Abstract
Malignant glioma is usually accompanied by vigorous angiogenesis to provide essential nutrients. An effective glioma targeting moiety should include excellent tumor-cell homing ability as well as good neovasculature-targeting efficiency, and should be highly resistant to enzyme degradation in the bloodstream. The phage display-selected heptapeptide, the glioma-initiating cell peptide (GICP), was previously reported as a ligand for the VAV3 protein (a Rho-GTPase guanine nucleotide exchange factor), which is mainly expressed on glioma cells; the stabilized heptapeptide DA7R has been shown to be the ligand of both vascular endothelial growth factor receptor 2 (VEGFR2) and neuropilin-1 (NRP-1), and has demonstrated good neovasculature-targeting ability. By linking DA7R and GICP, a multi-receptor targeting molecule was obtained. The stability of these three peptides was evaluated and their targeting efficiency on tumor-related cells and models was compared. The ability of these peptides to cross the blood--tumor barrier (BTB) was also determined. The results indicate that the coupled Y-shaped peptide DA7R–GICP exhibited improved tumor and neovasculature targeting ability and had higher efficiency in crossing the BTB than either individual peptide.
KEY WORDS: Glioma, Neovasculature, Target, Peptide, Stabilization, Blood–tumor barrier
Graphical abstract
This study demonstrates that a coupled Y-shape peptide DA7R–GICP exhibited enhanced tumor and neovasculature targeting ability with high efficiency of crossing blood-tumor barrier.
1. Introduction
Glioblastoma, the most common and aggressive type of brain tumor, accounts for 29% of all primary brain and central nervous system tumors and 80% of malignant tumors1, 2. It is highly fatal with a median survival time of 15 months, with only 10% of patients having a 5-year overall survival3, 4, 5. Conventional chemotherapy lacks selectivity after systemic administration, causing severe adverse effects6, 7. The current trend to develop glioma-targeting therapeutic strategies can minimize adverse effects, but most approaches focused only on killing tumor cells and decreasing systematic toxicity, ignoring the importance of inhibiting tumor angiogenesis, an essential component of tumor proliferation, evasion, and metastasis8, 9, 10. Moreover, the abundant proteases in blood are the first barriers for drug delivery, with the blood–tumor barrier (BTB) the second physiological and enzymatic barrier. The most severe disadvantage of a peptide-based targeting moiety is that it can be degraded by peptidases and proteases in blood, but it has been demonstrated that stabilized peptides can accumulate in tumor tissue to a greater extent than un-stabilized peptides11. In addition, because of BTB, circulating targeting molecules need to penetrate the neovasculature made up of vascular endothelia cells to reach the tumor. Therefore, targeting moieties must demonstrate efficient tumor-related cell and blood vessel uptake, as well as protease resistance12.
In a previous report, VAV3 (a Rho-GTPase guanine nucleotide exchange factor) was shown to be upregulated in glioma cells, and especially glioma stem cells13 and its expression in U87MG cells was greater than in HUVECs (regarded as tumor related vascular endothelial cells), suggesting that VAV3 could be a potential target for glioma-cell targeting14. The heptapeptide GICP (glioma-initiating cell peptide), consisting of all natural l-amino acids, was identified by phage display technique and shown to have a high affinity for the VAV3 protein. Due to its unique amino combination, GICP peptide cannot be optimized by a retro-inverse strategy that utilizes all d-amino acids in reverse sequence (unpublished results) but conjugation with a d-peptide could improve its enzymatic stability15. DA7R is a ligand of the vascular endothelial growth factor receptor 2 (VEGFR2) and neuropilin-1 (NRP-1), which is mainly and highly expressed by tumor neovasculature16, 17, 18, 19. Since A7R is a C-terminal peptide and the sequence of Pro-Arg plays a vital role in receptor recognition20, 21, 22, retro-inversed DA7R peptide was arranged such that the critical amino acids were exposed at the N-terminal when coupled to GICP to yield peptide DA7R–GICP. The new targeting peptide was achieved by linking GICP and DA7R by several glycines to form a Y-shaped multifunctional targeting moiety without affecting their individual affinity for their respective receptors19, 23
In the present work, the stability of the three peptides to enzyme degradation was investigated and their targeting efficiencies were compared in U87MG cells, HUVECs and an ex vivo neovasculature model formed by HUVECs. A lysosome colocalization assay and quantitative measurement of the amount of peptide able to cross the BTB model were also conducted. The results indicated that the new Y-shaped peptide DA7R--GICP exhibited better tumor and neovasculature targeting ability and had higher efficiency for crossing the BTB than GICP or DA7R individually. The DA7R–GICP may be a promising multi-receptor recognition peptide for glioma targeting.
2. Materials and methods
2.1. Materials
Boc-protected α-amino acids were purchased from GL Biochem Ltd. (Shanghai, China). 4-Methylbenzhydrylamine resin HCl salt (rink amide MBHA resin) was from Xi'an Innovision Bioscience Co., Ltd. O-Benzotriazole-N,N,N′,N′-tetramethyl-uronium hexafluorophosphate (HBTU) was purchased from American Bioanalytical Co., Ltd. (Natick, MA, USA). Diisopropylethylamine (DIEA) was supplied by Sigma–Aldrich. 4ʹ,6-Diamidino-2-phenylindole (DAPI) was supplied by Roche (Basel, Switzerland). 5-Carboxyfluorescein (FAM) was purchased from Sigma (St. Louis, MO, USA). Fluorescein-5-maleimide was purchased from FanboBiochemicals (Beijing, China). LysoTracker®Red DND-99 was from Invitrogen (Grand Island, NY, USA). Matrigel was obtained from BD Biosciences (San Diego, CA, USA). All chemicals were analytic reagent grade.
U87MG glioblastoma cells and HUVECs were obtained from ATCC. All cells were cultured in special Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (FBS, GE Healthcare's HyClone, USA) at 37 °C in a 5% CO2-humidified atmosphere.
