGXDTT, EXXR and FXXGXRXCXG motifs. These results are in accordance with previous reports [38, 41] where it was proposed that these P450s variations may be due to misaligned sequences or that the P450s are missing the invariant residues at these motifs. In our study, phylogenetic group 10 containing protein families of clan CYP65 showed highly conserved motifs, suggesting functional conservation of CYP65 clan in analysed Trichoderma species. All members of clan CYP65 are involved in secondary metabolism. CYP65 is reported to catalyze the epoxidation reaction during the synthesis of trichothecenes [56, 57] and radicicol [37]. Identification of conserved and variable CYP motif signatures among and within phylogenetic groups in the present study may provide us information on CYP evolution, structure, and function in Trichoderma and have application in classification of proteins in gene expression analysis [58].
Cyp56 clan, found to be unique to T. asperellum, T. harzianum and T. virens (mycoparasites) has been characterized earlier in yeast [59, 60]. Members of Cyp56 clan are involved in meiotic spore wall biogenesis, particularly in dityrosine biosynthesis [59–61]. Members of the clan CYP507, CYP530, CYP531, CYP532 and CYP548 are known to be involved in xenobiotics metabolism [36]. Abundance of these proteins in Trichoderma may be related to the ability of these fungi to metabolize a wide range of xenobiotics, including many fungicides. Similarly, ability of Trichoderma spp. to produce a plethora of secondary metabolites could be linked to the abundance of P450s belonging to the clan CYP574, CYP58 and CYP65 proteins that have been implicated in trichothecene biosynthesis [62]. In T. harzianum, three copies of Cyp504 protein are present as compared to single copy in other Trichoderma species. Expansion of Cyp504 proteins in T. harzianum suggest important role of Cyp504 protein in xenobiotic metabolism. Further, the family members of Cyp504 were also reported to be up-regulated during cuticle infection by insect pathogenic fungi Metarhizium anisopliae and M. acridum [63]. Cyp505 family was found to be expanded in T. asperellum, T. harzianum and T. virens where these species contained three Cyp505 proteins each. Cyp505 proteins are membrane-associated fatty acid hydroxylase [64]. Cyp528 family has only one protein entry in all Trichoderma species analysed except T. atroviride where family Cyp528 consisted of two proteins. Similarly, Cyp58 family has a single protein entry in all Trichoderma species analysed except in T. virens where family Cyp58 consisted of two proteins. Previous studies also showed expansion of clan Cyp58 proteins in fungi [36]. In Trichoderma, the increase in CYPome size of T. harzianum and T. virens may be due to the expansion of certain CYP gene families or the presence of novel genes that are essential for their lifestyle. Previous reports have associated expansions of the fungal CYP families with the evolution of various fungal traits including pathogenicity [65]. Our phylogenetic analysis showed uneven distribution of CYP group sizes in Trichoderma species, which are in concordance with extreme expansions and contractions of certain CYP families in the course of evolution. Expansion of CYP clans in different Trichoderma species could aid them in more competent survival in their respective habitats.
Trichoderma spp. are prolific producers of secondary metabolites, many with antimicrobial, anticancer and plant growth-promoting properties [45] Cyps are known to play central role in biosynthesis if many, if not most of the secondary metabolites of plant and microbial origin. Till date, however, only a handful of Trichoderma Cyps have been investigated for their role in biosynthesis of secondary metabolites [46, 66, 67]. Our present findings suggest that more than 20% of the catalogued Cyps from Trichoderma are part of putative secondary metabolism-related gene clusters. There is a need for systematic studies on the functions of these Cyps which would lead to the discovery of novel pathways, metabolites and intermediates with greater biotechnological significance.
Conclusion
Trichoderma CYPome described in our study is by combining information generated from existing databases, predicting conserved domains and identifying structural motifs in each hypothetical protein. By following internationally recognized nomenclature system, we have identified novel CYP clans and families unique to Trichoderma. Phylogenetic analysis elucidated distribution of Cyp families and clans in different evolutionary groups and their probable functions in metabolism or biosynthesis based on the comparisons with CYPomes of other organisms. The number of these proteins correlates with the genome size and many are species-specific. Unfortunately, the functions of none of these proteins are known. One reason being a lack of systematic studies and annotation of these proteins. Our CYPome annotation and evolutionary studies of seven Trichoderma species now provides opportunities for exploration of research-driven strategies to select Trichoderma species for various applications especially in relation to secondary metabolism and degradation of environmental pollutants. Several of these proteins could also have biotechnological applications like biotransformation and synthesis of pharmaceutically important drugs.