2.2. Synthesis of peptides
GICP (SSQPFWS), DA7R (DRDPDPDLDWDTDA) and DA7R–GICP (DRDPDPDLDWDTDA–GGG–C–GGG–SSQPFWS) were synthesized via active ester chemistry to couple Boc-protected amino acid to the de-protected resin. A cysteine was added to the C-terminal of GICP and DA7R to provide a thiol group in order to couple with imaging molecules. Crude products were precipitated with cold ether and purified by preparative C18 reversed-phase HPLC (RP-HPLC). After purification, GICP, DA7R and DA7R--GICP were labeled with fluorescein-5-maleimide via covalently conjugation. The products were purified by C18 RP-HPLC and molecular masses were ascertained by electrospray ionization mass spectrometry (ESI-MS).
2.3. Peptide stability assay
GICP, DA7R and DA7R–GICP were dissolved in distilled water to the concentration of 1 mg/mL and 150 μL of each solution was incubated with 1350 μL 50% rat serum. Incubation was at 37 °C for 0.25, 0.5, 1, 2, 4, 6, 8, 12 h, after which 100 μL of reaction solution was taken out to mix with 20 μL 15% (w/v) trichloroacetic acid aqueous solution to precipitate serum protein. The mixture was stored at 4 °C for 20 min and then centrifuged at 12,000 rpm (H1650-W, Xiangyi, Changsha, China) for 10 min. 20 μL of supernatant was analyzed via HPLC to quantify peptide hydrolysis.
2.4. Competitive inhibition assay
To determine if the addition of free DA7R and GICP would influence the targeting efficiency of DA7R–GICP peptides, a competitive inhibition assay was conducted on U87MG cells. Each peptide was tested for its ability to inhibit the cellular uptake of other fluorescein labeled peptides. U87MG cells were suspended in phosphate-buffered saline (PBS) and pre-incubated with peptide at 4 °C for 2 h, rinsed with PBS and further incubated with fluorescein-labeled peptide for 12 h, and then measured by flow cytometer.
2.5. Cellular uptake assay
To qualitatively assess the intracellular uptake of GICP, DA7R and DA7R–GICP peptides, U87MG cells and HUVECs were seeded in confocal dishes at a density of 5000 cells per well. Twenty-four hours after seeding, cells were incubated with FAM and fluorescein-labeled GICP, DA7R, DA7R–GICP for 2 h at the concentration of 5 μmol/L at 37 °C. The cells were rinsed with PBS and immobilized with 4% paraformaldehyde for 15 min. Intracellular fluorescence was visualized by a laser scanning confocal microscope.
For quantitative measurement, U87MG cells and HUVECs were seeded in 12-well plates at a density of 105 cells per well. Twenty-four hours after seeding, the cells were incubated with FAM and fluorescein-labeled GICP, DA7R and DA7R–GICP for 2 h at a concentration of 5 μmol/L at 37 °C. Fluorescence-positive cells were counted by flow cytometry.
2.6. Cellular uptake assay with serum pre-incubation
GICP, DA7R and DA7R–GICP peptides were pre-incubated with 50% rat serum for 1 h, and then incubated with U87 MG cells and HUVECs for 4 h. The other steps were as described in Section 2.4.
2.7. Lysosome colocalization assay
U87MG cells and HUVECs were seeded in confocal dishes at a density of 5000 cells per well. Twenty-four hours after seeding, cells were incubated with FAM and fluorescein-labeled GICP, DA7R and DA7R–GICP for 2 h at a concentration of 5 μmol/L at 37 °C. The solution was discarded, LysoTracker®Red DND-99 diluent was added to the dishes to incubate for another half-hour. The cells were rinsed with PBS and immobilized with 4% paraformaldehyde for 15 min. Lysosome colocalization was visualized by a laser scanning confocal microscope.
2.8. Neovasculature model uptake assay24
A HUVEC cell suspension was plated on 24-well plates precoated by Matrigel for 12 h. After tube formation, 10 μmol/L FAM, fluorescein-labeled GICP, fluorescein-labeled DA7R and fluorescein-labeled DA7R–GICP solution were given to the neovascular model for 1 h, then observed with a fluorescence microscope (DMI4000D, Leica, Germany). Semi-quantification was conducted with Image J software (National Institutes of Health, USA).
2.9. Tumor spheroid model penetration assay
U87MG cells were seeded in 48-well plates at the density of 4000 cells per well with the plates precoated with 150 μL of 2% (w/v) agarose to prepare the three-dimensional tumor spheroid model. The plates were gently shaken clockwise to make the cells clump and were incubated in a cell incubator for 7 days with 5% CO2 at 37 °C. The tumor spheroids were incubated with 20 μmol/L FAM and fluorescein-labeled GICP, DA7R and DA7R–GICP solution for 4 h. The tumor spheroids were then rinsed with PBS (pH 7.4), fixed with 4% paraformaldehyde for 15 min and observed under a laser scanning confocal microscope. Semi-quantification was conducted with Image J software.
2.10. Peptide crossing BTB assay
In order to evaluate the BTB transferability, a HUVECs/U87 co-culture model was established as described previously25. HUVECs were seeded in the apical chamber of a transwell and U87 cells were seeded into the basolateral chamber at a 1:5 HUVECs/U87 ratio. For quantitative measurement, three days after seeding, 30 μmol/L FAM and fluorescein-labeled GICP, DA7R and DA7R–GICP solution were added in the apical compartment and after 0.5, 1, 1.5, 2, 3 and 4 h of incubation at 37 °C, the transport ratio (%) was measured by microplate reader (Power Wave XS, Bio-TEK, USA). In order to better simulate the tumor environment, tumor spheroids were added into the basolateral chamber, 100 μL FAM and fluorescein-labeled GICP, DA7R and DA7R–GICP solution were added into the apical compartment at a concentration of 30 μmol/L in DMEM with 10% FBS. After a 4 h incubation, tumor spheroids were observed via laser scanning confocal microscope and semi-quantification was conducted by Image J software (NIH, MD, USA).
2.11. In vivo peptide distribution assay
GICP, DA7R and DA7R–GICP peptides were labeled with near-infrared dye Cy7. A subcutaneous tumor model was constructed by inoculating 5×106 U87MG cells (suspended in 100 μL of PBS) into the axilla of each mouse's right anterior limb. When the tumor size reached 0.8–1.0 cm3, 12 nude mice were randomly assigned into four groups. Free Cy7, GICP–Cy7, DA7R–Cy7 and DA7R–GICP–Cy7 at 0.01 μmol/L was injected through the tail vain. The fluorescent images were captured by an in vivo image system (VISUQE Invivo Elite, Gyeonggi Do Anyang, Korea) at 30, 60, 90, and 120 min after administration.
2.12. Statistical analysis
All data are presented as mean ± SD unless otherwise indicated. Statistically significant differences between the treatment groups was conducted by one-way ANOVA followed by Bonferroni test; a P value <0.05 was considered as significant.
3. Results and discussion
3.1. Characterization and serum stability of peptides
GICP, DA7R and DA7R–GICP were synthesized via a standard Boc-protected solid phase peptide synthesis strategy. After purification by preparative C18 RP-HPLC, analytical HPLC was used to confirm that purity was greater than 95% for each peptide. The results of electrospray ionization mass spectrometry (ESI-MS) indicated that the molecular weight of synthesized GICP, DA7R, DA7R–GICP peptide and fluorescein labeled GICP, DA7R, DA7R–GICP were 837.89 Da, 839.99 Da, 2105.31 Da, 1368.36 Da, 1370.26 Da and 2532.67 Da, respectively, which was consistent with the theoretical values, indicating that the peptides were correctly synthesized.
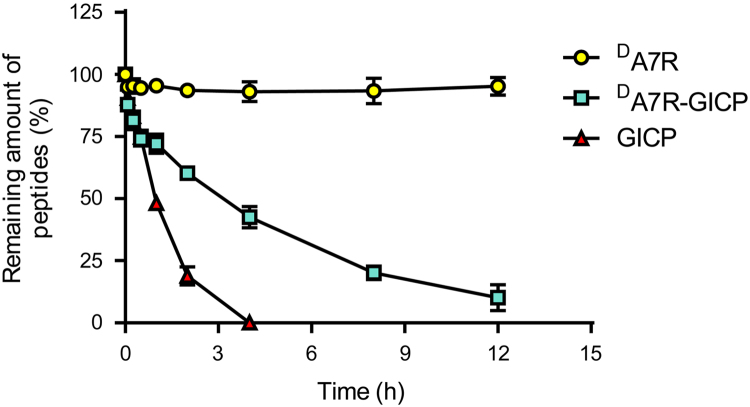
The serum stability assay was conducted in 50% rat serum at 37 °C to mimic the in vivo environment. As shown in Fig. 1, heptapeptide GICP containing natural l-amino acids was degraded in less than 4 h; stabilized peptide DA7R showed a great resistant to enzymatic degradation, and, surprisingly, the coupled peptide DA7R–GICP displayed a significant improvement of stability in serum compared with GICP, increasing its stability to greater than 12 h. The data implied that DA7R–GICP may have better tumor-targeting ability than GICP as a result of its longer circulating time in blood and the stability may be due to increased steric hindrance.
Figure 1.
Peptide stability in 50% sterile rat serum at 37 °C. Data are presented as Mean ± SD, n = 3.
3.2. Competitive inhibition of peptides
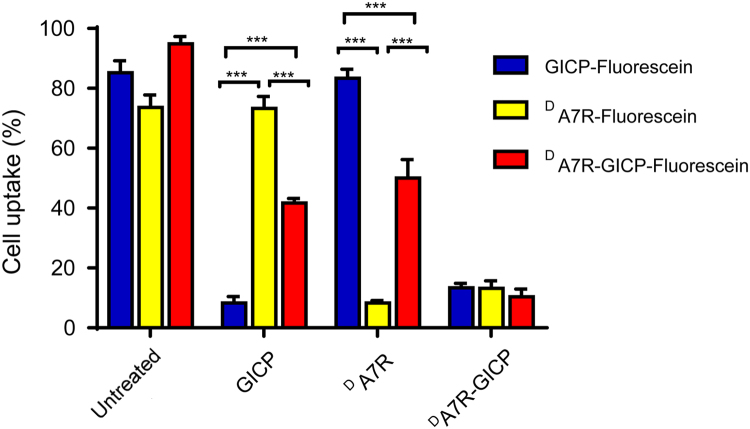
In order to determine whether GICP and DA7R each contribute to the process of DA7R–GICP internalization, a competitive inhibition experiment was carried out with U87MG cells. The fluorescein-labeled peptides could be efficiently internalized into U87MG cells. In contrast, after pre-incubation with GICP or DA7R peptide, the internalization of DA7R–GICP peptide was inhibited by up to 50%. Only pre-incubation with the coupled DA7R–GICP peptide itself was able to fully prevent internalization. The results shown in Fig. 2 indicate that GICP and DA7R in the DA7R–GICP peptide work individually.
Figure 2.
Cellular uptake of fluorescein-labeled peptides with or without treatment with GICP and DA7R in DA7R–GICP peptides in U87MG cells.