Methods
Sequence data
Sequences of Cytochrome P450s were retrieved from the Joint Genome Institute (JGI) fungal genome database MycoCosm (http://genome.jgi-psf.org/programs/fungi/index.jsf) for all the species of genus Trichoderma. The species included were T. asperellum (CBS 433.97) v1.0, T. atroviride (IMI 206040) v2.0, T. citrinoviride v4.0, T. harzianum (CBS 226.95) v1.0, T. longibrachiatum (ATCC 18648) v3.0, T. reesei (QM 6a) v2.0 and T. virens (Gv29-8) v2.0.
Annotation of CYPs
The annotation pipeline of the CYPome in the Trichoderma species was done in a two-step procedure of identification and annotation. The identification step of CYP family was performed by using Conserved Domain Database (CDD); the cut-off of positive hits was set at E-value of 10−2. Entries with incomplete sequences and domain were manually removed from the data. Cyp proteins with complete conserved cytochrome P450 domains were further subjected to the annotation procedure using the Nelson’s P450 database against all named fungal cytochrome P450s (http://blast.uthsc.edu) with the E-value of 10−4 [68]. For annotation, sequence similarity cut-off of 40% was used. For few entries, we have followed criteria of the phenomenon called family creep that allows sequences less than 40% to be included in a family. For such entries, we have used sequence similarity cut-off of 30% and above. These predicted CYPs were then assigned to the corresponding family and clan types based on their highest homology according to the International P450 Nomenclature Committee Databases used by Nelson (http://drnelson.uthsc.edu/CytochromeP450.html) [21] and the fungal cytochrome P450 database (http://p450.riceblast.snu.ac.kr) [27] respectively.
Structural feature analysis of CYP protein sequences
Presence of cytochrome P450 conserved domain was confirmed using conserved domain database [69]. To reveal phylogenetic group-specific conservation pattern of cytochrome P450 proteins, structural features were explored. To identify cyp conserved signature motifs, multiple protein sequence alignments for each phylogenetic group were built by MAFFT program [70] using E-INS-i iterative refinement method. Alignments were further refined and viewed using AliView [71]. Consensus logos of the alignments were automatically generated by WebLogo 3 program [72] and used for visualization of the conservation of signature motifs for each phylogenetic group. The generated logos were used for the analysis.
Phylogenetic reconstruction of CYPs
After removal of redundant and incomplete sequences, the protein sequences were aligned using MUSCLE [73]. The evolutionary history was inferred using the minimum evolution method [74]. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analysed [75]. The evolutionary distances were computed using the Poisson correction method [76] and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modelled with a gamma distribution (shape parameter = 1). The ME tree was searched using the close-neighbor-interchange (CNI) algorithm [77] at a search level of 1. The neighbor-joining algorithm [78] was used to generate the initial tree. Evolutionary analyses were conducted in MEGA5 [79]. Phylogenetic trees were visualized with FigTree v1.1.2 [80].
Identification of cytochrome P450s associated with secondary metabolism related gene clusters
A genome-wide survey was done to identify cytochrome P450s associated (presence in the vicinity) with secondary metabolism-related gene clusters, viz., NRPS, PKS, PKS/NRPS, NRPS-like and terpene cyclase clusters either manually (T. reesei, T. virens and T. atroviride [81, 82] or using automated pipeline on the respective genome pages (for T. citrinoviride, T. longibrachiatum, T. asperellum and T. harzianum).
Additional file
Additional file 1. Table S1: List of Cyp protein entries with incomplete cytochrome P450 domain. Table S2: Cytochrome P450s associated with predicted secondary metabolism-related gene clusters.