3.3. Cellular uptake of peptides
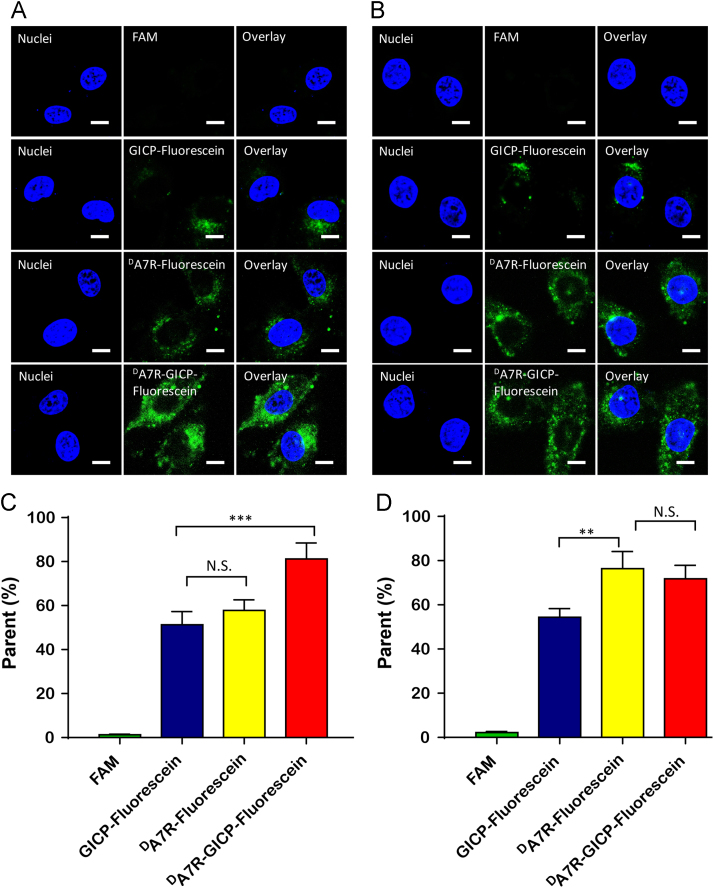
The cellular uptake efficiency of GICP, DA7R, DA7R–GICP molecules was evaluated with U87MG cells and HUVECs. The qualitative and the quantitative measurement of peptide uptake was conducted via flow cytometry and laser scanning confocal microscopy. The results are shown in Fig. 3; all three peptides can be taken up by U87MG cells and HUVECs, while FAM was barely absorbed. Because VAV3 is relatively overexpressed in tumor cells12, GICP peptide uptake was higher in U87MG cells than in DA7R (Fig. 3A and C), whereas DA7R was taken up more by HUVECs owing to higher expression of its receptor VEGFR and NRP-1 (Fig. 3B and D). However, the coupled GICP and DA7R peptide showed better tumor and neovasculature targeting ability, which may result from more receptors for DA7R–GICP peptide and its enhanced hydrophobicity caused by the seven amino-acids linker between DA7R and GICP.
Figure 3.
Cellular uptake of 5 μmol/L FAM, fluorescein-labeled GICP, DA7R and DA7R–GICP for 2 h. (A) Qualitative images of U87MG cells. (B) Qualitative images of HUVECs. (C) Quantitative data from U87MG cells. (D) Quantitative data for HUVECs. The scale bar indicates 10 μm. Data are presented as mean ± SD, n = 3; *P<0.05; **P<0.01.
3.4. Cellular uptake assay with serum pre-incubation
In order to further validate the influence of serum stability on cellular uptake, the three peptides were pre-incubated with 50% rat serum for 1 h and then incubated with U87MG cells to evaluate uptake efficiency. The results shown in Fig. 4 indicate that enhanced enzymatic stability of DA7R–GICP could maintain its uptake efficiency in both U87MG cells and HUVECs. In contrast, uptake of the non-stabilized peptide GICP was severely decreased by enzyme incubation.
Figure 4.
Cellular uptake of 5 μmol/L FAM, fluorescein-labeled GICP, DA7R and DA7R–GICP pre-incubated with 50% rat serum for 1 h and then incubated with cells for 4 h. (A) Qualitative images of U87MG cells. (B) Qualitative images of HUVECs. (C) Quantitative data from U87MG cells. (D) Quantitative data from HUVECs. The scale bar indicates 10 μm. Data are presented as mean ± SD, n = 3; *P<0.05; **P<0.01.
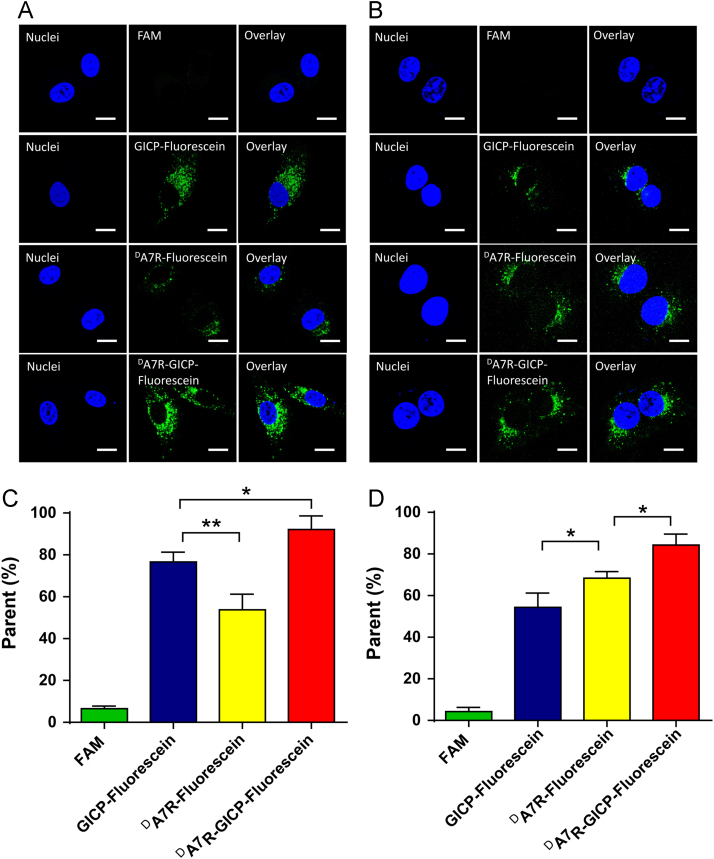
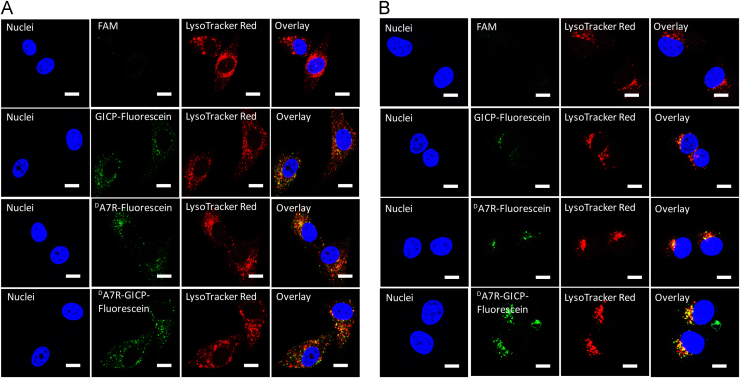
3.5. Peptide colocalization with lysosomes
FAM and fluorescein-labeled GICP, DA7R and DA7R–GICP were incubated with U87MG cells and HUVECs for 2 h at a concentration of 5 μmol/L at 37 °C. Lysosomes were marked with LysoTracker. The images of Fig. 5 demonstrate that three peptides can colocalize with lysosomes in both U87MG cells and HUVECs. The intracellular distribution of the endocytosed GICP, DA7R and DA7R–GICP demonstrate that they are in the same pathway for endocytosis. Since HUVECs were adopted as the neovasculature model cell line, colocalization of peptides and lysosomes in HUVEC indicates that enzymatic stability in trans-BTB process may play a key role in peptide distribution in vivo. As a consequence, DA7R–GICP, due to its multi-receptor and enhanced enzymatic stability, may lead to greater accumulation in tumors.
Figure 5.
Colocalization of endocytosed peptides and lysosomes. (A) Colocalization with U87MG cells. (B) Colocalization with HUVECs. The scale bar indicates 10 μm.
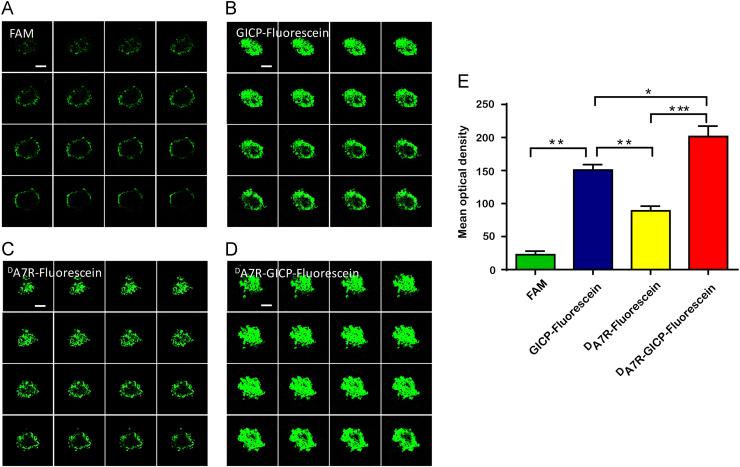
3.6. Tumor spheroid penetration
FAM and fluorescein-labeled GICP, DA7R and DA7R–GICP solution were added to the tumor spheroid incubations for 4 h. The images were captured under the same conditions by Z-stack mode of laser scanning confocal microscopy with an interval of 5 μm. As shown in Fig. 6, FAM could barely penetrate into the compact tumor cluster, only forming a slight fluorescent circle around tumor spheroid. GICP–fluorescein demonstrated slightly greater uptake than DA7R–fluorescein, which was consistent with the U87 cellular uptake results. The images (Fig. 6A–D) were analyzed by Image J software and the semi-quantitative results of mean optical density are shown in Fig. 6E. Fluorescein labeled DA7R–GICP exhibited better uptake and penetrating ability than the individual DA7R and GICP peptides, which might due to its dual-receptor targeting. The data indicate that DA7R–GICP could significantly enhance the penetration of single DA7R or GICP peptide into tumor spheroids.
Figure 6.
Penetration of (A) FAM, (B) GICP−fluorescein, (C) DA7R−fluorescein and (D) DA7R−GICP-fluorescein into U87MG tumor spheroids as observed by Z-stack mode laser scanning confocal microscopy with an interval of 5 μm. The scale bar indicates 100 μm. (E) Semiquantification of mean optical density (IOD/Area) in the tumor spheroid model. Data are presented as mean ± SD, n = 3; *P<0.05; **P<0.01, ***P<0.001.