Authors’ contributions
PKM, SC and ID conceptualized and framed the work. SC designed the study and performed CYP manual annotation, phylogenetics, function and motif predictions, analysed data, prepared figures and supplementary data. STM performed data mining and contributed to CYP function prediction. RB identified the association of the genes with secondary metabolism-related gene clusters. AK, AA and IVG at JGI did the whole genome sequencing and automated annotation. SC, PKM and ID wrote the manuscript. PKM coordinated this study. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Head, Nuclear Agriculture and Biotechnology Division, Bhabha Atomic Research Centre, Mumbai for encouragement and support. The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, was supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40694-018-0056-3) contains supplementary material, which is available to authorized users.
References
- 1.Druzhinina I, Seidl-Seiboth V, Herrera-Estrella A, Horwitz BA, Kenerley CM, Monte E, Mukherjee PK, Zeilinger S, Grigoriev IV, Kubicek CP. Trichoderma: the genomics of opportunistic success. Nat Rev Microbiol. 2011;9:749–759. doi: 10.1038/nrmicro2637. [DOI] [PubMed] [Google Scholar]
- 2.Bissett J, Gams W, Jaklitsch W, Samuels GJ. Accepted Trichoderma names in the year 2015. IMA Fungus. 2015;6:263–295. doi: 10.5598/imafungus.2015.06.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee PK, Horwitz BA, Singh US, Mukherjee M, Schmoll M. Trichoderma in agriculture, industry and medicine: an overview. In: Mukherjee PK, Horwitz BA, Singh US, Mukherjee M, Schmoll M, editors. Trichoderma: biology and applications. Oxfordshire: CABI; 2013. pp. 152–172. [Google Scholar]
- 4.Mukherjee PK, Horwitz BA, Herrera-Estrella A, Schmoll M, Kenerley CM. Trichoderma research in the genome era. Annu Rev Phytopathol. 2013;51:105–129. doi: 10.1146/annurev-phyto-082712-102353. [DOI] [PubMed] [Google Scholar]
- 5.Hatvani L, Manczinger L, Vágvölgyi C, Kredics L. Trichoderma as a human pathogen. In: Mukherjee PK, Horwitz BA, Singh US, Mukherjee M, Schmoll M, editors. Trichoderma: Biology and Applications. Oxfordshire: CABI; 2013. pp. 152–172. [Google Scholar]
- 6.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, Danchin EG, Grigoriev IV, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 7.Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, Zeilinger S, Casas-Flores S, Horwitz BA, Mukherjee PK, et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011;12:R40. doi: 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmoll M, Dattenböck C, Carreras-Villaseñor N, Mendoza-Mendoza A, Tisch D, Alemán MI, Baker SE, Brown C, Cervantes-Badillo MG, Cetz-Chel J, Cristobal-Mondragon GR, et al. The genomes of three uneven siblings: footprints of the lifestyles of three Trichoderma species. Microbiol Mol Biol Rev. 2016;80:205–327. doi: 10.1128/MMBR.00040-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, Smirnova T. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42(D1):D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Degtyarenko KN. Structural domains of P450-containing monooxygenase systems. Protein Eng. 1995;8:737–747. doi: 10.1093/protein/8.8.737. [DOI] [PubMed] [Google Scholar]
- 11.Klingenberg M. Pigments of rat liver microsomes. Arch Biochem Biophys. 1958;75:376–386. doi: 10.1016/0003-9861(58)90436-3. [DOI] [PubMed] [Google Scholar]
- 12.Han Y, Liu X, Benny U, Kistler HC, VanEtten HD. Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen Nectria haematococca. Plant J. 2001;25:305–314. doi: 10.1046/j.1365-313x.2001.00969.x. [DOI] [PubMed] [Google Scholar]