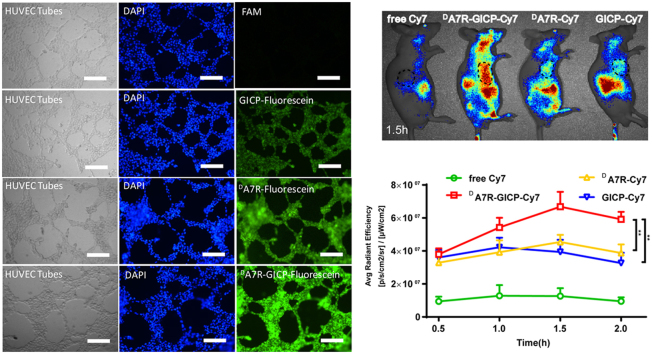
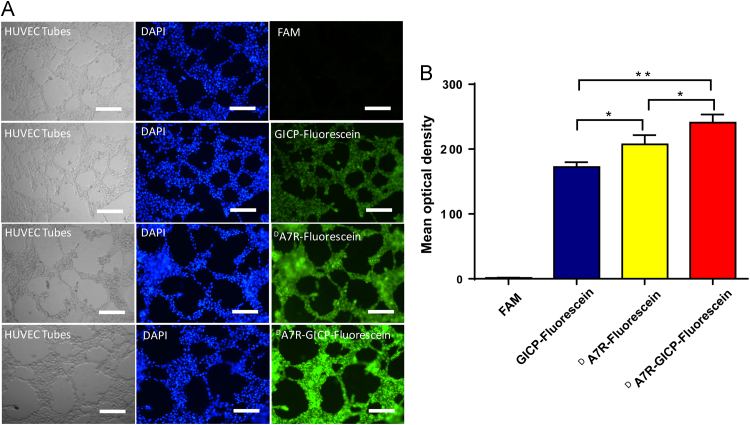
3.7. Neovasculature model uptake
The tubes formed by HUVECs on Matrigel were regarded as a neovasculature model, and 10 μmol/L FAM and fluorescein-labeled GICP, DA7R and DA7R–GICP solution were added for 1 h. The images of Fig. 7 demonstrate that the uptake of GICP, DA7R and DA7R–GICP were increased successively, which was consistent with HUVEC cell uptake. Fig. 7B shows the semi-quantification result of fluorescent pictures in Fig. 7A, analyzed by Image J software: the mean optical density of FAM, fluorescein-labeled GICP, DA7R and DA7R–GICP are 1.30 ± 0.64, 172.47 ± 7.29, 207.34 ± 14.30 and 241.0 ± 12.17, respectively. The results demonstrate that the coupled peptide may have increased neovasculature targeting ability and may serve as a potential tumor and tumor-related blood vessel co-targeted moiety.
Figure 7.
(A) Uptake ability of GICP, DA7R and DA7R−GICP peptides in a HUVEC-formed neovasculature model. The scale bar indicates 500 μm. (B) Semiquantification of mean optical density (IOD/Area) in neovasculature model. (Three random fields were observed and analyzed; *P<0.05; **P<0.01, ***P<0.001).
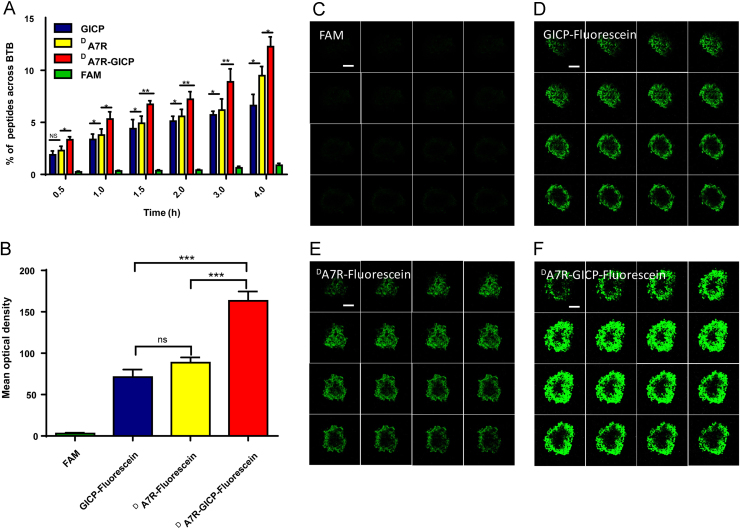
3.8. BTB crossing ability in a HUVEC/U87MG cell co-culture model
To quantitatively evaluate trans-BTB peptide transfer, the HUVEC/U87MG cells co-culture model was established as described in Section 2.8. Solutions of 30 μmol/L FAM and fluorescein-labeled GICP, DA7R and DA7R–GICP were added to the apical compartment, and after 0.5, 1, 1.5, 2, 3 and 4 h incubation at 37 °C samples were taken from the basolateral chamber and measured, with results shown in Fig. 8A. At each time point, DA7R–GICP peptide was much more effectively transferred through BTB model than GICP or DA7R individually, which may result from its dual-receptor recognition and increased hydrophobicity. In order to better simulate the tumor environment, tumor spheroids were placed under the BTB layer and 30 μmol/L FAM and fluorescein-labeled GICP, DA7R and DA7R–GICP solution was added to the apical compartment and the amount of peptide accumulating in tumor spheroid was observed using the Z-stack mode of the laser scanning confocal microscope. In contrast to the studies where the peptides were directly incubated with tumor cluster, FAM showed no fluorescence (Fig. 8C) whereas GICP and DA7R displayed almost the same fluorescent intensity as seen in ex vivo tumor model, which was far less than DA7R–GICP. The images (Fig. 8C–F) were analyzed by Image J software; the data (Fig. 8B) further demonstrate the advantage of DA7R–GICP.
Figure 8.
Qualitative and quantitative measurement of BTB crossing ability in a HUVEC/U87MG cell co-cultured model. (A) Transport ratios (%) of FAM, fluorescein-labeled GICP, DA7R and DA7R−GICP in the ex vivo BTB model. (B) Semiquantification of mean optical density (IOD/Area) in the trans-BTB tumor spheroid model. (C) FAM uptake in trans-BTB tumor spheroid model. (D) GICP−fluorescein uptake in the trans-BTB tumor spheroid model. (E) DA7R−fluorescein uptake in the trans-BTB tumor spheroid model. (F) DA7R−GICP−fluorescein uptake in the trans-BTB tumor spheroid model. The scale bar indicates 100 μm. Data are presented as mean ± SD, n = 3; *P<0.05; **P<0.01, ***P<0.001.