- 13.Siewers V, Viaud M, Jimenez-Teja D, Collado IG, Gronover CS, Pradier JM, Tudzynsk B, Tudzynski P. Functional analysis of the cytochrome P450 monooxygenase gene bcbot1 of Botrytis cinerea indicates that botrydial is a strain-specific virulence factor. MPMI. 2005;18:602–612. doi: 10.1094/MPMI-18-0602. [DOI] [PubMed] [Google Scholar]
- 14.Fan J, Urban M, Parker JE, Brewer HC, Kelly SL, Hammond-Kosack KE, Fraaije BA, Liu X, Cools HJ. Characterization of the sterol 14α-demethylases of Fusarium graminearum identifies a novel genus-specific CYP51 function. New Phytol. 2013;198:821–835. doi: 10.1111/nph.12193. [DOI] [PubMed] [Google Scholar]
- 15.Takaoka S, Kurata M, Harimoto Y, Hatta R, Yamamoto M, Akimitsu K, Tsuge T. Complex regulation of secondary metabolism controlling pathogenicity in the phytopathogenic fungus Alternaria alternata. New Phytol. 2014;202:1297–1309. doi: 10.1111/nph.12754. [DOI] [PubMed] [Google Scholar]
- 16.Zhang DD, Wang XY, Chen JY, Kong ZQ, Gui YJ, Li NY, Bao YM, Dai XF. Identification and characterization of a pathogenicity-related gene VdCYP1 from Verticillium dahliae. Sci Rep. 2016;6:27979. doi: 10.1038/srep27979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernhardt R. Cytochromes P450 as versatile biocatalysts. J Biotechnol. 2006;124:128–145. doi: 10.1016/j.jbiotec.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 18.Urlacher VB, Eiben S. Cytochrome P450 monooxygenases: perspectives for synthetic application. Trends Biotechnol. 2006;24:324–330. doi: 10.1016/j.tibtech.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Kizawa H, Tomura D, Oda M, Fukamizu A, Hoshino T, Gotoh O, Yasui T, Shoun H. Nucleotide sequence of the unique nitrate/nitrite inducible cytochrome P450 cDNA from Fusarium oxysporum. J Biol Chem. 1991;266:10632–10637. [PubMed] [Google Scholar]
- 20.Lamb DC, Skaug T, Song HL, Jackson CJ, Podust LM, Waterman MR, Kell DB, Kelly DE, Kelly SL. The cytochrome P450 complement (CYPome) of Streptomyces coelicolor A3 (2) J Biol Chem. 2002;277:24000–24005. doi: 10.1074/jbc.M111109200. [DOI] [PubMed] [Google Scholar]
- 21.Nelson DR. Cytochrome P450 and the individuality of species. Arch Biochem Biophys. 1999;369:1–10. doi: 10.1006/abbi.1999.1352. [DOI] [PubMed] [Google Scholar]
- 22.Nebert D, Adesnik M, Coon MJ, Estabrook RW, Gonzalez FJ, Guengerich FP, Gunsalus IC, Johnson EF, Kemper B, Levin W, et al. The P450 gene superfamily: recommended nomenclature. DNA. 1987;6:1–11. doi: 10.1089/dna.1987.6.1. [DOI] [PubMed] [Google Scholar]
- 23.Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenet. 1996;6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Nelson D, Werck-Reichhart DA. P450-centric view of plant evolution. Plant J. 2011;66(1):194–211. doi: 10.1111/j.1365-313X.2011.04529.x. [DOI] [PubMed] [Google Scholar]
- 25.Nelson DR. Metazoan cytochrome P450 evolution. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;121(1–3):15–22. doi: 10.1016/S0742-8413(98)10027-0. [DOI] [PubMed] [Google Scholar]
- 26.Deng J, Carbone I, Dean RA. The evolutionary history of cytochrome P450 genes in four filamentous Ascomycetes. BMC Evol Biol. 2007;7(1):10–30. doi: 10.1186/1471-2148-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park J, Choi J, Ahn K, Park B, Park J, Kang S, Lee Y-H. Fungal cytochrome P450 database. BMC Bioinform. 2008;9:402. doi: 10.1186/1471-2105-9-S3-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmeister D, Keller NP. Natural products of filamentous fungi, enzymes, genes and their regulation. Nat Prod Rep. 2007;24:393–416. doi: 10.1039/B603084J. [DOI] [PubMed] [Google Scholar]
- 29.Kelly DE, Kraševec N, Mullins J, Nelson DR. The CYPome (cytochrome P450 complement) of Aspergillus nidulans. Fungal Genet Biol. 2009;46:S53–S61. doi: 10.1016/j.fgb.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Kelly SL, Kelly DE. Microbial cytochromes P450: biodiversity and biotechnology. Where do cytochrome P450 come from, what do they do and what can they do for us? Philos Trans R Soc Lond B Biol Sci. 2013;368(1612):20120476. doi: 10.1098/rstb.2012.0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ichinose H. Metabolic diversity and cytochromes P450 of fungi. In: Yamazaki H, editor. Fifty years of cytochrome P450 research. Tokyo: Springer; 2014. pp. 187–205. [Google Scholar]
- 32.Syed K, Yadav JS. P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaete chrysosporium. Crit Rev Microbiol. 2012;38(4):339–363. doi: 10.3109/1040841X.2012.682050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newsome AW, Nelson D, Corran A, Kelly SL, Kelly DE. The cytochrome P450 complement (CYPome) of Mycosphaerella graminicola. Biotechnol Appl Biochem. 2013;60:52–64. doi: 10.1002/bab.1062. [DOI] [PubMed] [Google Scholar]
- 34.Lah L, Haridas S, Bohlmann J, Breuil C. The cytochromes P450 of Grosmannia clavigera: genome organization, phylogeny, and expression in response to pine host chemicals. Fungal Genet Biol. 2013;50:72–81. doi: 10.1016/j.fgb.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Chen W, Lee MK, Jefcoate C, Kim SC, Chen F, Yu JH. Fungal cytochrome p450 monooxygenases: their distribution, structure, functions, family expansion, and evolutionary origin. Genome Biol Evol. 2014;6(7):1620–1634. doi: 10.1093/gbe/evu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moktali V, Park J, Fedorova-Abrams ND, Park B, Choi J, Lee YH, Kang S. Systematic and searchable classification of cytochrome P450 proteins encoded by fungal and oomycete genomes. BMC Genom. 2012;13(1):525. doi: 10.1186/1471-2164-13-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sirim D, Widmann M, Wagner F, Pleiss J. Prediction and analysis of the modular structure of cytochrome P450 monooxygenases. BMC Struct Biol. 2010;10:34. doi: 10.1186/1472-6807-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sezutsu H, Le Goff G, Feyereisen R. Origins of P450 diversity. Philos Trans R Soc B. 2013;368:20120428. doi: 10.1098/rstb.2012.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werck-Reichhart D, Feyereisen R. Cytochromes P450: a success story. Genome Biol. 2000;1:6. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 41.Syed K, Mashele SS. Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: identification of evolutionarily conserved amino acid patterns characteristic of P450 Family. PLoS ONE. 2014;9(4):e95616. doi: 10.1371/journal.pone.0095616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey BA, Melnick RL. The endophytic Trichoderma. In: Mukherjee PK, Horwitz BA, Singh US, Mukherjee M, Schmoll M, editors. Trichoderma: biology and applications. Oxfordshire: CABI; 2013. pp. 152–172. [Google Scholar]
- 43.Komon-Zelazowska M, Bissett J, Zafari D, Hatvani L, Manczinger L, Woo S, Lorito M, Kredics L, Kubicek CP, Druzhinina IS. Genetically closely related but phenotypically divergent Trichoderma species cause green mold disease in oyster mushroom farms worldwide. Appl Environ Microbiol. 2007;73(22):7415–7426. doi: 10.1128/AEM.01059-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaidi NW, Singh US. Trichoderma in plant health management. In: Mukherjee PK, Horwitz BA, Singh US, Mukherjee M, Schmoll M, editors. Trichoderma: biology and applications. Oxfordshire: CABI; 2013. pp. 152–172. [Google Scholar]
- 45.Zeilinger S, Gruber S, Bansal R, Mukherjee PK. Secondary metabolism in Trichoderma—chemistry meets genomics. Fungal Biol Rev. 2016;30(2):74–90. doi: 10.1016/j.fbr.2016.05.001. [DOI] [Google Scholar]
- 46.Ramírez-Valdespino CA, Porras-Troncoso MD, Corrales-Escobosa AR, Wrobel K, Martínez-Hernández P, Olmedo-Monfil V. Functional characterization of TvCyt2, a member of the p450 monooxygenases from Trichoderma virens relevant during the association with plants and mycoparasitism. Mol Plant Microbe Interact. 2018;31:289–298. doi: 10.1094/MPMI-01-17-0015-R. [DOI] [PubMed] [Google Scholar]