3.9. Peptide distribution in vivo
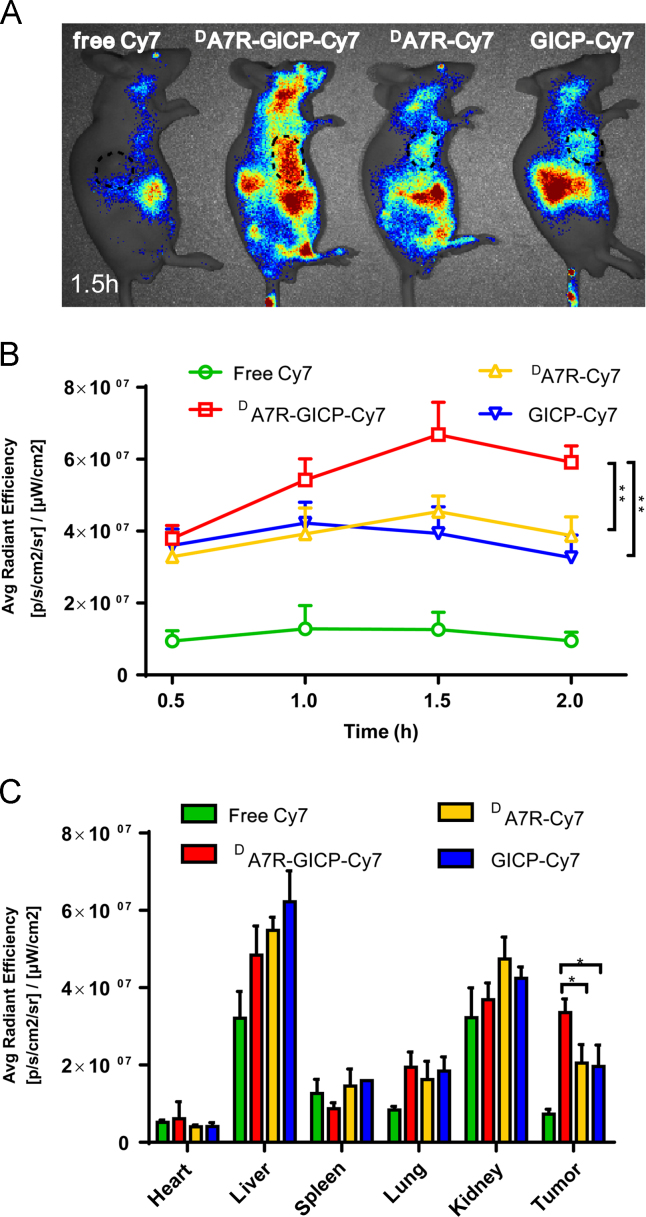
To evaluate the biodistribution of GICP, DA7R and DA7R–GICP peptide in nude mice bearing a subcutaneous U87MG tumor, twelve nude mice were randomly assigned into four groups. Free Cy7, GICP–Cy7, DA7R–Cy7 and DA7R–GICP–Cy7 were used as imaging probes and the mice were imaged with an IVIS system at different time points (Fig. 9). It appeared that DA7R–GICP peptide accumulated to a greater extent in the tumor region than the individual GICP and DA7R peptides, and reached a maximum at 1.5 h (Fig. 9B). After 2 h, the mice were killed and the organs were harvested for fluorescent semi-quantification (Fig. 9C). The results indicate that DA7R–GICP peptide could undergo significant accumulation in the tumors.
Figure 9.
(A)Biodistribution of free Cy7, GICP−Cy7, DA7R−Cy7 and DA7R−GICP−Cy7 in tumor-bearing nude mice at 1.5 h after injection; the tumor region is circled by a dashed line. (B) Semi-quantification of fluorescence intensity in the tumors at each time point. (C) Semi-quantification of the fluorescent intensity of organs harvested at 2 h after injection. Data are presented as mean ± SD, n = 3; *P<0.05; **P<0.01, ***P<0.001.
4. Conclusions
In summary, because of the different distributions of the VAV3, VEGFR2 and NRP-1 receptors in U87MG cells and HUVECs, the single receptor recognition peptides GICP and DA7R were inefficient in glioma cell targeting. In contrast, the coupled Y-shaped peptide DA7R–GICP exhibited higher tumor and tumor-related blood vessel accumulation, relatively improved the enzymatic stability compared with l-peptide GICP, and significantly increased the transport ratio in an ex vivo BTB model. These findings demonstrate that the DA7R–GICP peptide is a multifunctional peptide that should mediate effective tumor and tumor-related neovasculature targeting.