- 47.Ichinose H. Cytochrome P450 of wood-rotting basidiomycetes and biotechnological applications. Biotechnol Appl Biochem. 2013;60:71–81. doi: 10.1002/bab.1061. [DOI] [PubMed] [Google Scholar]
- 48.Bhattacharya SS, Syed K, Shann J, Yadav JS. A novel P450-initiated biphasic process for sustainable biodegradation of benzo[a]pyrene in soil under nutrient-sufficient conditions by the white rot fungus Phanerochaete chrysosporium. J Hazard Mater. 2013;261:675–683. doi: 10.1016/j.jhazmat.2013.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, Yamamoto R, Yamamoto Y, Tokumoto T, Dong J, Thomas P, Hirai H, Kawagishi H. Hydroxylation of bisphenol A by hyper lignin-degrading fungus Phanerochaete sordida YK-624 under non-ligninolytic condition. Chemosphere. 2013;93:1419–1423. doi: 10.1016/j.chemosphere.2013.07.026. [DOI] [PubMed] [Google Scholar]
- 50.Akiyama R, Kajiwara S, Shishido K. Catalytic reaction of basidiomycete Lentinula edodes cytochrome P450, Le. CYP1 enzyme produced in yeast. Biosci Biotechnol Biochem. 2004;68:79–84. doi: 10.1271/bbb.68.79. [DOI] [PubMed] [Google Scholar]
- 51.Ichinose H, Wariishi H, Tanaka H. Identification and heterologous expression of the cytochrome P450 oxidoreductase from the white-rot basidiomycete Coriolus versicolor. Appl Microbiol Biotechnol. 2002;59:658–664. doi: 10.1007/s00253-002-1083-8. [DOI] [PubMed] [Google Scholar]
- 52.Mgbeahuruike AC, Kovalchuk A, Ubhayasekera W, Nelson DR, Yadav JS. CYPome of the conifer pathogen Heterobasidion irregulare: inventory, phylogeny, and transcriptional analysis of the response to biocontrol. Fungal Biol. 2017;121:158–171. doi: 10.1016/j.funbio.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 53.Eschenfeldt WH, Zhang Y. H, Samaha H, Stols L, Eirich LD, Wilson CR. Donnelly MI. Transformation of fatty acids catalyzed by cytochrome P450 monooxygenase enzymes of Candida tropicalis. Appl Environ Microbiol. 2003;69:5992–5999. doi: 10.1128/AEM.69.10.5992-5999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skaggs BA, Alexander JF, Pierson CA, Schweitzer KS, Chun KT, Koegel C, Barbuch R, Bard M. Cloning and characterization of the Saccharomyces cerevisiae C-22 sterol desaturase gene, encoding a second cytochrome P-450 involved in ergosterol biosynthesis. Gene. 1996;169:105–109. doi: 10.1016/0378-1119(95)00770-9. [DOI] [PubMed] [Google Scholar]
- 55.Becher R, Wirsel SG. Fungal cytochrome P450 sterol 14 alpha-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl Microbiol Biotechnol. 2012;95:825–840. doi: 10.1007/s00253-012-4195-9. [DOI] [PubMed] [Google Scholar]
- 56.Keller G, Turner NP, Bennett JW. Fungal secondary metabolism-from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 57.Ward TJ, Bielawski JP, Kistler HC, Sullivan E, O’Donnell K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc Natl Acad Sci USA. 2002;99:9278–9283. doi: 10.1073/pnas.142307199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jensen S, Shen L, Liu J. Combining phylogenetic motif discovery and motif clustering to predict co-regulated genes. Bioinformatics. 2005;21:3832–3839. doi: 10.1093/bioinformatics/bti628. [DOI] [PubMed] [Google Scholar]
- 59.Melo NR, Moran GP, Warrilow AGS, Dudley E, Smith SN, Sullivan DJ, Lamb DC, Kelly DE, Coleman DC, Kelly SL. CYP56 (Dit2p) in Candida albicans: characterization and investigation of its role in growth and antifungal drug susceptibility. Antimicrob Agents Chemother. 2008;52:3718–3724. doi: 10.1128/AAC.00446-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crešnar B, Petrič S. Cytochrome P450 enzymes in the fungal kingdom. Biochim Biophys Acta. 2011;1814:29–35. doi: 10.1016/j.bbapap.2010.06.020. [DOI] [PubMed] [Google Scholar]
- 61.Briza P, Eckerstorfer M, Breitenbach M. The sporulation-specific enzymes encoded by the DIT1 and DIT2 genes catalyze a two-step reaction leading to a soluble LL-dityrosine-containing precursor of the yeast spore wall. Proc Natl Acad Sci USA. 1994;91(10):4524–4528. doi: 10.1073/pnas.91.10.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown DW, McCormick SP, Alexander NJ, Proctor RH, Desjardins AE. Inactivation of a cytochrome PJ450 is a determinant of trichothecene diversity in Fusarium species. Fungal Genet Biol. 2002;36:224–233. doi: 10.1016/S1087-1845(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 63.Gao Q, Jin K, Ying SH, Zhang Y, Xiao G, Shang Y, Duan Z, Hu X, Xie XQ, Zhou G, Peng G, Luo Z, Huang W, Wang B, Fang W, Wang S, Zhong Y, Ma LJ, St Leger RJ, Zhao GP, Pei Y, Feng MG, Xia Y, Wang C. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011;7(1):e1001264. doi: 10.1371/journal.pgen.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kitazume T, Takaya N, Nakayama N, Shoun H. Fusarium oxysporum fatty-acid subterminal hydroxylase (CYP505) is a membrane-bound eukaryotic counterpart of Bacillus megaterium cytochrome P450BM3. J Biol Chem. 2000;275:39734–39740. doi: 10.1074/jbc.M005617200. [DOI] [PubMed] [Google Scholar]
- 65.Soanes DM, Alam I, Cornell M, Wong HM, Hedeler C, Paton NW, Rattray M, Hubbard SJ, Oliver SG, Talbot NJ. Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PLoS ONE. 2008;3(6):e2300. doi: 10.1371/journal.pone.0002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malmierca MG, Cardoza RE, Alexander NJ, McCormick SP, Hermosa R, Monte E, Gutiérrez S. Involvement of Trichoderma trichothecenes in the biocontrol activity and induction of plant defense-related genes. Appl Environ Microbiol. 2012;78:4856–4868. doi: 10.1128/AEM.00385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bansal R, Sherkhane PD, Oulkar D, Khan Z, Banerjee K, Mukherjee PK. The viridin biosynthesis gene cluster of Trichoderma virens and its conservancy in the bat white-nose fungus Pseudogymnoascus destructans. Chem Select. 2018;3:1289–1293. [Google Scholar]
- 68.Nelson DR. The cytochrome P450 homepage. Hum Genom. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2017 doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Larsson A. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics. 2014;30:3276–3278. doi: 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crooks GE, Hon G, Chandoniam JM, Brennerm SE. WebLogo: a sequence logo generator. Genome Res. 2004;2004(14):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rzhetsky A, Nei MA. Simple method for estimating and testing minimum evolution trees. Mol Biol Evol. 1992;9:945–967. [Google Scholar]
- 75.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 76.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press; 1965. pp. 97–166. [Google Scholar]
- 77.Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- 78.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 79.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rambaut A. FigTree v1.4.3: Tree figure drawing tool. 2016. http://tree.bio.ed.ac.uk/software/figtree/. Accessed 5 Feb 2018.
- 81.Bansal R, Mukherjee PK. Identification of novel gene clusters for secondary metabolism in Trichoderma genomes. Microbiology. 2016;85:185–190. doi: 10.1134/S002626171602003X. [DOI] [Google Scholar]
- 82.Bansal R, Mukherjee PK. The terpenoid biosynthesis toolkit of Trichoderma. Nat Prod Commun. 2016;11:431–434. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Table S1: List of Cyp protein entries with incomplete cytochrome P450 domain. Table S2: Cytochrome P450s associated with predicted secondary metabolism-related gene clusters.