Acknowledgments
This work was supported by National Basic Research Program of China (973 Program, No. 2013CB932500), National Natural Science Foundation of China (Nos. 81773657, 81690263 and 81473149), Shanghai Education Commission Major Project (No. 2017-01-07-00-07-E00052) and Shanghai International Science and Technology Cooperation Project (No.16430723800).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Dolecek CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14:v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao X., Chen R., Liu M., Feng J., Chen J., Hu K. Remodeling the blood–brain barrier microenvironment by natural products for brain tumor therapy. Acta Pharm Sin B. 2017;7:541–553. doi: 10.1016/j.apsb.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei X., Chen X., Ying M., Lu W. Brain tumor-targeted drug delivery strategies. Acta Pharm Sin B. 2014;4:193–201. doi: 10.1016/j.apsb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson D.R., O'Neill B.P. Glioblastoma survival in the United States before and during the temozolomide era. J Neurooncol. 2012;107:359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 5.Purow B., Schiff D. Advances in the genetics of glioblastoma: are we reaching critical mass? Nat Rev Neurol. 2009;5:419–426. doi: 10.1038/nrneurol.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yakubov E., Buchfelder M., Eyüpoglu I.Y., Savaskan N.E. Selenium action in neuro-oncology. Biol Trace Elem Res. 2014;161:246–254. doi: 10.1007/s12011-014-0111-8. [DOI] [PubMed] [Google Scholar]
- 7.Hoang-Xuan K., Capelle L., Kujas M., Taillibert S., Duffau H., Lejeune J. Temozolomide as initial treatment for adults with low-grade oligodendrogliomas or oligoastrocytomas and correlation with chromosome 1p deletions. J Clin Oncol. 2004;22:3133–3138. doi: 10.1200/JCO.2004.10.169. [DOI] [PubMed] [Google Scholar]
- 8.van den Eynden G.G., van der Auwera I., van Laere S.J., Colpaert C.G., Turley H., Harris A.L. Angiogenesis and hypoxia in lymph node metastases is predicted by the angiogenesis and hypoxia in the primary tumour in patients with breast cancer. Br J Cancer. 2005;93:1128–1136. doi: 10.1038/sj.bjc.6602828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouhi P., Lee S.L., Cao Z., Hedlund E.M., Jensen L.D., Cao Y. Pathological angiogenesis facilitates tumor cell dissemination and metastasis. Cell Cycle. 2010;9:913–917. doi: 10.4161/cc.9.5.10853. [DOI] [PubMed] [Google Scholar]
- 10.Hendry S.A., Farnsworth R.H., Solomon B., Achen M.G., Stacker S.A., Fox S.B. The Role of the tumor vasculature in the host immune response: implications for therapeutic strategies targeting the tumor microenvironment. Front Immunol. 2016;7:621. doi: 10.3389/fimmu.2016.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei X., Zhan C., Shen Q., Fu W., Xie C., Gao J. A d-peptide ligand of nicotine acetylcholine receptors for brain-targeted drug delivery. Angew Chem Int Ed Engl. 2015;54:3023–3027. doi: 10.1002/anie.201411226. [DOI] [PubMed] [Google Scholar]
- 12.Ran D., Mao J., Shen Q., Xie C., Zhan C., Wang R. GRP78 enabled micelle-based glioma targeted drug delivery. J Control Release. 2017;255:120–131. doi: 10.1016/j.jconrel.2017.03.037. [DOI] [PubMed] [Google Scholar]
- 13.Liu J.K., Lubelski D., Schonberg D.L., Wu Q., Hale J.S., Flavahan W.A. Phage display discovery of novel molecular targets in glioblastoma-initiating cells. Cell Death Differ. 2014;21:1325–1339. doi: 10.1038/cdd.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M., Chen X., Ying M., Gao J., Zhan C., Lu W. Glioma-targeted drug delivery enabled by a multifunctional peptide. Bioconjugate Chem. 2017;28:775–781. doi: 10.1021/acs.bioconjchem.6b00617. [DOI] [PubMed] [Google Scholar]
- 15.Talluri R.S., Samanta S.K., Gaudana R., Mitra A.K. Synthesis, metabolism and cellular permeability of enzymatically stable dipeptide prodrugs of acyclovir. Int J Pharm. 2008;361:118–124. doi: 10.1016/j.ijpharm.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ying M., Shen Q., Liu Y., Yan Z., Wei X., Zhan C. Stabilized heptapeptide A7R for enhanced multifunctional liposome-based tumor-targeted drug delivery. ACS Appl Mater Interfaces. 2016;8:13232–13241. doi: 10.1021/acsami.6b01300. [DOI] [PubMed] [Google Scholar]
- 17.Starzec A., Ladam P., Vassy R., Badache S., Bouchemal N., Navaza A. Structure-function analysis of the antiangiogenic ATWLPPR peptide inhibiting VEGF165 binding to neuropilin-1 and molecular dynamics simulations of the ATWLPPR/neuropilin-1 complex. Peptides. 2007;28:2397–2402. doi: 10.1016/j.peptides.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Falcon B.L., Chintharlapalli S., Uhlik M.T., Pytowski B. Antagonist antibodies to vascular endothelial growth factor receptor 2 (VEGFR-2) as anti-angiogenic agents. Pharmacol Ther. 2016;164:204–225. doi: 10.1016/j.pharmthera.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Ma Y., Liang S., Guo J., Guo R., Wang H. 18F labeled RGD-A7R peptide for dual integrin and VEGF-targeted tumor imaging in mice bearing U87MG tumors. J Label Compd Radiopharm. 2014;57:627–631. doi: 10.1002/jlcr.3222. [DOI] [PubMed] [Google Scholar]
- 20.Giordano R.J., Cardo-Vila M., Salameh A., Anobom C.D., Zeitlin B.D., Hawked D.H. From combinatorial peptide selection to drug prototype (I): targeting the vascular endothelial growth factor receptor pathway. Proc Natl Acad Sci U S A. 2010;107:5112–5117. doi: 10.1073/pnas.0915141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamarulzaman E.E., Vanderesse R., Gazzali A.M., Barberi-Heyob M., Boura C., Frochot C. Molecular modelling, synthesis and biological evaluation of peptide inhibitors as anti-angiogenic agent targeting neuropilin-1 for anticancer application. J Biomol Struct Dyn. 2017;35:26–45. doi: 10.1080/07391102.2015.1131196. [DOI] [PubMed] [Google Scholar]
- 22.Teesalu T., Sugahara K.N., Kotamraju V.R., Ruoslahti E. C-End rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci U S A. 2009;106:16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belhadj Z., Ying M., Cao X., Hu X., Zhan C., Wei X. Design of Y-shaped targeting material for liposome-based multifunctional glioblastoma-targeted drug delivery. J Control Release. 2017;255:132–141. doi: 10.1016/j.jconrel.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 24.Gu G., Hu Q., Feng X., Gao X., Jiang M., Kang T. PEG--PLA nanoparticles modified with APTEDB peptide for enhanced anti-angiogenic and anti-glioma therapy. Biomaterials. 2014;35:8215–8226. doi: 10.1016/j.biomaterials.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 25.Khodarev N.N., Yu J., Labay E., Darga T., Brown C.K., Mauceri H.J. Tumour-endothelium interactions in co-culture: coordinated changes of gene expression profiles and phenotypic properties of endothelial cells. J Cell Sci. 2003;116:1013–1022. doi: 10.1242/jcs.00281. [DOI] [PubMed] [Google Scholar]